Abstract
Advances in genome sequencing technologies and genome-wide association studies (GWAS) have provided unprecedented insights into the molecular basis of microbial phenotypes and enabled the identification of the underlying genetic variants in real populations. However, utilization of genome sequencing in clinical phenotyping of bacteria is challenging due to the lack of reliable and accurate approaches. Here, we report a method for predicting microbial resistance patterns using genome sequencing data. We analyzed whole genome sequences of 1,680 Streptococcus pneumoniae isolates from four independent populations using GWAS and identified probable hotspots of genetic variation which correlate with phenotypes of resistance to essential classes of antibiotics. With the premise that accumulation of putative resistance-conferring SNPs, potentially in combination with specific resistance genes, precedes full resistance, we retrogressively surveyed the hotspot loci and quantified the number of SNPs and/or genes, which if accumulated would confer full resistance to an otherwise susceptible strain. We name this approach the ‘distance to resistance’. It can be used to identify the creep towards complete antibiotics resistance in bacteria using genome sequencing. This approach serves as a basis for the development of future sequencing-based methods for predicting resistance profiles of bacterial strains in hospital microbiology and public health settings.
Streptococcus pneumoniae, or the pneumococcus, is part of the normal bacterial flora of the human nasopharynx, but can occasionally infiltrate sterile sites of the body progressing to disease. An estimated 1.6 million deaths associated with S. pneumoniae are reported every year worldwide, mostly affecting children under five years. Despite the immunization efforts reducing pneumococcal disease, there is merely a marginal prospect of eliminating pneumococcal disease because the available pneumococcal conjugate vaccines (PCV) only protect against 13 of the over 97 circulating serotypes. Removal of vaccine-types also leads to rapid serotype replacement that consequently increases carriage prevalence and disease by non-vaccine serotypes with occasional increase in antibiotic resistance1.
Since the first case of penicillin-resistant pneumococcus was reported2, followed by outbreaks of disease caused by multidrug-resistant pneumococci3, the antibiotic resistance patterns of S. pneumoniae have drastically evolved and escalated worldwide. The pneumococcus is known to be highly recombinogenic allowing sequences that confer antimicrobial non-susceptibility to be readily introduced into the genome. Discovery of the genetic determinants underlying microbial phenotypes such as antimicrobial resistance and virulence is an important question in microbiology. Traditionally, changes in DNA which are associated with antibiotic resistance in S. pneumoniae were identified using sequence comparison, laboratory mutagenesis4, and identification of horizontally transferred sequences5. These techniques are limited in specificity and sensitivity necessary for applications in clinical laboratories. They only reveal common genomic regions where change has occurred in the so-called ‘mosaic’ genes, are narrow in their application to study actual populations, and may miss out on situations where multiple mutations occurring in different genomic loci are required for full antibiotic resistance5,6. Significant advances in high-throughput genome sequencing technologies and bacterial genome-wide association studies (GWAS) now allow identification of statistical association between plausible causal genetic variants and microbial phenotypes in real populations7. This approach was recently used to identify the single nucleotide polymorphisms (SNPs) in DNA that may confer beta-lactam resistance in S. pneumoniae6.
However, understanding how genetic variations contribute to antibiotics resistance remains underexplored. Here we report the use of genome sequencing and GWAS to investigate SNPs and genes associated with resistance to four essential classes of antibiotics; collectively referred to as antibiotic resistance hereafter. We name the cumulative effect of these resistance-conferring variants the “distance to resistance” for the pneumococcus. We analyzed 1,680 invasive and carriage pneumococcal isolates from Nijmegen, the Netherlands8, Massachusetts, USA9, Maela, Thailand6, and isolates from Sickle-cell anemic children (henceforth referred to as SCD; sickle cell disease) in the USA10. The genotypic and phenotypic diversity in these independent cohorts, whose draft genomes and phenotypes for antibiotic resistance are available, represent a unique dataset for identifying the contribution of each putative antibiotic resistance-conferring genetic variant to the pneumococcal resistance profile. With the premise that presence of particular genes or accumulation of specific SNPs precedes full drug resistance of a fit clone, whole genome sequencing and GWAS could be used to evaluate the rate of accumulation of candidate resistance-conferring variants and provide an early warning sign of increasing antibiotic resistance. We hypothesize that the SNPs and/or resistance associated genes separating the phenotypes of antibiotic resistance are the plausible maximum number of mutations, which if accumulated could render high antibiotic resistance to otherwise susceptible bacteria. As the clinics gradually embrace genome sequencing for microbiological analyses, the ability to use genomic sequencing data to predict relevant phenotypes such as antibiotic resistance will be essential and desirable. This study serves as a foundation for the development of future technologies that could utilize genomic sequencing to analyze the molecular epidemiological trends for bacterial strains reliably, and provide an early-warning measure for the edge towards antimicrobial resistance, crucially informing on clinical intervention strategies.
Results
Pneumococcal strains and phenotypes of antibiotic resistance
We analyzed 1,680 disease and carriage S. pneumoniae isolates systematically selected from diverse cohorts (see materials and methods). In carriage isolates from Maela and Massachusetts, 54 of the 1,012 were resistant to trimethoprim, penicillin, erythromycin and cotrimoxazole, representing about 0.054% resistance to four classes of essential antibiotics (multidrug resistance; MDR). Additionally, all 263 carriage isolates that showed full resistance to penicillin were also resistant to at least one other antibiotic of a different class (~26% resistance to penicillin and one other antibiotic). In contrast, only one isolate from Nijmegen (10208_2#41) showed resistance to all antibiotics tested. Three isolates from Nijmegen exhibited resistance to penicillin (Fig. 1; Supplementary Table S1).
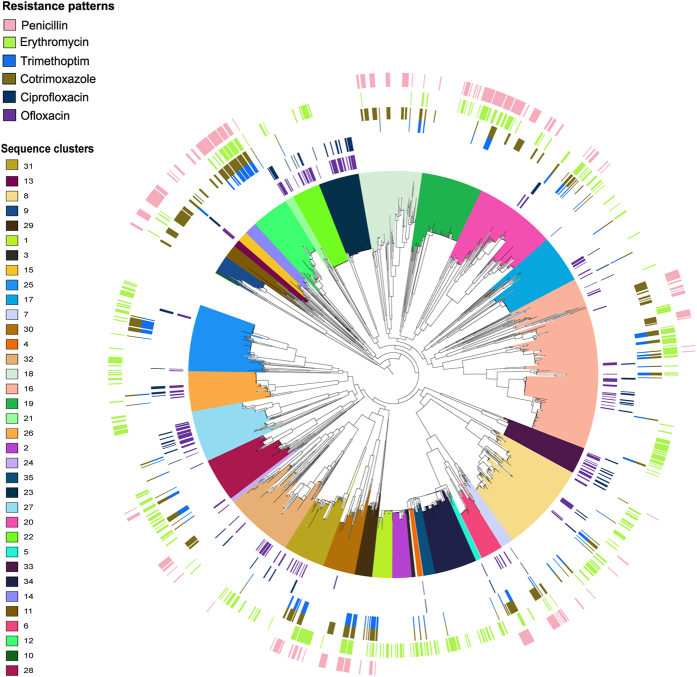
Figure 1. A maximum likelihood phylogeny of the concatenated variant regions from the core genome of 1682 pneumococcal isolates.
The clades are colored according to the sequence clusters. The circular stripes represent the antibiotic resistance phenotypes starting from the outermost: pink; penicillin, green; erythromycin, blue; trimethoprim, gold; cotrimoxazole, navy-blue; ciprofloxacin, and purple; ofloxacin.
Overall, the Nijmegen cohort had the lowest percentage proportion of isolates showing antibiotic resistance; penicillin 0.86% (3 isolates), trimethoprim 4.29% (15), erythromycin 2.28% (8), and cotrimoxazole 4.29% (15), as compared to the selection of Maela and Massachusetts isolates; penicillin 25.99% (263), trimethoprim 14.13% (143 isolates), erythromycin 35.57% (360), and cotrimoxazole 27.76% (281). However, the proportions of resistance to fluoroquinolones; ciprofloxacin 22.86% (80) and ofloxacin 50% (175) were remarkably high in the Nijmegen IPD isolates.
Specific polymorphisms and genes showing significant association with antibiotic resistance
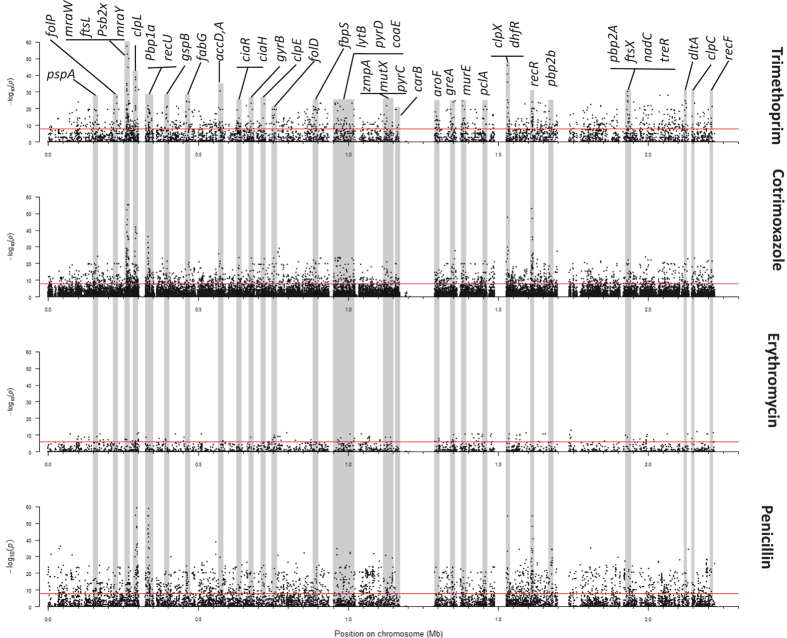
Candidate putative causal variants were selected as SNPs showing statistically significant associations with the resistance phenotype (p-values < 0.01, stratified for population substructure and Bonferroni-adjusted for multiple testing). Separate associations were tested for resistance to penicillin, trimethoprim, cotrimoxazole, erythromycin, ciprofloxacin, and ofloxacin. We identified, 4,317 SNPs that confer resistance to penicillin, 3,589 in coding or intragenic sequences and 548 in non-coding or intergenic sequences (Fig. 2; Supplementary Table S2). A q-q plot of the penicillin GWAS p-values showed a sharp deviation above an expected p-value indicating the presence of unusually high linkage disequilibrium (LD) and strong association of the phenotype with SNPs in heavily genotyped loci. To further control for inflation and increase confidence in verity, a more stringent p-value threshold cut-off was applied at the point of deviation of the observed p-values from the expected p-values. This new cut-off (p-value < 1.5E-20) produced a smaller subset of 426 SNPs associating to the penicillin resistance phenotype. The SNPs are localized primarily in genes previously reported to be involved in development of penicillin resistance, including genes involved in the peptidoglycan biosynthesis pathway such as penicillin binding proteins; PBPs (pbp1A, pbp1B, pbpX, pbp2A, penA), peptidoglycan biogenesis transferases (mraW, mraY) and synthesis of peptidoglycan precursors (murM, murN), pneumococcal surface proteins (pspA, pspC), recombination pathway (recU), cell division pathway (gpsB, ftsL), multi-drug resistance (MDR) proteins and drug efflux pumps (pmrA), drug antiporters6, heat shock proteins/chaperones (ClpL)11, and genes implicated in resistance to other essential classes of antibiotics like dihydrofolate reductase (dyr); involved in trimethoprim resistance12,13.
Figure 2. Manhattan plots summarizing the statistical significance of genome-wide associations between whole-genome SNPs and resistance to various antibiotics.
Specific loci that are significantly associated with resistance to antibiotic are shown in the top panel.
We employed a machine learning method, Random Forest (RF), to investigate the combinatorial effect of certain SNPs and or/genes and prioritize causal variants. From the 426 SNPs, the RF model identified 34 unique SNPs that are predictive of penicillin resistance (Table 1). A separate GWAS on the presence or absence of individual genes in each isolate also revealed that presence of variants of the cell division initiation protein, gpsB (og_2891 and og_1645; p-values 4.73E-44 and 4.06E-43 respectively; Bonferroni-adjusted for multiple testing) significantly correlates with penicillin resistance. Combining both the SNPs and the genes (represented by orthologous groups; OGs) in a RF model further determined that the gene gpsB (og_2891 and og_1645) is the only gene among the top 20 features (SNPS and genes) that are predictive of penicillin resistance.
Table 1. Single nucleotide polymorphisms that are associated with penicillin resistance.
| BP Position | RF importance | RF p-value | CMH p-value | Annotation | Gene |
|---|---|---|---|---|---|
| 1613086 | 20.62957785 | 0 | 3.308E-99 | penicillin-binding protein 2b | penA |
| 1612897 | 19.83961807 | 0 | 6.649E-85 | penicillin-binding protein 2b | penA |
| 333792 | 11.86365577 | 8.69E-98 | 1.234E-99 | Holliday junction-specific endonuclease | recU |
| 333282 | 11.70867819 | 1.55E-18 | 5.304E-55 | penicillin-binding protein 1 A | pbp1A |
| 334107 | 11.29654702 | 7.83E-162 | 9.151E-72 | Holliday junction-specific endonuclease | recU |
| 334639 | 10.42365322 | 0 | 1.203E-68 | hypothetical protein | — |
| 294991 | 10.24670847 | 0 | 5.276E-60 | phospho-N-acetylmuramoyl-pentapeptide-transferase | mraY |
| 335104 | 8.53526957 | 3.3E-17 | 2.659E-68 | DivIVA protein | — |
| 333345 | 7.379638353 | 6.46E-16 | 8.607E-24 | penicillin-binding protein 1 A | pbp1A |
| 1613422 | 7.320298527 | 9.51E-56 | 4.344E-55 | penicillin-binding protein 2b | penA |
| 292563 | 7.167180495 | 8.14E-08 | 1.294E-55 | penicillin binding protein 2x | pbpX |
| 332247 | 6.771832321 | 4.27E-26 | 3.671E-55 | penicillin-binding protein 1 A | pbp1A |
| 335955 | 6.631966798 | 8.25E-35 | 9.742E-45 | RNA methylase family protein | — |
| 1613770 | 6.108715366 | 7.27E-87 | 2.023E-78 | penicillin-binding protein 2b | penA |
| 333386 | 5.891738108 | 4.68E-13 | 8.607E-24 | penicillin-binding protein 1 A | pbp1A |
| 1532326 | 5.159428663 | 2.95E-11 | 4.143E-72 | dihydrofolate reductase | dyr |
| 1531915 | 5.154251817 | 1.21E-11 | 1.717E-20 | ATP-dependent protease ATP-binding subunit ClpX | clpX |
| 295737 | 4.619274035 | 1.67E-75 | 7.271E-49 | ATP-dependent protease ATP-binding subunit ClpL | clpL |
| 296748 | 4.473732826 | 4.15E-08 | 2.701E-36 | ATP-dependent protease ATP-binding subunit ClpL | clpL |
| 2193975 | 4.432372852 | 3.41E-61 | 8.905E-27 | elongation factor Ts | tsf |
Penicillin-resistant pneumococcus exhibit varying patterns of resistance to other β-lactams and are generally resistant to other classes of antibiotics that are usually active against pneumococci14. We evaluated resistance to trimethoprim and cotrimoxazole, erythromycin, as well as ofloxacin and ciprofloxacin. We identified SNPs associated with resistance to trimethoprim and cotrimoxazole in various genes encoding enzymes involved in folate metabolism, including dyr, folE, and folP (Fig. 2; Supplementary Tables S3–S4), and SNPs in genes implicated in resistance to other essential antibiotics like penicillin. RF analysis revealed that only mutations in genes involved in folate metabolism (dyr, folC, folE, folP, and 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase; HPPK (EC 2.7.6.3) encoded by folK or sulD), and chaperones/ATP-dependent proteases (clpL, clpX)15, as well as linked mutations in PBPs (pbp1A, pbpX, penA), recombination proteins (recR, recU), and peptidoglycan biogenesis transferases (mraW, mraY), were predictive of cotrimoxazole and trimethoprim resistance (Tables 2 and 3).
Table 2. Single nucleotide polymorphisms and genes that correlate with cotrimoxazole resistance.
| Feature | RF importance | RF p-value | CMH p-value | Annotation | Gene |
|---|---|---|---|---|---|
| 1531915 | 12.06447624 | 0 | 3.65E-32 | ATP-dependent protease ATP-binding subunit ClpX | clpX |
| 293661 | 10.58122979 | 2.05E-112 | 1.3E-75 | penicillin binding protein 2x | pbpX |
| 267233 | 10.09698603 | 0 | 5.656E-82 | GTP cyclohydrolase I | folE |
| 1532245 | 9.521815356 | 0 | 2.616E-81 | dihydrofolate reductase | dyr |
| 1613086 | 9.414234472 | 5.99E-237 | 8.051E-62 | penicillin-binding protein 2b | penA |
| 264912 | 8.905643954 | 0 | 1.032E-75 | dihydropteroate synthase | folP |
| 1531651 | 7.923116586 | 0 | 4.595E-60 | ATP-dependent protease ATP-binding subunit ClpX | clpX |
| 265536 | 7.247622768 | 5.56E-86 | 3.638E-67 | dihydropteroate synthase | folP |
| 291557 | 6.341493841 | 3.03E-43 | 3.097E-61 | cell division protein | ftsL |
| 292017 | 5.849575748 | 7.35E-31 | 4.103E-50 | penicillin binding protein 2x | pbpX |
| 1532054 | 5.822732316 | 0 | 1.203E-28 | hypothetical protein | — |
| 1612897 | 5.726042635 | 8.3E-102 | 1.213E-49 | penicillin-binding protein 2b | penA |
| 262352 | 5.671693703 | 0 | 1.208E-38 | — | — |
| 263885 | 5.610620269 | 9.89E-132 | 7.608E-41 | permease | — |
| 332799 | 5.455168203 | 5.73E-56 | 1.758E-48 | penicillin-binding protein 1A | pbp1A |
| 262539 | 5.429029061 | 0 | 2.304E-33 | — | — |
| 263190 | 5.333550128 | 9.15E-144 | 4.673E-26 | permease | — |
| 1613770 | 5.331345531 | 3.4E-65 | 2.497E-51 | penicillin-binding protein 2b | penA |
| 291982 | 5.096429153 | 4.18E-34 | 3.797E-47 | penicillin binding protein 2x | pbpX |
| 335104 | 4.434933551 | 0.000000165 | 1.249E-40 | DivIVA protein | — |
Table 3. Single nucleotide polymorphisms and genes that correlate with trimethoprim resistance.
| Feature | RF p-value | CMH p-value | Annotation | Gene |
|---|---|---|---|---|
| 267970 | 4.39E-262 | 2.51E-13 | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase | — |
| 291557 | 4.19E-57 | 6.03E-15 | Cell division protein | ftsL |
| 292017 | 4.99E-52 | 2.89E-16 | Penicillin binding protein 2x | pbpX |
| 291982 | 9.37E-47 | 1.34E-16 | Penicillin binding protein 2x | pbpX |
| cog_1652 | 3.01E-198 | 3.54E-08 | ImpB/MucB/SamB family protein | — |
| cog_1312 | 1.79E-58 | 0.00000173 | Macrolide efflux pump | — |
| cog_1379 | 4.39E-85 | 0.000000119 | YolD-like protein | — |
| cog_791 | 3.88E-48 | 0.00000049 | Ribose import ATP-binding protein rbsA | — |
| cog_2061 | 1.41E-90 | 0.000000119 | Hypothetical protein | — |
| 291286 | 3.39E-15 | 7.68E-17 | S-adenosyl-methyltransferase MraW | mraW |
| 290545 | 3.38E-26 | 6.7E-12 | — | — |
| 1558235 | 4.29E-19 | 4.69E-11 | Cytidylate kinase | cmk |
| cog_1645 | 0 | 0.001967 | Cell division initiation protein | — |
| cog_2891 | 0 | 0.000622 | Cell division initiation protein | — |
| cog_182 | 1.49E-25 | 0.001306 | Binding-protein-dependent transport systems inner membrane component | — |
| 2002234 | 2.83E-28 | 6.6E-15 | PTS system ascorbate-specific transporter subunit IIC | — |
| 1459029 | 5.58E-30 | 4.56E-12 | Asparaginyl-tRNA synthetase | asnC |
| 887794 | 1.4E-27 | 4.56E-12 | DNA polymerase III subunit epsilon | — |
| 1800425 | 0.0000387 | 1.73E-21 | DegT/DnrJ/EryC1/StrS family amino sugar synthetase | — |
Target site modification, characterized by presence of the ermB gene, and efflux from bacteria, mediated by the product of the mefA gene, are the most common mechanisms of bacterial resistance to macrolides. Before correcting for multiple testing, we observed a statistically significant association between the presence of ermB (og_1123) and resistance to erythromycin (p-value 3.137E-05). The OG comprises of two variants of the 23S rRNA (adenine [2058]-N6-)-methyltransferase (ermB): one variant is of the Staphylococcus aureus origin (ungapped protein blast - blastp - alignment with 100% sequence identity over 100% sequence coverage and an e-value of 3e-176). Surprisingly, this association significance diminishes after correcting for multiple testing (p-value 0.09; Bonferroni-corrected for multiple testing). Even so, we identified SNPs in the 16S rRNA; rsmE, 50S rRNA; rplM/S/B/T, rpmA/E2/F/H, rplE/L/I, and 30S rRNA; rpsA/M/N/P/D/H molecules that significantly associated with erythromycin resistance (Fig. 2; Supplementary Table S5). In the RF analysis, the presence of various genes was associated with resistance to erythromycin (Table 4). They include the macrolide efflux pump mefA (og_1312); ImpB/MucB/SamB family protein (og_1652): a family of error-prone DNA polymerases involved in DNA repair; the YolD-like protein (og_1379), a group of functionally uncharacterized proteins predicted to be functionally alike to the UmuD subunit of polymerase V from Gram-negative bacteria, and the ribose import ATP-binding protein (og_791; SP_1114).
Table 4. Single nucleotide polymorphisms and genes whose presence correlates with erythromycin resistance.
| Feature | RF importance | RF p-value | CMH p-value | Annotation | Gene |
|---|---|---|---|---|---|
| og_1652 | 7.180656298 | 0 | 3.701E-14 | ImpB/MucB/SamB family protein | — |
| og_1379 | 7.175992284 | 0 | 6.409E-15 | YolD-like protein | — |
| og_2061 | 7.097377018 | 0 | 6.409E-15 | hypothetical protein | — |
| og_1312 | 6.485953794 | 1.71E-217 | 2.683E-14 | Macrolide efflux pump | mefA |
| og_791 | 6.017699918 | 1.03E-210 | 6.671E-14 | Ribose import ATP-binding protein rbsA | — |
| 1741683 | 4.904707263 | 8.7E-52 | 3.01E-23 | Nudix-related transcriptional regulator NrtR | — |
| og_243 | 4.114250972 | 0.00000718 | 0.0002367 | IS1380-Spn1 transposase | — |
| og_10 | 3.24238261 | 0.000118 | 0.01343 | 3-ketoacyl-ACP reductase | — |
| 290545 | 3.240769624 | 1.2E-76 | 6.7E-12 | Tn916 ORF16 ATP/GTP-binding protein | — |
| 297653 | 3.016787219 | 4.01E-28 | 2.24E-16 | methyltransferase small domain superfamily | — |
| og_53 | 2.971456253 | 5.89E-142 | 4.8E-09 | UDP-glucose 6-dehydrogenase | — |
| og_129 | 2.962230108 | 0.000564 | 4.443E-07 | chlorohydrolase | — |
| 678325 | 2.695815692 | 1.6E-53 | 2.15E-15 | transposase, ISSmi4 | — |
| 136082 | 2.412872116 | 2.23E-15 | 7.73E-14 | cytidine deaminase | cdd |
| 637379 | 2.41015196 | 6.07E-44 | 1.36E-10 | nucleotidyl transferase WchZ | — |
| 987093 | 2.253968194 | 7.1E-12 | 4.43E-23 | Abi-alpha protein | — |
| 2058103 | 1.964208468 | 8.09E-10 | 5.51E-18 | transposase | — |
| og_4190 | 1.95393053 | 0.385 | 0.4235 | IS630-Spn1, transposase Orf1 | — |
| 1593956 | 1.93060019 | 8.68E-09 | 1.62E-10 | Ribose import ATP-binding protein rbsA | — |
| 794795 | 1.754564135 | 0.000000235 | 1.62E-10 | hypothetical protein | — |
Resistance in S. pneumoniae to fluoroquinolone is caused predominantly by mutations in DNA gyrase (gyrA) or DNA topoisomerase (parC and occasionally parE), which reduces binding of the drug to the site of activity. We did not observe any associations between SNPs in fluoroquinolone target proteins (DNA topoisomerase and DNA gyrase) and fluoroquinolone resistance (Supplementary Table S5). Nonetheless, we observed a statistically significant association between mutations in a multi-drug resistance efflux pump (pmrA) and resistance to ofloxacin (p-value 5.06E-05). We also observed that mutations in the heme exporter protein A (ccmA) and the recombination factor protein (rarA) associated with resistance to the fluoroquinolone ciprofloxacin.
Predicting antibiotic resistance profiles and the prospective value of the ‘distance to resistance’
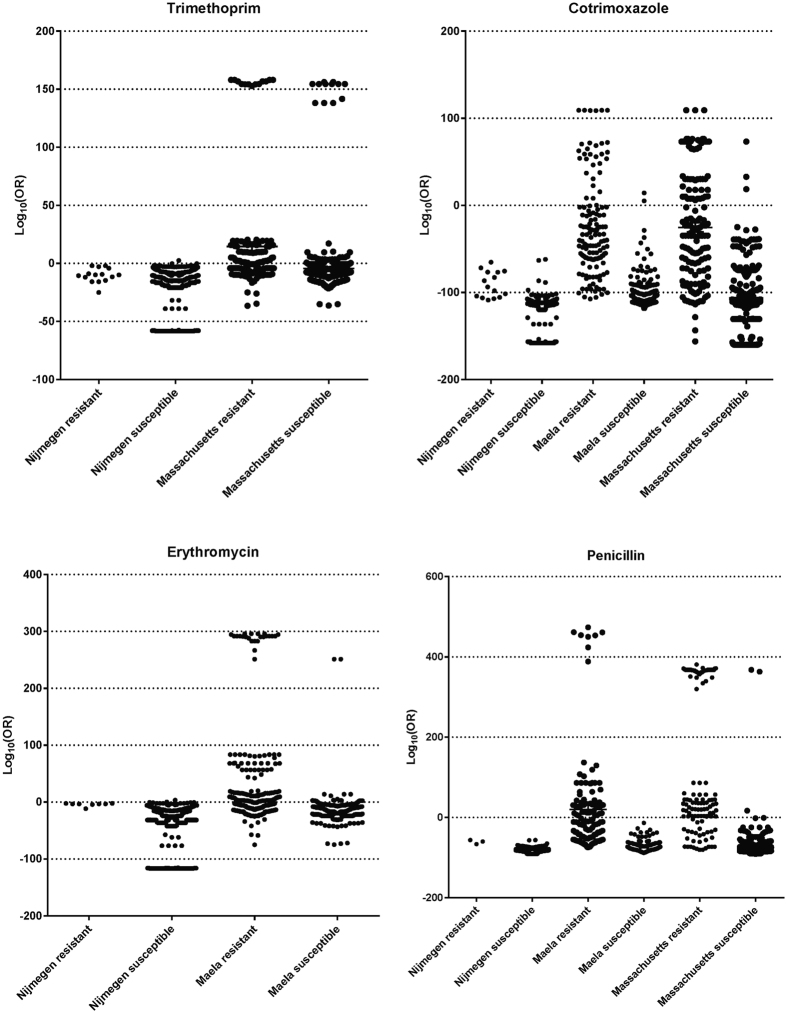
With a presupposition that accumulation of particular SNPs, or acquisition of certain genes leads to a creep towards antibiotic resistance and subsequently full-resistance, we sought to categorize the strains according to their phenotype of antibiotic resistance using the putative resistance-conferring SNPs and/or genes identified by a GWAS. We identified the partition between isolates by summing up the logarithmic derivatives of the odds ratios (sOR) for the putative antibiotic resistance-conferring SNPs (Fig. 3).
Figure 3. Penicillin, erythromycin, trimethoprim, and cotrimoxazole resistance profiles for isolates from individual geographical locations.
Each point represents the cumulative odds ratio effect of SNPs that significantly associate with resistance on each isolate.
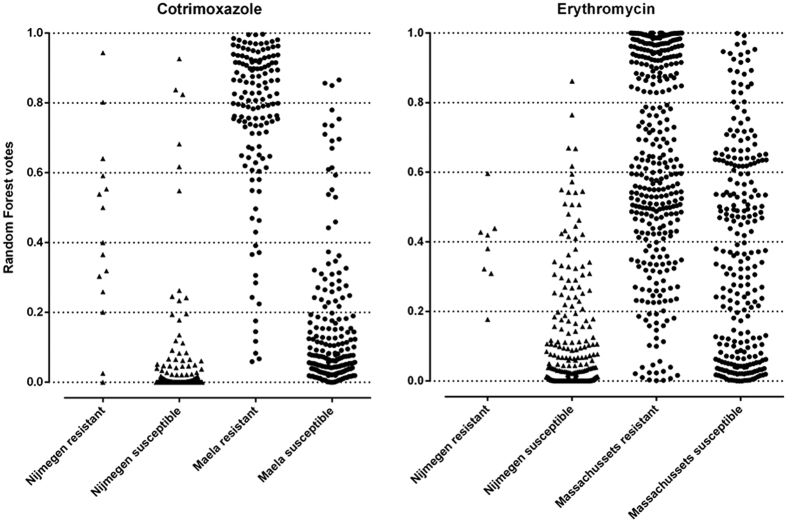
In response to environmental stress over time, bacteria evolve genetic adaptations such as the acquisition of resistance genes or accumulation of critical mutation that confer antibiotic resistance. Supplementary Figure 1 shows the resistance profiles for four antibiotics over time in different cohorts. There seems to be an increase, however subtle, in the number of isolates that have accumulated more resistance-conferring SNPs in each cohort every year. The IPD isolates from Nijmegen have particularly accumulated combinations of SNPs that confer resistance but are more close to susceptibility as compared to carriage isolates. From this data, it is possible to model the resistance profile of new clinical isolates and to predict strains that are approaching extreme antibiotic resistance using sequencing-based approach. Using the RF analysis, we observed that carriage (non-invasive) strains exhibited more resistance to antibiotics (through accumulating SNPs acquiring genes that are most likely to) than invasive strains (Fig. 4). They mostly contained resistance-associated SNPs with high odds ratios of conferring resistance. We also observed that increase in minimum inhibitory concentrations (MIC) against penicillin is characterized by an increase in high sORs SNPs are prioritized by the RF model (Fig. 5). Additionally, we observed that the antibiotics-susceptible isolates (MIC < 0.016 μg/ml) have also acquired some level of mutations that bring them closer to low-MIC penicillin resistance, which in clinical practice can be managed by increased penicillin dose.
Figure 4. The difference in resistance profiles between invasive and carriage isolates.
Each point represent the cumulative random forest vote for invasive (triangles) and carriage (circles) isolates.
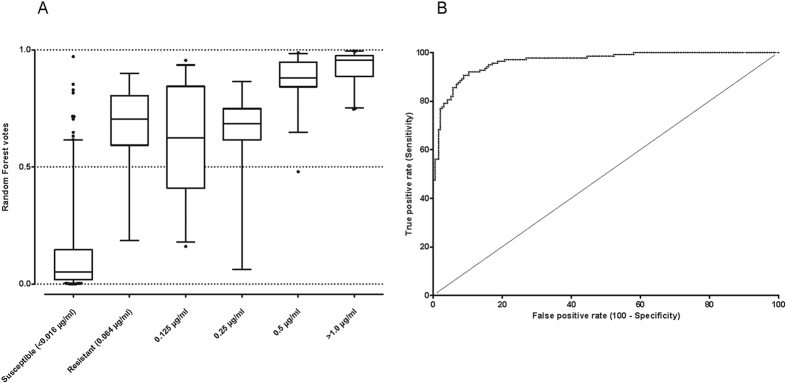
Figure 5.
(A) A box whisker plot of the random forest votes for the levels of resistance to penicillin. Increase in resistance (MIC >= 0.06 µg/ml), as measured by relative increase in the MIC per isolate, is coupled with increased accumulation of the resistance-conferring SNPs that in turn uplift the votes. (B) A Receiver Operating Characteristic (ROC) curve showing the discrimination power of this random forest model in predicting penicillin resistance (Area Under Curve; AUC min = 0.9417 and max = 0.9882 at 99.9% CI).
Discussion
This study includes a total of 1,680 S. pneumoniae isolates from diverse cohorts. Of these, a total of 1,012 carriage isolates (Massachusetts and Maela), and 350 IPD isolates (Nijmegen) were included in a GWAS to pre-screening for resistance-conferring variants. Another 318 SCD isolates were later included in the post-association evaluation of SNPs to determine the distance to resistance. Compared to the general population, SCD patients are usually at high risk of contracting potentially fatal IPD. Therefore, they receive long-term antibiotic prophylaxis and frequent empiric antibiotic treatment. In response to the antibiotic selective pressure, pneumococci isolated from SCD patients have been shown to exhibit high rates of antibiotic resistance10. As a result, isolates from these patients were expected to exhibit substantial resistance that could skew the statistical associations.
Antibiotic resistance profiles differed significantly between sampled populations, and between invasive and nasopharyngeal carriage isolates. Rates of antibiotic resistance were generally low in isolates from the Netherlands compared to those from Thailand and USA. The low levels of resistance in the Netherlands could be explained by two reasons. First, all Dutch isolates used in this study are from IPD cases whereas the Thai and US isolates are predominantly carriage isolates. There is ample opportunity for pneumococci in carriage to benefit from multiple co-colonizing strains of same or closely related species that provide new genetic material for homologous gene transformation. Rates of pneumococcal transformation are also much higher during colonization than during planktonic growth in sepsis16. Second, the Netherlands has implemented very stringent antibiotic control policies. The WHO provides defined daily dose (DDD) guidelines for antibiotics use in adults. Research has shown that aggregate hospital antibiotic use by DDD in the United States is discordant with the WHO stipulations17. Most studies established that majority of antibiotics were administered in primary care settings to treat infections for which antibiotic therapy is hardly indicated18,19. Similarly in Thailand, antibiotics are readily sold over-the-counter allowing for endemic misuse20. Such trends are likely to have accelerated the pneumococcal selection for resistance in the US and Thai populations, and could explain the observed higher levels of resistance as compared to, for example, the Dutch population where antibiotic use is vastly prudent.
Penicillin and other β-lactams have for long been the primary means of treating pneumococcal infections and are perhaps the most widely used group of antibiotics that work by inhibiting bacterial cell wall biosynthesis21. Resistance arises when the organism produces beta-lactamases, which are enzymes that cleave the beta-lactam ring, or modifies the drug targets; ‘penicillin binding proteins’; PBPs. Published research implicates changes in PBPs as the primary determinants of β-lactams resistance22,23,24. Variations in PBP2b and PBP2x modulate low-level (intermediate) β-lactams resistance with additional changes in PBP1a leading to extreme resistance. Growth inhibition assays show that β-lactams primarily kill the pneumococcus by inhibiting PBPs, particularly PBP2×25. PBP transpeptidase signatures are also significant indicators of resistance levels in various β-lactams26.
SNPs associated with beta-lactam resistance were previously reported using GWAS6. We identified 4,317 penicillin-resistance associated SNPs in the pneumococcus, which is more than Chewapreecha and colleagues observed (858 and 1,721 in Maela and Massachusetts cohorts respectively - 301 common SNPs). The difference could be explained by the fact that Chewapreecha et al. replicated their statistical associations in two independent cohorts and only collated candidate SNPs that were identified to be common between the two groups. In contrast, we selected all SNPs showing significant statistical association (p-value < 0.01) with the phenotype after correcting for multiple testing. Moreover, we potentially introduced more genotypic variance by analyzing a mixture of isolates, especially the disease isolates, from different geographical areas. The divergent genotypes in practice resulted into more sequence clusters (used for population substructure stratification) further partitioning the phylogenetic clusters/clades that may have been considered similar in the Chewapreecha study. Therefore, our approach allowed for identification of more SNPs. Applying a more stringent p-value threshold, however, reduced the number of SNPs allowing for further prioritization using RF. The onset of these SNPs is perhaps indicative of the development of antibiotic resistance. However, more studies will be required to validate these observations.
In the case of penicillin resistance, a single SNP or gene is not enough to confer full resistance. We used the RF analysis to evaluate the contribution of combinations of SNPs and/or genes to resistance to penicillin. The mixed RF model incorporating SNPs and genes showed that resistance to β-lactams is primarily driven by mutations in key genes and possibly by the presence of certain resistance genes. In this model, only the gpsB gene was among the top penicillin-resistance conferring features. GpsB is thought to be putatively essential27 and thus expected to be present in a single copy in all isolates. however, there appears to be two variants of the gpsB gene, og_1645; n = 1457 and og_2891; n = 215 in our isolates. GpsB is vital for peripheral and septal peptidoglycan synthesis in S. pneumoniae, particularly in the recruitment of PBP1 to the division complex and its removal from the cell pole soon after pole maturation is completed27. It shows overlapping although non-identical pattern of co-localization with FtsZ during cell division. Depletion of gpsB causes division defect characterized by significant cell elongation and enlargement, several unconstricted rings of division proteins Pbp2x, Pbp1a, FtsZ, and MreC, cessation of growth, and eventually cell lysis in S. pneumoniae D3927. These phenotypes are similar to those observed in Pbp2x depletion28 or inhibition of Pbp2x by the β-lactam antibiotic methicillin27. Therefore, the observed association is most likely a secondary effect of the functions of gpsB in peptidoglycan synthesis during cell division which may have fitness and pleiotropic consequences in maintaining cell integrity rather than a direct role in resistance.
Trimethoprim and cotrimoxazole (a combination of trimethoprim and sulfamethoxazole) reduce the ability of some bacteria to utilize folic acid for growing12 by blocking folate metabolism via dhfR or dyr (encoding dihydrofolate reductase), and folP/sulA (encoding dihydropteroate synthase) respectively. Although there is increasing prevalence of resistance of S. pneumoniae to these drugs, they are still deemed as excellent treatment particularly for pneumococcal infection as well as more generally for otitis, sinusitis, and acute exacerbation of chronic bronchitis. Mutational or recombinational changes on the target enzymes; dhfR and folP or their promoter regions have been reported to enhance resistance to trimethoprim and cotrimoxazole12,13,29. As expected, SNPs in various genes of the folate metabolism pathway correlated with resistance to trimethoprim and co-trimoxazole. Other significant associations for SNPs in genes implicated in resistance to other essential antibiotics, for example PBPs (pbp1A, pbpX, penA) implicated in penicillin resistance, indicating a strong co-selection for resistance to different classes of antibiotics by the pneumococcus. This could be due to the frequent simultaneous use of these antibiotics or an epistatic effect of linked compensatory pathways. More studies will be required to validate this observation.
Macrolides remain an important class of antibiotics for pneumococcal disease30. They inhibit protein synthesis by penetrating the bacterial cell membrane and binding to the ribosomal RNA molecules, particularly in the 50S subunit, of the bacterial ribosome blocking the exit of the growing peptide chain. Their prevalent use in other indications may be the primary driver of selection for macrolide resistance in pneumococci31,32,33. Pneumococcal resistance to macrolides is caused by drug efflux or alteration of the target site34,35. The phenotypic expression of target-site modification can be inducible or constitutive, and can be confirmed with the presence of mefA/E and ermB genes. MefA and mefE share >90% sequence homology and are carried in transposons which are comprised of additional open reading frames36. Perhaps due to the low prevalence of ermB (n = 124), the significance of statistical association waned after adjusting for multiple testing. Nonetheless, presence of other proteins like ImpB, YolD-like protein, SP_1114, and the macrolide efflux pump (encoded by mefA) was determining of resistance to erythromycin. The functional protein interaction network STRING shows that SP_1114 directly interacts with various efflux pumps associated with antibiotic resistance: the drug efflux ABC transporter ATP-binding protein/permease (SP_1342), the MATE efflux pump (SP2065), and the MATE family DinF transport system (SP1939)37, which may suggest a possible contribution to the efflux of drug. This may, however, need more studies to substantiate. YolD-like proteins, like their presumed equivalent (UmuD) in Gram-negative bacteria38, could be involved in modifying the DNA replication machinery to allow bypass synthesis across a damaged template. ImpB on the other hand copies undamaged DNA at stalled replication forks which arise in vivo from mismatched or misaligned primer ends. These misaligned primers could be extended by DNA polymerase IV subunit (polIV). The important role of these proteins in division and maintaining cellular integrity may perhaps explain their indirectly contribution to erythromycin resistance.
Fluoroquinolones (Ofloxacin and ciprofloxacin) are a family of related compounds that inhibit bacterial DNA synthesis by promoting cleavage of DNA in the DNA-enzyme complexes of DNA gyrase and type IV topoisomerase. Ciprofloxacin binds exclusively with topoisomerase IV whereas ofloxacin binds more avidly with topoisomerase IV and also binds the gyrase. Fluoroquinolone activity in Gram-positive bacteria, including S. pneumoniae, usually results from inhibition of DNA type IV topoisomerase whereas activity in Gram-negative bacteria corresponds with inhibition of DNA gyrase. Pneumococcal resistance to fluoroquinolone is thought to be a stepwise process; most intermediately resistant strains accumulate first-step mutations, which usually involve only a single mutation in the target genes39, although, these strains do tend to go on to develop subsequent second-step mutations which significantly diminishes the activity of most fluoroquinolones and renders the strains highly resistant40. This increasing mutational heterogeneity makes it harder to use GWAS in precisely identifying the role of each mutation in disseminating antibiotic resistance to fluoroquinolones. While there were no significant associations for mutations in the target proteins, mutations in other genes like pmrA, ccmA, and rarA were observed to be determining of resistance to fluoroquinolones. Mutations in correlated with resistance to ofloxacin. PmrA is homologous to other well-studied efflux pumps like NorA and Bmr, whose expression leads to reduced susceptibility against several diverse compounds41, and is associated with fluoroquinolone resistance in the pneumococcus42. The pneumococcus, like most pathogenic bacteria, mainly uses iron from hemoglobin and heme to support growth43. The direct role of CcmA in quinolone-resistance is unclear but in Group A streptococci (GAS), the heme exporter confers multi-drug resistance44. Apart from the ABC-transporter protein PmrA42, efflux mechanisms of fluoroquinolone resistance are poorly characterized in the pneumococcus. Nonetheless, efflux could reduce intracellular fluoroquinolone concentrations to sublethal levels facilitating resistance development45. More experiments are required to deduce the exact mechanisms involving these other efflux pumps.
Recombination is the primary source of genome plasticity in bacteria. Unlike in β-lactam resistance where recombination plays an important role46, fluoroquinolone resistance arises from very specific resistance-determining mutations within the target proteins47. Horizontal transfer of fluoroquinolone resistance loci between viridans group streptococci and the pneumococcus has been shown to occur in vitro but not in vivo48, with significantly higher rates during asymptomatic carriage than during invasive isolates49,50. Studies have reported a link between fluoroquinolone resistance and evolution of resistance to penicillin and macrolides51; both of which could be fostered by recombination. These studies suggest that the observed association between ciprofloxacin resistance and mutations in rarA could be an artifact of the linked resistance with other antibiotics. However, since we cannot rule out a novel mechanism involving these mutations, more studies are required to ascertain this observation.
Pneumococci exhibiting decreased susceptibility to regular penicillin doses are on the rise14. Many European guidelines recommend high-dose empirical treatments for systemic infections caused by such strains showing atypical susceptibility. Analyzing the strains for an effective dose, however, relies on laboratory assays. A recent publication reports the use of PBPs transpeptidase signatures (TPDs) from 2,528 clinical pneumococcal isolates to predict the MICs for various β-lactam antibiotics26. Li and colleagues constructed predictive models that link amino acid sequence variations in the TPDs of PBP1a, PBP2b, and PBP2x to β-lactam MIC levels among invasive pneumococcal isolates. They identified 68, 78, and 118 unique TPD amino acid sequences for PBP1a, PBP2b, and PBP2x, respectively. Using 307 unique combinations of these sequences which defined the PBP types, they observed that isolates whose PBP types exhibited more than 10% amino acid sequence divergence from a usual susceptible PBP type were associated with increased β-lactam MICs.
Using GWAS and RF, we have rapidly detected decreasing sensitivity of the pneumococcus to increasing doses of penicillin, and we have classified susceptible and resistant isolate using genome sequencing with the sensitivity and specificity comparable to the MICs determined using phenotypic susceptibility tests. The increase in MIC corresponded with accumulation of SNPs and acquisition of genes that we prioritized to be most determining of penicillin resistance. Moreover, we have determined how far the antibiotic susceptible strains are from acquiring the resistance-conferring features that will enable them attaining full-resistance, and what the creep pattern towards full-resistance is. The measure is dubbed the “distance to resistance”. Starting with a curated reference database of genes and SNPs/alleles that confer resistance in historical and contemporary isolates; it is possible to model a framework that predicts the antibiotic resistance profile of an isolate, and project the transition towards resistance over time. Although causality cannot always be drawn from statistical association, bacterial GWAS provide the means for identifying genomic variants underlying important microbial phenotypes like antibiotic resistance. We have demonstrated that prediction of advancing antimicrobial resistance could be achieved in silico using genomic sequencing data. Therefore, this study invokes a change of perspective for future research to focus on not only identify genetic variants underlying the resistance phenotype but also detecting how these variants herald the advancement towards full-resistance. Such sequencing-based frameworks are not only affordable and consistent but also allow for simultaneous discovery of other essential pneumococcal features such as serotype and sequence type. Altogether, this knowledge will greatly inform the choice of clinical intervention and improve public health surveillance thus precluding outbreaks caused by emerging multidrug resistant strains.
Materials and Methods
Strains and phenotypes in study
This study included 1,680 pneumococcal isolates and corresponding antibiotic resistance phenotypes; 349 from adults admitted with invasive pneumococcal disease between 2001 and 2011 in two hospitals in Nijmegen, The Netherlands8, a systematic selection (three from each “secondary BAPS” cluster) of published carriage isolates from Massachusetts, USA and Maela, Thailand6, and 318 isolates from children suffering from sickle-cell disease (SCD) in the USA10 which included isolates from the CDC ABC bacterial surveillance core and published collections52,53. Phenotypes of antimicrobial susceptibility were determined in vitro as previously described6,8,9. For the invasive Nijmegen isolates, the following breakpoints were used: penicillin susceptible (S) = <00.6; co-trimoxazole S = <1; erythromycin S = <0.25.
Determining SNPs and orthologous sequences
Bases were called from mapped sequences using kSNP v2 software54 against a single reference genome: multidrug-resistant S. pneumoniae ATCC 700669; Spain 23F ST8155. A total of 124,310 SNP calls were generated (Numerical feature IDs used in this study correspond to the SNP base-pair position in this reference genome). Filtering for SNPs present in more than ~90% of the isolates, (1,500 strains) resulted in 76,429 SNP calls that we used for further analysis. To determine clusters of orthologous sequences, all coding sequences (CDS) from the 1,680 isolates were predicted using Prodigal56. All coding sequences were analyzed using USEARCH57 and aligned in the ‘large-scale BLAST score ratio’ (LS-BSR) pipeline58 allowing 10% amino acid difference within clusters. The resulting representative sequences per group (“centroids”) were clustered through a Markov Clustering Algorithm (TRIBE-MCL)59 with an inflation factor of 2.5, resulting into 4687 orthologous groups (OGs). For each OG, we generated a binary metric of the presence (1) or absence (0) of a representative coding sequence(s) (CDS) from each strain. Each strain’s contribution of CDS to an OG was subsequently denoted by a single numeric value (1 or 0) to designate the presence or absence of a distinct gene, gene variants, or a group of paralogs. These groups were collated into a binary matrix and formatted for PLINK association analyses60.
Determining the population structure and statistical association
Resistance phenotypes were grouped according to antibiotic classes and analyzed separately for each population, and together in the final association analysis. The population clusters used to control for the effect of clonal inheritance of genetic variants and population stratification were determined using the Bayesian Analysis of Population Structure (BAPS) software61, and a phylogeny-based partitioning approach as proposed by Prosperi et al.62, which employs ‘ape, geiger, igraph, and phytools’ packages in R software63. An alignment of concatenated SNPs from the core or non-repetitive DNA of each of the isolates were analyzed in BAPS as previously described8. For the Prosperi clustering, a maximum-likelihood phylogenetic tree was constructed using RAxML version 8.2.064 and an alignment of all the concatenated SNPs from the core genome of all isolates as described before8. The general time-reversible model was used to calculate the maximum-likelihood ratios with a γ adjustment for site variation as the nucleotide substitution model. The support for nodes on the tree was tested using a hundred unsystematic bootstrap replicates. Resulting phylogenetic tree was visualized using iTOL version 2.165.
We used the Cochran-Mantel-Haenszel (CMH) correlation statistic to test for associations between antibiotic resistance phenotype and SNPs conditional on the population structure. Stratification for population structure minimized falsely positive associations that could be obtained merely by chance. We tested associations for resistance to penicillin, trimethoprim, cotrimoxazole, erythromycin, ofloxacin and ciprofloxacin, and corrected for population stratification using the genetic subpopulations (represented by the sequence clusters; SCs) determined using BAPS and/or the method proposed by Prosperi et al.62. The statistical associations were performed using PLINK software v1.960, and the results visualized as Manhattan and Q-Q plots in R using ‘qqman’ package.
Candidate resistance loci and a measure of the distance to resistance
We selected SNPs showing statistically significant associations (p-values < 0.01 at a minor allele frequency >0.01; Bonferroni-adjusted for multiple testing) as candidates for subsequent analysis. The percentage distribution of these candidate SNPs within resistance isolates relative to the susceptible isolates in each population was computed to determine how they vary in each cohort. For each SNP significantly associated with antibiotic resistance, we determined the odds ratio (OR) and nature (positive or negative) of the correlation. The accumulation of these significant SNPs in each isolate was also defined across all test cohorts. Each SNP present in an isolate was represented by the logarithmic derivative of the odds ratio; log10 (OR): The negative logarithmic values were used for SNPs negatively correlated with resistance. Q-Q plots were used to determine a more stringent p-value cut-off. The aggregate effect of the SNPs conferring antibiotic resistance is the sum of all the log10 (OR) values for SNPs above the p-value threshold. These represented a measure of the level of resistance of an isolate. These aggregate values were plotted in GraphPad Prism v6.05 software.
Random forest analysis for prioritizing variants and classifying isolates
To prioritize probable causal variants and investigate the effect of SNPs and/or gene combinations, a Random Forest (RF) classification using the Bioconductor randomForest package 4.6–10 was performed. The resulting candidates were used to discriminate resistant (R) and susceptible (S) isolates. This classification model, consisting of 5000 decision trees was trained on candidate genes and/or SNPs that were determined to be predictive of resistance through GWAS analysis. Statistical significance of the genes or SNPs that were able to discriminate between the classes were calculated by permuting the sample class labels. A normal distribution was determined for mean decrease of accuracy (MDA) values from the 300 feature-vector permutations using the pnorm package which is part of the R version 3.3.0 distribution63. Still using the pnorm package, a p-value was calculated comparing the average of mda values for RF 100 analyses (the out-of-box - OOB - error averaged over 100 RF runs) with the original sample classes (to account for slight differences between RF analyses) to the distribution of permuted MDA values.
Additional Information
How to cite this article: Mobegi, F. M. et al. Deciphering the distance to antibiotic resistance for the pneumococcus using genome sequencing data. Sci. Rep. 7, 42808; doi: 10.1038/srep42808 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This project was supported by a travel grant from The Royal Netherlands Academy of Arts and Sciences (Academy Ter Meulen Grant 2014) to FMM.
Footnotes
The authors declare no competing financial interests.
Author Contributions F.M., S.v.H. and A.Z conceived the study, designed the study and performed the bioinformatics analysis. F.M. wrote the manuscript. A.C. and M.d.J. were responsible for collecting and processing the clinical isolates from Nijmegen. S.B. provided meta-data for the Maela and Massachusetts isolates, and was responsible for sequencing the genome. S.v.H. M.d.J. S.B. and A.Z. were involved in supervising of the project. All authors have read and approved the manuscript.
References
- Pelton S. I. et al. Emergence of 19A as virulent and multidrug resistant Pneumococcus in Massachusetts following universal immunization of infants with pneumococcal conjugate vaccine. Pediatr Infect Dis J 26, 468–72 (2007). [DOI] [PubMed] [Google Scholar]
- Hansman D. & Bullen M. M. A Resistant Pneumococcus. The Lancet 290, 264–265 (1967). [Google Scholar]
- Appelbaum P. C. et al. Streptococcus pneumoniae resistant to penicillin and chloramphenicol. Lancet 2, 995–7 (1977). [DOI] [PubMed] [Google Scholar]
- Hakenbeck R., Bruckner R., Denapaite D. & Maurer P. Molecular mechanisms of beta-lactam resistance in Streptococcus pneumoniae. Future Microbiol 7, 395–410 (2012). [DOI] [PubMed] [Google Scholar]
- Sauerbier J., Maurer P., Rieger M. & Hakenbeck R. Streptococcus pneumoniae R6 interspecies transformation: genetic analysis of penicillin resistance determinants and genome-wide recombination events. Mol Microbiol 86, 692–706 (2012). [DOI] [PubMed] [Google Scholar]
- Chewapreecha C. et al. Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes. PLoS Genet 10, e1004547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. E. & Shapiro B. J. The advent of genome-wide association studies for bacteria. Curr Opin Microbiol 25, 17–24 (2015). [DOI] [PubMed] [Google Scholar]
- Cremers A. J. et al. The post-vaccine microevolution of invasive Streptococcus pneumoniae. Sci Rep 5, 14952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J. et al. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet 45, 656–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R. et al. Genomic analyses of pneumococci from children with sickle cell disease expose host-specific bacterial adaptations and deficits in current interventions. Cell Host Microbe 15, 587–99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T. D. et al. Decrease in penicillin susceptibility due to heat shock protein ClpL in Streptococcus pneumoniae. Antimicrob Agents Chemother 55, 2714–28 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huovinen P. Resistance to trimethoprim-sulfamethoxazole. Clin Infect Dis 32, 1608–14 (2001). [DOI] [PubMed] [Google Scholar]
- Pikis A., Donkersloot J. A., Rodriguez W. J. & Keith J. M. A conservative amino acid mutation in the chromosome-encoded dihydrofolate reductase confers trimethoprim resistance in Streptococcus pneumoniae. J Infect Dis 178, 700–6 (1998). [DOI] [PubMed] [Google Scholar]
- Perez-Trallero E. et al. Geographical and ecological analysis of resistance, coresistance, and coupled resistance to antimicrobials in respiratory pathogenic bacteria in Spain. Antimicrob Agents Chemother 49, 1965–72 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski A., Burghout P. & Morrison D. A. spr1630 is responsible for the lethality of clpX mutations in Streptococcus pneumoniae. J Bacteriol 191, 4888–95 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks L. R., Reddinger R. M. & Hakansson A. P. High levels of genetic recombination during nasopharyngeal carriage and biofilm formation in Streptococcus pneumoniae. MBio 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polk R. E., Fox C., Mahoney A., Letcavage J. & MacDougall C. Measurement of adult antibacterial drug use in 130 US hospitals: comparison of defined daily dose and days of therapy. Clin Infect Dis 44, 664–70 (2007). [DOI] [PubMed] [Google Scholar]
- Grijalva C. G., Nuorti J. P. & Griffin M. R. Antibiotic prescription rates for acute respiratory tract infections in US ambulatory settings. JAMA 302, 758–66 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh A. L., Shapiro D. J., Pavia A. T. & Shah S. S. Antibiotic prescribing in ambulatory pediatrics in the United States. Pediatrics 128, 1053–61 (2011). [DOI] [PubMed] [Google Scholar]
- Khamsarn S. et al. Epidemiology of Antibiotic Use and Antimicrobial Resistance in Selected Communities in Thailand. J Med Assoc Thai 99, 270–5 (2016). [PubMed] [Google Scholar]
- Elander R. P. Industrial production of beta-lactam antibiotics. Appl Microbiol Biotechnol 61, 385–92 (2003). [DOI] [PubMed] [Google Scholar]
- Grebe T. & Hakenbeck R. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of beta-lactam antibiotics. Antimicrob Agents Chemother 40, 829–34 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenbeck R. et al. Penicillin-binding proteins in beta-lactam-resistant Streptococcus pneumoniae. Microb Drug Resist 5, 91–9 (1999). [DOI] [PubMed] [Google Scholar]
- Smith A. M. & Klugman K. P. Alterations in PBP 1A essential-for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 42, 1329–33 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocaoglu O., Tsui H. C., Winkler M. E. & Carlson E. E. Profiling of beta-lactam selectivity for penicillin-binding proteins in Streptococcus pneumoniae D39. Antimicrob Agents Chemother 59, 3548–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Penicillin-Binding Protein Transpeptidase Signatures for Tracking and Predicting beta-Lactam Resistance Levels in Streptococcus pneumoniae. MBio 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land A. D. et al. Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol Microbiol 90, 939–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg K. H., Stamsas G. A., Straume D. & Havarstein L. S. Effects of low PBP2b levels on cell morphology and peptidoglycan composition in Streptococcus pneumoniae R6. J Bacteriol 195, 4342–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buwembo W., Aery S., Rwenyonyi C. M., Swedberg G. & Kironde F. Point Mutations in the folP Gene Partly Explain Sulfonamide Resistance of Streptococcus mutans. Int J Microbiol 2013, 367021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. C. & Jenney A. W. J. Macrolide resistance in pneumococci—is it relevant? Pneumonia 8, 1–3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda K. J., Hicks L. A., Roberts R. M., Hunkler R. J. & Taylor T. H. Trends and seasonal variation in outpatient antibiotic prescription rates in the United States, 2006 to 2010. Antimicrob Agents Chemother 58, 2763–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan J. D. et al. Evidence for clonal expansion after antibiotic selection pressure: pneumococcal multilocus sequence types before and after mass azithromycin treatments. J Infect Dis 211, 988–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare K. M. et al. Longitudinal nasopharyngeal carriage and antibiotic resistance of respiratory bacteria in indigenous Australian and Alaska native children with bronchiectasis. PLoS One 8, e70478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varaldo P. E., Montanari M. P. & Giovanetti E. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob Agents Chemother 53, 343–53 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler C. & Teuber M. The Macrolide Efflux Genetic Assembly of Streptococcus pneumoniae Is Present in Erythromycin-Resistant Streptococcus salivarius. Antimicrobial Agents and Chemotherapy 46, 3690–3691 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M. M., Doktor S., Flamm R. & Shortridge D. Characterization and prevalence of MefA, MefE, and the associated msr(D) gene in Streptococcus pneumoniae clinical isolates. J Clin Microbiol 42, 3570–4 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocci N. et al. Functional analysis of pneumococcal drug efflux pumps associates the MATE DinF transporter with quinolone susceptibility. Antimicrob Agents Chemother 57, 248–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuning P. J., Simon S. M., Godoy V. G., Jarosz D. F. & Walker G. C. Characterization of Escherichia coli Translesion Synthesis Polymerases and Their Accessory Factors. In Methods in Enzymology Vol. Volume 408 318–340 (Academic Press, 2006). [DOI] [PubMed] [Google Scholar]
- Brueggemann A. B. et al. Fluoroquinolone Resistance in Streptococcus pneumoniae in United States since 1994–1995. Antimicrobial Agents and Chemotherapy 46, 680–688 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletz M. W. et al. Prevalence of first-step mutants among levofloxacin-susceptible invasive isolates of Streptococcus pneumoniae in the United States. Antimicrob Agents Chemother 50, 1561–3 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I. T., Brown M. H. & Skurray R. A. Proton-dependent multidrug efflux systems. Microbiol Rev 60, 575–608 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M. J., Brenwald N. P. & Wise R. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 43, 187–9 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Espejel M. E., Gonzalez-Lopez M. A. & Olivares-Trejo Jde J. Streptococcus pneumoniae requires iron for its viability and expresses two membrane proteins that bind haemoglobin and haem. Metallomics 5, 384–9 (2013). [DOI] [PubMed] [Google Scholar]
- Sachla A. J. & Eichenbaum Z. The GAS PefCD exporter is a MDR system that confers resistance to heme and structurally diverse compounds. BMC Microbiol 16, 68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletz M. W. et al. Antihypertensives suppress the emergence of fluoroquinolone-resistant mutants in pneumococci: an in vitro study. Int J Med Microbiol 303, 176–81 (2013). [DOI] [PubMed] [Google Scholar]
- McGee L. et al. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol 39, 2565–71 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel L. M., Anderson G. J., Facklam R. R. & Tenover F. C. Genetic Analyses of Mutations Contributing to Fluoroquinolone Resistance in Clinical Isolates ofStreptococcus pneumoniae. Antimicrobial Agents and Chemotherapy 45, 3517–3523 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tankovic J., Perichon B., Duval J. & Courvalin P. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents and Chemother 40, 2505–2510 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre L., Ferrandiz M. J., Linares J., Tubau F. & de la Campa A. G. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob Agents Chemother 47, 2072–81 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletz M. W., McGee L., Beall B., Whitney C. G. & Klugman K. P. Interspecies recombination in type II topoisomerase genes is not a major cause of fluoroquinolone resistance in invasive Streptococcus pneumoniae isolates in the United States. Antimicrob Agents Chemother 49, 779–80 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton R., Morosini M., Enright M. C. & Morrissey I. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the global PROTEKT surveillance programme. J Antimicrob Chemother 52, 944–52 (2003). [DOI] [PubMed] [Google Scholar]
- McCavit T. L., Quinn C. T., Techasaensiri C. & Rogers Z. R. Increase in invasive Streptococcus pneumoniae infections in children with sickle cell disease since pneumococcal conjugate vaccine licensure. J Pediatr 158, 505–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw N. C. et al. Nasopharyngeal carriage of penicillin-resistant Streptococcus pneumoniae in children with sickle cell disease. Pediatrics 99, E7 (1997). [DOI] [PubMed] [Google Scholar]
- Gardner S. N. & Hall B. G. When whole-genome alignments just won’t work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8, e81760 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J. et al. Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniae Spain23F ST81. J Bacteriol 191, 1480–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–1 (2010). [DOI] [PubMed] [Google Scholar]
- Sahl J. W., Caporaso J. G., Rasko D. A. & Keim P. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ 2, e332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A. J., Van Dongen S. & Ouzounis C. A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30, 1575–84 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Connor T. R., Siren J., Aanensen D. M. & Corander J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30, 1224–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosperi M. C. et al. A novel methodology for large-scale phylogeny partition. Nat Commun 2, 321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team. R: a language and environment for statistical computing. 3.2.2 edn (Foundation for Statistical Computing, Vienna, Austria, 2010). [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I. & Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39, W475–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.