Abstract
A high-resolution structure of the eukaryotic ribosome has been determined and has led to increased interest in studying protein biosynthesis and regulation of biosynthesis in cells. The functional complexes of the ribosome crystals obtained from bacteria and yeast have permitted researchers to identify the precise residue positions in different states of ribosome function. This knowledge, together with electron microscopy studies, enhances our understanding of how basic ribosome processes, including mRNA decoding, peptide bond formation, mRNA, and tRNA translocation and cotranslational transport of the nascent peptide, are regulated. In this review, we discuss the crystal structure of the entire 80S ribosome from yeast, which reveals its eukaryotic-specific features, and application of X-ray crystallography of the 80S ribosome for investigation of the binding mode for distinct compounds known to inhibit or modulate the protein-translation function of the ribosome. We also refer to a challenging aspect of the structural study of ribosomes, from higher eukaryotes, where the structures of major distinctive features of higher eukaryote ribosome—the high-eukaryote–specific long ribosomal RNA segments (about 1MDa)—remain unresolved. Presently, the structures of the major part of these high-eukaryotic expansion ribosomal RNA segments still remain unresolved.
This article is part of the themed issue ‘Perspectives on the ribosome’.
Keywords: yeast ribosome, crystal structure, inhibitors
1. Introduction
The ribosome is a ribonucleoprotein assembly that is found in all living cells and translates the genetic code into proteins. Recent progress in ribosomal structural biology has included X-ray structure determinations and cryo-electron microscopy (EM) studies, which are based on previous knowledge of individual ribosomal components, such as ribosomal RNA, ribosomal proteins, ribosomal subunits and ribosome complexes in solution [1,2]. The shape of the bacterial ribosome and its non-symmetric ribosomal subunits was first reconstituted from negatively stained EM images in the laboratories of Vasiliev & Lake [3,4]. Ribosomes from bacteria and archaea consist of a large (50S) and a small (30S) subunit, which together constitute an approximately 2.5 megadalton (MDa) 70S ribosome. The eukaryotic counterparts are the 60S and 40S subunits and the 80S ribosome, which range in size from 3.5 MDa in lower eukaryotes to 4.0 MDa in higher eukaryotes. Many key components of the ribosome are conserved across the three kingdoms of life (Bacteria, Archaea and Eukarya), which highlights their importance in the fundamental process of protein biosynthesis [5]. Protein synthesis has been intensely studied during the last five decades, but for most of that time, the detailed three-dimensional structure of the ribosome remained unknown. Cryo-electron microscopy and single-particle analysis produced the first direct visualizations of the bacterial ribosome in different functional states [6–9]. However, it was not until the X-ray crystallographic structures of the entire 70S ribosome (as well as structures of the individual 30S and 50S subunits) began to emerge that accurate atomic models became available [10–16]. Efforts in ribosome crystallography started early with the crystallization of 50S ribosomal subunits isolated from Bacillus stearothermophilus and Haloarcula marismortui [17–19] and the 30S subunit as well as the full 70S ribosome isolated from Thermus thermophilus [20,21]. The first crystal structures of the 30S subunit from T. thermophilus and the 50S subunit from H. marismortui have been determined and were used to interpret X-ray electron density maps of the full ribosome from T. thermophilus [14–16]. Later, a 3.5 Å resolution crystal structure of the 70S ribosome from Escherichia coli was reported [22]. X-ray crystallography of individual ribosomal subunits and full ribosomes have been used for modelling and the study of ribosome function through complexes with functional ligands, ligand analogues and antibiotics, which has been summarized in several review articles [23,24].
Over the last decade, remarkable advances have been made in full-ribosome crystallography, to the extent that it is now possible to obtain a medium- or high-resolution structure of not only vacant ribosomes but also ribosomal complexes with key functional ligands, such as messenger RNA (mRNA), transfer RNAs (tRNA) and various protein translocation factors [16,25–31]. These structural studies can help to explain the mechanism of tRNA binding in the presence of elongation factor Tu [32], the processes of mRNA decoding [30,33–35] and the mechanism of GTP hydrolysis [36] as well as translocation [31,37], termination [38–45] and ribosomal recycling [46,47]. Crystallography of full ribosome complexes can also be used for cotranslational modification studies [48] and studies of translational regulation [49–51]. Until 2010, only studies concerning the X-ray crystal structures of the bacterial ribosome were available, because efforts to elucidate the structure of the eukaryotic ribosome remained unsuccessful.
Crystal structures of the eukaryotic ribosome from Saccharomyces cerevisiae were the first to be successfully determined at 4.2 Å and later at 3.0 Å resolution, and significantly increased our understanding of protein synthesis and regulation of protein synthesis in cells [52,53].
Later, the 40S and 60S ribosomal subunits from a eukaryotic organism (Tetrahymena thermophila) were successfully crystallized with their protein factors, and the complex structures were determined at 3.8 and 3.6 Å resolution, respectively [54,55].
Crystal structures of ribosome complexes also help researchers interpret lower-resolution data from cryo-EM image reconstructions and can provide a more thorough understanding of ribosomal complexes and their functions. For example, this approach has been used in investigations of the translocation mechanism [56–58] and protein transport [59,60].
2. Structure of the eukaryotic 80S ribosome
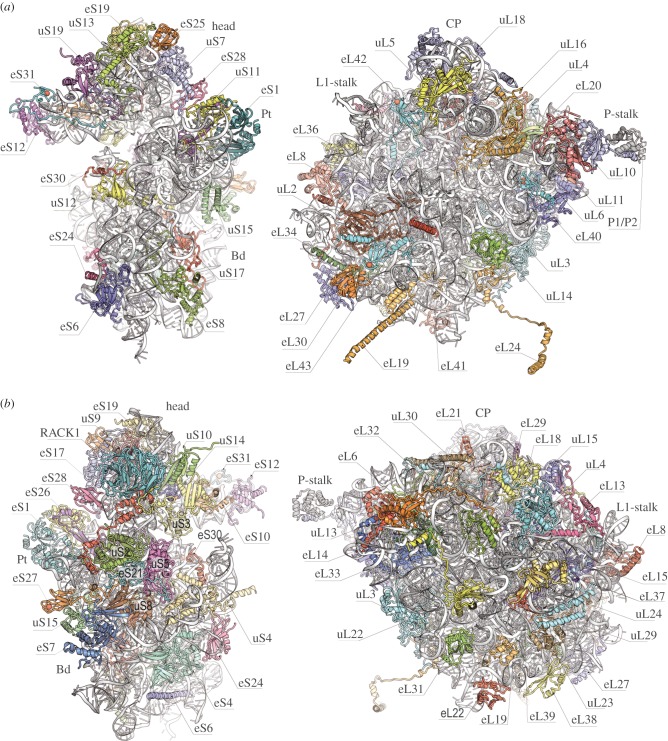
The 80S ribosome is an asymmetric assembly with 80 different proteins and four RNA chains (figure 1). Each ribosomal component has a single copy in the ribosome, except for P-stalk proteins, which have four copies. Experiments also demonstrated that the bacterial and eukaryotic ribosomes share a common structural core comprising 34 conserved proteins (15 in the small subunit and 19 in the large subunit) and approximately 4400 RNA bases, which together form the major functional centres of the ribosomes, including the decoding site, peptidyl transferase centre (PTC), and the tRNA-binding sites [61].
Figure 1.
Architecture of the ribosome subunits (40S subunit on the left site and 60S subunit on the right site) with new protein nomenclature. Ribosome proteins from all three domains (bacteria, archaea and eukaryotes) have the prefix ‘u’ (universal). Bacteria-specific proteins have the prefix ‘b’ (bacterial). Eukaryote-specific proteins have the prefix ‘e’ (eukaryote). (a) Interface view of the 60S and 40S subunits. Landmarks include the head, body (Bd) and platform (Pt) of 40S as well as the central protuberance (CP), L1-stalk and P-stalk of 60S. (b) Solvent-side view of the 60S and 40S subunits.
The 80S ribosome of yeast contains 46 eukaryote-specific proteins (18 in the 40S subunit, 28 in the 60S subunit) and extensions as well as insertions in most of the core proteins. The rRNA also contains several extensions in its conserved chains, with a total length of 900 bases or more [5]. Most of these rRNA and protein moieties envelop the core from the solvent side and are accessible for potential interactions with molecular partners, such as translation factors and chaperone proteins. In eukaryotes, the size of the 80S ribosome varies within the 0.5 MDa range, which is largely attributed to insertions in the RNA expansion segments ES7 L, ES15 L, ES27 L and ES39 L in 25S–28S rRNA (figure 2b). In a few cases, ribosomes may contain either fewer or additional ribosomal proteins [5].
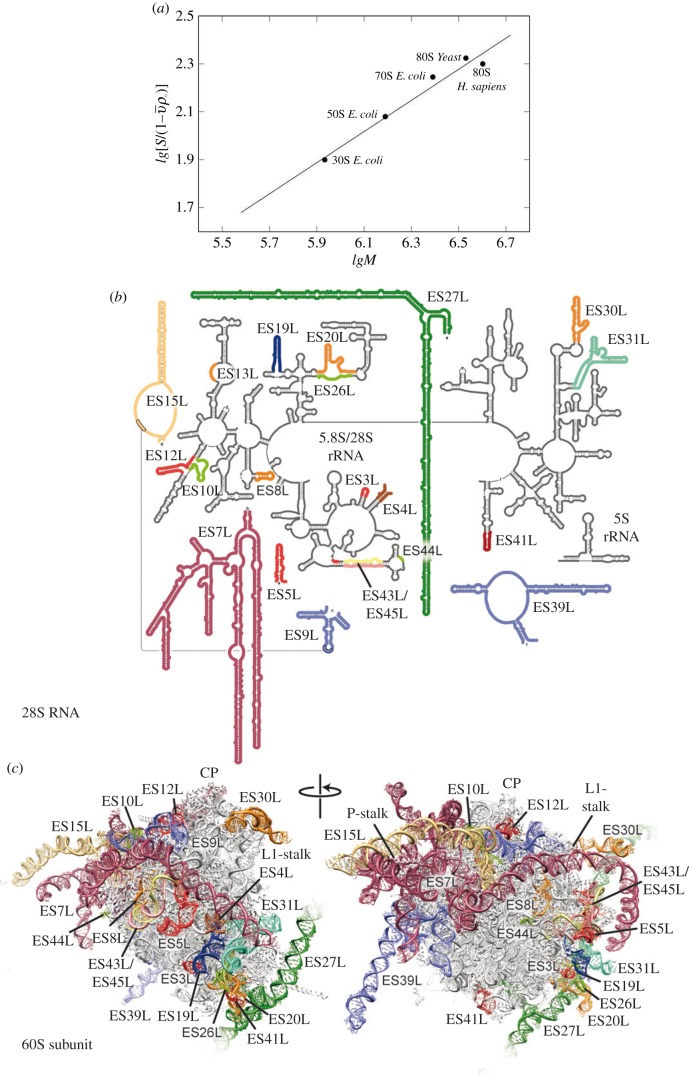
Figure 2.
Dependence of the sedimentation coefficient of the ribosome particle on molecular weight (a). Graphic kindly provided by S. Agalarov. Secondary structure of the human 28S RNA with coloured expansion segments (b). Molecular models of the 60S subunits from H. sapiens with ES coloured as in (b). Landmarks include the central protuberance (CP), L1-stalk and P-stalk. Figure kindly provided by D. Wilson.
The 40S ribosomal small subunit has structural landmarks known as the ‘head’, ‘body’, ‘platform’ and ‘beak’ (figure 1). The names of the structural domains and information about the ribosomal functional sites came from the crystal structure analysis of bacterial ribosome functional complexes [25,28]. The mRNA-binding sites and the three tRNA-binding sites (A, P and E) are located on the subunit interface. The structure showed that mRNA enters through a tunnel located between the head and the shoulder and wraps around the neck of the 40S subunit. The mRNA exit site (5′ end of the mRNA) is located between the head and the platform. The decoding centre of the small subunit, where the codon and anticodon are paired to ensure fidelity in mRNA decoding, is located on the surface of the interface. After comparing the overall structures, it was evident that there are extensive differences between eukaryotes and bacteria on the solvent side of the small ribosomal subunit. These differences are directly correlated with the considerably more complex pathway of translation initiation that exists in eukaryotic cells.
The large ribosomal subunit has an overall crown-like shape, which includes the central protuberance, L1-stalk and the P-stalk (figure 1). Located on the interface side of the large ribosomal subunit are the three (A, P and E) tRNA-binding sites and the PTC where peptide bond formation is catalysed. This PTC is adjacent to the entrance of a tunnel, along which nascent proteins progress before they emerge from the ribosome on the solvent side. The overall absence of bacteria- and eukaryote-specific moieties in the central regions of both the subunit's solvent and interface sides is consistent with the universally conserved functions of these areas. This conservation is seen at the PTC on the intersubunit surface, which is relatively devoid of bacteria- and eukaryote-specific moieties, as well as around the peptide tunnel on the solvent side, which is used for ribosomal association with membranes during protein synthesis. There are, however, important structural differences between the large subunits in bacteria and yeast, such as differences in the organization of the peptide tunnel and the surrounding area, that can be understood in terms of functional divergence.
3. New nomenclature for ribosomal proteins
To facilitate comparison between ribosomes from different species, a nomenclature system based on the names of protein families has been adopted (figure 1) [62]. Because the ribosomal proteins from Escherichia coli were the first to be isolated and fully sequenced, their archaeal and eukaryotic homologues were assigned E. coli names. Proteins found in ribosomes from all three domains (bacteria, archaea and eukaryotes) are given the prefix ‘u’ (for universal), which is followed by their E. coli names. Bacterial proteins without eukaryotic homologues are designated using the prefix ‘b’ (for bacterial). Eukaryotic proteins without bacterial homologues have the letter ‘e’ before the protein name [62].
4. Expansion segments
The ribosomal RNA expansion elements are located predominantly on the periphery of the solvent-exposed sides of both subunits of the eukaryotic ribosome. The interface between ribosomal subunits as well as the area around the mRNA entrance and the polypeptide exit tunnel is highly conserved and contains very few expansion segments and eukaryote-specific proteins. Structural information about expansion segments were reported in the crystal structure of the yeast 80S ribosome and the cryo-EM structure of the Drosophila and human 80S ribosomes [52,63]. All 30 expansion segments were interpreted as well as nine expansion segments of the 40S subunit and 21 expansion segments of the 60S subunit. The 28S RNA expansion segments are shown in figure 2b. Although human and Drosophila contain a set of expansion segments similar to yeast, their expansion segments are generally much longer. For example, ES3S, ES7 L, ES9 L, ES15 L, ES27 L and ES39 L in yeast are approximately 110, 200, 70, 20, 160 and 140 nucleotides, respectively, and the same segments in humans are longer by 50, 670, 40, 170, 550 and 100 nucleotides, respectively. Human ribosomal RNA expansion segments ES3S, ES6S, ES7 L, ES15 L, ES27 L, ES30 L and ES39 L could only be partially interpreted in the cryo-EM reconstructions owing to their unstructured flexibility (i.e. the tentacle-like shape of ES shown in figure 2c). Comparison of the sedimentation coefficients of the bacterial ribosomal 50S and 30S subunits, the bacterial 70S ribosome, the yeast 80S ribosome and the human ribosome showed a partially unfolded structure of the human ribosome. The experimental sedimentation coefficients of the human ribosome and the yeast ribosome were 78S and 80S, respectively (figure 2a). However, the molecular weight of the human ribosome is approximately 500 000 Da more than the yeast ribosome owing to the longer expansion segments of the 28S RNA. A partially unfolded structure of the human ribosome expansion segments made the crystallization of this ribosome difficult and made other structure functional studies challenging to accomplish.
The importance of the folded compact ribosome structure was shown in yeast ribosome expansion segments. ES6S is approximately 200 nucleotide long in the small ribosomal subunit [52]. Research showed that the expansion segment emerges on the solvent side of the platform, where several eukaryote-specific proteins, including a 60 amino acid–long alpha-helical extension of the C-terminus of protein eL19, envelop it. ES6S then extends one of its two long arms in the direction of the shoulder, where it interacts with protein uS8. The second long arm of this expansion segment runs down towards the bottom of the small subunit. The tip of the second arm is located approximately 120 Å away from the tip of the first arm. ES6S is in contact with the ribosomal components that form part of both the exit and entry sites of the mRNA. Therefore, it seems possible that ES6S is involved in translation initiation as a docking surface for factors that participate in activities on both the mRNA exit and entry sites [64].
5. Intersubunit bridges
The importance of intersubunit bridges is evident because they maintain communication pathways between the small and large subunits during protein synthesis. During translation, the ribosome undergoes global conformational rearrangements that are required for mRNA and tRNA translocation, termination and other processes. These changes involve intersubunit rotation and swiveling of the head domain of the small subunit. The interactions between the ribosomal subunits change with each of these rearrangements and have a dynamic composition. The 80S ribosome model derived from crystals captured the ribosome in a rotated state.
Several eukaryote-specific bridges were visualized in low-resolution cryo-EM studies of the yeast ribosome [65,66]. Our model, at 3.0 Å, provided a more accurate and detailed view of the molecular components involved in these contacts between ribosomal subunits. There are seven bridges in the ribosomal core as well as a few bacteria- and eukaryote-specific bridges [16,22,26,52]. In virtually all of the additional bridges, nearly all of the participating components on both subunits are eukaryote-specific. In contrast to bacteria, proteins play the dominant role in forming eukaryote-specific bridges [16]. Eukaryote-specific bridges are located on the periphery of the subunit interface and on the solvent sides of both subunits. The appearance of these numerous additional bridges on the outer edge of the eukaryotic subunit interface, which significantly increases the interaction surface between subunits, is potentially the reason for the preferential rotated state of eukaryotic ribosomes [52,66,67].
There is only one eukaryote-specific bridge positioned at the centre of the ribosome—bridge eB14. The bridge is formed by the smallest protein in yeast cells (25 amino acids) protein eL41 (figure 1), which consists of a single alpha-helix. eL41 protrudes from the 60S subunit into the 40S subunit in the proximity of the decoding centre and is nearly buried in a binding pocket composed of helices h27, h45 and h44. There are two remarkable aspects of this bridge. First, the binding pocket of eL41 in the small subunit is highly conserved in eukaryotes and bacteria. The second aspect is that, in the context of the full ribosome, eL41 is much more strongly associated with the 40S subunit than with the 60S subunit. Interestingly, although eL41 forms only minor contacts with the 60S subunit, it remains a part of the large subunit upon dissociation. In bacteria, there is only one example of this type of unusual bridge that is formed by a ribosomal protein of the large subunit and binds to the small subunit through substantial parts of their structures [29]. This unusual bridge is formed by protein bL31, which is conserved among bacteria and connects the central protuberance of the large subunit with the labile head domain of the small subunit.
The distinguishing features of the eukaryotic large subunit are two long protein helices extending from its left and right sides that are markedly distinct from the bridges of the core. These helices, which are eukaryote-specific additions to proteins eL19 and eL24 (figure 1), create the bridges eB12 and eB13, respectively, which are not buried within the intersubunit interface and are accessible from the solvent. Bridge eB12 below the mRNA exit tunnel is mainly formed through multiple interactions between several turns of the 60 residue–long α-helical extension at the C-terminus of eL19 and ES6S. Protein eL24 consists of an N-terminal domain that resides in the 60S subunit followed by a long flexible linker that protrudes deep into the side of the 40S body and a C-terminal domain that reaches the back of the 40S subunit. This architecture of eL24 should be considered in light of the finding that eL24 is a key player in re-initiation of the translation of polycistronic mRNAs [68–70].
6. Inhibition of the eukaryotic ribosome
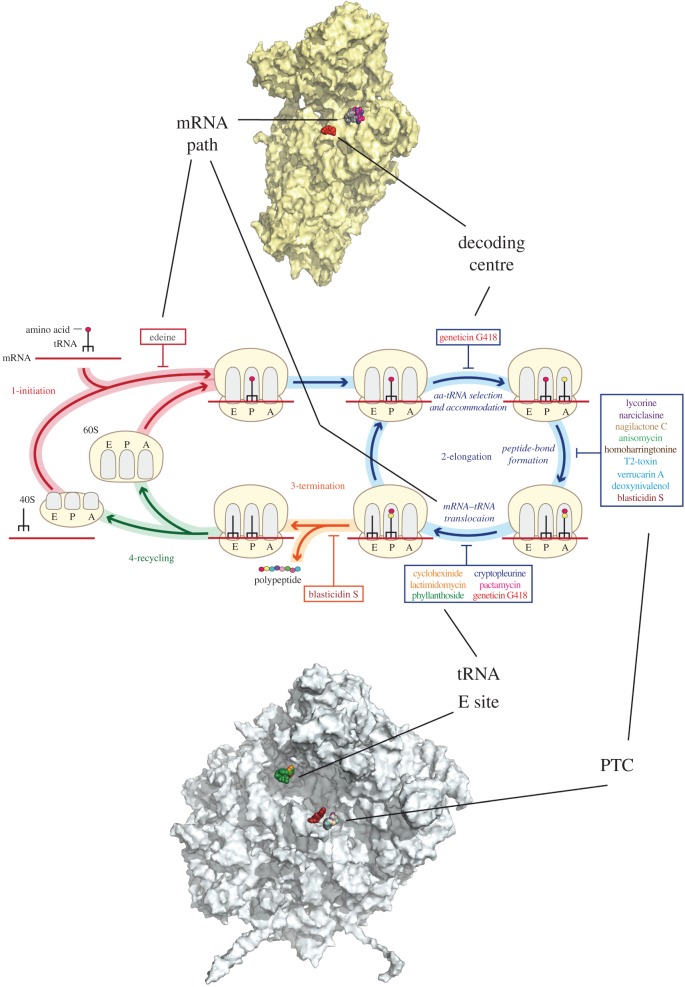
Decades of studies on antibacterial agents (antibiotics) have shown a diversity of molecular mechanisms for inhibiting protein synthesis [71]. The atomic structures of prokaryotic ribosomes provide the basis for the development of novel ribosome inhibitors that can serve as tools to study protein synthesis in bacteria. Similarly, the eukaryotic ribosome is a major target for eukaryote-specific inhibitors isolated from natural sources. Despite a limited understanding of their molecular mechanisms, eukaryote-specific inhibitors are increasingly used in research and have the potential to function as new therapeutics against a wide range of infectious diseases, cancers and genetic disorders [72–75]. Some eukaryote-specific inhibitors were investigated using crystals from the 50S subunit of the archaea Haloarcula marismortui owing to its similarity with some parts of the eukaryotic ribosome [76,77]. Recently, 16 eukaryote–specific inhibitors were investigated in the S. cerevisiae ribosome using X-ray analysis [78]. The broad-spectrum inhibitors target the PTC on the large subunit (blasticidin S), the decoding centre (geneticin G418) and the mRNA–tRNA-binding site on the small subunit (pactamycin, edeine). Eukaryote-specific inhibitors cycloheximide, lactimidomycin and phyllanthoside interact with the E tRNA-binding site, and T-2 toxin, deoxynivalenol, verrucarin, narciclasine, lycorine, nagilactone C, anisomycin, homoharringtonine and cryptopleurine interact with the PTC (figure 3). The glutarimide inhibitors cycloheximide and lactimidomycin were located in the tRNA E-site on the large subunit in a pocket formed by the universally conserved nucleotides of the 25S rRNA and a stretch of the eukaryote-specific protein eL42. Lactimidomycin bears an additional lactone ring that is positioned on top of eL42 and directed towards the subunit interface. Although chemically unrelated to glutarimides, phyllanthoside makes contact with the same rRNA nucleotides and interacts with eL42 in a manner resembling the tRNA CCA-end. The strict selectivity of E-site inhibitors towards eukaryotes is explained by the presence of two bacteria-specific rRNA residues that occlude the binding pocket. Although cycloheximide and lactimidomycin bind to the same site and likely compete with the E-tRNA, they affect translation in a different way. Lactimidomycin preferentially arrests ribosomes at the first peptide bond, whereas cycloheximide stalls ribosomes during ongoing translation [79–81].
Figure 3.
Scheme view of protein synthesis steps in eukaryotes impaired by inhibitors. Binding sites of the inhibitors shown on the interface of the ribosomal subunits and shown with connections to the protein synthesis steps.
The PTC is exclusively composed of rRNA nucleotides and is located on the large subunit. Peptide bond formation requires the two substrates, peptidyl-tRNA and aminoacyl-tRNA, to be properly aligned in the A- and P-sites, respectively. In contrast to blasticidin S, which binds to the P-site of the large subunit in the same way as in bacteria and archaea, numerous eukaryote-specific inhibitors were found to be associated with the A-site of the PTC (figure 3). Chemically different inhibitors share a similar mode of binding within the pocket. Upon binding, all A-site inhibitors induce a similar pattern of structural rearrangements in their direct vicinity that propagate up to 15 Å away from the PTC. It has been shown that nucleotides U2873 (U2504 in E. coli) and C 2824 (C 2452 in E. coli) are involved in interactions with inhibitors in yeast. Different orientations of these nucleotides in bacteria prevent ribosome binding to inhibitors.
The decoding centre of the ribosome forms a geometrically restricted pocket that accurately selects aminoacyl-tRNA in accordance with mRNA codons positioned in the A-site. In bacteria, aminoglycoside antibiotics alter translation accuracy and inhibit tRNA translocation by perturbing the conformation of the decoding centre nucleotides. In addition to their potent activity against Gram-negative bacteria, the aminoglycoside-induced suppression of premature termination holds the potential to treat inherited disorders caused by nonsense mutations [75,82]. The canonical aminoglycoside-binding site is located within the internal loop of helix 44 of the 18S rRNA, which is part of the decoding centre that contains the essential and universally conserved nucleotides A1755 (A1492 in E. coli) and A1756 (A1493 in E. coli). In close vicinity, two nucleotides differ between bacteria and eukaryotes but are identical in yeast and humans: G1645 (A1408 in E. coli) and A1754 (G1491 in E. coli) [83,84]. Geneticin binds to the aminoglycoside pocket and induces the flipping of A1755 and A1756. The structure highlights direct interactions between geneticin ring I and the eukaryote-specific residues G1645 and A1754.
Pactamycin, cryptopleurine and edeine are located exclusively in the 40S E-site and share the same binding pocket. These inhibitors are found in the mRNA channel in the E-site and interact only with the 18S rRNA. Pactamycin and edeine are broad-spectrum inhibitors. Pactamycin has a conserved binding site in bacteria and eukaryotes. Edeine in the yeast ribosome binds to the same region but adopts a different conformation from that in the bacterial small subunit. In contrast, cryptopleurine was described as a eukaryote-specific inhibitor [85]. The structure of cryptopleurine bound to the yeast ribosome does not provide the reason for its specificity. The location of edeine, pactamycin and cryptopleurines suggest that they act on translocation from the P- to the E-site and may also affect initiation in eukaryotes.
7. Conclusion
The development of new technologies in X-ray crystallography and cryo-EM has the possibility of facilitating better studies of the structure of ribosomes by modelling different steps of protein synthesis at atomic resolutions, where significant parts of the electron density maps allow researchers to interpret precise positions of chemical groups in the functional pockets of complexes and to suggest mechanisms for ribosome functions. However, a significant part of the ribosome, mainly the solvent side of subunits, is more flexible and does not allow interpreting of structure at high resolution. This problem becomes very significant in the case of the ribosomes from higher eukaryotes, where the biggest part of the high-eukaryote–specific long ribosomal RNA segments (approx. 1 MDa,) remain unresolved despite innovations in cryo-EM. Determining how these ribosome elements can be stabilized for structural studies remains an open area of research.
Acknowledgements
We thank Sultan Agalarov (Institute of Protein Research, Russia) and Daniel Wilson (Gene Center, Munich) for kindly providing figure 2.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the ERC Advanced grant 294312 (to M.Y.), the French National Research Agency grant ANR-15-CE11-0021-01 (to G.Y.) and the Russian Government Programme of Competitive Growth of Kazan Federal University (to M.Y.).
References
- 1.Wittmann HG. 1983. Architecture of prokaryotic ribosomes. Annu. Rev. Biochem. 52, 35–65. ( 10.1146/annurev.bi.52.070183.000343) [DOI] [PubMed] [Google Scholar]
- 2.Serdyuk IN, Agalarov SC, Sedelnikova SE, Spirin AS, May RP. 1983. Shape and compactness of the isolated ribosomal 16 S RNA and its complexes with ribosomal proteins. J. Mol. Biol. 169, 409–425. ( 10.1016/S0022-2836(83)80058-8) [DOI] [PubMed] [Google Scholar]
- 3.Vasiliev VD. 1974. Morphology of the ribosomal 30S subparticle according to electron microscopic data. Acta Biol. Med. Ger. 33, 779–793. [PubMed] [Google Scholar]
- 4.Lake JA. 1976. Ribosome structure determined by electron microscopy of Escherichia coli small subunits, large subunits and monomeric ribosomes. J. Mol. Biol. 105, 131–139. ( 10.1016/0022-2836(76)90200-X) [DOI] [PubMed] [Google Scholar]
- 5.Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567. ( 10.1038/nsmb.2313) [DOI] [PubMed] [Google Scholar]
- 6.Frank J, et al. 1995. A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem. Cell Biol. 73, 757–765. ( 10.1139/o95-084) [DOI] [PubMed] [Google Scholar]
- 7.Stark H, Orlova EV, Rinke-Appel J, Junke N, Mueller F, Rodnina M, Wintermeyer W, Brimacombe R, van Heel M. 1997. Arrangement of tRNAs in pre- and posttranslocational ribosomes revealed by electron cryomicroscopy. Cell 88, 19–28. ( 10.1016/S0092-8674(00)81854-1) [DOI] [PubMed] [Google Scholar]
- 8.Stark H, Rodnina MV, Rinke-Appel J, Brimacombe R, Wintermeyer W, van Heel M. 1997. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389, 403–406. ( 10.1038/38770) [DOI] [PubMed] [Google Scholar]
- 9.Agrawal RK, Penczek P, Grassucci RA, Frank J. 1998. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl Acad. Sci. USA 95, 6134–6138. ( 10.1073/pnas.95.11.6134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. 1999. X-ray crystal structures of 70S ribosome functional complexes. Science 285, 2095–2104. ( 10.1126/science.285.5436.2095) [DOI] [PubMed] [Google Scholar]
- 11.Clemons WM Jr, May JL, Wimberly BT, McCutcheon JP, Capel MS, Ramakrishnan V. 1999. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature 400, 833–840. ( 10.1038/23631) [DOI] [PubMed] [Google Scholar]
- 12.Ban N, Freeborn B, Nissen P, Penczek P, Grassucci RA, Sweet R, Frank J, Moore PB, Steitz TA. 1998. A 9 Å resolution X-ray crystallographic map of the large ribosomal subunit. Cell 93, 1105–1115. ( 10.1016/S0092-8674(00)81455-5) [DOI] [PubMed] [Google Scholar]
- 13.Ban N, Nissen P, Hansen J, Capel M, Moore PB, Steitz TA. 1999. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature 400, 841–847. ( 10.1038/23641) [DOI] [PubMed] [Google Scholar]
- 14.Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T. 2000. Structure of the 30S ribosomal subunit. Nature 407, 327–339. ( 10.1038/35030006) [DOI] [PubMed] [Google Scholar]
- 15.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920. ( 10.1126/science.289.5481.905) [DOI] [PubMed] [Google Scholar]
- 16.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896. ( 10.1126/science.1060089) [DOI] [PubMed] [Google Scholar]
- 17.Yonath A, Mussig J, Wittmann HG. 1982. Parameters for crystal growth of ribosomal subunits. J. Cell Biochem. 19, 145–155. ( 10.1002/jcb.240190205) [DOI] [PubMed] [Google Scholar]
- 18.Yonath A, Tesche B, Lorenz S, Mussig J, Erdmann VA, Wittmann HG. 1983. Several crystal forms of the Bacillus stearothermophilus 50 S ribosomal particles. FEBS Lett. 154, 15–20. ( 10.1016/0014-5793(83)80868-0) [DOI] [PubMed] [Google Scholar]
- 19.Harel M, Shoham M, Frolow F, Eisenberg H, Mevarech M, Yonath A, Sussman JL. 1988. Crystallization of halophilic malate dehydrogenase from Halobacterium marismortui. J. Mol. Biol. 200, 609–610. ( 10.1016/0022-2836(88)90547-5) [DOI] [PubMed] [Google Scholar]
- 20.Trakhanov SD, Yusupov M, Agalarov S, Garber M, Ryazantzev S, Tischenko SV, Shirokov VA. 1987. Crystallization of 70 S ribosomes and 30 S ribosomal subunits from Thermus thermophilus. FEBS Lett. 220, 319–322. ( 10.1016/0014-5793(87)80838-4) [DOI] [Google Scholar]
- 21.Yusupov MM, et al. 1987. Crystallization of the 30S subunits of Thermus thermophilus ribosomes. Dokl. Akad. Nauk (USSR) 292, 1271–1274. [Google Scholar]
- 22.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834. ( 10.1126/science.1117230) [DOI] [PubMed] [Google Scholar]
- 23.Steitz TA. 2008. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 9, 242–253. ( 10.1038/nrm2352) [DOI] [PubMed] [Google Scholar]
- 24.Schmeing TM, Ramakrishnan V. 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461, 1234–1242. ( 10.1038/nature08403) [DOI] [PubMed] [Google Scholar]
- 25.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. 2001. The path of messenger RNA through the ribosome. Cell 106, 233–241. ( 10.1016/S0092-8674(01)00435-4) [DOI] [PubMed] [Google Scholar]
- 26.Selmer M, Dunham CM, Murphy FVT, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942. ( 10.1126/science.1131127) [DOI] [PubMed] [Google Scholar]
- 27.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. 2006. Structural basis for messenger RNA movement on the ribosome. Nature 444, 391–394. ( 10.1038/nature05281) [DOI] [PubMed] [Google Scholar]
- 28.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. 2010. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17, 555–560. ( 10.1038/nsmb.1790) [DOI] [PubMed] [Google Scholar]
- 29.Jenner L, Demeshkina N, Yusupova G, Yusupov M. 2010. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat. Struct. Mol. Biol. 17, 1072–1078. ( 10.1038/nsmb.1880) [DOI] [PubMed] [Google Scholar]
- 30.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. 2012. A new understanding of the decoding principle on the ribosome. Nature 484, 256–259. ( 10.1038/nature10913) [DOI] [PubMed] [Google Scholar]
- 31.Dunkle JA, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JHD. 2011. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332, 981–984. ( 10.1126/science.1202692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V. 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694. ( 10.1126/science.1179700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmeing TM, Voorhees RM, Kelley AC, Ramakrishnan V. 2011. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat. Struct. Mol. Biol. 18, 432–436. ( 10.1038/nsmb.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozov A, Demeshkina N, Khusainov I, Westhof E, Yusupov M, Yusupova G. 2016. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 7, 10457 ( 10.1038/ncomms10457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2015. Structural insights into the translational infidelity mechanism. Nat. Commun. 6, 7251 ( 10.1038/ncomms8251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330, 835–838. ( 10.1126/science.1194460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Y-G, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699. ( 10.1126/science.1179709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petry S, Brodersen DE, Murphy FVT, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. 2005. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123, 1255–1266. ( 10.1016/j.cell.2005.09.039) [DOI] [PubMed] [Google Scholar]
- 39.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322, 953–956. ( 10.1126/science.1164840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin H, Kelley AC, Loakes D, Ramakrishnan V. 2010. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc. Natl Acad. Sci. USA 107, 8593–8598. ( 10.1073/pnas.1003995107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin H, Kelley AC, Ramakrishnan V. 2011. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc. Natl Acad. Sci. USA 108, 15 798–15 803. ( 10.1073/pnas.1112185108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl Acad. Sci. USA 105, 19 684–19 689. ( 10.1073/pnas.0810953105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. 2008. Structural basis for translation termination on the 70S ribosome. Nature 454, 852–857. ( 10.1038/nature07115) [DOI] [PubMed] [Google Scholar]
- 44.Korostelev A, Zhu J, Asahara H, Noller HF. 2010. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 29, 2577–2585. ( 10.1038/emboj.2010.139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J, Lancaster L, Trakhanov S, Noller HF. 2012. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA 18, 230–240. ( 10.1261/rna.031187.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pai RD, Zhang W, Schuwirth BS, Hirokawa G, Kaji H, Kaji A, Cate JHD. 2008. Structural insights into ribosome recycling factor interactions with the 70S ribosome. J. Mol. Biol. 376, 1334–1347. ( 10.1016/j.jmb.2007.12.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V. 2007. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat. Struct. Mol. Biol. 14, 733–737. ( 10.1038/nsmb1282) [DOI] [PubMed] [Google Scholar]
- 48.Bingel-Erlenmeyer R, et al. 2008. A peptide deformylase–ribosome complex reveals mechanism of nascent chain processing. Nature 452, 108–111. ( 10.1038/nature06683) [DOI] [PubMed] [Google Scholar]
- 49.Polikanov YS, Blaha GM, Steitz TA. 2012. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336, 915–918. ( 10.1126/science.1218538) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA. 2012. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science 335, 1370–1372. ( 10.1126/science.1217443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blaha G, Stanley RE, Steitz TA. 2009. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science 325, 966–970. ( 10.1126/science.1175800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529. ( 10.1126/science.1212642) [DOI] [PubMed] [Google Scholar]
- 53.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. 2010. Crystal structure of the eukaryotic ribosome. Science 330, 1203–1209. ( 10.1126/science.1194294) [DOI] [PubMed] [Google Scholar]
- 54.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1 . Science 331, 730–736. ( 10.1126/science.1198308) [DOI] [PubMed] [Google Scholar]
- 55.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334, 941–948. ( 10.1126/science.1211204) [DOI] [PubMed] [Google Scholar]
- 56.Fu J, Munro JB, Blanchard SC, Frank J. 2011. Cryoelectron microscopy structures of the ribosome complex in intermediate states during tRNA translocation. Proc. Natl Acad. Sci. USA 108, 4817–4821. ( 10.1073/pnas.1101503108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ratje AH, et al. 2010. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713–716. ( 10.1038/nature09547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. 2010. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 466, 329–333. ( 10.1038/nature09206) [DOI] [PubMed] [Google Scholar]
- 59.Seidelt B, et al. 2009. Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326, 1412–1415. ( 10.1126/science.1177662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Becker T, et al. 2009. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373. ( 10.1126/science.1178535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aaltonen T, et al. 2013. Search for a two-higgs-boson doublet using a simplified model in pp̄ collisions at √s = 1.96 TeV. Phys. Rev. Lett. 110, 121801 ( 10.1103/PhysRevLett.110.121801) [DOI] [PubMed] [Google Scholar]
- 62.Ban N, et al. 2014. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 24, 165–169. ( 10.1016/j.sbi.2014.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. 2013. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85. ( 10.1038/nature12104) [DOI] [PubMed] [Google Scholar]
- 64.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. 2009. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136, 447–460. ( 10.1016/j.cell.2009.01.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G,Frank J. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell 107, 373–386. ( 10.1016/S0092-8674(01)00539-6) [DOI] [PubMed] [Google Scholar]
- 66.Spahn CM, et al. 2004. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 23, 1008–1019. ( 10.1038/sj.emboj.7600102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CMT. 2011. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol. Cell 44, 214–224. ( 10.1016/j.molcel.2011.07.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park H-S, Himmelbach A, Browning KS, Hohn T, Ryabova LA. 2001. A plant viral ‘reinitiation’ factor interacts with the host translational machinery. Cell 106, 723–733. ( 10.1016/S0092-8674(01)00487-1) [DOI] [PubMed] [Google Scholar]
- 69.Nishimura T, Wada T, Yamamoto KT, Okada K. 2005. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17, 2940–2953. ( 10.1105/tpc.105.036533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thiebeauld O, Schepetilnikov M, Park HS, Geldreich A, Kobayashi K, Thiébeauld O, Keller M, Hohn T, Ryabova LA. 2009. A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation. EMBO J. 28, 3171–3184. ( 10.1038/emboj.2009.256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12, 35–48. ( 10.1038/nrmicro3155) [DOI] [PubMed] [Google Scholar]
- 72.Hobbie SN, Kaiser M, Schmidt S, Shcherbakov D, Janusic T, Brun R, Böttger EC. 2011. Genetic reconstruction of protozoan rRNA decoding sites provides a rationale for paromomycin activity against Leishmania and Trypanosoma. PLoS Negl. Trop. Dis. 5, e1161 ( 10.1371/journal.pntd.0001161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu W, Roongsawang N, Mahmud T. 2011. Biosynthetic studies and genetic engineering of pactamycin analogs with improved selectivity toward malarial parasites. Chem. Biol. 18, 425–431. ( 10.1016/j.chembiol.2011.01.016) [DOI] [PubMed] [Google Scholar]
- 74.Santagata S, et al. 2013. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science 341, 1238303 ( 10.1126/science.1238303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bidou L, Allamand V, Rousset J-P, Namy O. 2012. Sense from nonsense: therapies for premature stop codon diseases. Trends Mol. Med. 18, 679–688. ( 10.1016/j.molmed.2012.09.008) [DOI] [PubMed] [Google Scholar]
- 76.Gürel G, Blaha G, Steitz TA, Moore PB. 2009. Structures of triacetyloleandomycin and mycalamide a bind to the large ribosomal subunit of Haloarcula marismortui. Antimicrob. Agents Chemother. 53, 5010–5014. ( 10.1128/AAC.00817-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gürel G, Blaha G, Moore PB, Steitz TA. 2009. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J. Mol. Biol. 389, 146–156. ( 10.1016/j.jmb.2009.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garreau de Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, Yusupov M. 2014. Structural basis for the inhibition of the eukaryotic ribosome. Nature 513, 517–522. ( 10.1038/nature13737) [DOI] [PubMed] [Google Scholar]
- 79.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217. ( 10.1038/nchembio.304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee S, Liu B, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl Acad. Sci. USA 109, E2424–E2432. ( 10.1073/pnas.1207846109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. ( 10.1126/science.1168978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shulman E, Belakhov V, Wei G, Kendall A, Meyron-Holtz EG, Ben-Shachar D, Schacht J, Baasov T. 2014. Designer aminoglycosides that selectively inhibit cytoplasmic rather than mitochondrial ribosomes show decreased ototoxicity: a strategy for the treatment of genetic diseases. J. Biol. Chem. 289, 2318–2330. ( 10.1074/jbc.M113.533588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan-Minogue H, Bedwell DM. 2008. Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity. RNA 14, 148–157. ( 10.1261/rna.805208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Recht MI, Douthwaite S, Puglisi JD. 1999. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 18, 3133–3138. ( 10.1093/emboj/18.11.3133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dolz H, Vazquez D, Jimenez A. 1982. Quantitation of the specific interaction of [14a-3H] cryptopleurine with 80S and 40S ribosomal species from the yeast Saccharomyces cerevisiae. Biochemistry 21, 3181–3187. ( 10.1021/bi00256a023) [DOI] [PubMed] [Google Scholar]