Abstract
Hundreds of commensal bacterial species inhabit the gastrointestinal tract. This diverse microbial ecosystem plays a crucial role in the prevention and resolution of infectious diseases. In this review we will describe the major mechanisms by which the intestinal microbiota confers protection against infections, focusing on those caused by intestinal bacterial pathogens. These mechanisms include both non-immune- and immune-cell-mediated pathways, notably through bacterial production of inhibitory molecules and nutrient deprivation by the former and innate lymphoid cell-, myeloid cell- or lymphocyte-dependent stimulation by the latter. Finally, we will discuss novel therapeutic approaches based on commensal microbes and their products, which could potentially be used to combat infections.
Introduction
The intestinal tract is home to hundreds of bacterial species, referred to collectively as the intestinal microbiota. During the last decade, high-throughput technologies, including next-generation sequencing, metabolomics and proteomics, have allowed an in-depth study of the composition of the intestinal microbiota, the genes and functions expressed by commensal bacteria, and the metabolites and proteins derived from their biological activities.1 Use of these high-throughput techniques in combination with mouse models, such as germ free mice (GFM) or antibiotic-treated mice, has highlighted the extent to which commensal microbes are essential for conferring protection against pathogens, and how alterations of the microbiota, such as those induced by antibiotics, can promote infections.2, 3, 4, 5
Recent studies have started to thrown light on the mechanisms by which different commensal bacterial species confer resistance against infections. In this review, we will discuss both non-immune- and immune-mediated mechanisms used by commensal bacteria to confer pathogen resistance. These mechanisms range from direct inhibitory molecule production and nutrition competition (Figure 1 and Table 1) to indirect pathways through the stimulation of local innate lymphoid cells (ILCs), myeloid cells, or T- and B-cell responses (Figure 2 and Table 1). The review will mainly focus on the way the intestinal microbiota protects against gastrointestinal bacterial pathogens. Nevertheless, we will also review some studies that have demonstrated an effect of intestinal microbes on systemic immunity and defense against systemic infections. Finally, we will discuss novel therapeutic approaches for treating infections using commensal microbes and their products that have emerged from recent studies in the field.
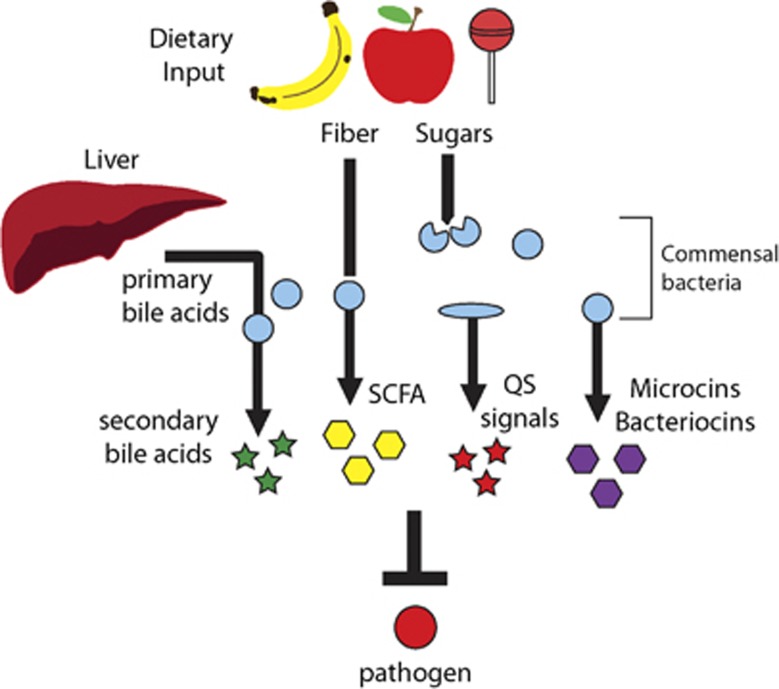
Figure 1.
Non-immune-derived mechanisms of protection. The intestinal microbiota can confer protection without the induction of the immune system. (i) Commensal microbes consume and deplete simple sugars that could be utilized by pathogens such as Escherichia coli 0157:H7, Salmonella typhimurium or Clostridium difficile. (ii) Anaerobic fermentation of dietary fiber by the microbiota generates short-chain fatty acids (SCFA), which can inhibit the growth of certain pathogens such as E. coli strain O157:H7. (iii) Clostridium scidens converts primary bile acids, synthesized in the liver, into secondary bile acids, which inhibit the growth of C. difficile. (iv) Ruminococcus obeum interferes with the expression of colonization factors expressed by Vibrio cholerae through production of the quorum-sensing (QS) signal AI-2. (v) Certain commensals such as Enterococcus faecalis, Staphylococcus lugdunensis or E. coli strain Nissle 1917 secrete small peptides (bacteriocins or microcins) that inhibit the growth of pathogens such as Staphylococcus aureus, vancomycin-resistant Enterococcus or S. typhimurium.
Table 1. Commensal bacterial species that confer protection against pathogens.
| Commensal | Pathogen | Mechanism | Reference |
|---|---|---|---|
| Staphylococcus lugdunensis | Staphylococcus aureus | Peptide antibiotic with bactericidal activity | 7 |
| Enterococcus faecalis with pPD1 plasmid | Vancomycin-resistant Enterococcus | Plasmid-encoded bacteriocin that inhibits pathogen growth | 6 |
| Bacillus thuringiensis | Clostridium difficile | Bacteriocin with bactericidal activity | 87 |
| Escherichia coli strain Nissle 1917 | Salmonella typhimurium | Microcins with antimicrobial activity | 8 |
| Clostridium scidens | C. difficile | Conversion of primary to secondary bile acids which inhibit pathogen growth | 4 |
| Ruminococcus obeum | Vibrio cholerae | Quorum-sensing signals that interfere with pathogen gene expression | 11 |
| E. coli strains HS and Nissle 1917 | E. coli O157:H7 | Competition for carbohydrates | 15 |
| E. coli, Bacteroides thetaiotaomicron | Citrobacter rodentium | Competition for carbohydrates | 16 |
| E. coli strain Nissle 1917 | S. typhimurium | Competition for iron | 86 |
| B. thetaiotaomicron | Candida albicans | LL-37 antimicrobial peptide induction | 32 |
| Bifidobacterium | E. coli O157:H7 | Inhibition of Shiga toxin dissemination | 33 |
| Lactobacillus reuteri | C. albicans | Induction of type 3 innate lymphoid cells expansion and interleukin 22 production through tryptophan conversion to an aryl hydrocarbon receptor ligand | 46 |
| Segmented filamentous bacterium | C. rodentium | Induction of T helper 17 cells differentiation and subsequent expression of antimicrobial peptides | 53 |
| E. coli | S. typhimurium | Systemic induction of IgG | 69 |
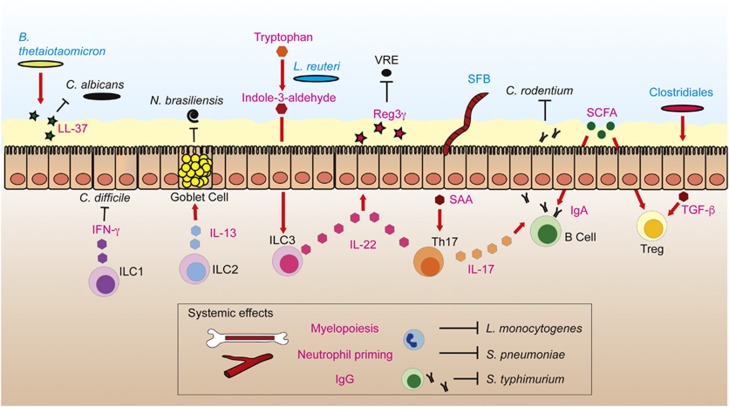
Figure 2.
Intestinal microbiota impacts local and systemic immunity against infection. Bacteroides thetaiotaomicron promotes the expression of LL-37 antimicrobial peptide by intestinal epithelial cells (IECs), which diminishes Candida albicans colonization. Commensal microbes affect the development and gene expression of innate lymphoid cells (ILCs). For example, tryptophan, an amino acid from the diet is metabolized by Lactobacillus reuteri to indole-3-aldehyde, which promotes interleukin (IL)-22 production by ILC3s. ILCs have been implicated in protection against different intestinal pathogens: (i) ILC1s protect against Clostridium difficile through interferon (IFN)-γ production, (ii) ILC2s protect against the helminth Nippostrongylus brasiliensis through production of IL-13, which stimulates mucus secretion in goblet cells, (iii) ILC3s produce IL-22, which induces Reg3γ expression in IECs, which in turn kills vancomycin-resistant Enterococcus (VRE). Segmented filamentous bacterium (SFB) induces differentiation of T helper 17 cells (Th17s) through production of serum amyloid A (SAA) by IECs. Th17s confer protection to pathogens through production of IL-22 and IL-17, a cytokine that enhances IgA responses. IgA confers protection against pathogens such as Citrobacter rodentium. Short-chain fatty acids (SCFA), commensal bacteria-derived products from the fermentation of dietary fiber, promote the differentiation of IgA+ B cells and T regulatory cells (Tregs). Clostridiales promote development of Tregs by inducing the synthesis of transforming growth factor-β (TGF-β) by IECs, probably through production of SCFA. In addition, the intestinal microbiota have systemic effects on the immune system: (i) it promotes myelopoiesis, conferring resistance against systemic infection by Listeria monocytogenes, (ii) it primes neutrophils, increasing their killing capabilities against Streptococcus pneumoniae and (iii) it induces systemic IgG responses against Salmonella typhimurium.
Non-immune-derived mechanisms of protection
Inhibitory molecules
The microbiota can confer resistance to pathogens in a direct manner, without the involvement of the immune system (Figure 1). Certain commensal strains can produce and secrete small molecules with bacteriostatic or bactericidal activity, such as bacteriocins or microcins produced by Gram-positive and gram-negative Enterobacteriaceae species, respectively.6, 7, 8 For example, an Enterococcus faecalis strain is capable of clearing vancomycin-resistant Enterococcus (VRE) from the intestinal tract of mice through the expression of a plasmid-encoded bacteriocin.6 Staphylococcus lugdunensis, a human nasal commensal, produces lugdunin, a recently discovered peptide antibiotic that diminishes both skin and nasal colonization by Staphylococcus aureus in rodents.7 On the other hand, production of microcins by the Escherichia coli strain Nissle 1917 reduces intestinal colonization by Salmonella enterica serovar Typhimurium.8 Interestingly, certain pathogens employ the same strategy to favor their own colonization. Listeria monocytogenes secretes a bacteriocin that alters the intestinal microbiota and allows this pathogen to colonize the intestinal tract and disseminate to other organs.9 On the other hand, the intestinal microbiota may confer resistance through the production of short-chain fatty acids (SCFA), a metabolic product derived from the bacterial fermentation of dietary fibers. Indeed, in vitro, SCFA markedly suppress the growth of the pathogenic E. coli strain O157:H7.10
Besides the production of molecules with an effect on pathogen growth, certain commensals produce small molecules that confer resistance to infection through interference with pathogen gene expression. For example, Ruminococcus obeum, a human intestinal commensal, diminishes intestinal colonization of Vibrio cholerae in mice through production of the quorum-sensing signal AI-2. This molecule interferes with the expression of the V. cholerae toxin co-regulated pilus operon, which is required to colonize the human intestinal tract.11 SCFA can also influence the expression of virulence factors. For example, butyrate and propionate, both present in high concentrations in the large intestine, downregulate the expression of genes encoded in S. typhimurium pathogenicity island 1 (SP1), required by this pathogen for invasion of intestinal epithelial cells (IECs).12, 13 In contrast, acetate, present in higher concentrations in the ileum (the primary site for Salmonella cell invasion), promotes the expression of SP1 genes. In this case, microbiota-derived products allow the pathogen to sense its location within the gut and regulate its gene expression accordingly.12
In addition to bacterial-synthetized molecules, host-derived molecules can be metabolized by commensals, resulting in the production of secondary metabolites that play a crucial role in defense against pathogens. This is the case of bile acids, which are synthesized in the liver and secreted as primary bile acids into the intestinal tract. In the presence of an intact microbiota, primary bile acids are converted into secondary bile acids, which can inhibit the growth of Clostridium difficile.4 Disruption of the microbiota by antibiotic treatment reduces the levels of secondary bile acids, allowing the growth of C. difficile vegetative forms. Conversely, after antibiotic treatment, there is an increase in levels of primary bile acids, including taurocholate, which promotes the germination of C. difficile spores.14 Thus, through two different mechanisms involving bile acid conversion, C. difficile infection is enhanced upon antibiotic disruption of commensal communities. Notably, a recent study has identified a key commensal bacterium that confers resistance to C. difficile through bile acid conversion.4 Clostridium scidens, a commensal associated with resistance to C. difficile infections in hospitalized patients, encodes the 7α-hydroxysteroid dehydrogenase enzyme, which is required for the conversion of primary to secondary bile acids. Colonization of antibiotic-treated mice with this commensal increases the level of intestinal secondary bile acids, which promotes mouse survival upon C. difficile challenge.4
Nutrient competition
Besides avoiding pernicious molecules, in order to colonize the intestinal tract a bacterial pathogen must compete for the same nutrient sources with commensal bacterial species that are highly adapted to the gut environment and very efficient at obtaining energy from the diet or utilizing host-derived nutrients. Not surprisingly, a major mechanism by which the microbiota inhibits intestinal colonization by bacterial pathogens is through nutrient competition. This is the case of infections produced by E. coli O157:H7, which can be prevented in mice that have been pre-colonized with two commensal E. coli strains (HS and Nissle 1917).15 Interestingly, when both strains are present, the pathogen is not able to colonize the gut; however, the presence of only one of these two strains is not sufficient to prevent pathogen colonization. Each of these commensal strains can utilize some but not all of the five most important sugars consumed by the pathogen in the gut. Thus, if only one strain is present, the pathogen can still exploit the remaining sugars that have not been consumed by the commensal. However, when both commensal strains are present, all five sugars are depleted, preventing gut colonization by E. coli O157:H7. Similarly, competition for the same nutrients is a mechanism by which the microbiota partially confers protection against Citrobacter rodentium, a natural mouse pathogen used as a model of Enterohemorrhagic E. coli (EHEC) disease. In GFM fed a regular chow, C. rodentium intestinal burden is reduced upon administration of E. coli, but not after inoculation of Bacteroides thetaiotaomicron.16 Like C. rodentium, E. coli consumes simple sugars in order to grow, while B. thetaiotaomicron can catabolize both mono- and polysaccharides. Thus, in mice receiving a regular diet containing both simple sugars and polysaccharides, B. thetaiotaomicron may preferentially use polysaccharides, allowing C. rodentium to obtain energy from available monosaccharides. However, if mice receive a diet consisting exclusively of simple sugars, B. thetaiotaomicron is forced to compete for the same nutrient sources as the pathogen, which diminishes C. rodentium levels.16 Thus, not only the composition of the microbiota, but also diet, influences the outcome of the infectious process.
Consistent with the effect that the microbiota has on resistance to infection through nutrient depletion, antibiotic clearance of commensal bacteria can lead to pathogen expansion through liberation of nutrient sources. For example, administration of cefoperazone to mice promotes C. difficile intestinal colonization.14 This antibiotic alters the murine intestinal microbiome and metabolome, increasing levels of sugars such as mannitol and sorbitol, which can serve as a carbon source for C. difficile.14 On the other hand, both C. difficile and S. typhimurium reach high levels in the intestine upon antibiotic treatment, in part due to a higher availability of the monosaccharide sialic acid.17 Interestingly, this sugar forms part of the glycosylated proteins mucins (the major components of the intestinal mucus layer), and is available to pathogens when liberated by certain commensal encoding sialidases, such as B. thetaiotaomicron.17 Thus, by selectively depleting commensal species that consume certain nutrient sources (i.e. sialic acid), but leaving intact microbes that provide those nutrients (i.e. sialidase encoders), antibiotics could promote the expansion of pathogens. This is also the case for C. difficile competition with commensals that consume succinate, a metabolite derived from the fermentation of dietary carbohydrates by primary fermenters such as B. thetaiotaomicron.18 During antibiotic treatment, succinate becomes available, which promotes intestinal colonization by C. difficile.18
Nutrients derived from the microbiota metabolism can also enhance pathogen colonization by regulating the expression of virulence factors. For example, EHEC can sense fucose, a sugar liberated from mucin by B. thetaiotaomicron.19 EHEC express virulence factors encoded by the LEE pathogenicity island to attach to epithelial cells.20 LEE expression is therefore required close to the epithelium but not on the mucus surface, where free fucose is available. When sensed by EHEC, fucose on the mucus surface represses LEE-encoded genes, therefore avoiding superfluous energy expenditure.19 In line with this, an EHEC mutant with a deletion in the fucose sensor is outcompeted by the wild-type strain.19
In summary, consumption of dietary nutrients by commensals can confer protection to infections, but certain pathogens exploit products derived from the microbiota metabolism to invade the gut.
Innate immune-derived mechanisms of protection
The mucus layer and IECs
The layer of mucus that covers the intestinal tract epithelium can be considered the first line of host defense against pathogens. The major component of the intestinal mucus is Muc2, a glycosylated protein, which is synthesized and secreted by goblet cells.21 Muc2 is vital for protection against enteric pathogens; indeed, deficiency has been shown to increase mortality in mice upon challenge with C. rodentium.22 The intestinal microbiota influences the quantity and properties of intestinal mucus. GFM have a thinner mucus layer in the small but not in the large intestine.23 In addition, the colonic mucus of conventional mice contains higher amounts of Muc2 and, in contrast to GFM mucus, is not penetrable by bacterial size beads.23 The effect that the intestinal microbiota has on mucus synthesis depends in part on the induction of Toll-like receptors (TLRs), which can recognize commensal-derived products.24 Indeed, mice with IECs deficient in myeloid differentiation primary response 88 gene (Myd88), an adaptor protein that mediates TLR signaling, synthetize less Muc2.24
Underneath the mucus lies the intestinal epithelium, which is composed of different cell types, such as enterocytes, paneth cells (cells specialized in producing antimicrobial peptides that reside at the base of the intestinal crypts) and goblet cells.21 The intestinal epithelium constitutes the second barrier separating the intestinal microbial ecosystem from the largely sterile underlying tissue. This layer of cells not only constitutes a physical barrier but is also able to synthetize and secrete antimicrobial peptides, which are essential for inhibiting pathogen colonization and for restraining commensal microbes from coming into direct contact with the epithelium.25, 26, 27, 28 Expression of these antimicrobial peptides is driven by the microbiota.26 Indeed, GFM exhibit diminished paneth cell expression of antimicrobial peptides RegIIIγ, RegIIIβ and defensin Defcr-rs-10.26 Moreover, expression of RegIIIγ, a bactericidal lectin that kills Gram-positive bacteria,29 is reduced upon microbiota depletion with antibiotics, which promotes murine intestinal colonization by the Gram-positive pathogen VRE.30 Microbiota induction of ReIIIγ and RegIIIβ depends on TLR signaling, while Defcr-rs-10 expression is driven by stimulation of nucleotide-binding oligomerization domain (NOD) receptors.26, 27 Besides conferring resistance to pathogens such as VRE, RegIIIγ also prevents overstimulation of the immune system by keeping commensal bacteria 50 μm apart from the small intestinal epithelial surface.28, 31 In addition to conferring protection against bacteria, microbiota induction of antimicrobial peptides also enhances resistance against fungi. Notably, certain components of the microbiota, including B. thetaiotaomicron, confer resistance to Candida albicans by promoting the expression of H1F-1α, a transcriptional regulator that induces the expression of the antimicrobial peptide LL-37, with anti-Candida activity.32
In addition to antimicrobial peptide induction, specific commensal strains can confer resistance to infection by decreasing intestinal permeability to bacterial toxins.33 This is the case of the resistance conferred to EHEC O157:H7, which colonizes the intestinal tract and produces Shiga toxin, which upon translocation to the bloodstream can induce a hemolytic uremic syndrome with fatal consequences.34 Certain Bifidobacterium strains, but not all, increase the survival rate of mice after EHEC infection.33 Notably, Bifidobacterium-protective strains encode an ABC-type sugar transporter that enhances carbohydrate consumption and subsequent production of acetate as a metabolic product.33 Higher acetate levels prevent the reduction in transepithelial electrical resistance induced by EHEC, decrease the translocation of the Shiga toxin from the apical to the basolateral side of colonic epithelial cells and diminish the dissemination of the toxin to the bloodstream and the associated mortality.33
ILCs and myeloid cells
Beneath the epithelium, several specialized innate immune cells are necessary to create an adequate response against intestinal pathogens. Within these cells, ILCs represent the most recently identified arm of the innate immune system, which is crucial for defense against intestinal pathogens.35 ILCs lack antigen-specific receptors, but, interestingly, their cytokine production and transcriptional factors that regulate their development mirror those of the three major T helper cell subsets (Th1, Th2, Th17).35 Based on these features, ILCs have been classified into three groups: ILC1s, which rely on transcriptional factor T-bet or eomesodermin and produce interferon(IFN)-γ ILC2s, which rely on the GATA-3 transcriptional factor and primarily produce interleukin (IL)-5 and IL-13; and ILC3s, which rely on RORγt and produce IL-17 and/or IL-22.35 Earlier studies reported that the gut microbiota influences the development of some but not all ILC types.36, 37, 38 Development of a subset of ILC3s that express NKp46 may be influenced by the microbiota, since it has been reported that GFM contain lower numbers of these cells.36 However, in another study, no differences in the number of murine NKp46+ ILCs were observed in GFM compared with conventional mice.39 Regarding ILC2s, one study reported higher numbers of these cells in the intestine of GFM than in that of conventional mice,37 while intraepithelial ILC1s can develop independently of the intestinal microbiota.38 A very recent study using state-of-the-art techniques such as single-cell RNA-Seq has expanded our knowledge of the diversity of ILCs and the contribution of the microbiota to the maintenance and development of different ILC types.40 Based on transcriptome profiles, this study identified several clusters within each of the ILC types (four ILC1 clusters, four ILC2 clusters and five ILC3 clusters) in mice. Importantly, marked differences were observed between different clusters belonging to the same ILC type. For example, IL-22 expression, a hallmark cytokine of ILC3s, was expressed almost exclusively by one of the five clusters identified within this ILC type. In addition, the authors of the study were able to identify two additional transcriptional ILC states that did not correspond with any of the three canonical ILC types. In order to study the impact of commensal microbes in the different ILC types, a similar transcriptome analysis was performed in GFM or in adult mice in which the microbiota had been depleted using broad-spectrum antibiotics. Notably, the general transcriptional profile of all ILC types was preserved upon antibiotic treatment; however, levels of hundreds of transcripts were significantly altered. When the authors focused on the relative abundance of different ILC types, both GFM and antibiotic-treated mice exhibited a higher proportion of ILC3s and ILC2s (categorized by their transcriptome), while the relative abundance of ILC1s was reduced. Moreover, the transcriptional profiles of ILC1s and ILC2s were seen to become more similar to that of ILC3s upon antibiotic administration, suggesting that ILC expression tends toward an ILC3 profile when microbial stimulation is removed. In summary, the results obtained in the study in question indicate that the microbiota has an impact on both the relative abundance of different ILC types and their expression.40
As indicated above, the three ILC types produce different cytokines and consequently have been implicated in protection against different pathogens. For example, while ILC1s and IFN-γ were found to be critical to the host's defense against C. difficile, ILC3s and IL-22 made a minor contribution to protection.41 On the other hand, ILC3s are important for conferring protection against C. rodentium infection in mice that lack B cells and T cells.42 C. rodentium is a pathogen that colonizes the surface of IECs. Therefore, ILC3s provided protection against this infection may depend on the ability of these cells to produce IL-22, a cytokine that confers protection against C. rodentium through induction of antimicrobial peptide synthesis by IECs.43 On the other hand, ILC2s, through production of IL-13, are key for protection against helminth infections (i.e. Nippostrongylus brasiliensis infections).44 IL-13 secreted by ILC2s stimulates the production of mucus by goblet cells and induces smooth muscle contraction, which is thought to facilitate helminth expulsion.45 Thus, different ILCs confer protection against different pathogens and the microbiota influences ILC functionality.
Recent studies have begun to elucidate the precise mechanisms by which the microbiota influence ILC expression and subsequent pathogen clearance. One such mechanism involves the production of secondary metabolites from diet components.46 ILC3s depend on the aryl hydrocarbon receptor (AhR) for IL-22 production.47 Interestingly, metabolites derived from tryptophan, an amino acid found in the diet, are ligands for AhR.48 Tryptophan's metabolism frequently depends on host-encoded enzymes;48 nevertheless, certain bacteria are also able to metabolize tryptophan and produce ligands for AhR.46 Indeed, a strain of Lactobacillus reuteri can metabolize tryptophan to indole-3-aldehyde (IAld), a molecule with AhR ligand activity.46 Production of IAld by L. reuteri promotes the expansion of ILC3s and production of IL-22, which diminish the burden of C. albicans in the stomach.46
The intestinal microbiota is also required for the proper development and functionality of myeloid cells.49, 50, 51, 52 A recent study demonstrated that GFM possess lower numbers of splenic macrophages, monocytes and neutrophils,49 which was attributed to reduced myelopoiesis in the absence of commensal microbes. Importantly, defects in myelopoiesis undermined protection against systemic L. monocytogenes infection.49 On the other hand, neutrophil activation by the microbiota plays a role in C. difficile infection,50 during which there is translocation of commensal bacteria due to intestinal damage. Notably, bacteria translocation promotes synthesis of the cytokine IL-1β, mainly by neutrophils, which then induces the production of CXCL1, a chemokine that attracts neutrophils to the site of infection.50 Thus, a positive loop occurs in which neutrophils promote their own recruitment to the site of infection, and this positive loop depends on the dissemination of commensal microbes. Furthermore, peptidoglycan, a component of the bacterial cell wall, translocates from the gut into the circulation, priming neutrophils through induction of the NOD1 receptor, which enhances their ability to kill bacteria.51 In other cases, such as Toxoplasma gondii infection, neutrophil activation during infection can be detrimental for the host due to exacerbation of tissue damage.52 In this case, however, the commensal microbiota plays a protective role through induction of a regulatory phenotype in Ly6Chi monocytes, characterized by the expression of prostaglandin E2.52 This molecule suppresses the activation of neutrophils and the pathology associated with T. gondii infection.52 In this way, contrary microbiota effects through induction of different innate immune cells (i.e. activation of pro-inflammatory neutrophils or regulatory monocytes) are required to counteract infections.
Adaptive immune-derived mechanisms of protection
T cells
Microbiota induction of the adaptive arm of the immune system, including B cells and T cells, plays a central role in the defense against intestinal pathogens in the gastrointestinal tract. Studies using GFM colonized with different commensal microbes have demonstrated that certain intestinal bacterial species play a major role in the differentiation of T cells into different subsets, including T helper cells Th1s, Th2s, Th17s and T regulatory cells (Tregs).53, 54, 55 Intestinal microbes can influence the local intestinal pool of T cells, but can also have systemic effects. For example, GFM contain lower numbers of splenic CD4+ cells and a higher ratio of Th2/Th1 cells.54 Reconstitution of GFM with Bacteroides fragilis, an anaerobic bacterium that exclusively colonizes the intestinal tract, is sufficient to restore the number of splenic CD4+ cells and correct the Th2/Th1 imbalance.54
Within the intestinal tract, two major T-cell subsets with very different functions are greatly influenced by commensal microbes: Th17s, which synthetize IL-17 and IL-22 cytokines and play a major role in pathogen protection but also in autoimmune disorders due to their pro-inflammatory potential; and Tregs, which synthetize immunosuppressive cytokines such as IL-10, TGF-β or IL-35, and play a key role in controlling inflammation to avoid excessive tissue damage upon infection.56
Th17s can confer protection against bacterial pathogens through production of IL-22 and subsequent synthesis and secretion of antimicrobial peptides by IECs.42, 43, 56 In this sense, Th17s play a similar protective role against infection to that of ILC3s, which are also a source of IL-22 during infection. Indeed, IL-22 production by ILC3s is not necessary to confer protection against C. rodentium infection when T cells are present.42 However, as previously indicated, in the absence of T cells, mice develop a more pronounced pathology upon infection if ILC3s are also depleted.42 Th17s may also confer protection against infection by increasing, in a IL-17-dependent manner, the intestinal levels of IgA and polymeric Ig receptor, which is required for the translocation of IgA into the intestinal lumen.57
Th17 numbers in the intestinal lamina propria depend on the composition of the intestinal microbiota. Ivanov et al.53 reported that the same mouse strain acquired from different vendors contained a different microbiota, which would influence the number of Th17s in the small intestine. Notably, a single bacterial species, segmented filamentous bacterium (SFB), was responsible for increasing Th17 intestinal numbers.53 Although the mechanisms by which SFB induces the development of Th17s are not fully understood, several studies have started to unravel key aspects of this process.53, 58, 59 SFB induces the production of IL-22 by ILC3s, which subsequently promotes the expression of serum amyloid A (SAA) by IECs.58 SAA promotes the expression of IL-17 by Rorγt+ T cells.58 In addition, SAA appear to condition CD11c+ myeloid cells to produce IL-1β to enhance Th17 differentiation.59
On the other hand, intestinal Tregs play an important role in diminishing the tissue damage caused by the immune response against pathogenic bacteria. For example, GPR15 is a receptor expressed by Tregs that is necessary for their migration to the lamina propria of the large intestine.60 After C. rodentium infection, most mice resolve inflammation and survive. However, mice defective in GPR15, which have lower numbers of Tregs in the gut, suffer increased weight loss, inflammation, tissue damage and mortality.60 Interestingly, the intestinal microbiota induces the expression of GPR15 by Tregs.60 Thus, in this case, the microbiota may protect by promoting migration of Tregs to the site of infection, therefore diminishing the collateral damage derived from the inflammatory response against the pathogen.
The development and functionality of Tregs in the intestinal tract depend on the presence of specific commensal microbes. GFM have a lower quantity of Tregs in the large intestine.55 Notably, administration of a cocktail of bacterial strains from the Clostridia clusters IV, XIVa and XVIII has been shown to restore numbers of Tregs to those observed in conventional mice.55, 61 One mechanism by which Clostridia species may enhance Treg differentiation is through production of SCFA, which several studies have shown to be crucial in Treg differentiation.62, 63, 64 Clostridia species produce SCFA, which, at least in vitro, enhance the expression by IECs of TGF-β, a key cytokine that contributes to the differentiation of Tregs.61 In addition, SCFA may influence Tregs differentiation by inhibiting histone deacetylases and enhancing histone H3 acetylation in the promoter of Foxp3, the master regulator involved in Treg development.63, 64 In addition to SCFA, bacterial-derived molecules that activate TLR receptors can also play a role in Tregs differentiation. Through TLR signaling in macrophages, commensal microbes induce the production of IL-1β, which acts on ILC3s to promote the release of the granulocyte–macrophage colony-stimulating factor (Csf2).65 This cytokine promotes recruitment to the colon of macrophages and DCs that produce factors involved in Treg differentiation, such as retinoic acid and IL-10.65 Consequently, mice deficient in Csf2 have lower number of Tregs in the intestine.65 Thus, different commensal-derived molecules, including TLR ligands and SCFA, are required for the adequate development of Tregs in the gut.
B cells
Production of IgA by B cells is key for the host to control infections on mucosal surfaces, including the gastrointestinal tract.66 Several studies have demonstrated that the intestinal microbiota influences B-cell development and antibody production. Early studies using GFM showed that the presence of gut commensal microbes increases the intestinal levels of plasma cells producers of IgA, but were unable to determine the exact mechanisms by which the microbiota influences IgA production.67 Notably, a recent study has shown a clear relation between SCFA derived from microbiota metabolism, B-cell differentiation and IgA production.68 Kim and co-workers demonstrated that SCFA directly affect the differentiation of B cells into IgA-producing plasma cells by increasing the expression of several genes related to B-cell differentiation, including genes necessary for IgA synthesis. Further in vitro experiments suggested that SCFA might influence IgA production by inhibiting histone deacetylases and enhancing histone acetylation of key regulatory regions, including Igα class-switch regions. In concordance with their in vitro results, mice receiving a diet with high levels of fiber (a major source of microbial production of SCFA) had higher numbers of intestinal IgA+ B cells than those receiving a low-fiber diet. In addition, upon C. rodentium infection, mice fed with the high-fiber diet produced more IgA pathogen-specific antibodies and were more resistant to pathogen intestinal colonization than those on a low-fiber diet. Moreover, supplementation of the low-fiber diet with SCFA increased the murine IgA response against the pathogen. Thus, taking into account the effects on both Treg and B-cell differentiation, SCFA derived from the microbiota's metabolism can be considered key molecules in the regulation of adaptive immune responses in the gut.
Besides influencing IgA production, certain members of the microbiota can also affect systemic IgG antibody responses.69 Zeng et al.69 showed that specific commensals, including members of the Enterobacteriaceae family such as E. coli, are able to disseminate from the gut in homeostatic conditions and induce the synthesis of IgG-specific antibodies. A major antigen recognized by IgG was murein lipoprotein, a highly conserved Gram-negative outer membrane protein. Importantly, IgG antibodies against commensal E. coli murein lipoprotein also targeted Salmonella, which conferred protection against systemic infection produced by this pathogen.69
Pathogen exploits immune responses
Although the host has developed numerous strategies to avoid infections, some pathogens, such as Salmonella, are able to outcompete commensal microbes by exploiting host immune responses. A pioneer study by Stecher et al.70 demonstrated that S. typhimurium promotes intestinal inflammation in a way that alters the microbiota's composition and enhances pathogen intestinal colonization. Stecher et al.70 showed that an avirulent S. typhimurium mutant is outcompeted by the microbiota unless inflammation is provided through co-infection with the wild-type strain or by infecting hosts that spontaneously develop colitis, such as IL-10−/− mice. Following this work, several other studies have thrown light on the specific mechanisms that allow pathogens to exploit inflammation to their own benefit. Some involve the consumption of nutrients made available during inflammation that can be utilized by the pathogen but not by the microbiota. For example, colonic bacteria produce high amounts of hydrogen sulfide, which is very toxic for the host and is therefore converted into thiosulfate in the intestinal mucosa.71 During inflammation, oxygen radicals are produced, which convert thiosulfate into tetrathionate.72 This molecule can be used as a terminal electron acceptor to obtain energy from ethanolamine by S. typhymurium, but not by competing commensals.72, 73 Thus, through generation of tetrathionate, Salmonella-induced inflammation confers the pathogen a metabolic advantage over the microbiota. Similarly, during inflammation, production of superoxide and nitric oxide radicals lead to the generation of nitrate.74 This molecule confers a growth advantage to E. coli, since it can utilize nitrate as a terminal electron acceptor for anaerobic respiration, as opposed to obligate anaerobic members of the microbiota.75 On the other hand, Salmonella benefits from the inflammatory state through competition for iron, which is crucial for bacterial growth. During Salmonella infection, IL-22 is highly expressed, which induces the synthesis of lipocalin, an antimicrobial peptide that sequesters siderophores expressed by bacteria to uptake iron.76, 77 For Enterobacteriaceae commensals, inflammation creates an environment that restricts their growth. However, Salmonella encodes salmochelin, a siderophore that cannot be bound by lipocalin.78 Thus, under limited iron conditions imposed by inflammation, Salmonella has a competitive advantage over other Enterobacteriaceae competitors.76 Consistently, this advantage is lost, and Salmonella colonization is restricted in mice lacking IL-22 that express low levels of lipocalin.77
Therapeutic approaches to limit infections based on the microbiota
In the previous sections, we have described some of the mechanisms by which the microbiota can confer resistance to infection. The accumulated knowledge can now be applied for designing novel therapeutic approaches based on commensal microbes or their products, in order to restrict intestinal colonization by pathogens. Indeed, therapies based on the microbiota have started to be applied in the clinical setting with extremely high success. This is the case of fecal transplants utilized for treating C. difficile infections. C. difficile colonizes the intestinal tract of patients with an altered microbiota due mainly to previous antibiotic exposure. Some of these patients, upon C. difficile treatment, develop recurrent infections due to major and permanent changes in their microbiota.79 Thus, reconstitution with an unaltered microbiota could in principle facilitate the elimination of the pathogen. This was shown to be the case in one trial in which administration of feces from healthy donors to patients with recurrent C. difficile infection cured the disease in 93% of subjects, while a 30% success rate was obtained with vancomycin administration.80 Following this pioneer study, multiple studies have confirmed the astonishing efficacy of fecal transplants for the treatment of recurrent C. difficile infection.79 Fecal transplant could also be used to treat other infectious pathogens, such as VRE, whose intestinal colonization is restricted by the microbiota. This has indeed been demonstrated in mice treated with antibiotics in which VRE was completely cleared after administration of fecal pellets from untreated mice.81 In addition, lower relative VRE abundance was observed upon fecal transplantation to a patient infected with C. difficile and heavily colonized with VRE.82 Nevertheless, further studies should be performed to confirm the capacity of fecal transplants to eliminate VRE. Although such transplants seem to be very efficient in eliminating certain pathogens, they are not free of risk. Indeed, taking into account that the microbiota has been associated with other diseases, including obesity and cancer,83 one cannot rule out negative effects upon introduction of hundreds of new commensal microbes into the intestinal tract, even if donors are carefully selected. Therefore, alternative approaches, including the administration of specific microbes or bacterial-derived molecules, are desirable in order to diminish the appearance of negative side effects.
Administration of specific microbes has been proven to confer protection against infections. A cocktail of 10 commensals, including anaerobic and facultative aerobic bacteria, was shown to be effective at curing recurrent C. difficile infection in patients.84 However, only five patients were included in the study in question, and so additional studies with larger cohorts are necessary in order to validate this approach. A cocktail of six phylogenetically diverse commensals was shown to re-establish a healthy microbiota and clear C. difficile in mice.85 Moreover, as described above, administration of C. scidens diminishes the intestinal C. difficile burden and increases survival in mice.4 Regarding other intestinal pathogens, administration of the E. coli strain Nissle 1917 diminishes murine intestinal levels of S. typhimurium through nutrient competition for iron.86 E. coli strain Nissle 1917 encodes a siderophore that acquires iron in the presence of lipocalin, therefore attenuating the competitive advantage of Salmonella during inflammation.86 Thus, administration of single or multiple bacteria could be an efficient method for conferring resistance against infections.
As an alternative to the ‘probiotic'-based approach, direct administration of bacterial-derived products is proven to be effective in restricting intestinal colonization by pathogens. For example, administration of thuricin CD, a narrow spectrum bacteriocin produced by Bacillus thuringiensis, considerably reduced C. difficile levels in an in vitro model of distal colon, without altering the microbiota.87 On the other hand, inoculation of bacterial-derived products that activate certain components of the immune system has also been tested with success. As an example, antibiotic therapy in mice diminishes Reg3γ expression and promotes VRE colonization of the intestinal tract.30 Oral administration of bacterial lipopolysaccharide restores Reg3γ intestinal levels through stimulation of TLR4, which enhances VRE killing.30 In addition, oral administration of resiquimod, a synthetic ligand for the TLR7 receptor, diminishes VRE intestinal density in antibiotic-treated mice.88 In this case, TLR7 stimulation on CD11c+ DCs induces IL-23 expression which promotes IL-22 synthesis by ILCs and subsequent induction of Reg3γ expression.88 Similarly, systemic stimulation of TLR5 by flagellin promotes Reg3γ expression and limits intestinal colonization by VRE and C. difficile in antibiotic-treated mice.89, 90 In this way, deficits in the immune response caused by antibiotic disruption of the microbiota can be restored by administration of bacterial products.
Conclusions
Commensal microbes that inhabit the gastrointestinal tract are essential for the proper development and functionality of multiple immune cell types. The intestinal microbiota can have a local effect on intestinal immune cell populations, including the production of cytokines by ILCs or Th17s or the differentiation of Tregs. Consequently, through its induction of the immune system, the microbiota is crucial for conferring protection against intestinal pathogens, including C. rodentium, C. difficile or VRE. Our knowledge about the immune populations that reside in the intestinal tract is rapidly expanding. For example, novel subtypes of ILCs have recently been discovered, although their role in defense against infections and how commensal microbes influence their functionality has yet to be defined. It is expected that novel interactions between the intestinal immune system, the microbiota and the pathogen will be discovered in the next few years.
Commensal microbes that colonize the gut can also have systemic effects on immunity, including the activation of neutrophils, induction of IgG responses and enhancement of myelopoiesis. Although these systemic effects have been observed in mice, a recent study has shown that the microbiome composition also influences the cytokine production potential of blood cells in humans.91
The key commensal bacteria involved in the induction of the different components of the immune system are now starting to be identified, including those that promote Th17 or Treg differentiation. One of the difficulties encountered by this area of research has been the impossibility of cultivating a major fraction of the microbiota. However, recent research has greatly expanded the number of cultivable human and murine intestinal microbiota species.92, 93, 94 Subsequently, future research using mouse models may elucidate the impact of these novel cultivable bacterial species on the immune system and defense against infections.
The molecular mechanisms involved in the interaction between commensals and immune responses against pathogens are starting to be understood. In some cases, bacterial-encoded molecules have a direct effect on the immune defense against pathogens through stimulation of innate immune receptors. In other cases, products derived from the microbiota metabolism of dietary components represent the effector molecules that influence immune responses, such as SCFA that promote the development of Tregs. In the latter case, the type of diet consumed by the host can play an important role in the defense against infections, a field that requires further investigation.
On the other hand, the microbiota can confer protection to pathogens through mechanisms that do not require the induction of the immune system. These mechanisms include the production of molecules that inhibit the growth of the pathogen or that interfere with their colonization capabilities. High-throughput sequencing techniques have led to the generation of a vast amount of sequencing data from commensal microbes. Careful analysis of a subset of this data has begun to reveal novel commensal-encoded antibiotics that are effective against pathogens.95 Further analysis of new subsets of metagenomic sequences, in combination with in vitro and in vivo experiments, should therefore expand our knowledge of bacterial-derived molecules that can directly influence pathogen colonization capabilities. Competition for nutrients has also been shown to play a major role in resistance against infections. In this case, a greater understanding of the biology of commensals and pathogens is required to identify novel mechanisms of protection. For example, little is known about the genes required for intestinal colonization and exploitation of nutrients by some of the pathogens that colonize the intestinal tract, such as VRE. Further studies in this area are necessary in order to fully clarify how certain pathogens are able to invade the intestinal tract and the mechanisms by which key members of the microbiota outcompete them.
From a translational point of view, it is expected that the acquired knowledge will be applied to prevent or cure infections in patients. Indeed, we have discussed a few examples of novel strategies based on commensal bacteria or derived products that appear to enhance resistance to infection. Further validation in human subjects of these new therapies, and of those that are sure to emerge from the knowledge being generated rapidly in this field, will facilitate the treatment of infections in the near future. These novel microbiota-based therapies will be especially relevant for multidrug-resistant pathogens like VRE, for which most available therapies have become ineffective.
Acknowledgments
This work was supported by the InfectERA-ERANET-Acciones complementarias grant (PCIN-2015-094) and grants from the Spanish Ministerio de Economía y Competitividad (SAF2014-60234-R and SAF2014-62369-EXP) and from the Generalitat Valenciana (PROMETEO/2016/122) to CU; and an F.P.I. postgraduate fellowship from the Spanish Ministerio de Economía y Competitividad to SI.
Footnotes
The authors declare no conflict of interest.
References
- Morgan XC, Huttenhower C. Meta'omic analytic techniques for studying the intestinal microbiome. Gastroenterology 2014; 146: 1437–1448.e1. [DOI] [PubMed] [Google Scholar]
- Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120: 4332–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac S, Scher JU, Djukovic A, Jiménez N, Littman DR, Abramson SB et al. Short- and long-term effects of oral vancomycin on the human intestinal microbiota. J Antimicrob Chemother 2016; 72: 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2014; 517: 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Pamer EG. Antibiotics, microbiota, and immune defense. Trends Immunol 2012; 33: 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 2015; 526: 719–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016; 535: 511–516. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi M, Nuccio S-P, Liu H, Hernandez D, Vu CT, Takahashi AA et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 2016; 540: 280–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quereda JJ, Dussurget O, Nahori M-A, Ghozlane A, Volant S, Dillies M-A et al. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc Natl Acad Sci USA 2016; 113: 5706–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Suzuki M, Morishita Y. Influence of intestinal anaerobes and organic acids on the growth of enterohaemorrhagic Escherichia coli O157:H7. J Med Microbiol 2002; 51: 201–206. [DOI] [PubMed] [Google Scholar]
- Hsiao A, Ahmed AMS, Subramanian S, Griffin NW, Drewry LL, Petri WA et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 2014; 515: 423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon SD, Maurer R, Suyemoto M, Altier C. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol 2002; 46: 1451–1464. [DOI] [PubMed] [Google Scholar]
- Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A et al. Butyrate specifically down-regulates Salmonella pathogenicity Island 1 gene expression. Appl Environ Microbiol 2006; 72: 946–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 2014; 5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS ONE 2013; 8: e53957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 2012; 336: 1325–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013; 502: 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 2014; 16: 770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG et al. Fucose sensing regulates bacterial intestinal colonization. Nature 2012; 492: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott SJ, Wainwright LA, McDaniel TK, Jarvis KG, Deng YK, Lai LC et al. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol 1998; 28: 1–4. [DOI] [PubMed] [Google Scholar]
- Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 2014; 14: 141–153. [DOI] [PubMed] [Google Scholar]
- Bergstrom KSB, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog 2010; 6: e1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Jakobsson HE, Holmén-Larsson J, Schütte A, Ermund A, Rodríguez-Piñeiro AM et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 2015; 18: 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz AL, Rogier EW, Weber CR, Shen L, Cohen DA, Fenton LA et al. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol 2012; 5: 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl K, Plitas G, Schnabl B, Dematteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII and protect mice against intestinal Listeria monocytogenes infection. J Exp Med 2007; 204: 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA 2008; 105: 20858–20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Núñez G et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 2005; 307: 731–734. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 2011; 334: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006; 313: 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 2008; 455: 804–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonen LMP, Stolte EH, Jaklofsky MTJ, Meijerink M, Dekker J, Van Baarlen P et al. REG3γ-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol 2014; 7: 939–947. [DOI] [PubMed] [Google Scholar]
- Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X et al. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med 2015; 21: 808–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011; 469: 543–547. [DOI] [PubMed] [Google Scholar]
- Karpman D, Loos S, Tati R, Arvidsson I. Haemolytic uraemic syndrome. J Intern Med 2016; 281: 123–148. [DOI] [PubMed] [Google Scholar]
- Bostick JW, Zhou L. Innate lymphoid cells in intestinal immunity and inflammation. Cell Mol Life Sci 2015; 73: 237–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol 2009; 10: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature 2014; 516: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 2013; 38: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol 2011; 13: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 2016; 166: 1231–1246.e13. [DOI] [PubMed] [Google Scholar]
- Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Sušac B et al. Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe 2015; 18: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin LC, Girard-Madoux MJH, Seillet C, Mielke LA, Kerdiles Y, Fenis A et al. Complementarity and redundancy of IL-22-producinginnate lymphoid cells. Nat Immunol 2015; 17: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 2008; 14: 282–289. [DOI] [PubMed] [Google Scholar]
- Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TKA et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 2010; 464: 1367–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med 2015; 21: 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013; 39: 372–385. [DOI] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo X, Chen Z-ME, Fish K, Fu Y-X et al. The aryl hydrocarbon receptor regulates gut immunity through modulation ofinnate lymphoid cells. Immunity 2012; 36: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011; 478: 197–203. [DOI] [PubMed] [Google Scholar]
- Khosravi A, Yáñez A, Price JG, Chow A, Merad M, Goodridge HS et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 2014; 15: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kamada N, Jiao Y, Liu MZ, Nùñez G, Inohara N. Protective role of commensals against Clostridium difficile infection via an IL-1-mediated positive-feedback loop. J Immunol 2012; 189: 3085–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 2010; 16: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger JR, Wohlfert EA, Fuss IJ, Bouladoux N, Askenase MH, Legrand F et al. Inflammatory monocytes regulate pathologic responses to commensals during acute gastrointestinal infection. Nat Med 2013; 19: 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005; 122: 107–118. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature 2016; 535: 75–84. [DOI] [PubMed] [Google Scholar]
- Cao AT, Yao S, Gong B, Elson CO, Cong Y. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol 2012; 189: 4666–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 2015; 163: 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S et al. Th17 cell induction by adhesion of microbes tointestinal epithelial cells. Cell 2015; 163: 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SV, Xiang WV, Kwak C, Yang Y, Lin XW, Ota M et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science 2013; 340: 1456–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H et al. induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500: 232–236. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504: 446–450. [DOI] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013; 504: 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 2014; 343: 1249288–1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity 2008; 28: 740–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MC, Ducluzeau R, Guy-Grand D, Muller MC. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun 1978; 21: 532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 2016; 20: 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M et al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 2016; 44: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. Plos Biol 2007; 5: 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt MD, Furne J, Springfield J, Suarez F, DeMaster E. Detoxification of hydrogen sulfide and methanethiol in the cecal mucosa. J Clin Invest 1999; 104: 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 2010; 467: 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA 2011; 108: 17480–17485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov 2007; 6: 662–680. [DOI] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013; 339: 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio S-P, Paixao TA et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 2009; 5: 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 2014; 40: 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Lin H, Zhou L, Yu Y, Abergel RJ, Liu DR et al. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc Natl Acad Sci USA 2006; 103: 16502–16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CR, Kahn S, Kashyap P, Laine L, Rubin D, Atreja A et al. Update on fecal microbiota transplantation 2015: indications, methodologies, mechanisms, and outlook. Gastroenterology 2015; 149: 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368: 407–415. [DOI] [PubMed] [Google Scholar]
- Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun 2013; 81: 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stripling J, Kumar R, Baddley JW, Nellore A, Dixon P, Howard D et al. Loss of vancomycin-resistant Enterococcus fecal dominance in an organ transplant patient with Clostridium difficile colitis after fecal microbiota transplant. Open Forum Infect Dis 2015; 2: ofv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell 2012; 148: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvede M, Rask-Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet 1989; 1: 1156–1160. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 2012; 8: e1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H et al. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 2013; 14: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea MC, Dobson A, O'Sullivan O, Crispie F, Fouhy F, Cotter PD et al. Effect of broad-and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci USA 2011; 108: 4639–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abt MC, Buffie CG, Sušac B, Becattini S, Carter RA, Leiner I et al. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med 2016; 8: 327ra25–327ra25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis 2010; 201: 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarchum I, Liu M, Lipuma L, Pamer EG. Toll-like receptor 5 stimulation protects mice from acute Clostridium difficile colitis. Infect Immun 2011; 79: 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 2016; 167: 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol 2016; 1: 16131. [DOI] [PubMed] [Google Scholar]
- Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD et al. Culturing of ‘unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature 2016; 533: 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J-C, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol 2016; 1: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Donia MS, Cimermancic P, Schulze CJ, Brown LCW, Martin J, Mitreva M et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 2014; 158: 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]