Abstract
Differential mRNA polyadenylation plays an important role in shaping the neuronal transcriptome. In C. elegans, several ankyrin isoforms are produced from the unc-44 locus through alternative polyadenylation. Here, we identify a key role for an intronic polyadenylation site (PAS) in temporal- and tissue-specific regulation of UNC-44/ankyrin isoforms. Removing an intronic PAS results in ectopic expression of the neuronal ankyrin isoform in non-neural tissues. This mis-expression underlies epidermal developmental defects in mutants of the conserved tumor suppressor death-associated protein kinase dapk-1. We have previously reported that the use of this intronic PAS depends on the nuclear polyadenylation factor SYDN-1, which inhibits the RNA polymerase II CTD phosphatase SSUP-72. Consistent with this, loss of sydn-1 blocks ectopic expression of neuronal ankyrin and suppresses epidermal morphology defects of dapk-1. These effects of sydn-1 are mediated by ssup-72 autonomously in the epidermis. We also show that a peptidyl-prolyl isomerase PINN-1 antagonizes SYDN-1 in the spatiotemporal control of neuronal ankyrin isoform. Moreover, the nuclear localization of PINN-1 is altered in dapk-1 mutants. Our data reveal that tissue and stage-specific expression of ankyrin isoforms relies on differential activity of positive and negative regulators of alternative polyadenylation.
KEY WORDS: DAPK-1/DAP kinase, SSUP-72/SSU72, PINN-1/PIN1 peptidyl-prolyl isomerase, Nuclear polyadenylation, SYDN-1
Summary: Selective repression of a neuronal isoform of ankyrin in epidermis depends on alternative polyadenylation at an intronic transcriptional termination site and is necessary for epidermal tissue integrity.
INTRODUCTION
Alternative processing of pre-mRNA is an important contributor to transcriptome diversity and cellular differentiation in multicellular organisms. Alternative processing of pre-mRNA 3′ ends, also known as alternative polyadenylation (APA), often involves coordinated action of transcriptional termination and polyadenylation using sites within introns, and is emerging as an important mechanism for spatiotemporal control of gene expression (Di Giammartino et al., 2011; Proudfoot, 2011). Tissue-specific APA has been documented in C. elegans (Blazie et al., 2015), Drosophila (Smibert et al., 2012), zebrafish (Ulitsky et al., 2012) and human (Lianoglou et al., 2013; Wang et al., 2008; Zhang et al., 2005), although its functional significance is only beginning to be understood.
The nervous system expresses specific protein isoforms that result from differential polyadenylation of mRNAs (Hilgers et al., 2012; Miura et al., 2013; Smibert et al., 2012; Ulitsky et al., 2012; Wang et al., 2008; Zhang et al., 2005). It has been hypothesized that brain-specific RNA-binding proteins, polyadenylation factors or RNA modifications play roles in brain-specific APA. For example, ELAV (embryonic-lethal abnormal visual system) binds proximal polyadenylation signals (PASs) and promotes 3′ UTR extension (Hilgers et al., 2012; Oktaba et al., 2015); brain-specific expression of a splice variant of polyadenylation factor CstF-64 (βCstF-64) also probably regulates APA of neural mRNAs (Shankarling et al., 2009); and higher proximal m6A density correlates with the use of distal PASs in the brain (Ke et al., 2015). Nevertheless, the mechanistic relationship of neuronal versus non-neuronal alternative polyadenylation remains poorly understood.
The nematode C. elegans has about 21,000 protein-coding genes. Similar to the human genome, more than half of C. elegans genes are thought to have alternative poly(A) sites (Blazie et al., 2015). Recent transcriptomic characterization of C. elegans 3′UTRs has revealed extensive use of alternative poly(A) sites, resulting in inclusion or exclusion of cis-regulatory elements within 3′UTRs (Jan et al., 2011; Mangone et al., 2010). Furthermore, most components of the pre-mRNA 3′ end processing complex are highly conserved between C. elegans and mammals (Cui et al., 2008). C. elegans is thus a simple and tractable model with which to dissect conserved mechanisms underlying tissue-specific APA.
The C. elegans unc-44 gene generates multiple ankyrin isoforms, of which the large isoform (UNC-44L) is generally thought to be neuron specific (Otsuka et al., 2002). We recently identified one mechanism that promotes UNC44L expression in neurons via nuclear alternative polyadenylation. A novel nuclear protein SYDN-1 inhibits SSUP-72, a Ser5 phosphatase for the C-terminal domain of RNA polymerase II. In neurons, this inhibition attenuates the use of an internal polyadenylation site (PAS) in unc-44, allowing downstream pre-mRNA polyadenylation and subsequent production of UNC-44L protein (Chen et al., 2015; Van Epps et al., 2010).
UNC-44L expression in the nervous system might simply require a neuron-specific pathway involving SYDN-1 and SSUP-72. However, based on expression and phenotypic data, both SYDN-1 and SSUP-72 likely function in non-neuronal cells. Here, by visualizing endogenous UNC-44 isoforms using GFP knock-ins, we show that deletion of the intronic unc-44 PAS causes ectopic expression of UNC-44L in the epidermis, disrupting epidermal morphology. The antagonistic interaction between SYDN-1 and SSUP-72 promotes the use of the internal PAS of unc-44 to repress the production of UNC-44L in the epidermis. The tumor suppressor death-associated protein kinase DAPK-1 counteracts the action of SYDN-1. We further identify PINN-1, the C. elegans member of the Pin1 family of peptidyl-prolyl isomerases, as a new partner of SSUP-72 and find that DAPK-1 can affect PINN-1 localization in epidermis. Our results demonstrate how tissue-specific expression of ankyrin isoforms is driven by the differential function of positive and negative regulators of alternative polyadenylation.
RESULTS
Expression of the ankyrin long isoform UNC-44L is temporally and spatially distinct from other UNC-44 proteins
Ankyrins, a highly conserved family of scaffold proteins, mediate attachment of integral membrane proteins to the spectrin-actin-based membrane cytoskeleton (Bennett and Baines, 2001). In C. elegans, the unc-44 ankyrin gene extends over 40 kb, and produces multiple mRNA isoforms that are terminated predominantly at three poly(A) sites (designated proximal PAS1, PAS2 and the distal PAS3) (Chen et al., 2015) (Fig. 1A, Fig. S1A). The N-terminal common domains of the major UNC-44 isoforms contain multiple ankyrin repeats and a spectrin-binding domain followed by a regulatory domain (Otsuka et al., 2002). Previous studies of UNC-44 expression used antibodies raised against the spectrin-binding domain, the conventional regulatory domain, or the extended C-terminal set of STEP domains specific to the large isoform. These studies showed that the common ankyrin proteins were widely expressed in multiple tissues, including oocytes, epidermis, the vulva and the nervous system, whereas the long isoform was restricted to the nervous system (Otsuka et al., 2002).
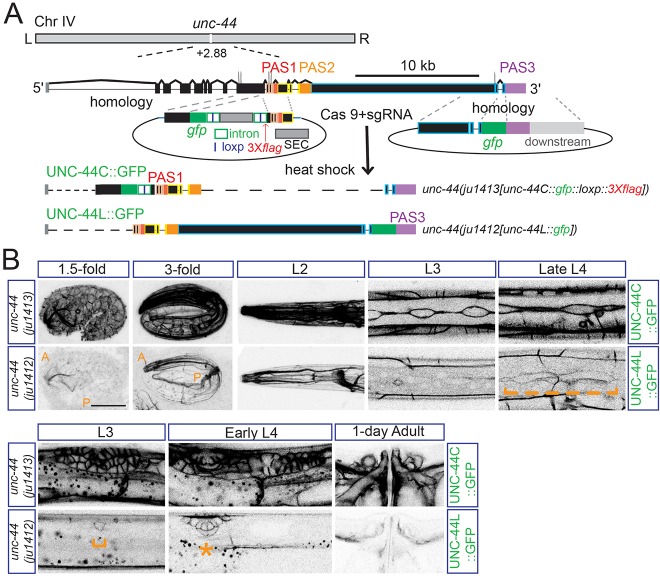
Fig. 1.
Expression of the neuronal ankyrin UNC-44L is temporally and spatially distinct from other UNC-44 proteins. (A) Experimental design for gfp knock-in lines of unc-44. The unc-44 gene locus produces multiple mRNA isoforms terminating at three major poly(A) sites: PAS1, PAS2 and PAS3. The use of PAS3 is reported to be neuron specific. Templates for recombination at different locations are indicated. Scale bar: 10 kb. (B) UNC-44L shows distinct temporal and tissue-specific expression from UNC-44C. Upper panels show GFP fluorescence of common UNC-44 isoforms (UNC-44C) and lower panels show GFP fluorescence of neuronal UNC-44 (UNC-44L). UNC-44C was strongly expressed in multiple tissues: nervous system (from 1.5-fold stage to adult), epidermis [from early embryo (Fig. S1A) to adult], seam cells (from L1 to L4), vulva (from L3 to adult), spermathecal/sheath cells (from L4 to adult). Starting from the 1.5-fold stage, UNC-44L was expressed abundantly in the nervous system, although low-level expression was also detected in seam cells at late L4 (orange broken bracket) and in vulva cells from L3 (lower panels, orange bracket and star). The anterior (A) and posterior (P) bodies are labeled at the 1.5-fold and 3-fold stages. Scale bar: 20 µm.
To examine the distribution of differentially polyadenylated isoforms of unc-44 in live animals, we used CRISPR/Cas9 genome editing to tag the endogenous unc-44 gene (see Materials and Methods). We inserted GFP after Leu1818 in an exon encoding the spectrin-binding domain, which is present in most isoforms of unc-44 [Fig. 1A, ju1413(unc-44::gfp::loxp::3Xflag), UNC-44C::GFP, with GFP inserted after L1818], and after His6987 in the last exon specific to the transcript encoding the large isoform [Fig. 1A, ju1412(unc-44::gfp(PAS3)), UNC-44L::GFP, GFP inserted after H6987]. Strong loss of function in unc-44 causes extremely uncoordinated movement (Otsuka et al., 1995). Neither insertion disrupted unc-44 function, as the knock-in animals superficially resembled the wild type in movement and growth rate. In adults, the pattern of UNC-44C::GFP(ju1413) fluorescence was generally consistent with that described for UNC-44 common isoforms by immunostaining (Otsuka et al., 2002); however, we also observed strong expression in muscles and in gonadal spermathecal/sheath cells (Fig. 1B, upper panel). UNC-44C::GFP(ju1413) expression was detected from early embryos to late larvae. Immediately before the bean stage of embryogenesis, UNC-44C::GFP proteins were expressed in epidermal cells, within which they localized to the cell periphery (Fig. S1, embryo). At postnatal L1 stage, UNC-44C::GFP was evident at the peripheries of epidermal seam cells and gut cells (Fig. S1, L1). In late larvae and adults, UNC-44C::GFP was seen in cells undergoing morphogenesis or fusion, such as vulval and seam cells.
In contrast to UNC-44C::GFP, we did not detect expression of UNC-44L::GFP(ju1412) before the bean stage of embryogenesis. By the 1.5-fold stage of embryogenesis, we observed expression of UNC-44L::GFP mainly in the developing nervous system (Fig. 1B, lower panel). UNC-44L::GFP became progressively more concentrated in the nervous system, predominantly localizing to neuronal processes and to the peripheries of neuronal cell bodies (Otsuka et al., 2002) (Fig. 1B, lower panel). UNC-44L has been previously thought to be exclusively neuronal, based on immunostaining experiments. However, we observed faint expression of UNC-44L::GFP in epidermal seam cells at the L4 stage, and in the gonadal anchor cell at L3 stage, as well as in neighboring vulval cells at L4 (Fig. 1B, lower panel). Thus, the regulation of UNC-44L is temporally and spatially distinct from the more widely expressed UNC-44C isoforms, and is predominantly but not exclusively confined to neurons.
SSUP-72 regulates tissue- and stage-specific expression of UNC-44L via use of the internal PAS2
Previously, we found that a novel nuclear protein SYDN-1 is required for UNC-44L expression in neurons, acting to inhibit SSUP-72 and a subset of pre-mRNA 3′ end processing factors (Chen et al., 2015). Consistent with these findings, sydn-1(0) mutants displayed greatly decreased expression of UNC-44L::GFP in the nervous system and in the adult vulva. In sydn-1(0); ssup-72(0) double mutants, expression of UNC-44L::GFP in neurons was restored to normal, consistent with SYDN-1 inhibiting SSUP-72 to allow production of UNC-44L (Fig. 2A). Intriguingly, in ssup-72(0) single mutants and sydn-1(0); ssup-72(0) double mutants, we also observed ectopic expression of UNC-44L::GFP in non-neuronal tissues such as the epidermis (Fig. 2A), suggesting that SSUP-72 normally inhibits UNC-44L expression in non-neuronal tissues.
Fig. 2.
SSUP-72 is required for the temporal and tissue-specific expression of unc-44 large isoform via the intronic PAS2 region. (A) SYDN-1 is required for the expression of UNC-44L in all tissues by negatively regulating SSUP-72. Images show GFP fluorescence of neuronal UNC-44 (UNC-44L) in unc-44(ju1412[unc-44L::gfp]) animals. In sydn-1(0), the expression of unc-44 large isoform is strikingly reduced in multiple tissues, especially in the nervous system. In sydn-1(0); ssup-72(0), expression of UNC-44L in the nervous system and vulva is recovered. Moreover, ectopic expression of UNC-44L in epidermis and other tissues was also observed. Scale bar: 20 µm. (B) unc-44(ju1378) contains a 180 bp deletion in the PAS2-containing intron, which affects the internal PAS2 but not the splice sites. The deleted region is outlined by the orange rectangle. The template for C-terminal recombination was injected into unc-44(ju1378) to tag the large isoform of unc-44 in unc-44(ju1378). (C) SSUP-72 functions through the internal PAS2 to regulate the spatiotemporal pattern of UNC-44L. Images show GFP fluorescence of neuronal UNC-44 (UNC-44L). Compared with UNC-44L expression in the wild type [1st row, unc-44(ju1412)], when PAS2 was deleted [2nd row, unc-44(ju1378 ju1412)], UNC-44L was mis-expressed in epidermis (red stars) from the embryonic stage in L3 seam cells (broken red bracket), L3 vulva cells (orange bracket and broken red bracket) and in L4/adult spermathecal/sheath cells (broken red bracket); UNC-44L was also upregulated in the nervous system, in L4 seam cells (broken orange and red brackets) and L4/adult vulva (orange stars). ssup-72(0) [3rd row, unc-44(ju1412); ssup-72(0)] shows similar expression of UNC-44L to the PAS2-deleted strain [unc-44(ju1378 ju1412)]. Upregulation and ectopic expression of UNC-44L was not further enhanced in unc-44(ju1378 ju1412); ssup-72(0) (4th row) compared with either unc-44(ju1378 ju1412) or unc-44(ju1412); ssup-72(0). Anterior (A) and posterior (P) bodies are labeled at the 1.5-fold and 3-fold stages. Scale bar: 20 µm. (D) Quantitation of the effect of SSUP-72 and internal PAS2 on upregulation of UNC-44L in the nervous system. GFP intensity along the dorsal cord was scored. Intensities from different animals were normalized to the average intensity from the control strain (unc-44(ju1412[unc-44L::gfp])). Data are mean normalized intensities±s.e.m. Numbers of animals analyzed are indicated. *P<0.05, **P<0.01 (one-way ANOVA and Bonferroni's multiple comparison post-test).
Our previous studies also showed that a 180 bp deletion in the intron containing PAS2, unc-44(ju1378), results in increased production of unc-44L mRNA in neurons (Chen et al., 2015). To test how PAS2 affects the tissue specificity of UNC-44L expression, we inserted GFP after His6987 in the unc-44(ju1378) deletion locus [Fig. 2B, allele designation unc-44(ju1378 ju1412)], together denoted as UNC-44(ju1378)L::GFP. We also detected UNC-44(ju1378)L::GFP in non-neuronal tissues, at higher levels and at earlier times than UNC-44L::GFP. At the 1.5-fold stage of embryogenesis, UNC-44(ju1378)L::GFP began to be expressed in the epidermis; this epidermal expression became more obvious at L2 and later stages (Fig. 2C, unc-44(ju1378 ju1412)). UNC-44(ju1378)L::GFP was expressed in seam cells at the L3 stage, earlier than UNC-44L::GFP (Fig. 2C, L3). Besides the L3 anchor cell, UNC-44L::GFP was also expressed in neighboring vulval cells (Fig. 2C, L3). We observed higher levels of UNC-44(ju1378)L::GFP expression in the adult vulva (Fig. 2C, 1-day adult) and in spermathecal/sheath cells in the somatic gonad from L4 stage (Fig. 2C, L4), which was never observed in PAS2-containing UNC-44L::GFP animals. Taken together, deletion of the internal PAS2 results in upregulation of UNC-44L::GFP in the cells and stages that it is normally expressed in, i.e. nervous system, L4 seam cells and adult vulva, as well as precocious or ectopic expression in the epidermis and other cells [Fig. 2C, unc-44(ju1412) vs unc-44(ju1378 ju1412)]. These results suggest that PAS2 use is a key determinant of the spatiotemporal pattern of UNC-44L expression.
As the expression of UNC-44(ju1378)L::GFP resembles that of UNC-44L::GFP in ssup-72(0), we next made double mutants of ssup-72(0); UNC-44(ju1378)L::GFP. Loss of function in ssup-72 did not result in further upregulation or misexpression of UNC-44(ju1378)L::GFP (Fig. 2D). As a control, neither sydn-1(0) nor ssup-72(0) affected UNC-44C::GFP expression in ju1447(unc-44C::gfp::loxp::SEC::loxp::3Xflag), suggesting the SYDN-1/SSUP-72 pathway does not affect transcription of common isoforms of unc-44 (Fig. S2, see Materials and Methods). It has been reported that genes with multiple UTRs tend to be transcribed ubiquitously and display tissue-specific changes in 3′UTR use (Lianoglou et al., 2013). Our data suggest that tissue-specific regulation of PAS2 use by SYDN-1 and SSUP-72 controls the ratio of UNC-44L to other UNC-44 isoforms both in neurons and in other tissues.
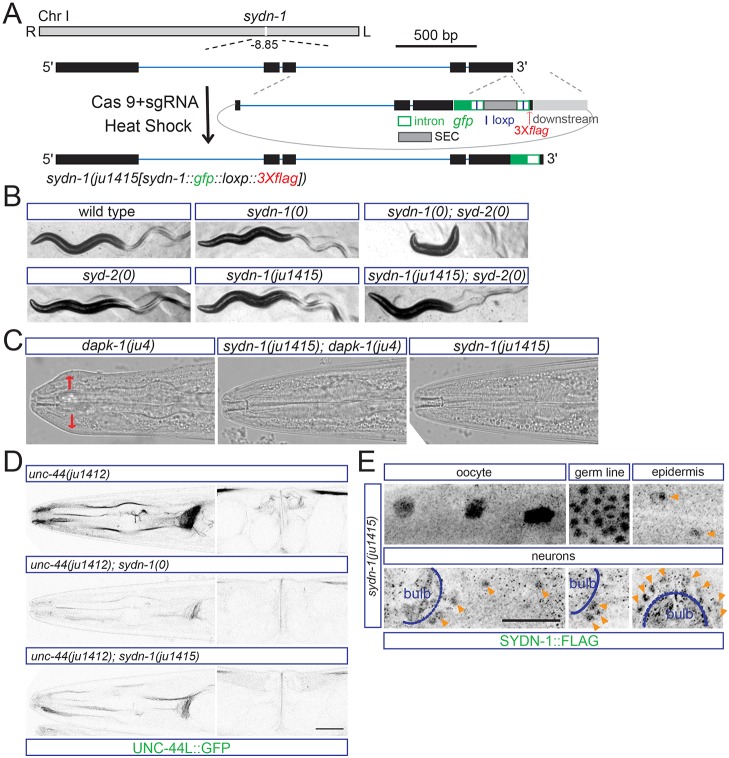
SYDN-1 is expressed at low levels in many tissues
Our previous studies showed that SYDN-1 and SSUP-72 act cell-autonomously in neurons (Chen et al., 2015). ssup-72 is expressed in many tissues, whereas the endogenous expression pattern of SYDN-1 has not yet been reported. As shown above, UNC-44L::GFP appears to be expressed at low levels in non-neuronal tissues (Fig. 1B), and sydn-1(0) mutants displayed decreased UNC-44L in both neurons and non-neuronal tissues (Fig. 2A), suggesting that SYDN-1 might regulate UNC-44L in multiple tissues. To visualize SYDN-1 expression from the endogenous chromosomal locus, we inserted gfp::3Xflag at the C terminus of SYDN-1 [Fig. 3A, ju1415(sydn-1::gfp::loxp::3Xflag)]. The sydn-1(ju1415) knock-in allele mildly disrupts SYDN-1 function as animals showed slightly uncoordinated movement. Unlike sydn-1(0) mutants, sydn-1(ju1415) did not strongly enhance syd-2(0) locomotor defects (Fig. 3B). Expression of UNC-44L::GFP was visibly reduced in sydn-1(ju1415) compared with sydn-1(+), although it was noticeably higher than in sydn-1(0) (Fig. 3D). Taken together, these observations suggest the sydn-1(ju1415) causes a partial loss of sydn-1 function. We did not detect GFP fluorescence in sydn-1(ju1415) knock-in animals, probably reflecting the low endogenous expression levels of SYDN-1. By immunostaining with FLAG antibody, we detected weak signals in multiple tissues, including oocytes, germ cells, neurons and epidermis (Fig. 3E). These findings suggest the SYDN-1 pathway can control UNC-44L expression in multiple tissues in a cell-autonomous manner.
Fig. 3.
SYDN-1 is expressed in multiple tissues. (A) Experimental design for making gfp::3Xflag knock-in worms of the sydn-1 gene by CRISPR/Cas9 editing. gfp is inserted at the C terminus of sydn-1. The template for C-terminal recombination is indicated. Scale bar: 500 bp. (B) The knock-in allele of sydn-1 mildly enhances syd-2(0) defects. syd-2/α-liprin is essential for synaptogenesis. sydn-1(0) or syd-2(0) single mutants display mildly uncoordinated movement, whereas double mutants display severe paralysis. sydn-1(ju1415) also shows mild uncoordinated movement similar to sydn-1(0). However, sydn-1(ju1415); syd-2(0) double mutants show a less severe locomotor phenotype compared with sydn-1(0); syd-2(0). (C) ju1415 is a loss-of-function allele of sydn-1. Images show head morphology of wild type and mutants. sydn-1(ju1415) suppresses the Mor phenotypes of dapk-1 to the same extent as sydn-1(0). Head region with epidermal defects is labeled by red arrows. (D) UNC-44L::GFP fluorescence from unc-44(ju1412) in sydn-1(ju1415) is reduced compared with wild type, and is noticeably higher than in sydn-1(0). (E) SYDN-1 is widely expressed. Images show immunostaining results with anti-FLAG in sydn-1(ju1415) animals. Immunostaining with anti-FLAG shows expression of SYDN-1::GFP::Flag in nuclei of oocytes, germline, epidermis and neurons. The pharyngeal bulb is labeled in the head image. Nuclear staining of SYDN-1 in epidermal cells or neurons is labeled (orange arrowheads). Scale bar: 20 µm.
SYDN-1-dependent inhibition of SSUP-72 is required for epidermal morphology defects of dapk-1 mutants
The C. elegans ortholog of the tumor suppressor death-associated protein kinase DAPK-1 regulates epidermal integrity (Tong et al., 2009). Loss of DAPK-1 function, as caused by the dominant-negative ju4 allele or by the gk219 deletion allele, results in morphological defects in the epidermis (Mor phenotype) and formation of progressive scar-like structures in the cuticle (Chuang et al., 2016). These defects are suppressed by sydn-1(0) (Tong et al., 2009), and this suppression is rescued by epidermal expression of SYDN-1, indicating SYDN-1 functions cell-autonomously in the epidermis. We observed that the sydn-1(ju1415) insertion also completely suppressed dapk-1 Mor phenotypes (Fig. 3C), comparable with suppression by sydn-1(0) (Fig. 4B), suggesting that, in dapk-1 mutants, epidermal development is sensitized to disruption of sydn-1 function. dapk-1 sydn-1(0) ssup-72(0) triple mutants displayed epidermal Mor defects resembling those of dapk-1 single mutants, indicating that negative regulation of SSUP-72 by SYDN-1 contributes to the Mor phenotype. Another sydn-1(0) suppressor, zfp-3(0), also restored epidermal Mor defects in dapk-1(ju4) sydn-1(0), consistent with DAPK-1 negatively regulating or antagonizing the SYDN-1-dependent pre-mRNA nuclear polyadenylation.
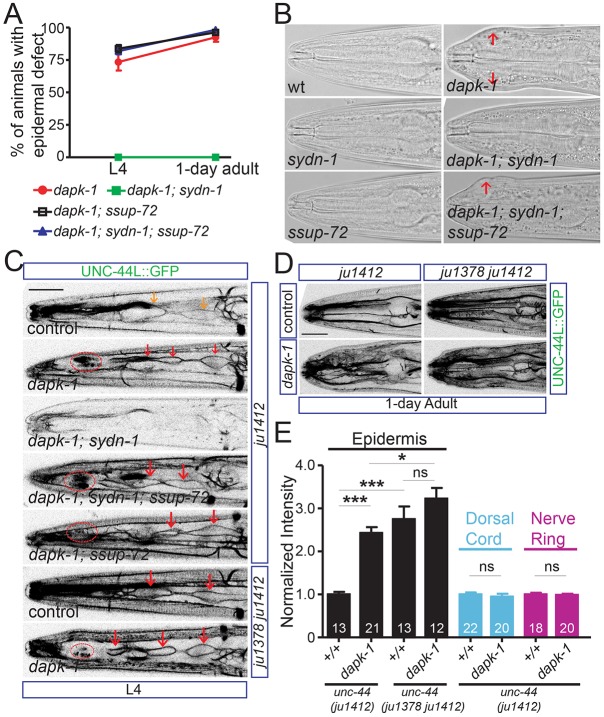
Fig. 4.
DAPK-1 inhibits ectopic expression of UNC-44L in epidermis by antagonizing SYDN-1 function. (A) DAPK-1 maintains epidermal integrity in part by negatively regulating the SYDN-1 and SSUP-72 pathway. Graph shows fraction of epidermal morphology (Mor) defects at L4 and 1-day-old adult stages. (B) sydn-1(0) completely suppresses the Mor defects of dapk-1(ju4), and ssup-72(0) recovers the Mor defect in dapk-1(ju4); sydn-1(0). Images show head morphology in wild type and mutants (epidermal defects are labeled by red arrows). (C) DAPK-1 inhibits UNC-44L::GFP expression in L4 seam cells by negatively regulating the SYDN-1/SSUP-72 pathway. Representative images show GFP fluorescence of UNC-44L (unc-44(ju1412[unc-44L::gfp])) in L4 seam cells in genotypes indicated. dapk-1(ju4) induced upregulation of UNC-44L in L4 seam cells, which is suppressed by sydn-1(0). ssup-72(0) recovered the upregulation of UNC-44L in L4 seam cells in dapk-1(ju4); sydn-1(0). dapk-1(ju4) does not further enhance the UNC-44L expression in L4 seam cells in the unc-44(ju1378) strain (lower panels). Orange arrows label seam cell expression of UNC-44L-GFP in control; red arrows label increased seam cell expression of UNC-44L-GFP; red ovals label scar-like structures, which show autofluorescence in the same channel as GFP. Scale bar: 20 µm. (D) DAPK-1 inhibits ectopic expression of UNC-44L in epidermis via the internal PAS2. Representative images show epidermal GFP fluorescence of UNC-44L (unc-44(ju1412[unc-44L::gfp])) in the genotypes indicated. In dapk-1(ju4), UNC-44L::GFP is ectopically expressed in epidermal cells. Ectopic expression of UNC-44L, caused by deletion of the internal PAS2, is not further enhanced in dapk-1(ju4). Scale bar: 20 µm. (E) DAPK-1 specifically inhibits UNC-44L expression in epidermis. dapk-1(ju4) mutants display elevated UNC-44L expression in the adult epidermis and normal neuronal expression. Ectopic expression of UNC-44L::GFP in adult epidermis is not further enhanced in dapk-1(ju4); unc-44(ju1378 ju1412) compared with unc-44(ju1378 ju1412). GFP intensity was scored. Intensities from different animals were normalized to the average intensity from the control strain (unc-44(ju1412[unc-44L::gfp])). Data are mean normalized intensities±s.e.m. Numbers of animals scored are indicated. *P<0.05, ***P<0.001 (one-way ANOVA and Bonferroni's multiple comparison post-test).
As SYDN-1 and SSUP-72 regulate the expression of UNC-44L, we asked whether UNC-44L expression might account for the cellular defects of dapk-1 mutants. We tested two alleles of dapk-1: ju4 and gk219 (Chuang et al., 2016). dapk-1(ju4) mutants displayed increased UNC-44L::GFP expression in L4 seam cells, and ectopic expression of UNC-44L::GFP in the head and tail epidermal cells at L4 and later stages (Fig. 4C-E). UNC-44L::GFP expression in neurons and vulva was not altered by dapk-1(ju4) (Fig. S3A), suggesting DAPK-1 only inhibits UNC-44L expression in the epidermis. dapk-1(gk219) also induced ectopic expression of UNC-44L only in adult head epidermis (Fig. S4A), although upregulation of UNC-44L was not seen in L4 seam cells. The expression of UNC-44C in seam cell or head epidermis was not altered in dapk-1(ju4) (Fig. S3B) or dapk-1(gk219) (Fig. S4B), implying that DAPK-1 normally decreases UNC-44L/UNC-44C ratio in the seam cells and epidermis.
Consistent with previous findings that sydn-1(0) suppresses the epidermal defects of dapk-1 (Tong et al., 2009), sydn-1(0) also suppressed the upregulation of UNC-44L::GFP in the seam cells of dapk-1 mutants (Fig. 4C). Upregulation of UNC-44L in seam cells was recovered in ssup-72(0); sydn-1(0); dapk-1(ju4) (Fig. 4C), suggesting that, as in neurons, expression of UNC-44L in seam cells depends on cell-autonomous inhibition of SSUP-72 activity by SYDN-1. Loss of function in dapk-1 did not further increase UNC-44L::GFP expression in seam cells and epidermis in unc-44(ju1378) ΔPAS2 animals (Fig. 4C, lower panel; 4D, right panel; and 4E), consistent with DAPK-1 negatively regulating SYDN-1/SSUP-72 function at the internal PAS2 region to decrease the UNC-44L/UNC-44C ratio.
Ectopic expression of UNC-44L in epidermis contributes to dapk-1 epidermal defects
We next asked whether ectopic expression of UNC-44L in the epidermis contributed to the epidermal defects of dapk-1 mutants. We used unc-44(tm349), which contains a deletion within the large isoform-specific exon and causes loss of function of the unc-44 large isoform (Chen et al., 2015). unc-44(tm349) completely suppressed the epidermal defects of dapk-1 mutants (Fig. 5A), consistent with DAPK-1 negatively regulating UNC-44L. Although unc-44(ju1378) and ssup-72(0) resulted in abundant ectopic expression of UNC-44L in epidermis and upregulation of UNC-44L in L4 seam cells (Fig. 2C), they neither induced epidermal defects in a wild-type background, nor enhanced epidermal defects of dapk-1(ju4) (Fig. 5B), suggesting elevated UNC-44L levels are insufficient to induce Mor defects. We infer that the epidermal defects of dapk-1 mutants are likely caused by ectopic expression of UNC-44L in the epidermis as well as other events downstream of loss of DAPK-1 activity. Interestingly, both unc-44(ju1378) and ssup-72(0) enhanced the lethality of dapk-1(ju4) in early adulthood (Fig. 5C). It is possible that dapk-1(ju4) is specifically sensitized to ectopic expression and upregulation of UNC-44L, leading to enhanced epidermal fragility and lethality. unc-44(ju1378) did not enhance the lethality of dapk-1(gk219) in early adulthood (Fig. S4C,D), likely due to the less pronounced change in UNC-44L levels induced by dapk-1(gk219).
Fig. 5.

Ectopic expression of UNC-44L causes epidermal defects in dapk-1 mutants. (A) DAPK-1 negatively regulates UNC-44L. Images show head morphology in wild type and mutants. unc-44(tm349), which contains a deletion in the large isoform-specific exon and causes an open reading frame shift, suppresses the Mor phenotypes of dapk-1(ju4) (arrows). (B) Dysregulation of UNC-44L does not enhance the Mor phenotype of dapk-1(ju4). Quantification shows the fraction of animals with epidermal defects in genotypes indicated at L4 and 1-day-old adult stages. The fraction of animals displaying Mor defects in ssup-72(0); dapk-1(ju4) or unc-44(ju1378 ju1412); dapk-1(ju4) is comparable with dapk-1 single mutants. (C) Dysregulated UNC-44L exacerbates lethality of dapk-1(ju4). Percentage of living animals in genotypes indicated at 1-day-old and 2-day-old adult stages. In ssup-72(0); dapk-1(ju4) or unc-44(ju1378 ju1412); dapk-1(ju4), fewer worms survive to early adulthood compared with dapk-1(ju4) single mutants. (D) Exclusion of UNC-44L in larval epidermis is required for maintaining epidermal integrity in dapk-1(ju4). Expression of SSUP-72 in larval epidermis driven by Pcol-10 or Pdpy-7 strongly reduces Mor phenotypes in dapk-1(ju4); sydn-1(0); ssup-72(0); unc-44(ju1412). Expression of SSUP-72 in adult epidermis driven by Pcol-19 modestly reduces Mor phenotypes in dapk-1(ju4); sydn-1(0); ssup-72(0); unc-44(ju1412). Expression of SSUP-72 in neurons driven by Prgef-1 does not reduce Mor phenotypes in dapk-1(ju4); sydn-1(0); ssup-72(0); unc-44(ju1412). Transgenic worms were picked and animals with epidermal defects were counted. Data are average ratio±s.e.m. from three independent experiments. Numbers of animals analyzed are indicated. *P<0.05, ***P<0.001 (one-way ANOVA and Bonferroni's multiple comparison post-test).
To address whether ectopic expression of UNC-44L in the epidermis contributes to morphological defects of dapk-1 mutants, we expressed SSUP-72 in the epidermis of dapk-1(ju4); sydn-1(0); ssup-72(0); unc-44(ju1412) animals. We used three epidermal cell-specific promoters (Fig. S5A): Pcol-10, which is active in larval and adult stages (Cox and Hirsh, 1985; Thein et al., 2003); Pcol-19, which is active from late L4 to adult (Thein et al., 2003); and Pdpy-7, which is active in embryonic and larval, but not adult, epidermis (Gilleard et al., 1997). Overexpression of SSUP-72 in larval epidermis (Pcol-10 or Pdpy-7) strongly suppressed Mor phenotypes in the dapk-1(ju4); sydn-1(0); ssup-72(0); unc-44(ju1412) background (Fig. 5D). Moreover, epidermal mis-expression of UNC-44L in L4 and adult dapk-1(ju4); sydn-1(0); ssup-72(0); unc-44(ju1412) animals was strikingly reduced in animals expressing SSUP-72 in larval epidermis (Fig. S5B,C, Tables S1, S2). By contrast, expression of SSUP-72 under the control of the adult-specific Pcol-19 promoter slightly reduced the Mor defects of dapk-1(ju4); sydn-1(0); ssup-72(0); unc-44(ju1412) animals (Fig. 5D) and did not significantly reduce epidermal UNC-44L::GFP at the L4 stage, although we observed a mild reduction of UNC-44L::GFP in adults (Fig. S5B,C). When SSUP-72 was driven by the pan-neuronal promoter (Prgef-1), neuronal expression of UNC-44L in dapk-1(ju4); sydn-1(0); ssup-72(0); unc-44(ju1412) was greatly decreased (Fig. S5B,C), but the fraction of animals with epidermal defects was unchanged (Fig. 5D). The above results suggest ectopic expression of UNC-44L in the larval epidermis, due to inappropriate activity of SSUP-72, contributes to epidermal defects in dapk-1 mutants.
The peptidyl-prolyl isomerase PINN-1 represses expression of UNC-44L
Next, we asked how SSUP-72 activity might be upregulated in the epidermis to prevent expression of UNC-44L. DAPK-1 is not localized to cell nuclei (Chuang et al., 2016) nor does loss of function in dapk-1 affect expression levels of SYDN-1, suggesting DAPK-1 might regulate the SYDN-1/SSUP-72 pathway indirectly.
The dephosphorylation of RNA polymerase II C-terminal domain (CTD) by the yeast homolog Ssu72 could be facilitated by Ess1/Pin1 peptidyl-prolyl cis-trans isomerase (Krishnamurthy et al., 2009; Werner-Allen et al., 2011). Moreover, murine Pin1 can be phosphorylated by death-associated protein kinase 1 (DAPK1), and this phosphorylation alters nuclear localization and enzymatic activity of Pin1 (Lee et al., 2011). The Ser residue phosphorylated by DAPK-1 is conserved in C. elegans PINN-1 (Fig. S6A, residue 66 in C. elegans PINN-1). We therefore asked whether pinn-1 might function in ssup-72-dependent regulation of unc-44 APA. pinn-1(tm2235) removes the first three exons of the gene, likely resulting in a null mutation (Fig. S6B). pinn-1(tm2235) suppressed the behavioral defects of sydn-1(0) and recovered neuronal expression of UNC-44L in sydn-1(0) (Fig. 6A). In pinn-1(0) single mutants and in sydn-1(0); pinn-1(0) double mutants, misexpression of UNC-44L was also observed in the epidermis (Fig. 6A). The spatiotemporal expression of UNC-44L in pinn-1(0) resembled that in ssup-72(0) mutants (Fig. 6B). These observations suggest PINN-1, like SSUP-72, might antagonize SYDN-1 function.
Fig. 6.

SYDN-1 and PINN-1 have antagonistic effects on UNC-44L expression. (A) SYDN-1 counteracts the function of PINN-1 in regulating UNC-44L expression. Images show GFP fluorescence of UNC-44L in unc-44(ju1412[unc-44L::gfp]) animals. The null allele of pinn-1, tm2235, recovered neuronal expression of UNC-44L in sydn-1(0). In sydn-1(0); pinn-1(0) double mutants and in pinn-1(0), UNC-44L is ectopically expressed in the epidermis (red stars) and upregulated in the vulva. Scale bar: 20 µm. (B) PINN-1 regulates spatiotemporal pattern of UNC-44L expression. Images show GFP fluorescence of UNC-44L in unc-44(ju1412[unc-44L::gfp]) animals. Compared with UNC-44L expression in wild type [upper panels, unc-44(ju1412)], UNC-44L was mis-expressed in epidermis from embryonic stage (red star), in L3 seam cells (broken red bracket), L3 vulva cells (orange bracket and broken red bracket) and in L4/adult spermathecal/sheath cells (broken red bracket). Anterior (A) and posterior (P) bodies are labeled at the 1.5-fold stage. Scale bar: 20 µm. (C) DAPK-1 promotes nuclear localization of PINN-1 in the epidermis. Images show GFP fluorescence of PINN-1 in the epidermal nuclei of pinn-1(ju1504[pinn-1::gfp::loxp::3Xflag]) animals. Endogenous tagging of PINN-1 reveals cytoplasmic and nuclear localization. dapk-1(ju4) mutants display reduced level of PINN-1 in epidermal nuclei of pinn-1(ju1504[pinn-1::gfp::loxp::3Xflag]) animals. Images are representative epidermal nuclei. Scale bar: 5 µm. (D) Quantification of epidermal nuclear intensity of PINN-1 in wild type and dapk-1(ju4) mutants. GFP intensity in nucleus was scored. Individual intensities from different animals were normalized to average intensity from control strain. Data are mean normalized intensities±s.e.m. Numbers of nuclei analyzed are indicated. ***P<0.001 (Mann–Whitney test).
To examine PINN-1 expression, we inserted GFP after Lys161 [Fig. S6B, pinn-1(ju1504[pinn-1::gfp::loxp::3Xflag]), PINN-1::GFP] at the endogenous locus. PINN-1::GFP was distributed in the cytoplasm and nucleus (Fig. 6C and Fig. S6C). Intriguingly, expression of PINN-1::GFP in epidermal nuclei was significantly reduced in L4 and adult dapk-1 mutant animals compared with wild-type animals (Fig. 6C,D and Fig. S6C,D), suggesting DAPK-1 normally represses UNC-44L expression in the epidermis by promoting nuclear localization of PINN-1.
DISCUSSION
Using the C. elegans unc-44/ankyrin as a model, we have dissected genetic and cellular mechanisms underlying tissue-specific APA. Our previous study revealed that in neuronal development the activity of a set of nuclear polyadenylation factors, including SSUP-72/Ssu72, is inhibited at the internal PAS2 of unc-44 to promote production of the UNC-44L isoform (Chen et al., 2015). Here, by examining tissue-specific expression of endogenous unc-44 isoforms, our results indicate that the same set of polyadenylation factors converge at the internal PAS2 of unc-44 to repress expression of unc-44L isoform in epidermis and other non-neuronal tissues. Thus, the tissue-specific pattern of differentially polyadenylated mRNA isoforms of unc-44 depends on the competition of positive and negative regulation on the usage of the same internal polyadenylation site (Fig. S7).
Pre-mRNA 3′ end processing involves a large, and highly conserved, protein machinery that executes cleavage and polyadenylation to produce the 3′UTRs of mature mRNAs (Colgan and Manley, 1997; Millevoi and Vagner, 2010). Biochemical studies that led to the initial identification of core factors involved in pre-mRNA 3′ end processing used viral pre-mRNAs as substrates (Shi et al., 2009); the composition of these factors likely reflects the state of the polyadenylation machinery at general or consensus 3′ end. Whether cleavage and polyadenylation of internal PAS shares the same 3′ end processing machinery has not been examined. Using CRISPR genome-editing mediated knock-ins, we monitored the expression of GFP-tagged unc-44 isoforms from endogenous locus in living animals. Although observations based on GFP fluorescence also potentially reflects regulation at post-transcriptional and translational levels, the following findings – that SYDN-1 and SSUP-72 interact with RNA polymerase II, that SSUP-72 belongs to pre-mRNA 3′ end processing machinery (Chen et al., 2015), and that ssup-72(0) and pinn-1(0) mutants display ectopic UNC-44L expression resembling that caused by the unc-44 internal PAS2 deletion – support our conclusion that our experimental design allows us to examine differential use of alternative PASs. Given that ssup-72(0) displays normal distal polyadenylation of unc-44, it is likely that the polyadenylation machinery at the internal 3′ end (PAS2) and the distal 3′ end (PAS3) differs in regulatory components. PINN-1, a conserved peptidyl prolyl isomerase, plays important roles in diverse cellular processes by mediating conformational changes of many proteins (Lu and Hunter, 2014). Our data reveal an in vivo role for PINN-1 in alternative polyadenylation. Like its yeast homologs, C. elegans PINN-1 could stimulate SSUP-72 enzymatic activity by promoting the cis configuration of the phospho-Ser5-Pro bond in RNA polymerase II CTD (Werner-Allen et al., 2011; Xiang et al., 2010). PINN-1 may also serve as a sensor to detect cytoplasmic changes and, by shuttling between cytoplasm and nucleus, to regulate 3′ end processing at alternative PAS sites.
Mammalian death-associated protein kinase (DAPK), a Ser/Thr kinase, mediates cell death by promoting autophagy or apoptosis (Bialik and Kimchi, 2006). The C. elegans ortholog of death-associated protein kinase, DAPK-1, maintains epidermal integrity and inhibits epidermal innate immune responses (Tong et al., 2009) and wound healing (Xu and Chisholm, 2011). Epidermal expression of DAPK-1 rescues dapk-1(0) epidermal defects, supporting cell-autonomous function of DAPK-1 (Tong et al., 2009). DAPK-1 represses ectopic expression of UNC-44L in the epidermis by antagonizing the SYDN-1/PINN-1/SSUP-72 pathway, suggesting PAS2 use in non-neuronal tissues ensures the predominantly neuron-specific expression of UNC-44L. DAPK-1 is a cytoplasmic protein that, to date, has no known role in RNA processing. Nuclear localization of PINN-1 is partially impaired in dapk-1 worms. We speculate that in the wild-type epidermis, phosphorylation of PINN-1 by DAPK-1 promotes its nuclear localization, thus upregulating SSUP-72 activity. It is notable that in mammalian cells phosphorylation of Pin1 by mammalian DAPK1 has been interpreted to inhibit its nuclear localization and catalytic activity. Moreover, in a C. elegans model of neurodegeneration, DAPK-1 and PINN-1 have been proposed to act antagonistically (Del Rosario et al., 2015). Future studies will be required to determine why the interactions of DAPK-1 and PINN-1 differ depending on cellular context.
Epidermal expression of neuronal ankyrin/UNC-44L causes epidermal fragility in dapk-1 mutants, highlighting the importance of tissue-specific APA in maintaining normal function of different tissues. Ankyrin proteins are components of the cortical cytoskeleton (Zhang and Rasband, 2016). Compared with other UNC-44 proteins, UNC-44L has a unique extended C terminus. Interestingly, the C terminus of AnkG, a mammalian ankyrin with an extended C terminus, radially extends towards the cytosol, likely interacting with other proteins (Leterrier et al., 2015), and plays important roles in the assembly of axon initial segment (Jenkins et al., 2015). We speculate that ectopic expression of the C terminus of UNC-44L in the epidermis may disrupt the cortical cytoskeleton, possibly by interfering with the activity of UNC-44C isoforms or by recruiting other proteins. Our results underscore the importance of precise spatiotemporal control of ankyrin isoform expression in tissue development.
MATERIALS AND METHODS
C. elegans genetics
Strains were maintained on NGM plates at 20-22.5°C as described previously (Brenner, 1974). Table S1 lists the strains used in this study. Double or triple mutants were constructed following standard procedures. We also followed standard procedures to generate extra-chromosomal arrays (Mello et al., 1991). Expression constructs were injected at 20 ng/µl with co-injection marker Pttx-3-RFP at 80 ng/µl. For most expression constructs, two or three independent transgenic lines were analyzed (Table S2).
Generation of GFP knock-in lines by CRISPR/Cas9-mediated genome editing
To make CRISPR knock-in lines, we generated plasmids carrying the sgRNAs and gfp sequence with 2 kb homology on both arms (Table S3). Peft-3-cas9-NLS-pU6-sgRNA vectors were constructed by site-directed mutagenesis of pDD162 (Addgene #47549) (Dickinson et al., 2013). Templates for recombination were made using Gibson Assembly (Gibson et al., 2009). Briefly, pDD282 (Addgene #66823) was digested overnight with SpeI and ClaI, which were heat inactivated afterwards. Left and right homology arms were amplified by primers with homology sequence from pDD282 and then assembled with digestion products of pDD282.
For unc-44(ju1412), in which gfp was inserted at the C terminus of the unc-44 long isoform, we generated the template in pCR8 backbone (pCZGY3125) and co-injected it with sgRNA vector (pCZGY3122) into 50 one-day-old adult animals (P0). Three days later, F1s with any GFP expression were singled out and maintained for one more generation. F2s with stable expression of GFP were kept and gfp insertion was verified by PCR (Table S3).
For other knock-in lines, we used self-excising drug selection cassette as described previously (Dickinson et al., 2015). Homology arms of each target site were cloned into pDD282 (Addgene #66823). Homology arms containing plasmid (50 ng/µl), sgRNA vector (50 ng/µl) and markers (10 ng/µl Prab-3::mcherry, 5 ng/µl Pmyo-3::mcherry and 2.5 ng/µl Pmyo-2::mcherry) were co-injected into 60 1-day-old adults (P0). Three days later, each plate was treated with 5 mg/ml hygromycin. Three days later, roller worms not expressing mCherry co-injection markers were singled out and screened for 100% roller progeny. L1-L2 worms were heat shocked for 4 h to excise the drug selection cassette. When the SEC cassette is not excised by heat shock, transcription from the inserted gene is assumed to stop after the gfp sequence. Each gfp insertion line after heat shock was verified by PCR (Table S3).
Fluorescence microscopy and image analyses
Worms were paralyzed in M9 buffer containing 0.5-1% 1-phenoxy-2-propanol (TCL America, Portland, OR, USA) for imaging under Zeiss LSM710 confocal microscope equipped with Chroma HQ filters. Images were taken from the head, vulva, dorsal cord and tail regions of GFP-expressing strains. Z-stack sections at 0.5 µm intervals were collected from the entire region of interest. We distinguished L3 from L4 stage worms using vulval morphology. One-day-old adult worms were imaged 1 day after L4 stage.
Images were processed using MetaMorph software with the observer blinded to genotypes. Maximum projection images from 12 sections of nerve ring, nine sections of head epidermis or the entire dorsal cord were used to calculate either the line average intensity or rectangle average intensity between genotypes after subtracting background. To calculate the nuclear intensity of PINN-1::GFP, maximum projection images from head epidermal nuclei were outlined and average intensity was calculated after subtracting background. All intensities were normalized to the average intensity of controls in the same group.
Analyses of mutant phenotypes
To measure lethality, we picked over 100 L4s. Moving worms were transferred to new plates consecutively for the next 2 days. Quantification was from at least three independent experiments. The fraction of animals with epidermal morphology defects was scored at L4 and adult stage following previous studies (Tong et al., 2009). Over 100 animals per experiment were counted and the average from three independent experiments is reported.
For epidermal and body morphology phenotypes with 100% penetrance, we score a large population (i.e. over 10 broods for multiple generations) of worms of different genotypes. We show representative images of mutant phenotypes, from a collection of images of more than 10 individual animals/genotype grown from at least three different parents.
Acknowledgements
We thank Zhiping Wang for sharing her CRISPR genome editing protocol, Marian Chuang and Tiffany Hsiao for sharing observations on dapk-1 genetic interactions, and Shohei Mitani (Japan National Bioresource Project) and the C. elegans gene knockout consortium for deletion alleles. We thank members of the Jin and Chisholm labs for discussion and comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
F.C., A.D.C. and Y.J. designed the study; F.C. performed experiments and analyzed the data; A.D.C. and Y.J. interpreted the data; F.C., A.D.C. and Y.J. wrote the manuscript.
Funding
This research was supported by the National Institutes of Health (R01 GM054657 to A.D.C. and R01 NS035546 to Y.J.). F.C. is an Associate of and Y.J. is an Investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.146001.supplemental
References
- Bennett V. and Baines A. J. (2001). Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol. Rev. 81, 1353-1392. [DOI] [PubMed] [Google Scholar]
- Bialik S. and Kimchi A. (2006). The death-associated protein kinases: structure, function, and beyond. Annu. Rev. Biochem. 75, 189-210. 10.1146/annurev.biochem.75.103004.142615 [DOI] [PubMed] [Google Scholar]
- Blazie S. M., Babb C., Wilky H., Rawls A., Park J. G. and Mangone M. (2015). Comparative RNA-Seq analysis reveals pervasive tissue-specific alternative polyadenylation in Caenorhabditis elegans intestine and muscles. BMC Biol. 13, 4 10.1186/s12915-015-0116-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhou Y., Qi Y. B., Khivansara V., Li H., Chun S. Y., Kim J. K., Fu X.-D. and Jin Y. (2015). Context-dependent modulation of Pol II CTD phosphatase SSUP-72 regulates alternative polyadenylation in neuronal development. Genes Dev. 29, 2377-2390. 10.1101/gad.266650.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang M., Hsiao T. I., Tong A., Xu S. and Chisholm A. D. (2016). DAPK interacts with Patronin and the microtubule cytoskeleton in epidermal development and wound repair. Elife 5, e15833 10.7554/elife.15833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan D. F. and Manley J. L. (1997). Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11, 2755-2766. 10.1101/gad.11.21.2755 [DOI] [PubMed] [Google Scholar]
- Cox G. N. and Hirsh D. (1985). Stage-specific patterns of collagen gene expression during development of Caenorhabditis elegans. Mol. Cell. Biol. 5, 363-372. 10.1128/MCB.5.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Allen M. A., Larsen A., MacMorris M., Han M. and Blumenthal T. (2008). Genes involved in pre-mRNA 3′-end formation and transcription termination revealed by a lin-15 operon Muv suppressor screen. Proc. Natl. Acad. Sci. USA 105, 16665-16670. 10.1073/pnas.0807104105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rosario J. S., Feldmann K. G., Ahmed T., Amjad U., Ko B. K., An J. H., Mahmud T., Salama M., Mei S., Asemota D. et al. (2015). Death Associated Protein Kinase (DAPK) -mediated neurodegenerative mechanisms in nematode excitotoxicity. BMC Neurosci. 16, 25 10.1186/s12868-015-0158-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino D. C., Nishida K. and Manley J. L. (2011). Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853-866. 10.1016/j.molcel.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J. and Goldstein B. (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028-1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D. and Goldstein B. (2015). Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200, 1035-1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. III and Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343-345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gilleard J. S., Barry J. D. and Johnstone I. L. (1997). cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol. Cell. Biol. 17, 2301-2311. 10.1128/MCB.17.4.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Lemke S. B. and Levine M. (2012). ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev. 26, 2259-2264. 10.1101/gad.199653.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C. H., Friedman R. C., Ruby J. G. and Bartel D. P. (2011). Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469, 97-101. 10.1038/nature09616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins P. M., Kim N., Jones S. L., Tseng W. C., Svitkina T. M., Yin H. H. and Bennett V. (2015). Giant ankyrin-G: a critical innovation in vertebrate evolution of fast and integrated neuronal signaling. Proc. Natl. Acad. Sci. USA 112, 957-964. 10.1073/pnas.1416544112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S., Alemu E. A., Mertens C., Gantman E. C., Fak J. J., Mele A., Haripal B., Zucker-Scharff I., Moore M. J., Park C. Y.. et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037-2053. 10.1101/gad.269415.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S., Ghazy M. A., Moore C. and Hampsey M. (2009). Functional interaction of the Ess1 prolyl isomerase with components of the RNA polymerase II initiation and termination machineries. Mol. Cell. Biol. 29, 2925-2934. 10.1128/MCB.01655-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Chen C.-H., Suizu F., Huang P., Schiene-Fischer C., Daum S., Zhang Y. J., Goate A., Chen R.-H., Zhou X. Z.. et al. (2011). Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol. Cell 42, 147-159. 10.1016/j.molcel.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier C., Potier J., Caillol G., Debarnot C., Rueda Boroni F. and Dargent B. (2015). Nanoscale architecture of the axon initial segment reveals an organized and robust scaffold. Cell Rep. 13, 2781-2793. 10.1016/j.celrep.2015.11.051 [DOI] [PubMed] [Google Scholar]
- Lianoglou S., Garg V., Yang J. L., Leslie C. S. and Mayr C. (2013). Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380-2396. 10.1101/gad.229328.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. and Hunter T. (2014). Prolyl isomerase Pin1 in cancer. Cell Res. 24, 1033-1049. 10.1038/cr.2014.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M., Manoharan A. P., Thierry-Mieg D., Thierry-Mieg J., Han T., Mackowiak S. D., Mis E., Zegar C., Gutwein M. R., Khivansara V. et al. (2010). The landscape of C. elegans 3′UTRs. Science 329, 432-435. 10.1126/science.1191244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D. and Ambros V. (1991). Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S. and Vagner S. (2010). Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 38, 2757-2774. 10.1093/nar/gkp1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P., Shenker S., Andreu-Agullo C., Westholm J. O. and Lai E. C. (2013). Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res. 23, 812-825. 10.1101/gr.146886.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktaba K., Zhang W., Lotz T. S., Jun D. J., Lemke S. B., Ng S. P., Esposito E., Levine M. and Hilgers V. (2015). ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol. Cell 57, 341-348. 10.1016/j.molcel.2014.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka A. J., Franco R., Yang B., Shim K. H., Tang L. Z., Zhang Y. Y., Boontrakulpoontawee P., Jeyaprakash A., Hedgecock E., Wheaton V. I.. et al. (1995). An ankyrin-related gene (unc-44) is necessary for proper axonal guidance in Caenorhabditis elegans. J. Cell Biol. 129, 1081-1092. 10.1083/jcb.129.4.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka A. J., Boontrakulpoontawee P., Rebeiz N., Domanus M., Otsuka D., Velamparampil N., Chan S., Vande Wyngaerde M., Campagna S. and Cox A. (2002). Novel UNC-44 AO13 ankyrin is required for axonal guidance in C. elegans, contains six highly repetitive STEP blocks separated by seven potential transmembrane domains, and is localized to neuronal processes and the periphery of neural cell bodies. J. Neurobiol. 50, 333-349. 10.1002/neu.10036 [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. (2011). Ending the message: poly(A) signals then and now. Genes Dev. 25, 1770-1782. 10.1101/gad.17268411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarling G. S., Coates P. W., Dass B. and MacDonald C. C. (2009). A family of splice variants of CstF-64 expressed in vertebrate nervous systems. BMC Mol. Biol. 10, 22 10.1186/1471-2199-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Di Giammartino D. C., Taylor D., Sarkeshik A., Rice W. J., Yates J. R. III, Frank J. and Manley J. L. (2009). Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33, 365-376. 10.1016/j.molcel.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Miura P., Westholm J. O., Shenker S., May G., Duff M. O., Zhang D., Eads B. D., Carlson J., Brown J. B.. et al. (2012). Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 1, 277-289. 10.1016/j.celrep.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein M. C., McCormack G., Winter A. D., Johnstone I. L., Shoemaker C. B. and Page A. P. (2003). Caenorhabditis elegans exoskeleton collagen COL-19: an adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Dev. Dyn. 226, 523-539. 10.1002/dvdy.10259 [DOI] [PubMed] [Google Scholar]
- Tong A., Lynn G., Ngo V., Wong D., Moseley S. L., Ewbank J. J., Goncharov A., Wu Y.-C., Pujol N. and Chisholm A. D. (2009). Negative regulation of Caenorhabditis elegans epidermal damage responses by death-associated protein kinase. Proc. Natl. Acad. Sci. USA 106, 1457-1461. 10.1073/pnas.0809339106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I., Shkumatava A., Jan C. H., Subtelny A. O., Koppstein D., Bell G. W., Sive H. and Bartel D. P. (2012). Extensive alternative polyadenylation during zebrafish development. Genome Res. 22, 2054-2066. 10.1101/gr.139733.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Epps H., Dai Y., Qi Y., Goncharov A. and Jin Y. (2010). Nuclear pre-mRNA 3′-end processing regulates synapse and axon development in C. elegans. Development 137, 2237-2250. 10.1242/dev.049692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P. and Burge C. B. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470-476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Allen J. W., Lee C.-J., Liu P., Nicely N. I., Wang S., Greenleaf A. L. and Zhou P. (2011). cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 286, 5717-5726. 10.1074/jbc.M110.197129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K., Nagaike T., Xiang S., Kilic T., Beh M. M., Manley J. L. and Tong L. (2010). Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467, 729-733. 10.1038/nature09391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. and Chisholm A. D. (2011). A Galphaq-Ca(2)(+) signaling pathway promotes actin-mediated epidermal wound closure in C. elegans. Curr. Biol. 21, 1960-1967. 10.1016/j.cub.2011.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. and Rasband M. N. (2016). Cytoskeletal control of axon domain assembly and function. Curr. Opin. Neurobiol. 39, 116-121. 10.1016/j.conb.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lee J. Y. and Tian B. (2005). Biased alternative polyadenylation in human tissues. Genome Biol. 6, R100 10.1186/gb-2005-6-12-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]