Abstract
A current goal of molecular biology is to identify transcriptional networks that regulate cell differentiation. However, identifying functional gene regulatory elements has been challenging in the context of developing tissues where material is limited and cell types are mixed. To identify regulatory sites during sex determination, we subjected Sertoli cells from mouse fetal testes to DNaseI-seq and ChIP-seq for H3K27ac. DNaseI-seq identified putative regulatory sites around genes enriched in Sertoli and pregranulosa cells; however, active enhancers marked by H3K27ac were enriched proximal to only Sertoli-enriched genes. Sequence analysis identified putative binding sites of known and novel transcription factors likely controlling Sertoli cell differentiation. As a validation of this approach, we identified a novel Sertoli cell enhancer upstream of Wt1, and used it to drive expression of a transgenic reporter in Sertoli cells. This work furthers our understanding of the complex genetic network that underlies sex determination and identifies regions that potentially harbor non-coding mutations underlying disorders of sexual development.
KEY WORDS: Genome-wide enhancer profiling, Sex determination, Sertoli cell development, Mouse
Summary: Chromatin profiling identifies numerous putative transcriptional enhancers of genes important for sex determination and Sertoli development, and validate a novel enhancer upstream of Wt1.
INTRODUCTION
During development, multipotent progenitor cell commitment to one cell fate versus another is driven by complex gene networks controlled by diverse regulatory factors, including transcription factors (TFs), chromatin remodelers and non-coding RNAs (ncRNAs). Although identification of the cohorts of genes that drive cell fate determination is an important focus of developmental studies, it is equally important to identify the regulatory sites through which these factors operate. Mutations of regulatory sites in enhancers and promoters that alter gene expression have been discovered serendipitously, but until recently, an unbiased genome-wide approach to identify regulatory elements in specific fetal cell types remained difficult. Thus, it has not been clear how the accessibility of chromatin sites and the availability of TFs act together to control cell fate decisions during fetal development from a pluripotent to a committed fate.
The developing gonad is a powerful model system for understanding the gene regulatory mechanisms that drive cell fate determination. The process of sex determination occurs in gonad progenitor cells whose differentiation controls the sexual fate of the organism. Although genetic sex (XY/XX) is established upon fertilization, these innate chromosomal differences do not produce phenotypic differences in male or female early embryos (Eggers and Sinclair, 2012). Instead, sexual dimorphism is initiated in the early gonad supporting cells, a single cell lineage that differentiates into Sertoli (XY) (Sekido et al., 2004) or pregranulosa (XX) cells (Albrecht and Eicher, 2001; Mork et al., 2012). Prior to sex determination, XX and XY supporting cells are bipotential and express nearly all genes at similar levels, with minor differences limited to genes on the X and Y chromosomes (Nef et al., 2003; Munger et al., 2013).
In the mouse, male sex determination is initiated around embryonic day 11.5 (E11.5) by expression of the Y-linked gene Sry, which diverts XY supporting cells to the Sertoli cell fate (Fig. 1A). Sry is transiently expressed and many SRY-binding sites are subsequently bound by its downstream target SOX9 (Li et al., 2014). SRY and SOX9 control the differentiation of Sertoli cells by triggering a dramatic transcriptional reprogramming of bipotential progenitor cells within just 24 h, leading to the upregulation of over 200 genes important for Sertoli cell development and downregulation of ∼100 pregranulosa cell-expressed genes that were expressed at the bipotential stage (Munger et al., 2013). In the absence of Sry, XX supporting cells differentiate to pregranulosa cells, but in contrast to the dramatic transcriptional changes that accompany Sertoli cell specification, pregranulosa cell differentiation proceeds with fewer transcriptional changes consistent with the finding that undifferentiated XX and XY supporting cells are initially transcriptionally biased toward ovarian fate (Jameson et al., 2012b).
Fig. 1.
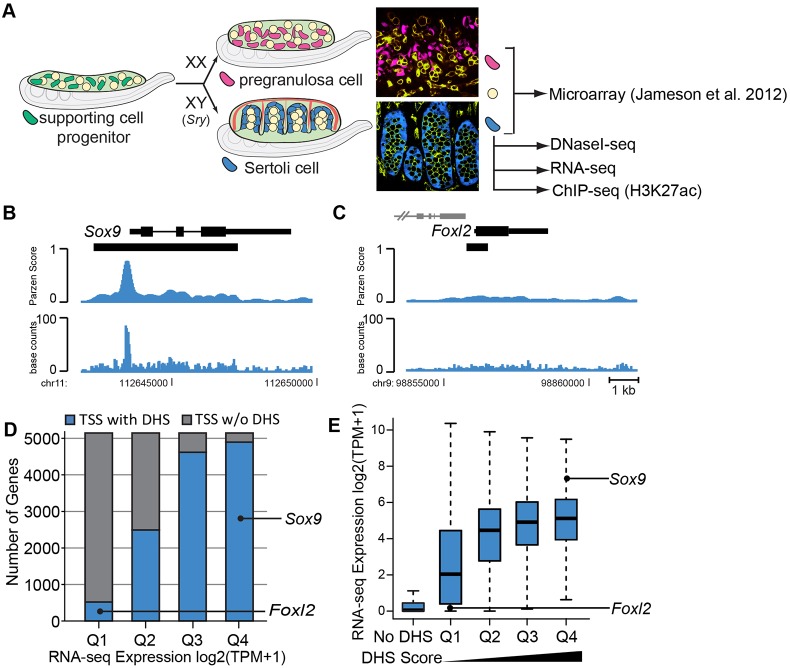
DNaseI-seq and RNA-seq analysis of Sertoli cells. (A) Overview of sex determination. At the bipotential stage (E11.5), XX and XY gonads are indistinguishable and contain a mixture of somatic and germ cells (yellow). The supporting cell lineage (green) gives rise to Sertoli cells of the testis (XY; blue) or pregranulosa cells of the ovary (XX; pink) upon sex determination. Confocal images show a whole-mount immunostained E13.5 ovary (top) and testis (bottom). XX pregranulosa cells express the Sry-eGFP transgene (Albrecht and Eicher, 2001) (pink), XY Sertoli cells express the Sox9-CFP transgene (blue; germ cells and vasculature are yellow/green). Microarray data were previously collected from FACS-isolated E13.5 pregranulosa cells, Sertoli cells and germ cells (Jameson et al., 2012b). DNaseI-seq was performed on FACS-isolated E13.5 and E15.5 Sertoli cells. RNA-seq was performed on E15.5 Sertoli cells. (B,C) DNaseI-seq data identified a strong DHS at (B) the Sox9 promoter (expressed in Sertoli cells) and a weaker DHS at (C) the Foxl2 promoter (repressed in Sertoli cells). Only peaks overlapping the TSS are shown and gene names are positioned adjacent to the TSS. Nearby genes are indicated in gray. Black bars under the gene indicate DHSs. The top track indicates the Parzen score, or DHS score, while the bottom track shows the smoothed base counts. (D,E) Comparative analysis of E15.5 DNaseI-seq and RNA-seq data. (D) RNA-seq data [log base 2 (transcripts per million (TPM+1))] from E15.5 Sertoli cells was divided into quartiles based on expression values [Q1, log2(TPM+1)=0-0.1; Q2, log2(TPM+1)=0.1-2.34; Q2, log2(TPM+1)=2.34-5.02; Q2, log2(TPM+1)=5.02-12.74]. The chart indicates the number of genes within each quartile that had an overlapping DHS (blue) or did not have an overlapping DHS (gray) at the TSS. (E) Genes with a DHS overlapping the TSS were divided into quartiles based on DHS scores (Q1-Q4, low to high DHS scores). The distribution of expression values for each group is shown as a boxplot (excluding outliers).
Although SRY acts as a pivotal switch in the sex-determining pathway, mutations in SRY account for only ∼10-15%, of XY disorders of sex development (DSDs) (Cameron and Sinclair, 1997). Over 35 other genes have been identified that contribute to DSDs (Arboleda and Vilain, 2011; Arboleda et al., 2013), suggesting that a complex genetic network controls fate commitment in the early gonad. It is evident from human DSD cases where gene dose is altered by duplications or haploinsufficiencies (Foster et al., 1994; Muscatelli et al., 1994; Zanaria et al., 1994; Jordan et al., 2001) that perturbations to the level or timing of individual genes within this network can lead to sex reversals. In the C57BL/6J inbred mouse strain, the altered expression of several sex-determining genes causes unique strain-dependent sex-reversal phenotypes in response to specific mutations (Colvin et al., 2001; Bouma et al., 2005; Kim et al., 2007b; Munger et al., 2009, 2013; Correa et al., 2012; Warr et al., 2012). These studies point to the importance of precise spatial and temporal gene regulation. However, our lack of knowledge regarding the locations of crucial gene-regulatory sites limits our ability to study the regulatory mechanisms that control gene expression in Sertoli cells.
To identify putative regulatory sites in Sertoli cells, we performed DNaseI-seq on purified Sertoli cells from fetal testes just after sex determination. Comparison with DNaseI-seq data from a variety of other mouse tissues and cell types revealed thousands of novel Sertoli cell-specific putative regulatory elements, which are enriched near genes expressed in Sertoli cells, as well as genes that are silent in Sertoli cells, but expressed in the alternate pregranulosa cell lineage. This suggests that DNaseI-seq identified sites involved in the activation and repression of genes required for Sertoli cell fate commitment. Using ChIP-seq for H3K27ac, we located active enhancer elements specifically enriched near genes expressed in Sertoli cells, and used this data to identify TF motifs that distinguish active from inactive enhancers. Finally, we validated the enhancer activity of an active regulatory element located 50 kb upstream of Wt1, which encodes a TF essential for early gonad development. This data will enable future experiments to characterize the regulatory network driving Sertoli cell development and may aid in the identification of non-coding human mutations underlying unexplained cases of DSDs, the majority of which cannot be attributed to mutation within the coding region of a known sex-determining gene (Arboleda and Vilain, 2011; Arboleda et al., 2013).
RESULTS
Adapting DNase1-seq to developmentally limited cell populations
We conducted a proof-of-principle experiment to establish the efficacy of DNaseI-seq in identifying regulatory elements in Sertoli cells that had been purified by fluorescence-activated cell sorting (FACS) followed by long-term storage at −80°C. E15.5 Sertoli cells were isolated by FACS based on the Sertoli cell-specific expression of the Sox9-CFP transgene (Fig. 1A). Significantly reducing the starting material required, we found that 5×106 cells routinely yielded enough chromatin to perform the DNaseI-seq assay.
DNaseI-seq identified over 81,000 DNaseI hypersensitive sites (DHSs) in E15.5 Sertoli cells. Using previously published methods (Song et al., 2011), DHSs were determined to be in gene promoters (23%), intergenic (31%) and genic regions (exons 10%, introns 36%), and displayed a median length of 330 bp (Fig. S1). Over 13,000 genes (∼60%) had a DHS peak overlapping a transcriptional start site (TSS), whereas over 9000 TSSs (∼40%) lacked a DHS peak. Two loci representing a gene expressed in Sertoli cells (Sox9) and a granulosa-specific gene (Foxl2) displayed DNase signals of differing magnitudes at their TSS (Fig. 1B,C), suggesting that the DHS signal at the TSS may correlate with gene expression levels. To examine this possibility, RNA-seq was performed on E15.5 Sertoli cells (Table S1). Consistent with prior studies (Boyle et al., 2008a), the presence and magnitude of a DHS peak at the promoter was associated with increased gene expression values (Fig. 1D,E).
Further validating the quality of our data, we identified the only known distal enhancer for a Sertoli cell-expressed gene: Sox9. Although the Sox9 enhancer TES is likely over-represented in our data set due to the multi-copy nature of transgene insertions in the Sox9 (Tes)-CFP line, our DHS data identifies only the smaller functional element, TESCO (Sekido and Lovell-Badge, 2008), as having putative enhancer activity (Fig. 2A).
Fig. 2.
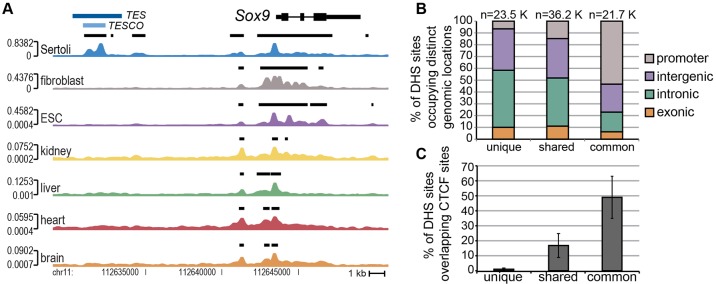
Identification of DHSs unique to the Sertoli cell lineage. (A) Comparison of the genomic region around Sox9 in E15.5 Sertoli cells versus other cell types (fibroblast, ESC) and tissues (kidney, liver, heart, brain). The Sox9-CFP transgene is driven by the 3 kb TES enhancer (blue bar) located 13 kb upstream from the Sox9 TSS. Enhancer activity was previously narrowed to a subregion of TES: the 1.3 kb TESCO element (Sekido and Lovell-Badge, 2008) (light-blue bar). DNaseI-seq identifies a DHS (black bar) spanning the TESCO element that is unique to the Sertoli cell lineage, consistent with the expression pattern of the transgene. Black bars above the track denote DHSs; Parzen scores are shown to the left of each track. (B) Based on their cell type specificity, E15.5 Sertoli cell DHSs were divided into unique, shared or common. Each group was subcategorized based on genomic location (promoter, gray; intergenic, purple; intronic, green; exonic, orange). (C) The percentage of DHSs overlapping CTCF sites based on the average overlap with eight previously generated CTCF ChIP-seq data sets (Shen et al., 2012).
Identification of DHSs important for gene regulation in Sertoli cells
Given the success of this experiment, we performed DNaseI-seq on E13.5 Sertoli cells, a stage of development for which we have extensive expression data (Nef et al., 2005; Bouma et al., 2007a, Jameson et al., 2012b) (Table S2). Our E13.5 DNaseI-seq data showed similar properties to the data from E15.5 Sertoli cells; ∼71% of the DHSs were shared between both data sets and had nearly identical genomic distributions (Fig. S2A,B).
To identify the DHSs most relevant for gene expression during testis development, we conducted DNaseI-seq on six other mouse samples [primary skin fibroblasts, embryonic stem cells (ESCs), adult kidney, liver, heart and brain]. To identify putative enhancers important for gene expression in Sertoli cells, we used the six other somatic cell types to classify E13.5 and E15.5 Sertoli cell DHSs as unique (present in only Sertoli cells), shared (in Sertoli cells and in one to five additional cell types) or common (in all cell types). For example, at the Sox9 promoter, all cell types had a DHS (common DHS); however, the TES enhancer was found only in the E13.5 and E15.5 Sertoli cell datasets (unique DHS) (Fig. 2A). Twenty-seven percent (21,743) of all E15.5 Sertoli cell DHSs and 25% (21,863) of E13.5 Sertoli cell DHSs were common, with promoters accounting for over half of the common sites (Fig. 2B; Fig. S2C). CTCF binding sites are highly conserved across different cell types (Kim et al., 2007a; Xi et al., 2007; Song et al., 2011) and, consistent with this, CTCF-binding sites found in eight other cell types (Shen et al., 2012) were enriched in the common DHSs (Fig. 2C). In fact, of the common sites not overlapping a promoter, ∼75% overlapped a CTCF binding site. The remaining DHSs were unique (E15.5: 29%; 23,548, E13.5: 32%, 28,133) to the Sertoli cell lineage or shared (E15.5: 44%; 36,176, E13.5: 43%, 38,111). A high proportion of unique (85%) and shared (73%) sites localized to intergenic or intronic regions (Fig. 2B; Fig. S2B), and are likely to be important regulators of gene expression in Sertoli cells.
Sertoli cell DHSs associate with Sertoli and pregranulosa cell-expressed genes, whereas active enhancers are enriched near Sertoli cell genes
The DHSs identified by DNaseI-seq correlate with several genomic features, including promoters, enhancers, repressors and insulators (Heintzman et al., 2007, 2009; Boyle et al., 2008a). To focus on regions of the genome driving Sertoli cell-specific gene expression, we conducted chromatin profiling for H3K27ac, a strong predictor of active enhancer activity (Creyghton et al., 2010; Rada-Iglesias et al., 2011). H3K27ac ChIP-seq on E13.5 Sertoli cells identified >28,000 H3K27ac peaks, ∼70% of which overlapped a DHS in our E13.5 Sertoli cell dataset (Fig. 3A, Fig. S4).
Fig. 3.
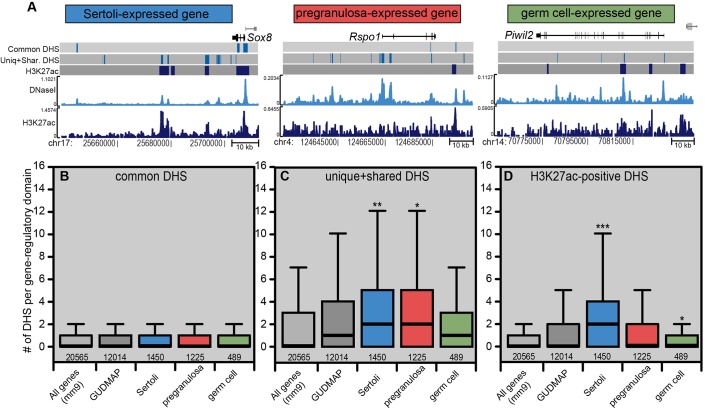
Enrichment of DHSs near Sertoli and pregranulosa cell-expressed genes. (A) Using GREAT (McLean et al., 2010) each gene was assigned a gene-regulatory domain extending to the nearest 3′ and 5′ gene, but not overlapping the proximal promoter of either neighboring gene. Regulatory domains are shown for three examples: a Sertoli cell-expressed gene (Sox8), a pregranulosa cell-expressed gene (Rpso1) and a germ cell-expressed gene (Piwil2). Using data from E13.5 Sertoli cells, the three bars under the gene show the locations of common DHSs (top) and cell type-specific DHSs (middle) (vertical light-blue lines), or H3K27ac-positive DHSs (bottom; vertical dark-blue lines) for each region. The light-blue track shows the DNaseI-Seq Parzen score (autoscaled) and the dark-blue track shows H3K27 signal. Gene names are positioned adjacent to the TSS and other genes in the region are indicated in gray. (B-D) The correlation between E13.5 DHS sites and genes expressed in male or female supporting cells was plotted. The DHSs that mapped to each gene's regulatory domain were counted and the distribution of peaks per domain is shown as a box plot (outliers excluded). The results are shown for common DHSs (B), cell-type specific DHSs (C) and H3K27ac-positive DHSs (D). mm9 refers to all RefSeq genes (light gray); GUDMAP is the subset of mm9 genes that were analyzed in our previous microarray study [gray (Jameson et al., 2012b)]. Sertoli cell genes (blue), pregranulosa cell genes (red) and germ cells genes (green) categories are described in the supplementary Materials and Methods. Statistical significance was calculated using a two-sample Mann–Whitney (two-sided) test to compare each gene set to the GUDMAP reference set. *P<1×10−5, **P<1×10−10, ***P<2.2×10−16.
We predicted that active enhancers would be enriched around Sertoli cell-expressed genes. To test this, the enrichment of DHSs, or H3K27ac-positive DHSs, was examined around Sertoli cell-expressed genes and compared against genes expressed in pregranulosa and germ cells (Jameson et al., 2012b). Using a nearest-neighbor approach (McLean et al., 2010), we assigned DHSs to one or two neighboring genes (Fig. 3A) and the number of DHSs (Fig. 3B,C) or H3K27ac-positive DHSs (Fig. 3D) was counted around genes enriched in these cell types.
As expected, common DHSs were not preferentially enriched around Sertoli-, pregranulosa- or germ cell-expressed genes (Fig. 3B). However, there was a significant increase in the number of cell type-specific (unique+shared) DHSs neighboring both Sertoli and pregranulosa cell-expressed genes (Fig. 3C). The similar enrichment of DHSs around Sertoli and pregranulosa cell-expressed genes may reflect the shared developmental origin of the supporting cell lineages, which have similar expression profiles just a few days prior to our analysis (E10.5) (Nef et al., 2005; Munger et al., 2013). DHSs around pregranulosa cell-expressed genes may define sites where repression complexes are bound to silence the alternative female pathway upon Sertoli cell differentiation.
H3K27ac-positive DHSs, which are indicative of active enhancers, were significantly enriched specifically in neighboring Sertoli cell-expressed genes (Fig. 3D), as expected. Additionally, we observed no enrichment of cell type-specific DHSs or H3K27ac-positive DHSs near germ cell-expressed genes, consistent with the unrelated origin of this cell lineage. Thus, using DNaseI-seq, we identified thousands of regulatory sites that are likely to be critical regulators of gene expression during Sertoli cell differentiation. Additionally, ChIP-seq for H3K27ac classified these into active enhancers (associated with actively-expressed genes; H3K27ac-positive DHSs) or inactive enhancers (DHS-positive only) (Table S2).
Sertoli cell DHSs are enriched for TFs important for supporting cell development
To identify the TFs that bind putative Sertoli cell regulatory sites, we conducted computational analysis of the sequences in the Sertoli cell unique DHSs that did not overlap a promoter. We counted 6-mers in these DHSs, and used these as features in an L1 logistic regression classifier to classify these regions from length-matched sites flanking these DHSs (Koh et al., 2007) (Fig. 4A). A similar approach was shown to aid in the identification of sequence features underlying enhancer function in other mammalian systems (Lee et al., 2011). Our method is designed to identify enriched 6-mers found in a substantial fraction of DHSs. However, similar to other motif enrichment approaches, it is possible that it does not identify TFs that play a significant role in Sertoli cell biology, but bind to only a few locations.
Fig. 4.
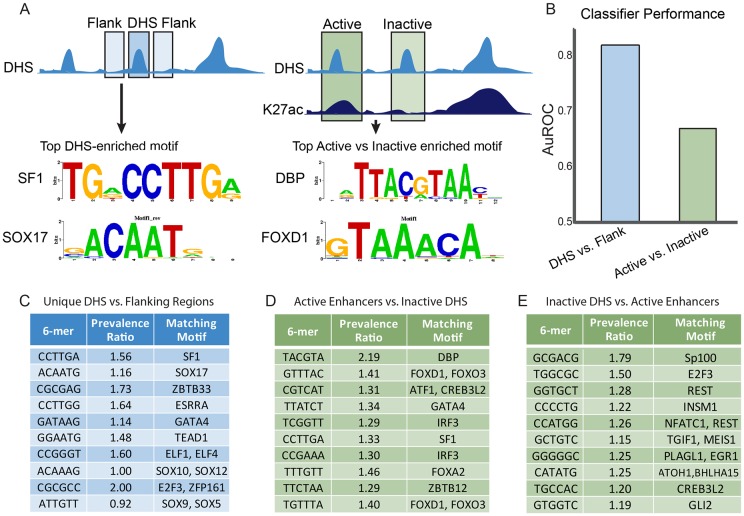
TF-binding sites in active enhancers and inactive DHSs. (A) Schematic representation of the classification approach to identify motifs and TFs predictive of Sertoli cell DHSs. The top enriched motifs with known binding factors for sites predictive of DHS versus flank (left) and active versus inactive DHSs (right) are shown. (B) AuROC for different L1-logistic regression classifiers. Classifying active enhancers and inactive DHS against flanking regions shows high classifier performance (∼0.82), while classifying active enhancers against inactive DHS shows lower performance (∼0.67). (C-E) Top motifs driving predictive performance for the different classification tasks. TFs shown are best hits from the MEME suite's TOMTOM tool matching to the different 6-mers (Gupta et al., 2007). The prevalence ratio is defined as: the ratio of the average length normalized frequency of that 6-mer in DHSs unique to Sertoli cells relative to the flanking regions (C); the ratio of the average length normalized frequency of that 6-mer in active enhancers to inactive DHSs (D); the ratio of the average length normalized frequency of that 6-mer in inactive DHSs to active enhancers (E). The top 10 6-mers are shown in order of the absolute value of their regression coefficients. Only the first 6-mer matching to a particular TF is shown. TFs identified in D and E were identified in the same classifier, with those TFs shown in D more highly prevalent in active enhancers and those in E more prevalent in inactive DHSs. Table S3 contains the complete results for the analyses in C-E.
Our analysis revealed 305 6-mers identified within Sertoli cell unique DHSs that are highly predictive of whether a region will be open specifically in Sertoli cells compared with flanking regions [area under receiver operating characteristic curve (AuROC)=0.82] (Fig. 4B). To rule out the possibility of high classification accuracy due to repeats in flanking regions, we trained a classifier to distinguish Sertoli cell DHSs from a union set of unique DHSs from other cell types, including ES and fibroblast cells, and heart, kidney, liver and brain tissue. The classifier again showed high classification accuracy (AuROC=0.87). Furthermore, the 6-mers identified are similar between the two classification tasks, indicating that the features identified are robust predictors of Sertoli cell-specific DHSs (Table S3A,D).
We used TOMTOM (Bailey et al., 2009) to identify TFs that match these 6-mer motifs and found 75 6-mers matching known motifs. As multiple TFs have similar motifs, we used Sertoli cell gene expression (Jameson et al., 2012b) to identify the most likely candidates. The 6-mer most predictive of open chromatin in Sertoli cells matched the TF-binding motif for SF1 (Fig. 4C, Table S3), a factor required for early gonad formation in both sexes (Luo et al., 1994; Baba et al., 2014). Other prominent motifs identified include the SOX motif, which reflects the crucial role of SOX9 and SOX8 in the divergence of the Sertoli cell lineage from the undifferentiated progenitor cells (Chaboissier et al., 2004; Barrionuevo and Scherer, 2010), and the GATA motif, likely representing GATA4 (which is involved in early gonadal differentiation in both sexes) (Bouma et al., 2007b; Manuylov et al., 2008). Matches to SF1, SOX9 and GATA4 motifs were also found to be statistically enriched in Sertoli cell-specific DHSs, supporting the validity of our method (Table S3E). Although less predictive of open chromatin, the FOX and TCF/LEF motifs were also identified (Table S3). FOXL2 (Schmidt et al., 2004; Ottolenghi et al., 2005) and Wnt signaling (which modulates gene expression through β-catenin binding with TCF/LEF factors) (Vainio et al., 1999; Chassot et al., 2008; Liu et al., 2009) are female-specific factors that are important for pregranulosa cell differentiation; enrichment of these motifs in Sertoli cell open chromatin may indicate sites requiring repression during Sertoli cell differentiation. Also identified was ELF1, an Ets-related TF. Ets family TFs can act downstream of FGF signaling (Sharrocks, 2001), which is crucial for repressing Wnt signaling to reinforce testis development (Kim et al., 2006; Jameson et al., 2012a). Additionally, we identified 6-mers matching TFs that are as yet not known to play a role in Sertoli cells, including TEAD1 and ZBTB33, as well as 6-mers that show no similarity to any known TF-binding motif (Table S3).
To determine whether distinct sequence features could differentiate active from inactive enhancers, we again used the L1-logistic regression classifier to classify the active enhancers (H3K27ac-positive DHS) from inactive DHSs (H3K27ac-negative DHS). The performance of the classifier was significantly reduced compared with our previous classifiers (AuROC=0.67), indicating that, despite the fact that active enhancers drive expression in Sertoli cells, sequence features between active enhancers and inactive DHSs are more similar than those between DHS and flanking regions (Fig. 4B). Inspection of the 6-mers that facilitate prediction of one class versus the other revealed 80 6-mers predictive of active enhancers and 64 6-mers predictive of inactive DHSs, many of which are not associated with a known TF. However, of the 6-mers predictive of an active enhancer, 25 did match a known motif. DBP, GATA4, FOXL1 and other FOX factor motifs were among the most predictive motifs for active enhancers (Fig. 4D). Although GATA4 is a well-known driver of Sertoli cell development (Bouma et al., 2007b), the role of DBP is unknown. Our identification of FOX motifs is consistent with a role of FOXL2 in repression of a Sox9 enhancer (TESCO) in granulosa cells after birth (Uhlenhaut et al., 2009) and suggests that FOXL2 may play a similar role earlier in fetal development (see Discussion). Alternatively, several other FOX factors are expressed in both Sertoli and pregranulosa cells (Jameson et al., 2012b) that have yet to be characterized and could bind these sites.
In addition, the same analysis identified 20 6-mers with matching motifs that were more predictive of inactive DHSs compared with active enhancers. Although we did not identify motifs associated with known pregranulosa cell-specific TFs, the H3K27ac-negative DHSs likely represent a diversity of open chromatin sites, potentially associated with many genes that are repressed or poised in Sertoli cells, with or without sex-specific functions (Fig. 4E). Interestingly, some of the highest scoring 6-mers in this group were not associated with known binding factors.
The motif for another TF important for Sertoli cell development, DMRT1, did not come up as predictive of open chromatin in this analysis. However, we used available ChIP-seq data from E13.5 whole testes to analyze the enrichment of DMRT1-bound sites in Sertoli cell open chromatin (Matson and Zarkower, 2012). We found that 70% of DMRT1 sites overlapped a DHS in our Sertoli cell dataset. However, the majority of DMRT1 sites overlapped promoters, with only 30% overlapping Sertoli cell unique and shared sites (Fig. S3). The exclusion of promoters from our 6-mer motif analysis may account for the absence of DMRT1 enrichment in DHSs analyzed.
DNaseI-Seq identifies a novel enhancer of Sertoli cell gene expression near Wt1
With the exception of the TESCO element, few distal regulatory elements have been identified for sex-determining genes. Our dataset provides an excellent resource with which to identify enhancers driving the expression of genes important in Sertoli cell differentiation. To demonstrate this, we performed a transient transgenic assay with a putative active enhancer of the Wt1 gene (Fig. 5A,B). Wt1 is initially expressed in multiple embryonic tissues, including the mesonephros and XX and XY gonadal somatic cells. Mutation of Wt1/WT1 in mice and humans leads to kidney and genitourinary defects, including disorders of sexual development (Kreidberg et al., 1993; Eggers and Sinclair, 2012). After sex determination, Wt1 expression becomes restricted to Sertoli cells in the testis and remains active in most somatic cells in XX gonads (Pelletier et al., 1991; Armstrong et al., 1993; Maatouk et al., 2012). We identified a DHS peak located 50 kb upstream of Wt1 that was unique to E13.5 and E15.5 Sertoli cells compared with the other cell types we examined, and marked by H3K27ac, suggesting that it functions as an active enhancer. Finally, this region contained matches to several of the 6-mers identified in our motif analysis, including binding sites for SF1, SOX and FOX motifs.
Fig. 5.
Transient transgenic analysis of a DHS unique to Sertoli cells identifies a new Sertoli cell enhancer upstream of Wt1. (A) DNaseI-seq and H3K27ac ChIP-Seq data around the Wt1 locus. Gene names indicate the TSS of the transcript. Nearby genes are indicated in gray. DNaseI-Seq data (Parzen score) and peak calls (boxes above track) are in light blue; H3K27ac ChIP-seq data and peak calls are in dark blue. A box surrounds the DHS chosen for in vivo analysis. (B) Detailed view of the DHS showing the DNaseI-seq Parzen score, the smoothed base counts (light blue) and the H3K27ac data (dark blue). The light- and dark-blue bars indicate the DNaseI-seq and H3K27ac peaks, respectively. The black bar marks the region cloned upstream of an hsp68-LacZ reporter cassette. (C) An E13.5 testis from a transgenic embryo showed β-galactosidase expression (TgWt1, green) specifically in Sertoli cells. Sertoli cells were labeled by AMH immunostaining (red) and germ cells by CDH1 (blue). The confocal image was taken using a 20× objective. Scale bar: 100 µm. (D) Confocal image of the sample in C taken with a 40× objective. Scale bar: 12.5 µm. All three panels show germ cells (blue); the left panel shows Sertoli cells (red); the middle panel shows β-galactosidase expression (green); the right panel shows the merge (yellow indicates co-expression of β-galactosidase and AMH).
Analysis of in vivo enhancer activity using a transgenic β-galactosidase reporter showed that the putative Wt1 enhancer was active in Sertoli cells of the testis (6/7 transient transgenic XY embryos; Fig. 5C,D and Fig. S5), where it exclusively localized in cells expressing the Sertoli cell marker AMH. Expression was also detected in somatic cells of the ovary (11/12 XX embryos; Fig. S5). However, no expression was detected in the mesonephros, embryonic kidney, heart or brain (Fig. 5C, Fig. S5 and data not shown) consistent with the cell-type specificity of this enhancer in our Sertoli cell datasets. These results demonstrate the power of our approach for the identification of distal enhancers that activate transcription in Sertoli cells.
DISCUSSION
Identification of Sertoli cell gene regulatory sites
Understanding the genetic networks that underlie developmental processes requires a comprehensive understanding of the genes involved as well as the regulatory mechanisms that modulate the expression of these genes. This is among a small number of studies to adapt epigenetic approaches, including DNaseI-seq and ChIP-seq, to address the resolution of cell fate in a cell type isolated from a complex developing tissue.
To discover cis-regulatory sequences during sex determination, we used DNaseI-seq to identify open chromatin sites and H3K27ac ChIP-seq to identify active enhancers in Sertoli cells. Using these data, we discovered TFs that might bind to these sites. We identified a novel active enhancer unique to Sertoli cells upstream of Wt1, and showed that it functions as a specific Sertoli cell enhancer in transgenic animals, as predicted. These data will enable future studies of cis-regulatory elements in Sertoli cells.
Although we were able to reduce the number of cells required for DNaseI-seq to 5 million cells, collection required more than 1 year, considering that a pair of E13.5 fetal testes contains only 10-20,000 Sertoli cells. Unfortunately, our attempts to perform DNaseI-seq on pregranulosa cells failed, possibly owing to the longer storage times required to collect sufficient numbers of cells (our pregranulosa cell reporter enabled purification of only 2-4000 cells per embryo). Rather than identifying sites of open chromatin based on DNaseI-seq, a new technique, ATAC-seq, uses a transposome-based method where the transposome preferentially integrates into sites of open chromatin (Buenrostro et al., 2013) and is highly correlative with DNaseI-seq data. Working on as few as 500 cells, this method presents an attractive alternative to enable studies of chromatin during cellular differentiation. Future studies will address whether a common chromatin architecture is maintained in differentiated XX and XY supporting cells, or whether these sites are eventually remodeled in a sexually dimorphic manner as differentiation proceeds.
DHSs adjacent to pregranulosa cell genes might indicate TF-mediated repression in Sertoli cell fate maintenance
In multipotent progenitor cell types, many developmentally regulated genes exist in an epigenetically poised state. Cis-regulatory elements of poised genes are marked by H3K4me1 and open chromatin (Heintzman et al., 2007). Upon differentiation, regulatory sites for active genes gain H3K27ac (Creyghton et al., 2010; Rada-Iglesias et al., 2011) and retain open chromatin, while repressed genes either remain poised or exhibit a loss of H3K4me1 and open chromatin (Whyte et al., 2012; Lara-Astiaso et al., 2014), perhaps rendering regulatory sites refractory to access by TFs.
In bipotential progenitor cells of the gonad, it is currently unknown whether genes that become sex-specific exist in an epigenetically poised state prior to sex determination. However, we hypothesized that, by E13.5, when sex-specific gene expression patterns are well-established, open chromatin would be enriched near genes actively expressed in Sertoli cells. Although the enrichment of H3K27ac-positive active enhancers was associated with only Sertoli cell-expressed genes as expected, an intriguing finding was the widespread enrichment of DHSs around both Sertoli and pregranulosa cell-expressed genes in E13.5 differentiated Sertoli cells. There is evidence that many genes associated specifically with ovarian development are expressed in both sexes at the bipotential stage, suggestive of lineage priming (Jameson et al., 2012b), and initiation of the male pathway is characterized by extensive upregulation of the testis-pathway genes with simultaneous downregulation of ovarian-pathway genes. In contrast, transcriptional priming of pregranulosa progenitors towards the female pathway results in fewer transcriptional changes upon differentiation (Munger et al., 2009; Jameson et al., 2012b). One possible explanation for the retention of open chromatin near repressed pregranulosa cell genes in Sertoli cells is that loss of open chromatin sites occurs slowly, downstream of faster transcriptional changes. At a later developmental stage, fewer open chromatin sites might be associated with repressed gene loci. During ES cell differentiation, reduced H3K4me1 and open chromatin domains were observed near repressed genes within 48 h of differentiation (Whyte et al., 2012; Buecker et al., 2014). However, in multiple hematopoietic lineage descendants, the loss of open chromatin near repressed genes did not occur until late differentiation stages (Lara-Astiaso et al., 2014). We observed a similar enrichment of DHSs around both Sertoli and pregranulosa cell genes at E13.5 and E15.5 (Fig. S2), up to 4 days after peak Sry expression. Although it remains possible that a loss of open chromatin near repressed pregranulosa cell genes occurs later in adult Sertoli cells, an alternative possibility is that open chromatin regions persist near granulosa-expressed genes, perhaps reflecting the original bipotential progenitor capable of differentiating as a Sertoli or granulosa cell. Open chromatin near granulosa genes could be bound either by TFs that function to preserve a poised chromatin state or bound by repressive TFs that maintain the Sertoli cell fate. It is unclear whether this is a generalizable feature of cell populations that derive from a common bipotential progenitor.
An intriguing characteristic of gonadal supporting cells is that long after sex determination, some mutations cause transdifferentiation to the alternate cell fate. In the testis, Dmrt1 is required to prevent postnatal Sertoli to granulosa cell transdifferentiation by repression of several pregranulosa cell genes, including Foxl2, Esr2 and β-catenin (Matson et al., 2011; Minkina et al., 2014). In a reciprocal manner, mutation of Foxl2 causes granulosa-to-Sertoli cell transdifferentiation in the adult ovary (Schmidt et al., 2004; Ottolenghi et al., 2005). Our finding that Sertoli cells maintain open chromatin near many repressed granulosa genes might suggest that continued repression by TFs represents a mechanism for long-term maintenance of the Sertoli cell fate, and that these repressive mechanisms are established early during Sertoli cell development. Consistent with this, the TESCO enhancer of Sox9 is bound by FOXL2 in adult granulosa cells (Uhlenhaut et al., 2009), where Sox9 is silenced; however, it is bound by SRY/SOX9 and SF1 in Sertoli cells (Sekido and Lovell-Badge, 2008), where Sox9 is active. The use of the same cis-regulatory element to repress expression (via FOXL2 binding) in granulosa cells and activate expression (via SRY/SOX9 and SF1 binding) in Sertoli cells suggests that at least some cis-regulatory sites serve a dual purpose during sex-specific differentiation.
If the same regulatory sites act as enhancers in one cell type and repressors in the opposing cell type, then motifs for both male-promoting and female-promoting TFs would be located within the active enhancers of Sertoli cell genes, available for competitive occupancy by male- or female-promoting TFs. Consistent with this, our motif analysis discriminating active versus inactive enhancers identified both the SOX and FOX motifs as predictive of active enhancers in Sertoli cells (Fig. 4 and Table S3). Furthermore, the lack of high classification accuracy from simple sequence features in active enhancers and inactive DHSs points to a complex and combinatorial regulatory code. Alternatively, differences in protein concentrations or post-translational modifications to regulators might explain the different transcriptional programs. The dual use of enhancers, such as TESCO, may underlie the ability of gonadal supporting cells to switch fates; however, it is currently unclear how extensive this mechanism is. Future studies mapping open chromatin sites in granulosa cells will reveal whether shared cis-regulatory sites represent a subset of gene regulatory mechanisms, or whether open chromatin sites are extensively shared between these functionally and morphologically distinct cell lineages.
Regulatory sites may harbor non-coding mutations that cause DSDs
The inability to provide a genetic diagnosis for the majority of individuals with a DSD has led to the development of more robust diagnostic tests to screen large regions of the genome for potential disease-causing mutations, including exome sequencing and copy number variation arrays (White et al., 2011; Arboleda et al., 2013; Baxter et al., 2015). This approach has revealed non-coding mutations upstream of SOX9 that result in DSDs (Kim et al., 2015). However, identifying non-coding mutations on a genome-wide scale remains difficult, and classification of non-coding mutations as potentially disease-causing is challenging, as these mutations do not directly alter the protein sequence. Recent studies have attempted to identify human enhancers for this purpose, using publicly available data from the NIH Roadmap Epigenomics Program (Roadmap Epigenomics Consortium et al., 2015). DNaseI-seq datasets for human fetal ovary and testis tissue have been used to identify putative enhancers (Ohnesorg et al., 2016). These tissues were presumably collected well after 4-6 weeks of gestation, the stage at which sex determination occurs in humans, and it is currently unclear whether enhancers that operate at the sex-determining stage are maintained in the late fetal stages. These issues can be addressed in mice; however, future studies will need to determine the extent of functional conservation between mouse and human enhancers. The genome-wide identification of regulatory sites that function during sex differentiation will enable more detailed studies on the regulation of sex-determining genes and will identify candidate regions that might harbor disease-causing mutations in humans with DSDs.
MATERIALS AND METHODS
Mouse strains, matings and cell/tissue isolation
To isolate Sertoli cells, Sox9-ECFP homozygous transgenic males (Kim et al., 2007b) were bred to CD-1 (Charles River) females in timed matings to generate E13.5 and E15.5 embryos. Following sorting by FACS, the cell pellet was snap-frozen and stored at −80°C for RNA-seq. For DNaseI-seq, cell pellets from the CFP-positive and CFP-negative fractions were resuspended in 250 µl of recovery-cell culture freezing media (Gibco) and slowly frozen to −80°C.
Mouse tissues (kidney, liver, heart, and brain) were collected from adult B6 mice. The B6 strain mouse fibroblast cell line was commercially available (Jackson Labs) and B6 strain ESCs were kindly provided by Ute Hochgeschwender (Duke University). For further details see the supplementary Materials and Methods.
Mice were housed in accordance with National Institutes of Health guidelines, and experimental protocols were approved by the Institutional Animal Care and Use Committee of Duke University Medical Center.
DNaseI-seq assay and data processing
DNaseI-seq assay was performed as previously described (Boyle et al., 2008a; Song and Crawford, 2010) with few modifications. DNaseI-seq libraries were prepared and data processed as previously described (Boyle et al., 2008b; Li and Durbin, 2009, Song and Crawford, 2010; Song et al., 2011). For further details see the supplementary Materials and Methods.
RNA preparation and RNA-seq data processing
E15.5 Sox9CFP-positive cells were pooled into three independent biological replicates containing ∼1 million cells. RNA isolation was performed using the RNeasy Micro Kit according to the manufacturer's protocol (Qiagen). Poly-A enriched mRNA libraries were generated from 0.8-1 µg of RNA using the standard Illumina Tru-Seq V2 protocol and sequencing was carried out by the Duke Genome Sequencing and Analysis Core.
RNA-seq data was processed using Bowtie version 0.12.7 (Langmead et al., 2009) and RSEM version 1.2.0 (Li and Dewey, 2011) as described in the supplementary Materials and Methods. RSEM expected counts were quartile normalized and square-root transformed.
TSS and regulatory domain assignments
Transcriptional start and end sites were downloaded from the UCSC Genome browser and used to generate regulatory domains using GREAT (McLean et al., 2010). For further details see the supplementary Materials and Methods.
Genomic location assignments
DHSs were assigned to specific genomic locations using previously published methods (Song et al., 2011). For further details see the supplementary Materials and Methods.
Cell type specificity categorization
Cell type specificity was determined by comparing DHSs from DNaseI-seq datasets from other cell types using the BedTools Software suite (Quinlan et al., 2010). For further details see the supplementary Materials and Methods.
CTCF overlap
We used previously published CTCF ChIP-seq datasets (Shen et al., 2012) and compared these against Sertoli-unique, Sertoli-specific and Sertoli-common DHSs using BedTools. For further details see the supplementary Materials and Methods.
DMRT1-binding site enrichment analysis
We used previously published DMRT1 ChIP-seq data (Krentz et al., 2013) and assessed overlap with Sertoli DHSs using BedTools. For further details see the supplementary Materials and Methods.
Enrichment of DHSs in Sertoli cell-expressed genes
To calculate enrichment, we assessed the number of DHSs that were assigned to each gene's regulatory domain. DHSs were divided as common, unique+shared or active enhancers (H3K27ac-positive). Significance was calculated using a two-sided Mann-Whitney test. For further details see the supplementary Materials and Methods.
Transient transgenics, immunocytochemistry and imaging
A putative regulatory region upstream of Wt1 was amplified and cloned into the Hsp68-LacZ reporter vector (Addgene; Plasmid #33351). DNA was prepared for zygote injection, and resuspended in EmbryoMax Injection Buffer (Millipore, MR-095-10F). Pronuclear injections into B6SJLF1/J zygotes were performed by the Duke Transgenic Core Facility to generate transient transgenics. Embryos were dissected at E13.5 and genotyped for the LacZ gene For immunostaining, fixed gonads were processed as previously described (Maatouk et al., 2012). Antibodies were used at the following dilutions: rat-anti-CDH1, 1:250 (Zymed, 13-1900); rabbit-anti-β-galactosidase, 1:10,000 (MP Biomedicals, 55976); goat-anti-MIS/AMH, 1:250 (Santa Cruz, sc-6886); Alexa Fluor 488-anti-rat, 1:500 (Molecular Probes, A21208); Cy3-anti-rabbit, 1:500 (Jackson ImmunoResearch Laboratories, 711-165-15); Alexa Fluor 647-anti-goat, 1:500 (Molecular Probes, A21447). For further details see the supplementary Materials and Methods.
ChIP-seq assay and data processing
For ChIP-seq, FACS-purified Sertoli cells were pelleted, resuspended, crosslinked and stored at −80°C. Briefly, 1 million cells were pooled from multiple sorts, washed, lysed, sonicated and incubated with bead-antibody complexes with 2.5 µg antibody (Rabbit-anti-H3K27ac; Abcam ab4729). Library preparation was performed on immunoprecipitated chromatin using the Rubicon ThruPLEX FD kit according to the manufacturer's protocol. Sequencing was performed at Duke's Genome Sequencing and Analysis core facility.
ChIP-seq reads were uniquely aligned with Bowtie (Langmead et al., 2009); peaks were called with SICER (Zang et al., 2009) using the input track as the control. For further details see the supplementary Materials and Methods.
Sequence analysis to identify predictive 6-mers and matching motifs
Enriched motifs were identified by incorporating previous methods (Lee et al., 2011; Natarajan et al., 2012). 6-mers and their reverse complements, were counted in each region, normalized by the length of the region, and used as features for an L1-norm sparse logistic regression classifier (Koh et al., 2007). Ten randomized iterations of fourfold cross-validation were performed to generate 40 different partitions of the data. 6-mers that showed non-zero regression coefficients in over 75% of the partitions were deemed to be significant and are shown in Table S3A-D. 6-mers were matched to known TF PWMs with TOMTOM (Gupta et al., 2007) using the JASPAR core vertebrate motifs, the UNIPROBE database and those generated by Jolma et al. (Bryne et al., 2007; Jolma et al., 2013; Hume et al., 2015). The SF1 (Baba et al., 2014) and DMRT1 (Krentz et al., 2013) motifs were included. For further details see the supplementary Materials and Methods.
Analysis of significance of motif matches
We used FIMO from the MEME Suite (Grant et al., 2011) to scan different regions of the genome. Length normalized motif match scores were used in a one-sided Mann-Whitney U-test to test for significance. For further details see the supplementary Materials and Methods.
Acknowledgements
We are grateful to the following people who assisted with this project: Olivier Fedrigo, Lisa Bukovnik and Fangfei Ye at Duke's Genome Sequencing and Analysis core facility; Chris Futtner, Cheryl Bock and Meilang Flowers at the Transgenic Core Facility for their work deriving the transient transgenic lines; and Mike Cook and Lynn Martinek at the Duke Comprehensive Cancer Center Flow Cytometry Shared Resource facility. The labs of Vivian Bardwell and David Zarkower (University of Minnesota) kindly shared DMRT1 ChIP-seq data prior to publication. We thank Steve Munger (Jackson Laboratories) for his guidance regarding RNA-seq and Ute Hochgeschwender (Duke University) for providing ESC material. Iordan Batchvarov and Allison Navis were very helpful in assisting with matings and genotyping. This article is dedicated to the memory of Danielle Maatouk, whose knowledge of mouse sex determination and exceptional laboratory skills made this study possible.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
D.M.M., A.N., G.E.C., U.O. and B.C. designed the experiments. D.M.M., A.N., L.S. and Y.S. carried out the experiments. D.M.M. and A.N. prepared the manuscript with input from all authors.
Funding
Funding was provided by the Duke University School of Medicine for use of the sequencing facility and the National Institutes of Health (5R01-HD039963-13 to B.C.). Deposited in PMC for release after 12 months.
Data availability
Datasets (including DNaseI-seq, ChIP-seq and RNA-seq) are publicly available in GEO (Accession Number GSE53076).
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.142554.supplemental
References
- Albrecht K. H. and Eicher E. M. (2001). Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev. Biol. 240, 92-107. 10.1006/dbio.2001.0438 [DOI] [PubMed] [Google Scholar]
- Arboleda V. A. and Vilain E. (2011). The evolution of the search for novel genes in mammalian sex determination: from mice to men. Mol. Genet. Metab. 104, 67-71. 10.1016/j.ymgme.2011.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleda V. A., Lee H., Sanchez F. J., Delot E. C., Sandberg D. E., Grody W. W., Nelson S. F. and Vilain E. (2013). Targeted massively parallel sequencing provides comprehensive genetic diagnosis for patients with disorders of sex development. Clin. Genet. 83, 35-43. 10.1111/j.1399-0004.2012.01879.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J. F., Pritchard-Jones K., Bickmore W. A., Hastie N. D. and Bard J. B. L. (1993). The expression of the Wilms’ tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 40, 85-97. 10.1016/0925-4773(93)90090-K [DOI] [PubMed] [Google Scholar]
- Baba T., Otake H., Sato T., Miyabayashi K., Shishido Y., Wang C.-Y., Shima Y., Kimura H., Yagi M., Ishihara Y. et al. (2014). Glycolytic genes are targets of the nuclear receptor Ad4BP/SF-1. Nat. Commun. 5, 3634 10.1038/ncomms4634 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W. and Noble W. S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202-W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrionuevo F. and Scherer G. (2010). SOX E genes: SOX9 and SOX8 in mammalian testis development. Int. J. Biochem. Cell Biol. 42, 433-436. 10.1016/j.biocel.2009.07.015 [DOI] [PubMed] [Google Scholar]
- Baxter R. M., Arboleda V. A., Lee H., Barseghyan H., Adam M. P., Fechner P. Y., Bargman R., Keegan C., Travers S., Schelley S.. et al. (2015). Exome sequencing for the diagnosis of 46,XY disorders of sex development. J. Clin. Endocrinol. Metab. 100, E333-E344. 10.1210/jc.2014-2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma G. J., Albrecht K. H., Washburn L. L., Recknagel A. K., Churchill G. A. and Eicher E. M. (2005). Gonadal sex reversal in mutant Dax1 XY mice: a failure to upregulate Sox9 in pre-Sertoli cells. Development 132, 3045-3054. 10.1242/dev.01890 [DOI] [PubMed] [Google Scholar]
- Bouma G. J., Affourtit J. P., Bult C. J. and Eicher E. M. (2007a). Transcriptional profile of mouse pre-granulosa and Sertoli cells isolated from early-differentiated fetal gonads. Gene Expr. Patterns 7, 113-123. 10.1016/j.modgep.2006.05.011 [DOI] [PubMed] [Google Scholar]
- Bouma G. J., Washburn L. L., Albrecht K. H. and Eicher E. M. (2007b). Correct dosage of Fog2 and Gata4 transcription factors is critical for fetal testis development in mice. Proc. Natl. Acad. Sci. USA 104, 14994-14999. 10.1073/pnas.0701677104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A. P., Davis S., Shulha H. P., Meltzer P., Margulies E. H., Weng Z., Furey T. S. and Crawford G. E. (2008a). High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311-322. 10.1016/j.cell.2007.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A. P., Guinney J., Crawford G. E. and Furey T. S. (2008b). F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics 24, 2537-2538. 10.1093/bioinformatics/btn480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne J. C., Valen E., Tang M.-H. E., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B. and Sandelin A. (2007). JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 36, D102-D106. 10.1093/nar/gkm955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C., Srinivasan R., Wu Z., Calo E., Acampora D., Faial T., Simeone A., Tan M., Swigut T. and Wysocka J. (2014). Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14, 838-853. 10.1016/j.stem.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y. and Greenleaf W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213-1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron F. J. and Sinclair A. H. (1997). Mutations in SRY and SOX9: testis-determining genes. Hum. Mutat. 9, 388-395. [DOI] [PubMed] [Google Scholar]
- Chaboissier M.-C., Kobayashi A., Vidal V. I. P., Lutzkendorf S., van de Kant H. J. G., Wegner M., de Rooij D. G., Behringer R. R. and Schedl A. (2004). Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 131, 1891-1901. 10.1242/dev.01087 [DOI] [PubMed] [Google Scholar]
- Chassot A.-A., Ranc F., Gregoire E. P., Roepers-Gajadien H. L., Taketo M. M., Camerino G., de Rooij D. G., Schedl A. and Chaboissier M.-C. (2008). Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum. Mol. Genet. 17, 1264-1277. 10.1093/hmg/ddn016 [DOI] [PubMed] [Google Scholar]
- Colvin J. S., Green R. P., Schmahl J., Capel B. and Ornitz D. M. (2001). Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell 104, 875-889. 10.1016/S0092-8674(01)00284-7 [DOI] [PubMed] [Google Scholar]
- Correa S. M., Washburn L. L., Kahlon R. S., Musson M. C., Bouma G. J., Eicher E. M. and Albrecht K. H. (2012). Sex reversal in C57BL/6J XY mice caused by increased expression of ovarian genes and insufficient activation of the testis determining pathway. PLoS Genet. 8, e1002569 10.1371/journal.pgen.1002569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., Steine E. J., Hanna J., Lodato M. A., Frampton G. M., Sharp P. A.. et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931-21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers S. and Sinclair A. (2012). Mammalian sex determination-insights from humans and mice. Chromosome Res. 20, 215-238. 10.1007/s10577-012-9274-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W., Dominguez-Steglich M. A., Guioli S., Kowk C., Weller P. A., Stevanović M., Weissenbach J., Mansour S., Young I. D., Goodfellow P. N.. et al. (1994). Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372, 525-530. 10.1038/372525a0 [DOI] [PubMed] [Google Scholar]
- Grant C. E., Bailey T. L. and Stafford Noble W. (2011). FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017-1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J. A., Bailey T. L. and Noble W. S. (2007). Quantifying similarity between motifs. Genome Biol. 8, R24 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A. et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311-318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., Ye Z., Lee L. K., Stuart R. K., Ching C. W.. et al. (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108-112. 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume M. A., Barrera L. A., Gisselbrecht S. S. and Bulyk M. L. (2015). UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 43, D117-D122. 10.1093/nar/gku1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson S. A., Lin Y.-T. and Capel B. (2012a). Testis development requires the repression of Wnt4 by Fgf signaling. Dev. Biol. 370, 24-32. 10.1016/j.ydbio.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson S. A., Natarajan A., Cool J., DeFalco T., Maatouk D. M., Mork L., Munger S. C. and Capel B. (2012b). Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 8, e1002575 10.1371/journal.pgen.1002575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A., Yan J., Whitington T., Toivonen J., Nitta K. R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G.. et al. (2013). DNA-binding specificities of human transcription factors. Cell 152, 327-339. 10.1016/j.cell.2012.12.009 [DOI] [PubMed] [Google Scholar]
- Jordan B. K., Mohammed M., Ching S. T., Délot E., Chen X.-N., Dewing P., Swain A., Rao P. N., Elejalde B. R. and Vilain E. (2001). Up-regulation of WNT-4 signaling and dosage-sensitive sex reversal in humans. Am. J. Hum. Genet. 68, 1102-1109. 10.1086/320125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kobayashi A., Sekido R., DiNapoli L., Brennan J., Chaboissier M.-C., Poulat F., Behringer R. R., Lovell-Badge R. and Capel B. (2006). Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 4, e187 10.1371/journal.pbio.0040187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Abdullaev Z. K., Smith A. D., Ching K. A., Loukinov D. I., Green R. D., Zhang M. Q., Lobanenkov V. V. and Ren B. (2007a). Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231-1245. 10.1016/j.cell.2006.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Bingham N., Sekido R., Parker K. L., Lovell-Badge R. and Capel B. (2007b). Fibroblast growth factor receptor 2 regulates proliferation and Sertoli differentiation during male sex determination. Proc. Natl. Acad. Sci. USA 104, 16558-16563. 10.1073/pnas.0702581104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.-J., Sock E., Buchberger A., Just W., Denzer F., Hoepffner W., German J., Cole T., Mann J., Seguin J. H. et al. (2015). Copy number variation of two separate regulatory regions upstream of SOX9 causes isolated 46,XY or 46,XX disorder of sex development. J. Med. Genet. 52, 240-247. 10.1136/jmedgenet-2014-102864 [DOI] [PubMed] [Google Scholar]
- Koh K., Kim S.-J. and Boyd S. (2007). An interior-point method for large-scale l1-regularized logistic regression. J. Machine Learn. Res. 8, 1519-1555. [Google Scholar]
- Kreidberg J. A., Sariola H., Loring J. M., Maeda M., Pelletier J., Housman D. and Jaenisch R. (1993). WT-1 is required for early kidney development. Cell 74, 679-691. 10.1016/0092-8674(93)90515-R [DOI] [PubMed] [Google Scholar]
- Krentz A. D., Murphy M. W., Zhang T., Sarver A. L., Jain S., Griswold M. D., Bardwell V. J. and Zarkower D. (2013). Interaction between DMRT1 function and genetic background modulates signaling and pluripotency to control tumor susceptibility in the fetal germ line. Dev. Biol. 377, 67-78. 10.1016/j.ydbio.2013.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. and Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D. A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S.. et al. (2014). Chromatin state dynamics during blood formation. Science 345, 943-949. 10.1126/science.1256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Karchin R. and Beer M. A. (2011). Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res. 21, 2167-2180. 10.1101/gr.121905.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. and Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. and Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754-1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng M. and Lau Y.-F. C. (2014). The sex-determining factors SRY and SOX9 regulate similar target genes and promote testis cord formation during testicular differentiation. Cell Rep. 8, 723-733. 10.1016/j.celrep.2014.06.055 [DOI] [PubMed] [Google Scholar]
- Liu C.-F., Bingham N., Parker K. and Yao H. H.-C. (2009). Sex-specific roles of beta-catenin in mouse gonadal development. Hum. Mol. Genet. 18, 405-417. 10.1093/hmg/ddn362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Ikeda Y. and Parker K. L. (1994). A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 77, 481-490. 10.1016/0092-8674(94)90211-9 [DOI] [PubMed] [Google Scholar]
- Maatouk D. M., Mork L., Hinson A., Kobayashi A., McMahon A. P. and Capel B. (2012). Germ cells are not required to establish the female pathway in mouse fetal gonads. PLoS ONE 7, e47238 10.1371/journal.pone.0047238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuylov N. L., Smagulova F. O., Leach L. and Tevosian S. G. (2008). Ovarian development in mice requires the GATA4-FOG2 transcription complex. Development 135, 3731-3743. 10.1242/dev.024653 [DOI] [PubMed] [Google Scholar]
- Matson C. K. and Zarkower D. (2012). Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163-174. 10.1038/nrg3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson C. K., Murphy M. W., Sarver A. L., Griswold M. D., Bardwell V. J. and Zarkower D. (2011). DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101-104. 10.1038/nature10239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M. and Bejerano G. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495-501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkina A., Matson C. K., Lindeman R. E., Ghyselinck N. B., Bardwell V. J. and Zarkower D. (2014). DMRT1 protects male gonadal cells from retinoid-dependent sexual transdifferentiation. Dev. Cell 29, 511-520. 10.1016/j.devcel.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mork L., Maatouk D. M., McMahon J. A., Guo J. J., Zhang P., McMahon A. P. and Capel B. (2012). Temporal differences in granulosa cell specification in the ovary reflect distinct follicle fates in mice. Biol. Reprod. 86, 37 10.1095/biolreprod.111.095208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger S. C., Aylor D. L., Syed H. A., Magwene P. M., Threadgill D. W. and Capel B. (2009). Elucidation of the transcription network governing mammalian sex determination by exploiting strain-specific susceptibility to sex reversal. Genes Dev. 23, 2521-2536. 10.1101/gad.1835809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger S. C., Natarajan A., Looger L. L., Ohler U. and Capel B. (2013). Fine time course expression analysis identifies cascades of activation and repression and maps a putative regulator of mammalian sex determination. PLoS Genet. 9, e1003630 10.1371/journal.pgen.1003630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscatelli F., Strom T. M., Walker A. P., Zanaria E., Récan D., Meindl A., Bardoni B., Guioli S., Zehetner G., Rabl W.. et al. (1994). Mutations in the DAX-1 gene give rise to both X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism. Nature 372, 672-676. 10.1038/372672a0 [DOI] [PubMed] [Google Scholar]
- Natarajan A., Yardimci G. G., Sheffield N. C., Crawford G. E. and Ohler U. (2012). Predicting cell-type-specific gene expression from regions of open chromatin. Genome Res. 22, 1711-1722. 10.1101/gr.135129.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nef S., Verma-Kurvari S., Merenmies J., Vassalli J.-D., Efstratiadis A., Accili D. and Parada L. F. (2003). Testis determination requires insulin receptor family function in mice. Nature 426, 291-295. 10.1038/nature02059 [DOI] [PubMed] [Google Scholar]
- Nef S., Schaad O., Stallings N. R., Cederroth C. R., Pitetti J.-L., Schaer G., Malki S., Dubois-Dauphin M., Boizet-Bonhoure B., Descombes P.. et al. (2005). Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev. Biol. 287, 361-377. 10.1016/j.ydbio.2005.09.008 [DOI] [PubMed] [Google Scholar]
- Ohnesorg T., Croft B., Tan J. and Sinclair A. H. (2016). Using ROADMAP data to identify enhancers associated with disorders of sex development. Sex. Dev. 10, 59-65. 10.1159/000445398 [DOI] [PubMed] [Google Scholar]
- Ottolenghi C., Omari S., Garcia-Ortiz J. E., Uda M., Crisponi L., Forabosco A., Pilia G. and Schlessinger D. (2005). Foxl2 is required for commitment to ovary differentiation. Hum. Mol. Genet. 14, 2053-2062. 10.1093/hmg/ddi210 [DOI] [PubMed] [Google Scholar]
- Pelletier J., Schalling M., Buckler A. J., Rogers A., Haber D. A. and Housman D. (1991). Expression of the Wilms’ tumor gene WT1 in the murine urogenital system. Genes Dev. 5, 1345-1356. 10.1101/gad.5.8.1345 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R. and Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A. and Wysocka J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279-283. 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317-330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Ovitt C. E., Anlag K., Fehsenfeld S., Gredsted L., Treier A.-C. and Treier M. (2004). The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 131, 933-942. 10.1242/dev.00969 [DOI] [PubMed] [Google Scholar]
- Sekido R. and Lovell-Badge R. (2008). Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453, 930-934. 10.1038/nature06944 [DOI] [PubMed] [Google Scholar]
- Sekido R., Bar I., Narváez V., Penny G. and Lovell-Badge R. (2004). SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev. Biol. 274, 271-279. 10.1016/j.ydbio.2004.07.011 [DOI] [PubMed] [Google Scholar]
- Sharrocks A. D. (2001). The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2, 827-837. 10.1038/35099076 [DOI] [PubMed] [Google Scholar]
- Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V. V.. et al. (2012). A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116-120. 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L. and Crawford G. E. (2010). DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010, pdb prot5384 10.1101/pdb.prot5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Zhang Z., Grasfeder L. L., Boyle A. P., Giresi P. G., Lee B.-K., Sheffield N. C., Graf S., Huss M., Keefe D.. et al. (2011). Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 21, 1757-1767. 10.1101/gr.121541.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenhaut N. H., Jakob S., Anlag K., Eisenberger T., Sekido R., Kress J., Treier A.-C., Klugmann C., Klasen C., Holter N. I.. et al. (2009). Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139, 1130-1142. 10.1016/j.cell.2009.11.021 [DOI] [PubMed] [Google Scholar]
- Vainio S., Heikkilä M., Kispert A., Chin N. and McMahon A. P. (1999). Female development in mammals is regulated by Wnt-4 signalling. Nature 397, 405-409. 10.1038/17068 [DOI] [PubMed] [Google Scholar]
- Warr N., Carre G.-A., Siggers P., Faleato J. V., Brixey R., Pope M., Bogani D., Childers M., Wells S., Scudamore C. L.. et al. (2012). Gadd45gamma and Map3k4 interactions regulate mouse testis determination via p38 MAPK-mediated control of Sry expression. Dev. Cell 23, 1020-1031. 10.1016/j.devcel.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S., Ohnesorg T., Notini A., Roeszler K., Hewitt J., Daggag H., Smith C., Turbitt E., Gustin S., van den Bergen J. et al. (2011). Copy number variation in patients with disorders of sex development due to 46,XY gonadal dysgenesis. PLoS ONE 6, e17793 10.1371/journal.pone.0017793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte W. A., Bilodeau S., Orlando D. A., Hoke H. A., Frampton G. M., Foster C. T., Cowley S. M. and Young R. A. (2012). Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221-225. 10.1038/nature10805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H., Shulha H. P., Lin J. M., Vales T. R., Fu Y., Bodine D. M., McKay R. D. G., Chenoweth J. G., Tesar P. J., Furey T. S. et al. (2007). Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 3, e136 10.1371/journal.pgen.0030136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanaria E., Muscatelli F., Bardoni B., Strom T. M., Guioli S., Guo W., Lalli E., Moser C., Walker A. P., McCabe E. R. B.. et al. (1994). An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature 372, 635-641. 10.1038/372635a0 [DOI] [PubMed] [Google Scholar]
- Zang C., Schones D. E., Zeng C., Cui K., Zhao K. and Peng W. (2009). A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952-1958. 10.1093/bioinformatics/btp340 [DOI] [PMC free article] [PubMed] [Google Scholar]