Abstract
Both bipolar disorder (BD) and major depressive disorder (MDD) have high morbidity and share a genetic background. Treatment options for these mood disorders are currently suboptimal for many patients; however, specific genetic variables may be involved in both pathophysiology and response to treatment. Agents such as the glutamatergic modulator ketamine are effective in treatment-resistant mood disorders, underscoring the potential importance of the glutamatergic system as a target for improved therapeutics. Here we review genetic studies linking the glutamatergic system to the pathophysiology and therapeutics of mood disorders. We screened 763 original genetic studies of BD or MDD that investigated genes encoding targets of the pathway/mediators related to the so-called tripartite glutamate synapse, including pre- and post-synaptic neurons and glial cells; 60 papers were included in this review. The findings suggest the involvement of glutamate-related genes in risk for mood disorders, treatment response, and phenotypic characteristics, although there was no consistent evidence for a specific gene. Target genes of high interest included GRIA3 and GRIK2 (which likely play a role in emergent suicidal ideation after antidepressant treatment), GRIK4 (which may influence treatment response), and GRM7 (which potentially affects risk for mood disorders). There was stronger evidence that glutamate-related genes influence risk for BD compared with MDD. Taken together, the studies show a preliminary relationship between glutamate-related genes and risk for mood disorders, suicide, and treatment response, particularly with regard to targets on metabotropic and ionotropic receptors.
INTRODUCTION
In the general population, bipolar disorder (BD) and major depressive disorder (MDD) have a lifetime prevalence of ~4 and 16%, respectively (Kessler et al, 2005), and both disorders share a genetic vulnerability (McGuffin et al, 2003). Despite the prevalence of mood disorders, available first-line treatment options for these illnesses have shown limited efficacy. The Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study found that one-third of MDD patients do not achieve remission even after four antidepressant trials (Rush et al, 2006). Similarly, BD patients remain symptomatically ill about half of the time and experience mostly depressive symptoms despite the use of standard treatments (Judd et al, 2002).
In this context, developing novel treatments for mood disorders is crucial. However, a major obstacle to developing such treatments is our lack of understanding of the pathophysiology of these disorders and the mechanism of action of effective interventions. Although the literature has focused on the role of monoamines in mood disorders, monoamine-related theories are limited in their ability to explain the underlying pathophysiology of these disorders. For instance, it takes about 2 weeks for antidepressants to improve mood, despite the fact that they affect monoamine levels immediately after treatment begins (Machado-Vieira et al, 2008).
A large body of preclinical evidence has implicated the glutamatergic system in the pathophysiology of mood disorders (Skolnick et al, 1996), including in the antidepressant effects of N-methyl-D-aspartate (NMDA) receptor antagonists in animal models (Papp and Moryl, 1994). Evidence also exists for the role of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA), supported by the knockout of the GluA1 subunit as a successful animal model of depression (Chourbaji et al, 2008) and the antidepressant properties of genetic ablation of kainate receptor subunit GluK4, also in animal models (Catches et al, 2012). Metabotropic glutamate receptors (mGluRs) have also been shown to regulate mood in several animal models (Witkin et al, 2007).
In addition to this preclinical evidence, human studies have shown that the glutamatergic modulator ketamine effectively reduces depressive symptoms in individuals with MDD or BD, including treatment-resistant patients (Zarate et al, 2006, 2012). Further evidence from postmortem studies shows that mood disorders are associated with expression of the glutamatergic genes GRIA1, GRIA3, GRM3, and GRIK1 (Sequeira et al, 2009). Identifying genes involved in the etiology and pathophysiology of mood disorders may reveal relevant mechanistic pathways that could, in turn, inform novel therapeutic targets. Although some genes have consistently been associated with BD (Craddock and Sklar, 2013), only two associations have been replicated in large MDD samples, namely SIRT1 and LHPP (CONVERGE Consortium, 2015). The lack of consistent replicated results poses both a challenge and an opportunity for investigation.
The extant evidence implicating glutamate genes in mood disorders—drawn largely from genome-wide association studies (GWASs) as well as from candidate gene studies—is inconsistent. For instance, no gene has been replicated in all the available studies to date, and associations in opposite directions have been reported for some single nucleotide polymorphisms (SNPs; Menke et al, 2008; Pu et al, 2013). Furthermore, genetic studies of glutamate-related genes have never been systematically reviewed, which hampers a synthesized view of the field and the identification of replicable targets on the glutamatergic pathway.
This paper provides a critical and systematic review of genetic studies investigating the tripartite glutamatergic synapse in patients with BD and MDD, encompassing specific mood symptoms and phenotypic characteristics, treatment response, and pathophysiology.
The Function of the Tripartite Glutamate Synapse: Pathways and Mediators
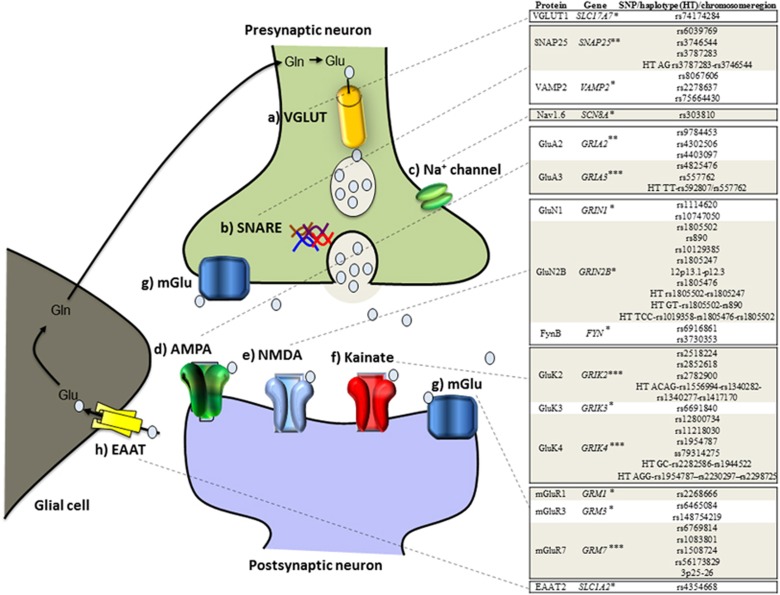
Glutamate is ubiquitous in the brain and, because it is crucial to promoting excitatory synaptic transmission (Orrego and Villanueva, 1993), plays a key role in synaptic plasticity and memory. In presynaptic neurons, glutamate is loaded into vesicles by vesicular glutamate transporter (VGLUT) proteins (Figure 1). These vesicles fuse with the presynaptic neuronal membrane by interacting with soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE), and are then released into the synaptic cleft. Sodium channels located in the presynaptic neuronal membrane depolarize the membrane, which also facilitates glutamate release (Lingamaneni et al, 2001). Glutamate acts on ionotropic AMPA, NMDA, and kainate receptors as well as mGluRs, mostly in the postsynaptic neuron. The ionotropic receptors are ligand-gated ion channels that open when an agonist binds to them. Notably, ketamine is thought to act by increasing glutamatergic throughput at the AMPA receptor relative to the NMDA receptor (Maeng et al, 2008).
Figure 1.
Glutamatergic pathway (on the left) and gene variants, haplotypes, and chromosome regions positively associated with mood disorders (on the right). Glutamate (Glu) is loaded into vesicles by vesicular glutamate transporter (VGLUT) proteins (a). These Glu vesicles are fused with the presynaptic neuronal membrane through interactions with soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins to be released to the synaptic cleft (b). Sodium channels located in the presynaptic neuron membrane play a role in regulating Glu release (c). In the postsynaptic neuron, Glu acts on several receptors: ionotropic receptors α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors (d), N-methyl-D-aspartate (NMDA) receptors (e), and kainate receptors (f). Glu also acts on metabotropic glutamate receptors (mGluRs) (g), which are G-protein coupled receptors attached to both the postsynaptic and the presynaptic neurons. The Glu released in the synaptic cleft is cleared by excitatory amino acid transporters (EAAT) (h), to be transformed into glutamine (Gln) in the glial cell. The Gln produced will be transformed into Glu in the neuron. The genes on the right are ranked according to the evidence as high interest (***), moderate interest (**), or low interest (*). High interest (***): at least one study with supporting evidence from a large sample (>1000 patients) and two studies with medium samples (200–1000 patients); moderate interest (**): evidence from one study with a large sample size (>1000 patients) but no more than one study with a medium sample size; low interest (*): findings from studies with medium or small sample sizes, but no study with a large sample size.
MATERIALS AND METHODS
This systematic review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) statement. Included studies had enrolled participants diagnosed with BD and/or MDD as established by a validated structured diagnostic interview such as the 10th revision of the International Statistical Classification of Diseases (ICD-10) or the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV). GWASs, linkage studies, and candidate gene studies in BD or MDD were included that investigated chromosomal regions or SNPs of genes involved in pathways/mediators related to the so-called tripartite glutamate synapse, comprising pre- and post-synaptic neurons and glial cells. The following specific terms were selected for inclusion from recently published non-systematic reviews (Duman, 2014; Machado-Vieira et al, 2009; Mathews et al, 2012; Rudy et al, 2015): ‘Vesicular Glutamate Transport Proteins', ‘SNARE Proteins', ‘Voltage-Gated Sodium Channels', ‘Receptors, Metabotropic Glutamate', ‘Postsynaptic Density Proteins', ‘Receptors, AMPA', ‘Receptors, NMDA', ‘Receptors, Kainic Acid', ‘Glutamate Plasma Membrane Transport Proteins', and ‘Glutamic Acid'. A literature search was performed on PubMed and Web of Science databases in March 2016 using the medical subject heading (MeSH) ‘Mood Disorders' associated with the other aforementioned MeSH terms.
Studies published in English, French, Spanish, German, or Portuguese were considered for inclusion. Studies with non-original data, postmortem analyses, and case reports were excluded. Studies of samples comprising individuals with multiple psychiatric diagnoses were also excluded from this review unless data for MDD or BD were reported separately.
RESULTS
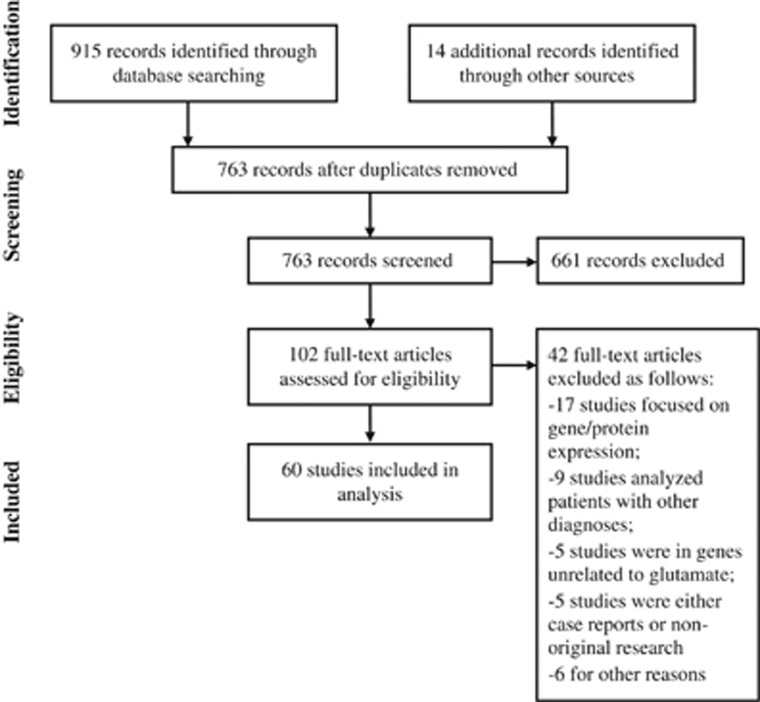
Two of the authors (RTDS and AAL) screened 763 abstracts for eligibility and reviewed 102 full texts of potentially eligible articles (Figure 2); reference lists in these publications were also reviewed and provided 14 other potential articles of interest. Disagreements regarding inclusion were resolved through consensus with an additional author (RMV). Sixty unique articles met inclusion criteria. Of these, 33 (55%) evaluated only BD, 23 (38%) evaluated only MDD, and four (7%) evaluated both BD and MDD.
Figure 2.
Flowchart showing the identification, screening, election, and inclusion of papers in the review.
Glutamate-Related Genes
Table 1 provides extensive information for all 60 studies, including study design, demographics, diagnosis, genes and proteins identified, and main study findings. Of the 60 papers included in this review, 34 had positive results—that is, the study implicated glutamate-related genes in mood disorders. Eight of these 34 studies (23.5%) were of large samples (>1000 patients), including two GWASs; 19 (56%) were conducted in medium-sized samples (200–1000 patients); and seven (20.5%) were conducted in small samples (<200 patients).
Table 1. Genetic Studies on Glutamate-Related Genes in Major Depressive Disorder (MDD) and Bipolar Disorder (BD): (a) Genes Related to Glutamate Pathways/Mediators Showing Positive Results; (b) Genes Related to Glutamate Pathways/Mediators with Negative Results.
| Gene | Protein | Study | Study design | MDD/BD | N | Population | Assessment | Main outcome |
|---|---|---|---|---|---|---|---|---|
|
(a)a | ||||||||
| VGLUT | ||||||||
| SLC17A7* | VGLUT1 | Li et al, 2014 | Candidate gene | MDD | 290 patients | Chinese Han | Treatment response | SLC17A7 rs74174284 allele C was associated with SSRI treatment response at week 6 in MDD patients (OR=0.57, 95 % CI=0.38–0.87, corrected P=0.032) |
| SNARE proteins | ||||||||
| SNAP25** | SNAP25 | Wang et al, 2015 | Candidate gene | MDD | 1045 patients and 1520 healthy controls | Chinese Han | Risk | SNAP25 rs3787283 and rs3746544 were associated with MDD (adjusted P=0.00387 and adjusted P=0.0485, respectively). The haplotype AG rs3787283-rs3746544 was also significantly associated with MDD (corrected P=6 × 10−4) |
| Etain et al, 2010 | Candidate gene | BD | 197 patients with early-onset BD, 202 patients with late-onset BD, and 136 healthy controls | French | Risk and phenotypic characteristics | SNAP25 rs6039769 allele A was associated with early-onset BD (OR=0.62, P=0.005, corrected P=0.03) but not with late-onset BD. SNAP25 rs363006 allele A was associated with late onset BD (OR=1.57, P=0.04), but did not survive correction. Homozygosity for allele C of SNAP25 rs6039769 was associated with a higher SNAP25b expression levels in prefrontal cortex (P=0.04) | ||
| VAMP2* | VAMP2 | Abou Jamra et al, 2008 | Candidate gene | BD | 409 patients and 407 healthy controls; a replication sample of 378 patients and 384 healthy controls | German | Risk | In the initial sample, there was an association of BD and VAMP2 rs2278637 (P=0.005), rs75664430 (P=0.033), and rs8067606 (P=0.007), which was not observed in the replication sample (all P>0.45) |
| Saito et al, 2006 | Candidate gene | MDD | 106 patients | Japanese | Treatment response | There was no association between VAMP2 rs1061032, VAMP2 rs8067606, or VAMP2 HT rs1061032-rs8067606 and response to fluvoxamine in MDD | ||
| Sodium channels | ||||||||
| SCN8A* | Nav1.6 | Wang et al, 2008 | Candidate gene | BD | 506 patients and 507 healthy controls | Chinese Han | Risk | SCN8A rs1601012 and rs303810 showed significant differences between BD patients and controls in both allele and genotype distribution, but only SCN8A rs303810 allele distribution remained significant after correction for multiple comparisons (P=0.0164). No linkage disequilibrium or haplotypes were observed among these SNPs |
| AMPA receptor | ||||||||
| GRIA2** | GRIA2 | Perlis et al, 2009 | GWAS | BD | 1177 patients, including 458 individuals treated with lithium carbonate or citrate. A second cohort of 359 patients | Caucasian | Treatment response | GRIA2 rs9784453 had a non-significant low p-value (P=4 × 10−4) in a STEP-BD analysis of association of the ability of lithium to prevent recurrence. The same GRIA2 rs9784453 was found to be associated with lithium's ability to prevent recurrence in a confirmatory cohort (P=0.03, alpha set at 0.05) |
| Chiesa et al, 2012b | Candidate gene | MDD | 145 patients and 170 healthy controls | Korean | Risk, treatment response, and phenotypic characteristics | GRIA2 rs4302506 and rs4403097 showed an association with age of onset in patients with MDD (P=0.003 and P=0.005, respectively; after Bonferroni, P set at 0.005), but no association with MDD or treatment response. Other GRIA2 SNPs (rs6536221, rs4260586, rs4441804, and rs3813296) were not associated with MDD and clinical characteristics or outcomes | ||
| Chiesa et al, 2012a | Candidate gene | BD | 132 patients and 170 healthy controls | Korean | Risk and phenotypic characteristics | GRIA2 rs6536221, rs4260586, rs4302506, rs4441804, rs3813296, and rs4403097 were not associated with BD diagnosis or treatment outcomes | ||
| Chiesa et al, 2013 | Candidate gene | MDD | 145 patients | Korean | Treatment response | GRIA2 rs4260586 was not associated with improvement on depression rating scale scores or other clinical/sociodemographic variables. There was no interaction between GRIA2 rs4260586, GRIA4 rs10736648, and improvement in depressive symptoms | ||
| GRIA3*** | GRIA3 | Laje et al, 2007 | Candidate gene | MDD | 1915 patients | Caucasian (78%) and other (22%) | Phenotypic characteristics | Suicidal ideation following citalopram treatment (treatment-emergent suicidal ideation) was associated with GRIA3 rs4825476 (corrected odds ratio=1.94; corrected P<0.01) |
| Menke et al, 2008 | Candidate gene | MDD | 397 inpatients | German | Phenotypic characteristics | Following treatment initiation, suicidal ideation was associated with GRIA3 rs4825476 (OR=2.7, uncorrected P=0.041), but with a different allele than that found in Laje et al, 2007) | ||
| Myung et al, 2012 | Candidate gene | MDD | 241 patients | Korean | Phenotypic characteristics | Guilt feelings in females were associated with GRIA3 rs557762 (corrected P=0.01) and GRIA3 TT HT rs592807-rs557762 (corrected P=5.0 × 10−3) | ||
| Gécz et al, 1999 | Linkage study | BD | 373 patients from 40 pedigrees | Caucasian | Risk | There was no evidence of linkage between GRIA3 and BD | ||
| Pu et al, 2013* | Candidate gene | MDD | 281 patients | Chinese Han | Treatment response | SNPs in GRIA3 (rs502434 and rs3761555) showed interactions with early-onset adverse events and recent negative life stress that influence treatment response, but the analyses did not withstand correction for multiple comparisons | ||
| NMDA receptor | ||||||||
| GRIN1* | GluN1 | Mundo et al, 2003 | Candidate gene | BD | 276 patients and their parents | European Caucasian (97%) | Risk | For both GRIN1 rs1114620 (P=0.03) and 6608-G/C (P=0.004) polymorphisms, a preferential transmission of the G allele to BD patients was observed |
| Georgi et al, 2006 | Candidate gene | BD | 306 patients and 319 healthy controls | German | Risk | No association was found between GRIN1 and BD | ||
| Hammer et al, 2014 | Candidate gene | MDD and BD | 88 MDD patients, 60 BD, and 1250 controls | Caucasian | Risk | In a study focusing on psychosis, a sub-analysis on patients with mood disorders found no association between mood disorders and GRIN1 rs524991 (P=0.13) | ||
| GRIN2B* | GluN2B | Martucci et al, 2006 | Candidate gene | BD | 318 trios (of which 158 probands had psychotic symptoms) | Caucasian | Risk | There was an association between GRIN2B rs1805502 (corrected P=0.04) and BD and between GRIN2B A5806C and BD with psychotic symptoms (corrected P=0.008). The HT TCC-rs1019358-A5806C-rs1805502 was transmitted more frequently in BD (P=0.015) |
| Avramopoulos et al, 2007 | Linkage study | BD | 41 patients with their families | Ashkenazi Jewish | Risk | Chromosome region 12p13.1–p12.3, which harbors the GRIN2B gene region, showed an increased association with BD in a fine-mapping analysis of regions nominally associated with BD | ||
| Zhao et al, 2011 | Candidate gene | BD | 480 patients and 480 healthy controls | Chinese Han | Risk | BD diagnosis was associated with GRIN2B rs1805247 allelic distribution (corrected P=0.018) and the haplotype consisting of rs1805502 and rs1805247 (uncorrected P=3.55 × 10−9) | ||
| Kuswanto et al, 2013 | Candidate gene | BD | 14 patients and 22 healthy controls | Chinese Han (>92% of the patients and the controls) | Phenotypic characteristics | T allele of GRIN2B rs890 in BD patients was associated with lower brain fractional anisotropy values in left frontal (corrected P<0.001), right frontal (corrected P<0.001), left parietal lobe (corrected P=0.001), left occipital (corrected P=0.006), right occipital region (corrected P<0.001), and left cingulate gyrus (corrected P<0.001), when compared with G allele of GRIN2B rs890 carriers | ||
| Zhang et al, 2014 | Candidate gene | MDD | 178 TRD patients, 612 non-TRD patients, and 779 healthy controls | Chinese Han | Treatment response | GRIN2B HT GT-rs1805502-rs890 was more frequent in the TRD group than in the controls (corrected P=0.007). Regarding GRIN2B rs1805502, there was an excess of the G allele in the TRD group compared with the non-TRD group (OR=1.55, 95 % CI=1.18−2.05, corrected P=0.008) | ||
| Fallin et al, 2005 | Linkage study | BD | 337 trios from 323 families | Ashkenazi Jewish | Risk | GRIN2B was associated with BD (uncorrected P<0.01), but this association did not survive correction for multiple comparisons | ||
| Szczepankiewicz et al, 2009d | Candidate gene | BD | 105 patients | Polish | Treatment response | No association was found between treatment response to lithium in BD and the three GRIN2B polymorphisms analyzed | ||
| Szczepankiewicz et al, 2009b | Candidate gene | BD | 419 patients and 487 healthy subjects | Polish | Risk | No association was found between BD and GRIN2B polymorphisms | ||
| Dalvie et al, 2010 | Candidate gene | BD | 191 patients and 188 healthy controls | Mixed ancestry (54%) and Caucasians (46%) | Phenotypic characteristics | The number of hospitalizations for mania in BD was influenced by an interaction between DAOA rs701567 and GRIN2B rs10129385 (P=0.011) | ||
| FYN* | FynB | Szczepankiewicz et al, 2009a | Candidate gene | BD | 425 patients and 518 healthy subjects | Polish | Risk | BD was associated with FYN rs6916861 T/G (uncorrected P=0.0004, significance set at 0.016) and rs3730353 T/C (uncorrected P=0.016, significance set at 0.016) |
| Szczepankiewicz et al, 2009c | Candidate gene | BD | 101 patients | Polish | Treatment response | No significant association was found between lithium response and FYN polymorphisms. There was a trend toward an association between the TT genotype and T allele of rs3730353 FYN T/C polymorphism and decreased response to lithium | ||
| Kainate receptor | ||||||||
| GRIK2*** | GluK2 | Laje et al, 2007 | Candidate gene | MDD | 1915 patients | Caucasian (78%) and other (22%) | Phenotypic characteristics | Suicidal ideation following citalopram treatment (treatment-emergent suicidal ideation) was associated with GRIK2 rs2518224 (odds ratio=8.23; corrected P<0.003) |
| Menke et al, 2008 | Candidate gene | MDD | 397 inpatients | German | Phenotypic characteristics | Following treatment initiation, suicidal ideation was associated with 15 GRIK2 SNPs; the SNPs with the best association were GRIK2 rs2852618 (OR=9.0, uncorrected P=0.005) and GRIK2 rs2782900 (OR=4.3, uncorrected P=0.007) | ||
| Myung et al, 2012 | Candidate gene | MDD | 241 patients | Korean | Phenotypic characteristics | GRIK2 HT ACAG rs1556994-rs1340282-rs1340277-rs141717 was associated with somatic anxiety (corrected P=5.9 × 10−4) | ||
| GRIK3* | GluK3 | Schiffer and Heinemann, 2007 | Linkage analysis | MDD and BD | 153 families with at least one sib-pair affected by MDD or BD and replication in 81 trios (early-onset MDD) | > 90% European ancestry | Risk | GRIK3 rs6691840 was preferentially transmitted to MDD patients (P=0.01), but not to BD type I patients in the first sample. In the replication sample GRIK3 rs6691840 did not show an association with maternal transmission (P=0.07) |
| GRIK4*** | GluK2 | Pickard et al, 2006 | Candidate gene | BD | 368 patients and 458 controls | Scottish | Risk | GRIK4 HT GC—rs2282586/rs1944522 was protective against BD (OR=0.62, P=0.0002), which remained significant after correction for multiple testing |
| Paddock et al, 2007 | Candidate gene | MDD | 1199 patients for discovery and 617 patients for replication | Caucasian (78%) and other (22%) | Treatment response | GRIK4 rs1954787 was associated with both response and remission after citalopram treatment in the discovery and replication groups | ||
| Pickard et al, 2008 | Candidate gene | BD | 356 patients and 286 controls (sample set overlapping with Pickard et al, 2006) and replication in 274 patients and 376 controls | Scottish | Risk | A deletion in the GRIK4 ss79314275 was associated with BD when analyzing genotype (P=2.73 × 10−6) and allele frequencies (P=1.9 × 10−7, OR=0.462). The replication confirmed the results for genotype (P=0.0166) and allele (P=0.0107, OR=0.694). Also, the combination of datasets showed an additive/dose-dependent protection: the association of deletion heterozygosity with BD was higher (OR=0.471) than deletion homozygosity (OR=0.325) | ||
| Horstmann et al, 2010 | Candidate gene | MDD | 275 patients | Caucasian | Treatment response and phenotypic characteristics | GRIK4 SNPs analyzed were not significantly associated with response and remission to five weeks of antidepressant treatment. Although GRIK4 rs12800734 association with remission did not survive correction for multiple comparisons, a sub-analysis in MDD patients with the GG genotype of GRIK4 rs12800734 showed a significant downregulation of hypothalamic-pituitary-adrenal (HPA) axis hyperactivity | ||
| Pu et al, 2013 | Candidate gene | MDD | 281 patients | Chinese Han | Treatment response | Antidepressant response in MDD patients receiving different SSRIs or SNRIs for 6 weeks was associated with GRIK4 rs1954787 (adjusted P=0.016, OR=1.87, CI 95% 1.17–2.98) and the A-G-G haplotype of GRIK4 (rs1954787-rs2230297-rs2298725) (adjusted P=0.004, OR=0.14, CI 95% 0.04–0.54) | ||
| Milanesi et al, 2015 | Candidate gene | MDD | 380 patients with treatment-resistant depression and 247 patients without treatment resistant depression | Caucasian of Italian descent | Treatment response and phenotypic characteristics | GRIK4 rs11218030 was associated with treatment resistance (corrected P=0.025) and GRIK4 rs1954787 was associated with the presence of psychotic symptoms (uncorrected P=0.005) | ||
| Perlis et al, 2010 | Candidate gene | MDD | 250 patients | 78% white, 10% African, 12% Hispanic or other descent | Treatment response | GRIK4 was not associated with treatment response to a six-week trial of duloxetine | ||
| Serretti et al, 2012 | Candidate gene | MDD | 223 MDD patients and 76 healthy controls | Caucasian | Risk, treatment response, and phenotypic characteristics | GRIK4 rs195478 was not associated with treatment response to antidepressants, with risk for MDD, or with characteristics of MDD | ||
| Drago et al, 2013 | Candidate gene | BD | 470 mania patients | Caucasian | Treatment response | GRIK4 rs2298723 was nominally associated with decreased manic symptoms after treatment, but did not survive correction for multiple testing | ||
| mGlu receptors | ||||||||
| GRM1* | mGluR1 | Menke et al, 2012 | Candidate gene | MDD | 350 patients and 370 controls and replication sample of 904 patients and 1012 controls | German (>89%) | Risk, treatment response, and phenotypic characteristics | 22 GRM1 SNPs were associated with MDD, of which six SNPs remained associated after correction for multiple testing (rs2268666 with best allelic P=7.0 × 10−5; corrected P set at 0.0002). In the replication sample, GRM1 rs2268666 was again associated with MDD in the genotypic and carrier-based tests (P=0.02/0.04). GRM1 rs2268666 genotype was also associated with brain hippocampal Glu, with regulation of the HPA-axis, and with treatment response at discharge. |
| GRM3* | mGluR3 | Tsunoka et al, 2009 | Candidate gene | MDD and BD | 325 MDD patients, 155 BD patients, and 802 controls | Japanese | Risk and treatment response | An association was found between GRM3 rs6465084 and MDD (corrected P=0.0371), but no association between GRM3 and fluvoxamine response in MDD. Also, no association was found between GRM2/GRM3 and BD, or between GRM2 and MDD. GRM2 was not associated with fluvoxamine response in MDD. |
| Dalvie et al, 2010 | Candidate gene | BD | 191 patients and 188 healthy controls | Mixed ancestry (54%) and Caucasians (46%) | Risk and phenotypic characteristics | GRM3 rs6465084 G-allele increased the risk of psychosis in BD (OR=3.9, P=0.004) | ||
| Kandaswamy et al, 2013 | Candidate gene | BD | One sample of 506 patients and 510 controls and another sample of 593 patients and 642 controls | British or Irish descent | Risk | An analysis on the discovery and the replication samples combined showed an association between BD and GRM3 rs148754219 (OR=4.20, 95% CI, 1.43–12.37, uncorrected P=0.005; corrected P=0.047). The study also shows evidence that GRM3 rs148754219 affects gene expression. | ||
| Sklar et al, 2008 | GWAS and linkage analysis | BD | 1461 patients and 2008 controls; replication on 409 trios (from 256 nuclear families) and 365 patients and 351 controls | Caucasian | Risk | GRM3 rs2237554 was nominally associated with BD in the initial scan (121st variant most associated, uncorrected P=0.001) and was associated with BD in the replication analysis (uncorrected P=0.035), though the results in the replication sample could be found by chance | ||
| Martí et al, 2002 | Candidate gene | BD | 283 BD patients and 162 healthy controls | German | Risk | There was no association between BD and the GRM3 rs2228595 | ||
| Fallin et al, 2005 | Linkage study | BD | 337 trios from 323 families | Ashkenazi Jewish | Risk | GRM3 was associated with BD (all uncorrected P<0.01), but did not survive correction for multiple comparisons | ||
| Jia et al, 2014 | Candidate gene | MDD | 409 patients and 619 healthy controls | Chinese Han | Risk | The GRM3 polymorphisms were not associated with MDD | ||
| GRM7*** | mGluR7 | Breen et al, 2011 | GW linkage study | MDD | 971 sibling pairs concordant for recurrent depression | European descent | Risk | Significant linkage of MDD to chromosome 3p25–26 (GRM7) when restricting diagnoses by severity, with a maximum LOD score of 4.0 centered at the linkage marker D3S1515 (corrected P=0.015). However, a fine mapping of the region in a case-control replication study could not replicate the finding |
| Pergadia et al, 2011 | Linkage study | MDD | 220 sibling pairs with history of heavy smoking | Australian | Risk | On a region that harbors GRM7 on chromosome 3 at 24.9 cM (3p26–3p25), there was a genome-wide significant multipoint LOD score of 4.14 for MDD (corrected P=0.004) | ||
| Fabbri et al, 2013 | Candidate gene | MDD | 1541 patients for discovery and 1109 patients for replication | White non-Hispanic (72%), white Hispanic (12%), and African-American (16%) on first sample. Second sample of white non-Hispanic | Treatment response | GRM7 GG genotype of rs1083801 was associated with early response in comparison with late response (P=2.03e−06) and to non-response (P=1.82e−05) to citalopram in a white and African-American sample. The results were confirmed in a white non-Hispanic sample, with GRM7 GG genotype of rs1083801 associated with early response when compared with late response (P=6.7 × 10−7) and non-response (P=2.1 × 10−5) (after Bonferroni, alpha set at 2.5 × 10−5) | ||
| Kandaswamy et al, 2014 | Candidate gene | BD | 506 patients and 510 healthy controls for discovery and 593 BD patients and 642 controls for replication | British or Irish descent | Risk | There was no association between BD and GRM7 SNPs in the replication sample. However, the analysis of the discovery and replication samples combined showed an association between BD and GRM7 rs1508724, rs56173829, and rs6769814. | ||
| Muglia et al, 2010 | GWAS | MDD | 926 patients and 866 controls and 494 and 1052 controls for replication | Caucasian | Risk | No significant variants were associated with MDD. In a sub-analysis on candidate genes, GRM7 rs162209 had the lowest P-value (P=0.0001) in the first sample, but was not significant in the replication (P=0.1); an analysis of the two samples found the lowest P-value for GRM7 rs 162209 (P=0.0002) | ||
| Alliey-Rodriguez et al, 2011 | GWAS | BD | 944 patients evaluated with Cloninger's Temperament and Character Inventory and 1007 patients with the Zuckerman-Kuhlman Personality Questionnaire | European | Phenotypic characteristics | The Zuckerman-Kuhlman Personality Questionnaire subscale evaluating Neuroticism-Anxiety was associated with GRM7 rs13080594 (uncorrected P=7.68 × 10−7), which did not survive correction for multiple comparisons | ||
| Shyn et al, 2011 | GWAS | MDD | 1221 patients and 1636 healthy controls | European descent | Risk | No genome-wide evidence for an association was found. A GRM7 SNP was the third most associated with MDD (P=1.11 × 10−6) | ||
| Verbeek et al, 2013 | Candidate gene | MDD and BD | 1738 patients and 1802 healthy controls | Dutch | Risk | No variants of GRM7 were associated with MDD in GAIN-MDD cohort | ||
| EAATs | ||||||||
| SLC1A2* | EAAT2 | Dallaspezia, et al, 2012 | Candidate gene | BD | 110 patients | Caucasian | Treatment response | SLC1A2 rs4354668 T/T genotype in BD patients was associated with a lower frequency of episodes (p<0.001). An interaction between lithium treatment and SLC1A2 genotype influenced the frequency of episodes in BD patients (P=0.026) |
| Poletti et al, 2014 | Candidate gene | BD | 86 patients | Caucasian | Phenotypic characteristics | SLC1A2 rs4354668 affected only BD patients exposed to fewer adverse childhood experiences, with T/T homozygotes showing the lowest, and G/G the highest right hippocampal volume (corrected P=0.001) and left hippocampal volume (corrected P=0.001) | ||
| (b) | ||||||||
| SNARE proteins | ||||||||
| SNAP29 | SNAP29 | Saito et al, 2001 | Candidate gene | BD | 124 patients and 107 healthy controls | Caucasian | Risk | There was no association between BD and four SNPs of SNAP29 |
| VAMP3 | VAMP3 | Abou Jamra et al, 2008 | Candidate gene | BD | 409 patients and 407 healthy controls; a replication sample of 378 patients and 384 healthy controls | German | Risk | No association was found between VAMP3 rs707455, rs2071987,rs2301489, or rs228729 and BD |
| VAMP7 | VAMP7 | Saito et al, 2000 | Candidate gene | BD | 110 patients not selected forsex-linked transmission and 119 control subjects. | Caucasian | Risk | There was a trend for association of one VAMP7 SNP in males with BD (P=0.06) but not females (P=0.66) |
| Müller et al, 2002 | Candidate gene | BD | 164 patients and 267 controls | German | Risk | There was an association of homozygosity between one VAMP7 SNP in females and BD compared with controls (uncorrected P=0.017), which became a trend after correction for multiple comparisons (corrected P=0.051) | ||
| AMPA receptor | ||||||||
| GRIA1 | GluA1 | Chiesa et al, 2012a | Candidate gene | BD | 132 patients and 170 healthy controls | Korean | Risk | GRIA1 rs707176 and rs6875572 were not associated with BD diagnosis or treatment outcomes |
| Chiesa et al, 2012b | Candidate gene | MDD | 145 patients and 170 healthy controls | Korean | Risk and treatment response | GRIA1 rs707176 and rs6875572 were not associated with MDD diagnosis or treatment response | ||
| Drago et al, 2013 | Candidate gene | BD | 470 BD mania patients | Caucasian | Treatment response | GRIA1 rs1461224 was nominally associated with a decrease in manic symptoms after treatment, but did not survive correction for multiple testing | ||
| GRIA4 | GluA4 | Chiesa et al, 2012a | Candidate gene | BD | 132 patients and 170 healthy controls | Korean | Risk and phenotypic characteristics | GRIA4 rs11226805, rs2166318, rs11822168, rs1938956, rs10736648, rs528205, rs11226867, rs667174, and rs641574 were not associated with BD diagnosis or treatment outcomes |
| Chiesa et al, 2012b | Candidate gene | MDD | 145 patients and 170 healthy controls | Korean | Risk and treatment response | GRIA4 rs11226805, rs2166318, rs11822168, rs1938956, rs10736648, rs528205, rs11226867, rs667174, and rs641574 were not associated with MDD diagnosis or treatment response | ||
| Chiesa et al, 2013 | Candidate gene | MDD | 145 patients | Korean | Treatment response and phenotypic characteristics | GRIA4 rs10736648 was not associated with improvement on depression rating scale scores or other clinical/sociodemographic variables. There was no interaction between GRIA2 rs4260586, GRIA4 rs10736648, and improvement in depressive symptoms | ||
| NMDA receptor | ||||||||
| GRIN3A | GluN3A | Takata et al, 2013 | Candidate gene | BD | 865 patients and 2781 controls | Japanese | Risk | In a sub-analysis of 865 Japanese BD patients compared with 2781 controls, GRIN3A rs149729514 was not associated with BD (P=0.14). |
| GRIN2D | GluN2D | Dorval et al, 2009 | Linkage study | MDD | 370 nuclear families with 450 children | Hungarian | Risk | Only GRIN2D rs276713 was associated with childhood-onset mood disorders (uncorrected P=0.04), but the association did not remain significant after correction for multiple testing |
| mGlu receptors | ||||||||
| GRM4 | mGluR4 | Fallin et al, 2005 | Linkage study | BD | 337 trios from 323 families | Ashkenazi Jewish | Risk | GRM4 was associated with BD (all uncorrected P<0.01), but did not survive correction for multiple comparisons |
Abbreviations: BD, bipolar disorder; GW, genome-wide; GWAS, genome-wide association study; MDD, major depressive disorder; SNRI, serotonin-norepinephrine reuptake inhibitor; SSRI, selective serotonin reuptake inhibitor; TRD, treatment resistant depression.
The studies retrieved in the systematic search were reviewed based on the following proteins related to the glutamatergic pathway: vesicular glutamate transport proteins (VGLUTs), soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor, N-methyl-D-aspartate (NMDA) receptor, kainate receptor, metabotropic glutamate receptor (mGluR), excitatory amino acid transporters (EAATs).
Most studies evaluated several genes; the study is located in the line(s) that correspond(s) to the most relevant results on glutamate-related genes.
Studies are underlined when they demonstrated a positive association between glutamate-related genes and mood disorders. The genes are ranked according to the following system: high interest***: at least one study with a large sample (>1000 patients) and two studies with medium-sized samples (200–1000 patients); moderate interest**: evidence from one study with a large sample size (>1000 patients) but no more than one study with a medium-sized sample; low interest*: findings from studies with medium or small sample sizes, but no study with a large sample size.
Interestingly, most of the positive findings (18 of 34 studies (53%)) came from studies that analyzed risk for mood disorders, particularly in BD. Twelve of the 34 positive studies (35%) showed an association between risk for BD and glutamate-related genes, and 7 (21%) showed an association between glutamate-related genes and risk for MDD. The influence of the glutamatergic system on treatment response was supported by 10 of the 34 positive studies (29% eight in MDD, two in BD). Eleven of the 34 positive studies (32% seven in MDD, four in BD) explored the links between phenotypic characteristics and glutamate-related genes.
The variants and regions of those genes showing a positive association with mood disorders are depicted in Figure 1 and Table 1. Specifically, positive evidence was obtained for 16 genes. The importance of these individual findings is highlighted throughout the manuscript, and in the tables and figures, via the following ranking system: high interest (***), moderate interest (**), and low interest (*). Evidence of high interest (***) was supported by at least one study with a large sample size (>1000 patients) and by two studies with medium sample sizes (200–1000 patients). Evidence of moderate interest (**) was supported by at least one study with a large sample size but no more than one study of medium sample size. Evidence of low interest (*) was supported by findings from studies with medium or low sample sizes (<200 patients), but no studies with large sample sizes.
Although to date no specific glutamate-related gene has consistently been associated with mood disorders, below we discuss four genes of high interest. As noted above, an additional 12 genes of moderate and low interest were identified; these are described only in Figure 1 and Table 1. Corroborating evidence from preclinical or postmortem studies is noted throughout the manuscript when extant.
AMPA Receptor
The ionotropic AMPA receptor encompasses several subunits encoded by the GRIA1–4 genes.
GRIA3***
GRIA3 rs4825476 of the gene encoding GluA3 was associated with emergent suicidal ideation following citalopram treatment in a mostly Caucasian sample (78%) of 1915 MDD patients drawn from the STAR*D study (Laje et al, 2007). The same GRIA3 rs4825476 was further associated with suicidal ideation following antidepressant treatment in a German sample of 397 MDD inpatients drawn from the Munich Antidepressant Response Signature (MARS) project; however, these findings implicated a different allele (AA; Menke et al, 2008).
Interestingly, another study found that GRIA3 rs557762 and GRIA3 TT HT rs592807-rs557762 were associated with guilt feelings in a subanalysis of 183 Korean MDD females (Myung et al, 2012). In a Chinese Han sample of 281 patients with MDD, the GRIA3 rs502434 and rs3761555 SNPs were also linked to early-onset adverse events and recent negative life stress that influenced treatment outcome, though the analyses did not withstand correction for multiple comparisons (Pu et al, 2013). In addition, a family study of 373 Caucasian individuals from 40 BD pedigrees from the US and Israel found no linkage between GRIA3 and BD (Gécz et al, 1999).
It should also be noted that a postmortem study found that levels of the GluA3 subunit of the glutamate receptor encoded by GRIA3 was significantly downregulated in the hippocampus of MDD subjects (Duric et al, 2013).
Kainate Receptor
GRIK2***
GRIK2 encodes a kainate receptor subunit that alters the structure and function of the GluK2 kainate receptor. GRIK2 rs2518224 was associated with suicidal ideation following citalopram treatment in the aforementioned sample of MDD patients (n=1915) drawn from the STAR*D study that analyzed 68 candidate genes (odds ratio (OR)=8.23; corrected P<0.003; Laje et al, 2007). In a subsequent replication study conducted in a German sample of 397 MDD inpatients who were part of the MARS project, 15 GRIK2 SNPs were further associated with suicidal ideation following antidepressant treatment; the SNPs with the strongest association with MDD were GRIK2 rs2852618 (OR=9.0, uncorrected P=0.005) and GRIK2 rs2782900 (OR=4.3, uncorrected P=0.007; Menke et al, 2008). Another study found that GRIK2 HT ACAG rs1556994-rs1340282-rs1340277-rs141717 was associated with somatic anxiety in a sample of 241 Korean MDD patients (Myung et al, 2012).
GRIK4***
GRIK4 encodes a protein that belongs to the kainate acid-type glutamate receptor GluK4. In a mostly (78%) Caucasian sample (divided into 1199 MDD patients for discovery and 617 for replication), the G allele of GRIK4 rs1954787 was directly associated with both response and remission after citalopram treatment (Paddock et al, 2007). In another Caucasian sample of 627 MDD patients, GRIK4 rs11218030 was associated with treatment resistance (corrected P=0.025), while the G allele of GRIK4 rs1954787 was linked to the presence of psychotic symptoms (Milanesi et al, 2015). Consistent with the study by Paddock and colleagues (Paddock et al, 2007), antidepressant response in a Chinese Han sample of 281 MDD patients receiving different SSRIs or serotonin norepinephrine reuptake inhibitors (SNRIs) for 6 weeks was directly and inversely associated with the G allele of the GRIK4 rs1954787 SNP and with the A-G-G HT of GRIK4 (rs1954787-rs2230297-rs2298725), respectively (Pu et al, 2013). GRIK4 SNPs analyzed in a Caucasian sample of 275 MDD patients were not significantly associated with response and remission within up to 5 weeks of treatment with several types of antidepressants, but the GG genotype of GRIK4 rs12800734 was associated with downregulation of hypothalamic–pituitary–adrenal (HPA) axis hyperactivity (Horstmann et al, 2010). In contrast, GRIK4 was not associated with treatment response to 6 weeks of duloxetine in a sample of 250 mostly Caucasian MDD patients (Perlis et al, 2010). GRIK4 was also not associated with response to antidepressants, risk for MDD, or phenotypic characteristics in a sample of 223 Caucasian MDD patients and 76 healthy controls (Serretti et al, 2012).
As regards BD, an association was observed between decreased manic symptoms and GRIK4 rs2298723 in a sample of 470 manic BD patients in the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD) Study who were treated with several antipsychotics, but this result did not survive correction for multiple testing (Drago et al, 2013). Notably, a Scottish sample of 368 BD patients and 458 controls found that the GRIK4 HT GC-rs2282586-rs1944522 protected against BD (OR=0.62, P=0.0002), an association that remained significant after correction for multiple testing (Pickard et al, 2006). In another Scottish sample of 356 patients and 286 controls (which overlapped with the 2006 study by Pickard and colleagues), a deletion in GRIK4 ss79314275 was associated with BD when analyzing genotype (P=2.73 × 10−6) and allele frequencies (P=1.9 × 10−7, OR=0.462); an independent replication sample of 274 patients and 376 controls confirmed this association for genotype (P=0.0166) and allele (P=0.0107, OR=0.694) with BD (Pickard et al, 2008). Interestingly, the combined discovery and replication samples showed an additive/dose-dependent protection: the association of deletion heterozygosity with BD was higher (OR=0.471) than with deletion homozygosity (OR=0.325) (Pickard et al, 2008). Thus, it appears that both of these kainate receptors—GRIK2 and GRIK4—are potentially implicated in mood disorders.
mGluRs
GRM7***
GRM7 encodes mGluR7. Linkage analyses encompass extensive regions, and GRM7 is one of the genes that could explain the signal associating the 3p25-26 region with MDD. One family study of 187 Australian sibling pairs, all them heavy smokers with a history of MDD, found that chromosome 3 at 24.9 cM (3p26–3p25) showed a genome-wide significant multipoint LOD score of 4.14 for MDD (corrected P=0.004; Pergadia et al, 2011). Another genome-wide study that replicated the finding showed significant linkage between severe MDD and chromosome 3p25–26, with a maximum LOD score of 4.0 centered at linkage marker D3S1515 (corrected P=0.015; Breen et al, 2011); the latter study analyzed 971 concordant sibling pairs for recurrent MDD, 118 discordant sibling pairs, and 12 unaffected sibling pairs, all of European descent.
A GWAS evaluating 926 Caucasian MDD patients and 866 controls in the discovery phase and 494 Caucasian MDD patients and 1052 controls in the replication phase obtained mixed results regarding GRM7. Although no significant variants were associated with MDD in the main analysis (Muglia et al, 2010), a sub-analysis of candidate genes found that GRM7 rs162209 had the lowest P-value (P=0.0001) in the first sample, but this finding was not significant in the replication study (P=0.1); when the two samples were analyzed together via meta-analysis, GRM7 rs162209 again had the lowest P-value, and the association occurred in the same direction (P=0.0002; Muglia et al, 2010). Another study of 1541 Caucasian and African-American MDD patients found that the GRM7 GG genotype of rs1083801 was associated with early response to citalopram compared with late response (P=2.03 × 10−6) and non-response (P=1.82 × 10−5; Fabbri et al, 2013). The result was confirmed in a Caucasian, non-Hispanic sample that found that the GRM7 GG genotype of rs1083801 was associated with early response to citalopram compared with late response (P=6.75 × 10−7) and non-response (P=2.12 × 10−5; after Bonferroni correction, alpha set at 2.51 × 10−5; Fabbri et al, 2013). Notably, after stratification by gender, the association between GRM7 rs1083801 and response to citalopram was shown to be significant only in females (females, uncorrected P=1.54 × 10−5; males, uncorrected P=0.0003). Finally, an analysis of 1221 MDD patients and 1636 controls of European ancestry drawn from the STAR*D study found no genome-wide evidence of an association between MDD and GRM7 (Shyn et al, 2011); however, the GRM7 rs9870680 SNP showed one of the lowest P-values (uncorrected P=1.1 × 10−6). Nevertheless, another study of 1738 MDD patients drawn from the Dutch GAIN-MDD sample found that none of the 204 GRM7 SNPs were associated with MDD (Verbeek et al, 2013).
As regards BD, a case–control study in BD patients of Irish or British descent found that three GRM7 SNPs were associated with BD in the discovery sample, a finding not replicated in a second sample (Kandaswamy et al, 2014). After combining the genotype data for the two samples (1099 BD patients and 1235 healthy controls), BD was significantly associated with GRM7 rs1508724 (OR=1.15, corrected P=0.043) and GRM7 rs6769814 (OR=1.15, corrected P=0.045; Kandaswamy et al, 2014). Another analysis of SNPs selected based on increased frequency in BD cases detected an association between GRM7 rs56173829 and BD in the two samples combined (OR=0.4829, P=0.035; Kandaswamy et al, 2014). Another study found that the GRM7 rs13080594 SNP was associated with Neuroticism-Anxiety (uncorrected P=7.68 × 10−7) as assessed by the Zuckerman-Kuhlman Personality Questionnaire in 1007 BD patients of European ancestry, but this finding did not survive correction for multiple comparisons (Alliey-Rodriguez et al, 2011). Interestingly, a GWAS in individuals of European ancestry with BD found one of the strongest P-values (uncorrected P=0.0001) for the glutamatergic SNP GRM7 rs1485171; however, it should be noted that 15% of the sample was diagnosed with schizoaffective disorder, BD subtype (CONVERGE Consortium, 2015).
Preclinical data also support the association between GRM7 and mood disorders. Specifically, mice with the GRM7 deletion had substantially less behavioral immobility in both the forced swim and tail suspension tests than their wild-type littermates (Cryan et al, 2003).
DISCUSSION
Here we reviewed the links between glutamate-related genes and mood disorders risk, treatment response, and phenotypic characteristics such as emergent suicidal ideation. As the evidence reviewed above demonstrates, more evidence exists linking glutamate-related genes to BD than MDD, but no specific glutamate-related gene has been consistently associated with mood disorders. However, several genes appear worthy of further exploration, including GRIA3, GRIK2, GRIK4, and GRM7.
Most of the studies reviewed here were candidate gene studies; only six GWASs were included. GWASs have advantages over other study designs because they use larger samples and analyze variants over the entire genome, thus providing more robust results. Indeed, in this review, four-fifths of the studies that found evidence of a relationship between glutamate-related genes and mood disorders were of medium or large samples. However, of the large studies evaluating glutamate-related genes, no analysis of a gene associated with mood disorders has yet been replicated.
The multiple glutamate-related gene targets of small relevance reviewed above are consistent with the overall sparse findings in genetic studies of mood disorders (Craddock and Sklar, 2013). As noted above, genetic studies in BD have been more conclusive than in MDD (13 studies in BD vs six in MDD), which could be explained by factors such as the much higher lifetime prevalence of MDD, which is over 16% (Kessler et al, 2005), the higher heritability of BD vs MDD (roughly 85% compared with 40% Lohoff, 2010; McGuffin et al, 2003), or the more diffuse phenotype of MDD compared with BD. Studies that focus on specific clinical characteristics of MDD, such as age of onset, could be more successful in this context, as could research focused on groups with specific endophenotypes, such as neuroimaging abnormalities.
Taken together, the studies reviewed above implicate several glutamate-related genes of high interest (***) in mood disorders: GRIA3, GRIK2, GRIK4, and GRM7. Specifically, several MDD (n=7) and BD (n=2) studies support the link between glutamate-related genes and treatment response. For instance, two large studies in MDD found evidence of a relationship between GRIK4 (Paddock et al, 2007) and GRM7 (Fabbri et al, 2013) and treatment response to SSRIs. Phenotypic characteristics were associated with both MDD (six studies) and BD (four studies). Notably, emergent suicidal ideation after SSRI treatment in MDD was linked to GRIA3 and GRIK2 in one large (Laje et al, 2007) and one medium-sized (Menke et al, 2008) study. In addition, a region spanning GRM7 was significantly associated with risk for MDD in one GWAS (Breen et al, 2011), and GRM7 was one of the best candidate genes emerging from two GWASs that evaluated risk for MDD (Muglia et al, 2010; Shyn et al, 2011). GRM7 was also linked to BD in one large study (Kandaswamy et al, 2014) and to MDD in one medium-sized study (Pergadia et al, 2011). Finally, GRIK4 influenced treatment response in one large and two medium-sized studies in MDD (Horstmann et al, 2010; Paddock et al, 2007; Pu et al, 2013) and was associated with risk for BD in two medium-sized samples (Pickard et al, 2006, 2008). All four of these genes (GRIA3, GRIK2, GRIK4, and GRM7) stand out as highly interesting candidates for further study. In addition, further clinical or preclinical evidence exists for all of these genes of functional involvement such as altered mRNA expression or induction of depressive-like behaviors (Beneyto et al, 2007; Catches et al, 2012; Cryan et al, 2003; Duric et al, 2013).
Though preliminary, evidence also suggests an association between glutamate genes with neuroimaging correlates in mood disorders. In MDD, GRM1 was associated with brain hippocampal glutamate levels (Menke et al, 2012). In BD patients, GRIN2B was associated with white matter integrity (Kuswanto et al, 2013), and SLC1A2 was found to modulate gray matter volume (Poletti et al, 2014). However, the results reported across studies have also obtained inconsistent results, which might be due to the high prevalence, relatively moderate heritability, and phenotypic heterogeneity of mood disorders. Another potential reason for inconsistencies is that common variants with small effects likely combine to make a large contribution to risk for mood disorders (Craddock and Sklar, 2013; Flint and Kendler, 2014), which leads to low power to detect effects for the SNPs studied as well as lack of replication.
Importantly, the availability of less costly exome sequencing or whole-genome sequencing techniques has improved the field; both methods are especially useful at finding rare variants that may have larger effects on mood disorders. Successful strategies for future studies could include focusing on more severe cases of MDD to retrieve a clearer signal (CONVERGE Consortium, 2015); using very large sample sizes to study depressive symptoms (Okbay et al, 2016); or studying families with a dense prevalence of mood disorders (Collins et al, 2013).
Pathway analyses are a powerful tool for overcoming limitations associated with studies that explain only a small proportion of phenotypic variance because they use previous knowledge of molecular and cellular processes to detect associations between genes and disorders (Wang et al, 2007). Interestingly, pathway analyses of large datasets support the involvement of the glutamatergic system—and specific glutamatergic pathways—in risk for MDD (Lee et al, 2012) and BD (Nurnberger et al, 2014; Torkamani et al, 2008). In addition, postsynaptic density was found to be related to risk for mood disorders (as well as schizophrenia; Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium, 2015), and the long-term potentiation pathway encompassing several glutamate genes was involved with response to citalopram in MDD (Hunter et al, 2013).
Another pathway analysis also found an association between BD and the glia-astrocyte pathway (Duncan et al, 2014); glial cells are central to glutamatergic uptake and recycling. Although the glutamatergic modulator ketamine is consistently associated with an effective antidepressant response (Zarate et al, 2006, 2012), research into ketamine is still in its infancy and we found no studies specifically investigating the genetics of response to ketamine, perhaps because no cohorts to date have been large enough to perform adequately powered GWASs. The lack of genetic studies on ketamine and other glutamatergic modulators could partly explain the low-interest evidence for the NMDA receptor in our review.
Limitations of this review include a methodology limited to searching for articles focused on glutamate-related genes and the diversity of the study designs. However, the inclusion of all genetic studies on the glutamate pathway in mood disorders is a strength of the present review. In future analyses, it might be valuable to explore the glutamatergic pathway in the context of patients enrolled in clinical trials with glutamatergic drugs.
Despite the absence of consistent results, these findings suggest directions that could help decipher the etiology and pathogenesis of mood disorders. The body of evidence suggests that glutamatergic genes are indeed involved in the pathophysiology and treatment of mood disorders. In particular, several targets identified by the present review—including GRIA3, GRIK2, GRIK4, and GRM7—are worthy of further exploration in future studies.
FUNDING AND DISCLOSURE
Funding for this work was supported by the Intramural Research Program at the National Institute of Mental Health, National Institutes of Health (IRP-NIMH-NIH; ZIA MH002857), by a NARSAD Independent Investigator toDr Zarate, and by a Brain & Behavior Mood Disorders Research Award to Dr Zarate. Dr Zarate is listed as aco-inventor on a patent for the use of ketamine and its metabolites in major depression. He has assigned his rights in the patent to the US government but will share a percentage of any royalties that may be received by the government. The remaining authors declare no conflict of interest.
Acknowledgments
We thank the 7SE research unit and staff for their support.
References
- Abou Jamra R, Gobina CM, Becker T, Georgi A, Schulze TG, Schmael C et al (2008). Association study between genetic variants at the VAMP2 and VAMP3 loci and bipolar affective disorder. Psychiatr Genet 18: 199–203. [DOI] [PubMed] [Google Scholar]
- Alliey-Rodriguez N, Zhang D, Badner JA, Lahey BB, Zhang X, Dinwiddie S et al (2011). Genome-wide association study of personality traits in bipolar patients. Psychiatr Genet 21: 190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramopoulos D, Lasseter VK, Fallin MD, Wolyniec PS, McGrath JA, Nestadt G et al (2007). Stage II follow-up on a linkage scan for bipolar disorder in the Ashkenazim provides suggestive evidence for chromosome 12p and the GRIN2B gene. Genet Med 9: 745–751. [DOI] [PubMed] [Google Scholar]
- Beneyto M, Kristiansen LV, Oni-Orisan A, McCullumsmith RE, Meador-Woodruff JH (2007). Abnormal glutamate receptor expression in the medial temporal lobe in schizophrenia and mood disorders. Neuropsychopharmacology 32: 1888–1902. [DOI] [PubMed] [Google Scholar]
- Breen G, Webb BT, Butler AW, van den Oord EJ, Tozzi F, Craddock N et al (2011). A genome-wide significant linkage for severe depression on chromosome 3: the depression network study. Am J Psychiatry 168: 840–847. [DOI] [PubMed] [Google Scholar]
- Catches JS, Xu J, Contractor A (2012). Genetic ablation of the GluK4 kainate receptor subunit causes anxiolytic and antidepressant-like behavior in mice. Behav Brain Res 228: 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa A, Crisafulli C, Porcelli S, Balzarro B, Han C, Patkar AA et al (2012. a). Case-control association study of GRIA1, GRIA2 and GRIA4 polymorphisms in bipolar disorder. Int J Psychiatry Clin Pract 16: 18–26. [DOI] [PubMed] [Google Scholar]
- Chiesa A, Crisafulli C, Porcelli S, Han C, Patkar AA, Lee SJ et al (2012. b). Influence of GRIA1, GRIA2 and GRIA4 polymorphisms on diagnosis and response to treatment in patients with major depressive disorder. Eur Arch Psychiatry Clin Neurosci 262: 305–311. [DOI] [PubMed] [Google Scholar]
- Chiesa A, Lia L, Lia C, Lee SJ, Han C, Patkar AA et al (2013). Investigation of possible epistatic interactions between GRIA2 and GRIA4 variants on clinical outcomes in patients with major depressive disorder. J Int Med Res 41: 809–815. [DOI] [PubMed] [Google Scholar]
- Chourbaji S, Vogt MA, Fumagalli F, Sohr R, Frasca A, Brandwein C et al (2008). AMPA receptor subunit 1 (GluR-A) knockout mice model the glutamate hypothesis of depression. FASEB J 22: 3129–3134. [DOI] [PubMed] [Google Scholar]
- Collins AL, Kim Y, Szatkiewicz JP, Bloom RJ, Hilliard CE, Quackenbush CR et al (2013). Identifying bipolar disorder susceptibility loci in a densely affected pedigree. Mol Psychiatry 18: 1245–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONVERGE Consortium (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523: 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, Sklar P (2013). Genetics of bipolar disorder. Lancet 381: 1654–1662. [DOI] [PubMed] [Google Scholar]
- Cryan JF, Kelly PH, Neijt HC, Sansig G, Flor PJ, van Der Putten H (2003). Antidepressant and anxiolytic-like effects in mice lacking the group III metabotropic glutamate receptor mGluR7. Eur J Neurosci 17: 2409–2417. [DOI] [PubMed] [Google Scholar]
- Dallaspezia S, Poletti S, Lorenzi C, Pirovano A, Colombo C, Benedetti F (2012). Influence of an interaction between lithium salts and a functional polymorphism in SLC1A2 on the history of illness in bipolar disorder. Mol Diagn Ther 16: 303–309. [DOI] [PubMed] [Google Scholar]
- Dalvie S, Horn N, Nossek C, van der Merwe L, Stein DJ, Ramesar R (2010). Psychosis and relapse in bipolar disorder are related to GRM3, DAOA, and GRIN2B genotype. Afr J Psychiatry (Johannesbg) 13: 297–301. [DOI] [PubMed] [Google Scholar]
- Dorval KM, Burcescu I, Adams J, Wigg KG, King N, Kiss E et al (2009). Association study of N-methyl-D-aspartate glutamate receptor subunit genes and childhood-onset mood disorders. Psychiatr Genet 19: 156–157. [DOI] [PubMed] [Google Scholar]
- Drago A, Giegling I, Schäfer M, Hartmann AM, Friedl M, Konte B et al (2013). AKAP13, CACNA1, GRIK4 and GRIA1 genetic variations may be associated with haloperidol efficacy during acute treatment. Eur Neuropsychopharmacol 23: 887–894. [DOI] [PubMed] [Google Scholar]
- Duman RS (2014). Pathophysiology of depression and innovative treatments: remodeling glutamatergic synaptic connections. Dialogues Clin Neurosci 16: 11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Holmans PA, Lee PH, O'Dushlaine CT, Kirby AW, Smoller JW et al (2014). Pathway analyses implicate glial cells in schizophrenia. PLoS One 9: e89441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duric V, Banasr M, Stockmeier CA, Simen AA, Newton SS, Overholser JC et al (2013). Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. Int J Neuropsychopharmacol 16: 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etain B, Dumaine A, Mathieu F, Chevalier F, Henry C, Kahn JP et al (2010). A SNAP25 promoter variant is associated with early-onset bipolar disorder and a high expression level in brain. Mol Psychiatry 15: 748–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri C, Drago A, Serretti A (2013). Early antidepressant efficacy modulation by glutamatergic gene variants in the STAR*D. Eur Neuropsychopharmacol 23: 612–621. [DOI] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA et al (2005). Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet 77: 918–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Kendler KS (2014). The genetics of major depression. Neuron 81: 484–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gécz J, Barnett S, Liu J, Hollway G, Donnelly A, Eyre H et al (1999). Characterization of the human glutamate receptor subunit 3 gene (GRIA3), a candidate for bipolar disorder and nonspecific X-linked mental retardation. Genomics 62: 356–368. [DOI] [PubMed] [Google Scholar]
- Georgi A, Jamra RA, Schumacher J, Becker T, Schmael C, Deschner M et al (2006). No association between genetic variants at the GRIN1 gene and bipolar disorder in a German sample. Psychiatr Genet 16: 183–184. [DOI] [PubMed] [Google Scholar]
- Hammer C, Stepniak B, Schneider A, Papiol S, Tantra M, Begemann M et al (2014). Neuropsychiatric disease relevance of circulating anti-NMDA receptor autoantibodies depends on blood-brain barrier integrity. Molecular Psychiatry 19: 1143–1149. [DOI] [PubMed] [Google Scholar]
- Horstmann S, Lucae S, Menke A, Hennings JM, Ising M, Roeske D et al (2010). Polymorphisms in GRIK4, HTR2A, and FKBP5 show interactive effects in predicting remission to antidepressant treatment. Neuropsychopharmacology 35: 727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter AM, Leuchter AF, Power RA, Muthen B, McGrath PJ, Lewis CM et al (2013). A genome-wide association study of a sustained pattern of antidepressant response. J Psychiatr Res 47: 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Zhang R, Wu B, Dai ZX, Zhu YS, Li PP et al (2014). Metabotropic glutamate receptor 3 is associated with heroin dependence but not depression or schizophrenia in a Chinese population. PLoS One 9: e87247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd LL, Akiskal HS, Schettler PJ, Endicott J, Maser J, Solomon DA et al (2002). The long-term natural history of the weekly symptomatic status of bipolar I disorder. Arch Gen Psychiatry 59: 530–537. [DOI] [PubMed] [Google Scholar]
- Kandaswamy R, McQuillin A, Curtis D, Gurling H (2014). Allelic association, DNA resequencing and copy number variation at the metabotropic glutamate receptor GRM7 gene locus in bipolar disorder. Am J Med Genet B Neuropsychiatr Genet 165B: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandaswamy R, McQuillin A, Sharp SI, Fiorentino A, Anjorin A, Blizard RA et al (2013). Genetic association, mutation screening, and functional analysis of a Kozak sequence variant in the metabotropic glutamate receptor 3 gene in bipolar disorder. JAMA Psychiatry 70: 591–598. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE (2005). Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 62: 593–602. [DOI] [PubMed] [Google Scholar]
- Kuswanto CN, Sum MY, Thng CR, Zhang YB, Yang GL, Nowinski WL et al (2013). GRIN2B gene and associated brain cortical white matter changes in bipolar disorder: a preliminary combined platform investigation. Biomed Res Int 2013: 635131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laje G, Paddock S, Manji H, Rush AJ, Wilson AF, Charney D et al (2007). Genetic markers of suicidal ideation emerging during citalopram treatment of major depression. Am J Psychiatry 164: 1530–1538. [DOI] [PubMed] [Google Scholar]
- Lee PH, Perlis RH, Jung JY, Byrne EM, Rueckert E, Siburian R et al (2012). Multi-locus genome-wide association analysis supports the role of glutamatergic synaptic transmission in the etiology of major depressive disorder. Transl Psychiatry 2: e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yu T, Huang X, Cao Y, Liu B, Yang F et al (2014). SLC17A7 gene may be the indicator of selective serotonin reuptake inhibitor treatment response in the Chinese Han population. J Clin Psychopharmacol 34: 331–336. [DOI] [PubMed] [Google Scholar]
- Lingamaneni R, Birch ML, Hemmings HC (2001). Widespread inhibition of sodium channel-dependent glutamate release from isolated nerve terminals by isoflurane and propofol. Anesthesiology 95: 1460–1466. [DOI] [PubMed] [Google Scholar]
- Lohoff FW (2010). Overview of the genetics of major depressive disorder. Curr Psychiatry Rep 12: 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Vieira R, Manji HK, Zarate CA (2009). The role of the tripartite glutamatergic synapse in the pathophysiology and therapeutics of mood disorders. Neuroscientist 15: 525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Vieira R, Salvadore G, Luckenbaugh DA, Manji HK, Zarate CA (2008). Rapid onset of antidepressant action: a new paradigm in the research and treatment of major depressive disorder. J Clin Psychiatry 69: 946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeng S, Zarate CA, Du J, Schloesser RJ, McCammon J, Chen G et al (2008). Cellular mechanisms underlying the antidepressant effects of ketamine: role of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors. Biol Psychiatry 63: 349–352. [DOI] [PubMed] [Google Scholar]
- Martí SB, Cichon S, Propping P, Nöthen M (2002). Metabotropic glutamate receptor 3 (GRM3) gene variation is not associated with schizophrenia or bipolar affective disorder in the German population. Am J Med Genet 114: 46–50. [DOI] [PubMed] [Google Scholar]
- Martucci L, Wong AH, De Luca V, Likhodi O, Wong GW, King N et al (2006). N-methyl-D-aspartate receptor NR2B subunit gene GRIN2B in schizophrenia and bipolar disorder: Polymorphisms and mRNA levels. Schizophr Res 84: 214–221. [DOI] [PubMed] [Google Scholar]
- Mathews DC, Henter ID, Zarate CA (2012). Targeting the glutamatergic system to treat major depressive disorder: rationale and progress to date. Drugs 72: 1313–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A (2003). The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry 60: 497–502. [DOI] [PubMed] [Google Scholar]
- Menke A, Lucae S, Kloiber S, Horstmann S, Bettecken T, Uhr M et al (2008). Genetic markers within glutamate receptors associated with antidepressant treatment-emergent suicidal ideation. Am J Psychiatry 165: 917–918. [DOI] [PubMed] [Google Scholar]
- Menke A, Sämann P, Kloiber S, Czamara D, Lucae S, Hennings J et al (2012). Polymorphisms within the metabotropic glutamate receptor 1 gene are associated with depression phenotypes. Psychoneuroendocrinology 37: 565–575. [DOI] [PubMed] [Google Scholar]
- Milanesi E, Bonvicini C, Congiu C, Bortolomasi M, Gainelli G, Gennarelli M et al (2015). The role of GRIK4 gene in treatment-resistant depression. Genet Res (Camb) 97: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ et al (2010). Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry 15: 589–601. [DOI] [PubMed] [Google Scholar]
- Müller DJ, Schulze TG, Jahnes E, Cichon S, Krauss H, Kesper K et al (2002). Association between a polymorphism in the pseudoautosomal X-linked gene SYBL1 and bipolar affective disorder. Am J Med Genet 114: 74–78. [DOI] [PubMed] [Google Scholar]
- Mundo E, Tharmalingham S, Neves-Pereira M, Dalton EJ, Macciardi F, Parikh SV et al (2003). Evidence that the N-methyl-D-aspartate subunit 1 receptor gene (GRIN1) confers susceptibility to bipolar disorder. Mol Psychiatry 8: 241–245. [DOI] [PubMed] [Google Scholar]
- Myung W, Song J, Lim SW, Won HH, Kim S, Lee Y et al (2012). Genetic association study of individual symptoms in depression. Psychiatry Res 198: 400–406. [DOI] [PubMed] [Google Scholar]
- Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium (2015). Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 18: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI Jr, Koller DL, Jung JY, Edenberg HJ, Foroud T, Guella I et al (2014). Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry 71: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA et al (2016). Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet 48: 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrego F, Villanueva S (1993). The chemical nature of the main central excitatory transmitter: a critical appraisal based upon release studies and synaptic vesicle localization. Neuroscience 56: 539–555. [DOI] [PubMed] [Google Scholar]
- Paddock S, Laje G, Charney D, Rush AJ, Wilson AF, Sorant AJ et al (2007). Association of GRIK4 with outcome of antidepressant treatment in the STAR*D cohort. Am J Psychiatry 164: 1181–1188. [DOI] [PubMed] [Google Scholar]
- Papp M, Moryl E (1994). Antidepressant activity of non-competitive and competitive NMDA receptor antagonists in a chronic mild stress model of depression. Eur J Pharmacol 263: 1–7. [DOI] [PubMed] [Google Scholar]
- Pergadia ML, Glowinski AL, Wray NR, Agrawal A, Saccone SF, Loukola A et al (2011). A 3p26-3p25 genetic linkage finding for DSM-IV major depression in heavy smoking families. Am J Psychiatry 168: 848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH, Fijal B, Dharia S, Heinloth AN, Houston JP (2010). Failure to replicate genetic associations with antidepressant treatment response in duloxetine-treated patients. Biol Psychiatry 67: 1110–1113. [DOI] [PubMed] [Google Scholar]
- Perlis RH, Smoller JW, Ferreira MA, McQuillin A, Bass N, Lawrence J et al (2009). A genomewide association study of response to lithium for prevention of recurrence in bipolar disorder. Am J Psychiatry 166: 718–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard BS, Knight HM, Hamilton RS, Soares DC, Walker R, Boyd JK et al (2008). A common variant in the 3'UTR of the GRIK4 glutamate receptor gene affects transcript abundance and protects against bipolar disorder. Proc Natl Acad Sci USA 105: 14940–14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard BS, Malloy MP, Christoforou A, Thomson PA, Evans KL, Morris SW et al (2006). Cytogenetic and genetic evidence supports a role for the kainate-type glutamate receptor gene, GRIK4, in schizophrenia and bipolar disorder. Mol Psychiatry 11: 847–857. [DOI] [PubMed] [Google Scholar]
- Poletti S, Locatelli C, Radaelli D, Lorenzi C, Smeraldi E, Colombo C et al (2014). Effect of early stress on hippocampal gray matter is influenced by a functional polymorphism in EAAT2 in bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry 51: 146–152. [DOI] [PubMed] [Google Scholar]
- Pu M, Zhang Z, Xu Z, Shi Y, Geng L, Yuan Y et al (2013). Influence of genetic polymorphisms in the glutamatergic and GABAergic systems and their interactions with environmental stressors on antidepressant response. Pharmacogenomics 14: 277–288. [DOI] [PubMed] [Google Scholar]
- Rudy CC, Hunsberger HC, Weitzner DS, Reed MN (2015). The role of the tripartite glutamatergic synapse in the pathophysiology of Alzheimer's disease. Aging Dis 6: 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D et al (2006). Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry 163: 1905–1917. [DOI] [PubMed] [Google Scholar]
- Saito S, Takahashi N, Ishihara R, Ikeda M, Suzuki T, Kitajima T et al (2006). Association study between vesicle-associated membrane protein 2 gene polymorphisms and fluvoxamine response in Japanese major depressive patients. Neuropsychobiology 54: 226–230. [DOI] [PubMed] [Google Scholar]
- Saito T, Guan F, Papolos DF, Rajouria N, Fann CS, Lachman HM (2001). Polymorphism in SNAP29 gene promoter region associated with schizophrenia. Mol Psychiatry 6: 193–201. [DOI] [PubMed] [Google Scholar]
- Saito T, Parsia S, Papolos DF, Lachman HM (2000). Analysis of the pseudoautosomal X-linked gene SYBL1in bipolar affective disorder: description of a new candidate allele for psychiatric disorders. Am J Med Genet 96: 317–323. [DOI] [PubMed] [Google Scholar]
- Schiffer HH, Heinemann SF (2007). Association of the human kainate receptor GluR7 gene (GRIK3) with recurrent major depressive disorder. Am J Hum Genet B Neuropsychiatr Genet 144B: 20–26. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V et al (2009). Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS One 4: e6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serretti A, Chiesa A, Crisafulli C, Massat I, Linotte S, Calati R et al (2012). Failure to replicate influence of GRIK4 and GNB3 polymorphisms on treatment outcome in major depression. Neuropsychobiology 65: 70–75. [DOI] [PubMed] [Google Scholar]
- Shyn SI, Shi J, Kraft JB, Potash JB, Knowles JA, Weissman MM et al (2011). Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol Psychiatry 16: 202–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K et al (2008). Whole-genome association study of bipolar disorder. Mol Psychiatry 13: 558–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick P, Layer RT, Popik P, Nowak G, Paul IA, Trullas R (1996). Adaptation of N-methyl-D-aspartate (NMDA) receptors following antidepressant treatment: implications for the pharmacotherapy of depression. Pharmacopsychiatry 29: 23–26. [DOI] [PubMed] [Google Scholar]
- Szczepankiewicz A, Rybakowski JK, Skibinska M, Dmitrzak-Weglarz M, Leszczynska-Rodziewicz A, Wilkosc M et al (2009. a). FYN kinase gene: another glutamatergic gene associated with bipolar disorder? Neuropsychobiology 59: 178–183. [DOI] [PubMed] [Google Scholar]
- Szczepankiewicz A, Skibinska M, Rybakowski J, Leszczynska-Rodziewicz A, Tomaszewska M, Twarowska-Hauser J (2009. b). Lack of association of three GRIN2B polymorphisms with bipolar disorder. World J Biol Psychiatry 10: 469–473. [DOI] [PubMed] [Google Scholar]
- Szczepankiewicz A, Skibinska M, Suwalska A, Hauser J, Rybakowski JK (2009. c). The association study of three FYN polymorphisms with prophylactic lithium response in bipolar patients. Hum Psychopharmacol 24: 287–291. [DOI] [PubMed] [Google Scholar]
- Szczepankiewicz A, Skibińska M, Suwalska A, Hauser J, Rybakowski JK (2009. d). No association of three GRIN2B polymorphisms with lithium response in bipolar patients. Pharmacol Rep 61: 448–452. [DOI] [PubMed] [Google Scholar]
- Takata A, Iwayama Y, Fukuo Y, Ikeda K, Okochi T, Maekawa M et al (2013). A population-specific uncommon variant in GRIN3A associated with schizophrenia. Biol Psychiatry 73: 532–539. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Topol EJ, Schork NJ (2008). Pathway analysis of seven common diseases assessed by genome-wise association. Genomics 92: 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoka T, Kishi T, Ikeda M, Kitajima T, Yamanouchi Y, Kinoshita Y et al (2009). Association analysis of group II metabotropic glutamate receptor genes (GRM2 and GRM3) with mood disorders and fluvoxamine response in a Japanese population. Prog Neuropsychopharmacol Biol Psychiatry 33: 875–879. [DOI] [PubMed] [Google Scholar]
- Verbeek EC, Bevova MR, Bochdanovits Z, Rizzu P, Bakker IM, Uithuisje T et al (2013). Resequencing three candidate genes for major depressive disorder in a Dutch cohort. PLoS One 8: e79921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M (2007). Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet 81: 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wang Y, Ji W, Zhou G, He K, Li Z et al (2015). SNAP25 is associated with schizophrenia and major depressive disorder in the Han Chinese population. J Clin Psychiatry 76: e76–e82. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang J, Li X, Ji J, Yang F, Wan C et al (2008). SCN8A as a novel candidate gene associated with bipolar disorder in the Han Chinese population. Prog Neuropsychopharmacol Biol Psychiatry 32: 1902–1904. [DOI] [PubMed] [Google Scholar]
- Witkin JM, Marek GJ, Johnson BG, Schoepp DD (2007). Metabotropic glutamate receptors in the control of mood disorders. CNS Neurol Disord Drug Targets 6: 87–100. [DOI] [PubMed] [Google Scholar]
- Zarate CA, Brutsche NE, Ibrahim L, Franco-Chaves J, Diazgranados N, Cravchik A et al (2012). Replication of ketamine's antidepressant efficacy in bipolar depression: a randomized controlled add-on trial. Biol Psychiatry 71: 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate CA, Singh JB, Carlson PJ, Brutsche NE, Ameli R, Luckenbaugh DA et al (2006). A randomized trial of an N-methyl-D-aspartate antagonist in treatment-resistant major depression. Arch Gen Psychiatry 63: 856–864. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li Z, Wu Z, Chen J, Wang Z, Peng D et al (2014). A study of N-methyl-D-aspartate receptor gene (GRIN2B) variants as predictors of treatment-resistant major depression. Psychopharmacology (Berl) 231: 685–693. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Che R, Zhang Z, Wang P, Li J, Li Y et al (2011). Positive association between GRIN2B gene and bipolar disorder in the Chinese Han Population. Psychiatry Res 185: 290–292. [DOI] [PubMed] [Google Scholar]