Abstract
Prostate cancer antigen 3 (PCA3) is a biomarker for diagnosing prostate cancer (PCa) identified in the Caucasian population. We evaluated the effectiveness of urinary PCA3 in predicting the biopsy result in 500 men undergoing initial prostate biopsy. The predictive power of the PCA3 score was evaluated by the area under receiver operating characteristic (ROC) curve (AUC) and by decision curve analysis. PCA3 score sufficed to discriminate positive from negative prostate biopsy results but was not correlated with the aggressiveness of PCa. The ROC analysis showed a higher AUC for the PCA3 score than %fPSA (0.750 vs 0.622, P = 0.046) in patients with a PSA of 4.0–10.0 ng ml−1, but the PCA3-based model is not significantly better than the base model. Decision curve analysis indicates the PCA3-based model was superior to the base model with a higher net benefit for almost all threshold probabilities, especially the threshold probabilities of 25%–40% in patients with a PSA of 4.0–10.0 ng ml−1. However, the AUC of the PCA3 score (0.712) is not superior to %fPSA (0.698) or PSAD (0.773) in patients with a PSA >10.0 ng ml−1. Our results confirmed that the RT-PCR-based PCA3 test moderately improved diagnostic accuracy in Chinese patients undergoing first prostate biopsy with a PSA of 4.0–10.0 ng ml−1.
Keywords: diagnosis, long noncoding RNA, prostate cancer antigen 3, prostatic neoplasms
INTRODUCTION
Prostate cancer is the most prevalent cancer in males in Western countries with an estimated 233 000 new cases and 29 480 estimated deaths in 2014 in the United States.1 Although its incidence is much lower in Asian countries, such as China,2 it is increasing at a tremendous rate. According to the Chinese Malignancy Report (2004, 2012), the incidence of PCa almost tripled from 2001 to 2009, ranking as the fastest growing malignancy.3,4 Although the lack of a systematic prostate cancer screening system in Asian countries may offer a partial explanation, the racial difference between Asian and Caucasian population may also have a substantial impact.5 This racial difference suggests that there may be different strategies for the diagnosis and treatment of PCa.
Diagnosis of PCa is currently based on digital rectal examination and prostate-specific antigen (PSA) tests. However, there are two major challenges with PSA. On the one hand, the high false positive rate of PSA tests leads to an unnecessary biopsy rate as high as 78% for initial biopsies and 90% for repeat biopsies with a PSA level of 4.0–10.0 ng ml−1; 6 on the other hand, there is not a safe PSA level below which men would not harbor PCa. A total of 26.9% of men with a PSA level of 3.1–4.0 were diagnosed with PCa in a US prevention study,7 making it difficult to set a lower limit. Although PSA derivatives, such as PSA density (PSAD), percent free PSA (%fPSA), and PSA velocity, have been demonstrated to be complementary tools to moderately improve the sensitivity and selectivity of PSA tests,8 there remains a need to improve the accuracy in the detection of PCa with new biomarkers.
In recent decades, new biomarkers to improve the detection of prostate cancer have emerged and shown promising results. Prostate cancer antigen 3 (PCA3), initially known as differential display clone 3 (DD3), has been extensively studied. Haese et al.9 showed that PCA3 was clinically effective in improving the detection of prostate cancer in patients with one or two previous negative biopsies. It was also confirmed that PCA3 is more effective than %fPSA, independent of prostate volume, age, and total PSA level.9 A meta-analysis illustrated that PCA3 had more discriminatory ability than total prostate-specific antigen, with 18 of 20 included studies demonstrating an improvement of AUC of 0.11 over PSA.10
Two studies in a Japanese population demonstrated an improvement in the diagnostic accuracy by applying the PCA3 test,11,12 which was confirmed in a Chinese study with a limited sample size of 64 benign prostatic hyperplasia patients and 35 PCa patients.13 However, these patients were not consecutive or in a clinical setting, making it difficult to predict the exact performance of PCA3 in clinical application in the Chinese population. As our previous RNA-seq study indicated, there are differences in the long noncoding RNAs among different population;14 this Western population-validated long noncoding RNA should be tested in a clinical setting before initiating a large-scale implication in Chinese population.
We performed the real-time RT-PCR-based PCA3 test in 500 patients who underwent initial prostate biopsies at Shanghai Changhai Hospital and compared the effectiveness of PCA3 with other PSA-based parameters.
METHODS
Patients and clinical specimens
This study was approved by the Institutional Review Board of Shanghai Changhai Hospital. Informed consent was obtained from all patients. Subjects were recruited between May 2012 and November 2013. All urine samples were collected from patients scheduled for initial prostate biopsy because of elevated serum PSA levels (≥4 ng ml−1) and/or suspicious DRE. We enrolled individuals visiting the out-patient department for various reasons, including health check-ups and voiding symptoms, regardless of digital rectal examination findings. Patients with other known tumors, recent instrumentation or catheterization of the urethra and those receiving finasteride or hormonal treatment were excluded.
Study design
The discriminative power of the PCA3 score between positive biopsy and negative biopsy was evaluated in two groups of patients with different PSA levels. Prostate volume (PV) was calculated using the equation D1 × D2 × D3 × (π/6), and the three dimensions of the prostate as measured by transrectal ultrasound (TRUS) performed by sonographers before biopsy. However, the PV of 22 patients was missing in the data collection. The diagnostic performance of the PCA3 score was assessed in these two groups. The diagnostic performance of the PCA3 score and other parameters was evaluated and compared in patients with all of these parameters available. Then, the combination of the PCA3 score and other parameters was evaluated by combining these parameters in logistic regression. The diagnostic performance of the PCA3 score and other parameters, the base model (PSA, age, %fPSA, and PV), and the PCA3-based model (PSA, age, %fPSA, PV, and PCA3 score) were evaluated using ROC curve analysis. Decision curve analysis was applied to test the clinical application of the base model and the PCA3-based model, calculating the net benefit and the net reduction in unnecessary biopsies at different thresholds (Figure 1).
Figure 1.
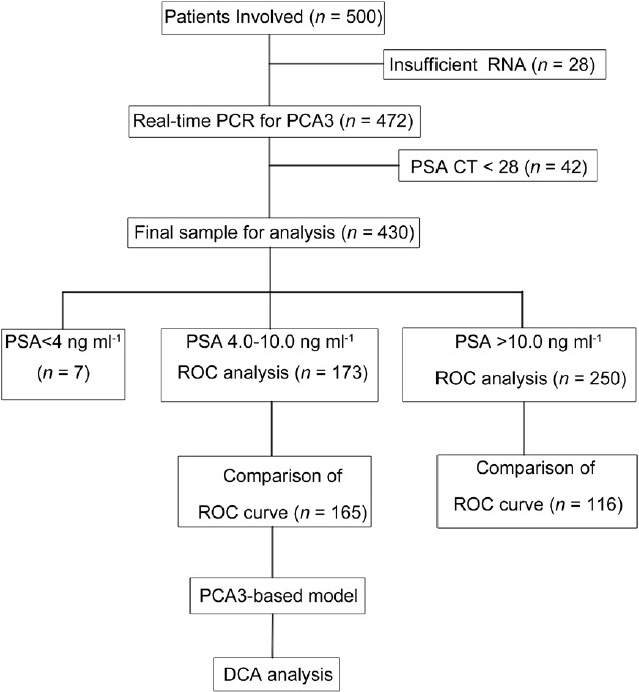
Flowchart of the study design.
Specimen collection and sample preparation
Biopsies were performed using an end-fire ultrasound transducer (Falcon 2101; B-K Medical, Inc.) and an automatic 18-gauge needle (Bard, Inc.). In all men, 10–12 core systematic, laterally directed, TRUS-guided biopsy was performed. First catch urine samples were collected following an attentive DRE (three strokes per lobe) before the biopsy was performed. The urine samples were immediately cooled on ice and processed within 2 h of collection. Urine samples were centrifuged at 2500 ×g for 15 min at 4°C, and then the pellets were washed twice with cold PBS (1×). The sediments were homogenized in TRIzol and were used for RNA extraction or stored at −80°C until use.
Quantitative RT-PCR analysis
Total RNA from urine sediments was extracted using TRIzol reagent (Invitrogen: No. 15596-026, USA). A total of 50 ng total RNA were treated with DNase I (TaKaRa: D2215, TaKaRa, Japan) prior to cDNA synthesis and then amplified with a TransPlex Complete Whole Transcriptome Amplification Kit (WTA2Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's instructions. qRT-PCR was performed using SYBR® Premix Ex Taq™ (TaKaRa: DRR081A TaKaRa, Japan) with an Applied BioSystems StepOne Plus according to the manufacturer's recommended cycling conditions. The gene-specific sequence information for the qRT-PCR primers is as follows: PSA-forward primer GTCTGCGGCGGTGTTCTG, PSA-reverse primer TGCCGACCCAGCAAGATC; PCA3 forward primer TGGTGGGAAGGACCTGATGATACAG, and PCA3 reverse primer TCTCCCAGGGATCTCTGTGCTTCC. Briefly, 2 μl of the cDNA solution was amplified using 10 μl SYBR® Premix Ex Taq™ (Perfect Real Time) (2×) (TaKaRa: DRR081A TaKaRa, Japan), 2 μl primers, 0.4 μl ROX Reference Dye (50×), and nuclease-free H2 O in a final volume of 20 μl. The data were analyzed with StepOne Software version v2.1 (Applied BioSystems, USA). A melt-curve analysis was performed at the end of the amplification. Samples with PSA Ct values of >2815 were excluded to ensure sufficient prostate cell collection. The PCA3 score was calculated as PCA3 mRNA/PSA mRNA × 1000 = 2Ct (PSA)−Ct (PCA3) ×1000. All experiments were performed in triplicate. No amplification of the signal was obtained when nuclease-free water was added instead of cDNA. The data were analyzed with StepOne Software version v2.1 (Applied BioSystems, USA).
Statistical analysis
The Mann–Whitney U-test, the Student's t-test, Pearson's Chi-square test, and Fisher's exact tests were used for statistical comparisons of the continuous and categorical variables. Univariate and multivariate logistic regressions were performed to identify independent predictors of PCa on biopsy. Receiver operating characteristic (ROC) curves were constructed to discriminate among different groups of patients. The area under the ROC curve (AUC) was used to assess the predictive power. The significant difference between the AUC of each pair of parameters, which includes the PCA3, PSA, %PSA, and PSAD, was calculated by a Z-test. A base model (age, PSA, %fPSA, and PV) was constructed using logistic regression, and the PCA3-based model was constructed by incorporating the PCA3 score into the base model (PCA3 score, age, PSA, %fPSA, and PV). Decision curve analysis was used to evaluate the clinical effects of the calculations. All of the statistical calculations were performed using MedCalc v. 10.4.7.0 (MedCalc Software bvba, Mariakerke, Belgium) and Stata 12.0 (Stata Corp., College Station, TX, USA). The P values were two-sided, and P < 0.05 was considered statistically significant.
RESULTS
Patient characteristics
Initially, 500 patients were included in this study, from which 28 samples were excluded because of insufficient RNA extraction from the sediments. After quantitative RT-PCR analysis, another 42 patients with PSA Ct values above 28 were excluded15 (insufficient prostate cell collection). A total of 430 patients were included in the final analysis, of which 173 patients had a PSA of 4.0–10.0 ng ml−1, 250 patients had a PSA >10.0 ng ml−1 and 7 patients had a PSA <4 ng ml−1 (Figure 1). The clinical characteristics are summarized in Table 1. There were no patients with a positive family history or familial cases in this study.
Table 1.
Clinical characteristics of men with positive and negative biopsy
PCA3 score sufficed to discriminate positive from negative prostate biopsy results
The PCA scores of patients with positive and negative biopsy are characterized in Figure 2. The median PCA3 score of positive and negative biopsy were 161.1 and 61.8, respectively (P < 0.0001). The median PCA3 score was also higher in patients with positive biopsy compared to negative biopsy with a PSA > 10.0 ng ml−1 (119.5 vs 60.1, P < 0.001).
Figure 2.
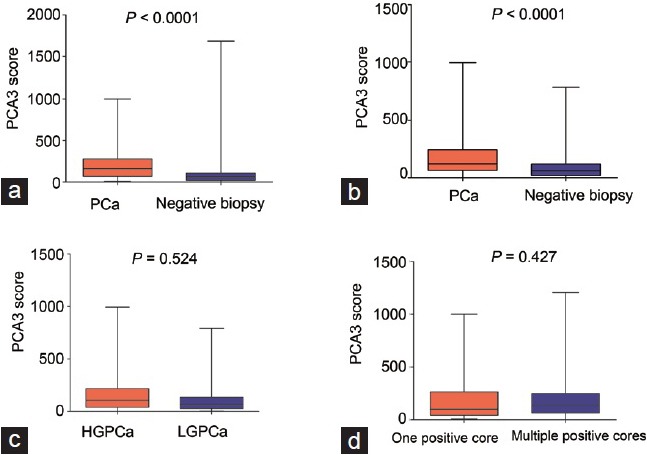
PCA3 score of prostate cancer patients and patients with negative biopsy in patients with (a) PSA 4.0–10.0 ng ml−1; (b) PSA > 10.0 ng ml−1; (c) patients with high-grade prostate cancer (HGPCa) versus low-grade prostate cancer; (d) prostate cancer patients with only 1 positive biopsy core and more than 1 positive biopsy cores.
PCA3 score is not correlated with the aggressiveness of prostate cancer
The median PCA3 score was higher in PCa patients with a Gleason score ≥7 (high-grade prostate cancer, HGPCa) than patients with a Gleason score ≤7 (low-grade prostate cancer, LGPCa) (P = 0.523, Figure 2c). There was no significant difference in the PCA3 score between patients with one or multiple positive cores (P = 0.427, Figure 2d).
PCA3 score in the diagnosis of prostate cancer
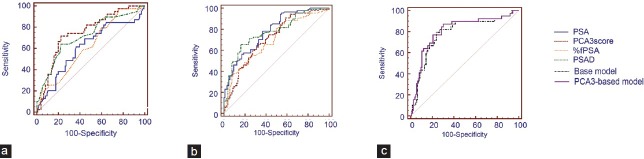
The ROC analysis indicated that the PCA3 score is effective in predicting biopsy results in both groups. In patients with a PSA of 4.0–10.0 ng ml−1, the AUC of PSA is 0.614, and the AUC of the PCA3 score is higher than that of %fPSA (0.750 vs 0.622, P = 0.046) but is not higher than that of PSAD (0.750 vs 0.718, P = 0.590) (Table 2, Figure 3). In patients with a PSA >10.0 ng ml−1, the AUC of PSA, PCA3 score, %fPSA, and PSAD was 0.780, 0.712, 0.698, and 0.773, respectively. No significant differences were observed between the PCA3 score and %fPSA or PSAD. The statistical optimal cut-off value of the PCA3 score was 114.8 with a sensitivity of 71.8% and specificity of 77.0%. At a cut-off PCA3 score of 23, 95% of PCa patients would be detected with a specificity of 27.0%. In patients with a PSA of 4.0–10.0 ng ml−1, the PCA3-based model achieves a higher AUC than the base model, but the advantage did not meet the common level of statistical significance (0.833 vs 0.825, P = 0.552).
Table 2.
Comparison of the AUC of the PCA3 score, PSA, PSAD, and %fPSA in patients with a PSA of 4.0–10.0 ng ml−1, 10.1–20.0 ng ml−1 and >20.0 ng ml−1
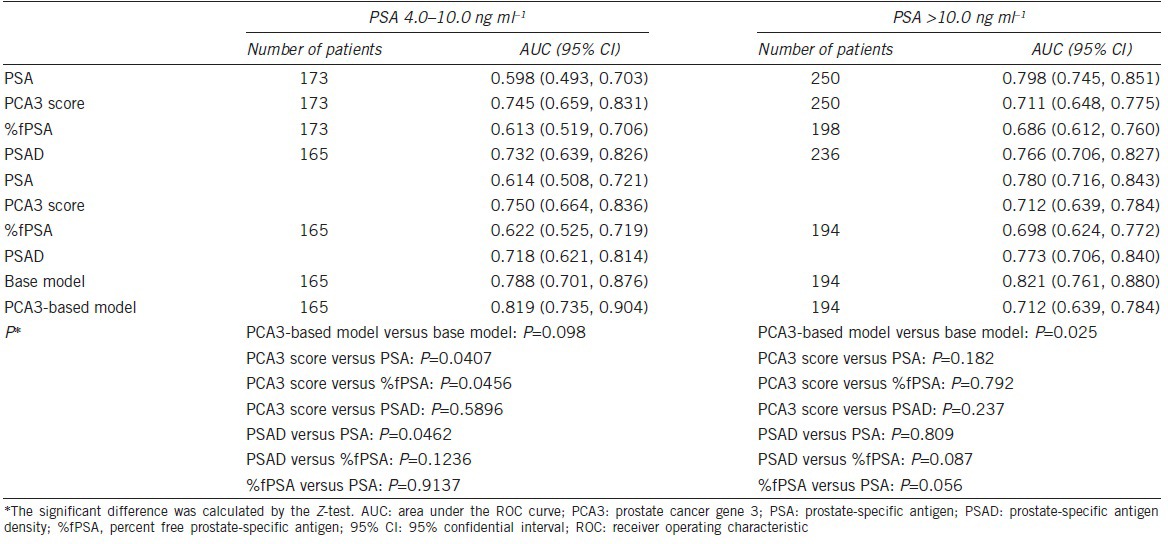
Figure 3.
(a) ROC curve analysis of the PSA, PCA3 score, %fPSA and PSAD in patients with PSA levels of 4.0–10.0 ng ml−1 and (b) ROC curve analysis of the PSA, PCA3 score, %fPSA and PSAD in patients with PSA levels of over 10.0 ng ml−1; (c) ROC curve analysis of the base model and the PCA3-based model in patients with a PSA of 4.0–10.0 ng ml−1.
PCA3 score in the diagnosis of high-grade prostate cancer
The PCA3 score, PSA, %fPSA, and PSAD have been tested for the ability to predict HGPCa in two groups of patients. In patients with a PSA of 4.0–10.0 ng ml−1, the AUC of PSAD (0.774, 95% CI: 0.665–0.884) is higher than the PSA (0.701, 95% CI: 0.587–0.815) and PCA3 score (0.709, 95% CI: 0.609–0.808); however, the differences did not meet the common level of statistical significance (PSAD vs PSA, P = 0.185 and PSAD vs PCA3 score, P = 0.312, respectively). In patients with a PSA of >10 ng ml−1, the AUC of PSA is higher than that of the PCA3 score (P = 0.086).
Logistic regression analysis of the diagnostic accuracy of the PCA3 score and other clinical parameters
In patients with a PSA of 4.0–10.0 ng ml−1, the four variables in the base model, age, PSA, %fPSA, and PV were identified to be associated with the biopsy result in the multivariable logistic regression analysis (Supplementary Table 1 (560KB, tif) ). All five variables in the PCA3-based model, including the PCA3 score, age, PSA, %fPSA, and PV, were associated with the biopsy result (Supplementary Table 1 (560KB, tif) ). The AUC of the PCA3-based model is slightly higher than that of the base model, but the benefit is not statically significant (0.819 vs 0.788, P = 0.098) in patients with PSA levels of 4.0–10.0 ng ml−1. The PCA3-based model does not perform better than the base model (AUC 0.712 in the PCA3-based model and 0.821 in the base model).
Multivariate logistic analysis for predicting prostate cancer in patients with PSA levels of 4.0-10.0 ng/ml
Decision curve analysis of using the PCA3-based model in patients with PSA 4.0–10.0 ng ml−1
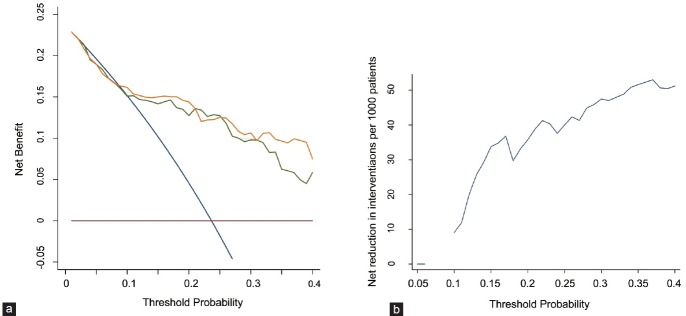
The decision curve analysis focused on patients with a PSA of 4.0–10.0 ng ml−1 because the diagnosis of prostate cancer in these patients is of particular clinical interest. As the decision curve indicates, the PCA3-based model was superior to the base model with a higher net benefit for almost all threshold probabilities, especially in threshold probabilities of 25%–40% (Figure 4a, Supplementary Table 2 (394.2KB, tif) ). For example, the net benefit of the PCA3-based model is moderately higher than that of the base model at a probability of 30% (9.784 vs 10.649) and 35% (6.247 vs 9.650). More than 40% of unnecessary biopsies would be spared using the PCA3-based model at the threshold probabilities of 25%–40% (Figure 4b, Supplementary Table 3 (674.7KB, tif) ). For example, slightly more biopsies would be prevented using the PCA3-based model than the base model (60.3% vs 57.1%), missing 4 PCa patients, 2 of which were HGPCa at the threshold probability of 30%.
Figure 4.
Decision curve analysis of the base model and the PCA3-based model in patients with a PSA of 4.0–10.0 ng ml−1. (a) The green line indicates the base model; the orange line indicates the prediction model that includes only age, PSA, %fPSA, prostate volume and PCA3 score. The horizontal line along the x-axis assumes that no patient will have PCa (no patient should undergo a prostate biopsy), whereas the solid gray line assumes that all patients will have PCa (all patients will need to undergo a prostate biopsy). (b) Net reduction in biopsies per 100 patients by threshold probability.
Net benefit and reduction in avoidable biopsies for the base model and PCA3-based model compared to the “treat all” strategy to biopsy every patient for different threshold probabilities in the same range for patients with a PSA of 4.0-10.0 ng/ml
Number of total and HGPCa missed and reduction in biopsies according to threshold probability in the range of 15–40% for the base model and PCA3.based model for patients with a PSA of 4.0–10.0 ng/ml
DISCUSSION
The PCA3 gene was first identified using differential display technology as a prostate-specific noncoding gene16 and was further explored in several studies.17,18 The initial method for measuring PCA3 mRNA in urine was quantitative real-time RT-PCR-based assay.19 The application of PCA3 was expanded20 after the approval of the third-generation PCA3 assay (PROGENSA®) by the European Committee (CE) in 200621 and the United States Food and Drug Administration approval of the PROGENSA® PCA3 Assay to help determine the need for repeat prostate biopsies in men who had a previous negative biopsy in 2012.
This study assessed the application of PCA3 and PCA3-based models in the diagnosis of PCa in Chinese men who underwent biopsy in a clinical setting. Our results indicated that PCA3 has a higher diagnostic accuracy than %fPSA in men with a PSA of 4.0–10.0 ng ml−1 in the ROC analysis, and the PCA3-based model has a higher net benefit than the base model in the decision curve analysis in men with a PSA of 4.0–10.0 ng ml−1. However, the PCA3-based model is only slightly better than the base model in the ROC analysis in men with PSA levels of 4.0–10.0 ng ml−1, but the benefit is not statistically significant. Moreover, PCA3 is not more accurate than %fPSA, PSAD, or PSA in men with PSA levels over 10.0 ng ml−1 in the ROC analysis. Our results suggested that PCA3 could moderately increase the diagnostic performance in men with PSA of 4.0–10.0 ng ml−1.
The AUC of the PCA3 test in our study is in accordance with a systematic review that reported an AUC of 0.58–0.83.8 However, the AUC in this study was slightly lower than in some studies in Western populations, and our result failed to illustrate a significant benefit of adding the PCA3 tests to the base model. As previously proposed, the variability among published studies might be the result of different rates of PCa.22 In this study, the positive rate of men with a PSA level of 4.0–10.0 ng ml−1 was 26%, which is much lower than that in most Western reports.23 This situation may lead to lower specificity of the PCA3 score in this study. Second, there may be racial differences between the Chinese and Western populations in the diagnostic accuracy of PCA3. However, as the result of our RNA-seq study, the ratio of mRNA expression of PCA3 in tumor tissue/adjacent normal tissue ranks as one of the highest among hundreds of long noncoding RNAs.14 The influence of race on the effectiveness of the PCA3 test should be further evaluated.
Chinese investigators have previously tested PCA3 expression in the PCa tissue of Chinese patients, and PCA3 mRNA was identified in 42/42 prostate cancer tissues and was not present in 70 nonprostate neoplastic tissues and nonprostate normal tissues.24 Further investigation in 35 PCa and 64 negative biopsies showed that the AUC of the PCA3 score was 0.79 (95% CI: 0.68–0.89).13 The sensitivity and specificity reached 62.9% and 90.6%, respectively, with a PCA3/PSA mRNA ratio cut-off of 0.107. Likewise, a study in Hong Kong validated these findings in a relatively small set of patients.25 The cut-off they used was a “PCA3 ratio” (ratio of the Ct value of PCA3/PSA mRNA) of 1.127. Due to the insufficiency of the involved cases and the fact that these patients were not enrolled in a prospective and consecutive manner, these studies may not reflect the real performance of the PCA3 test in Chinese patients.
The introduction of PCA3 into the current diagnostic strategy was not successful because of the limited diagnostic accuracy and the high cost of the current commercial assay. Readily available PCR methods may overcome the price problem once these assays are sold in accordance with the Freedom to Operate (FTO) policy. Other biomarkers, such as p2PSA and prostate health index (PHI), have also been validated to be useful in PCa diagnosis. PHI had an AUC of approximately 0.70 in two multi-center prospective studies in Western populations.26,27 The effectiveness of PHI has also been validated in a multi-center study in a Chinese population. The AUC of PHI is 0.73 in men with PSA levels of 2.0–10.0 ng ml−1, 28 similar to the AUC of the PCA3 tests in this study. However, the AUC of PHI is higher than that of PSA in men with PSA of 10.0–20.0 ng ml−1 or over 20.0 ng ml−1 because total PSA is integrated in PHI. Direct comparison of PHI and PCA3 is planned because both urine and serum were collected in the participating hospitals of the Chinese Prostate Cancer Consortium.
Several limitations of this study must be acknowledged. First, a manual real-time RT-PCR-based PCA3 assay was used instead of the commercial PROGENSA® PCA3 test. However, Filella et al.29 showed that there is no significant difference between the real-time RT-PCR-based manual assay and the commercial assay in terms of diagnostic accuracy and effectiveness. A large prospective study confirmed the effectiveness of the PCR-based PCA3 test.30 Second, the manual test has a lower informative rate of samples than the commercial assay. Our study illustrated an informative rate of 86.0%, with samples excluded for insufficient RNA and lower concentrations of prostate cell collection. As a validation study, these limitations may not substantially affect the effectiveness of the biomarker itself. Third, most of the patients received a biopsy due to elevated PSA levels; thus, it is unfair to compare the performance of PCA3 with that of total PSA. It is unknown which biomarker would perform best if of other biomarkers were used for biopsy indication. Fourth, there were limited participants in this study, and all of the patients were from the same institute. Multi-center studies with more cases are needed to validate the effectiveness of this test.
CONCLUSION
PCA3 moderately improved the diagnostic accuracy in Chinese patients undergoing first prostate biopsy. The PCA3-based model is not more effective than the base model in ROC analysis, but it demonstrates higher net benefit in decision curve analysis in patients with a PSA of 4.0–10.0 ng ml−1. However, there is not enough evidence supporting its utility in patients with a PSA >10.0 ng ml−1.
AUTHOR CONTRIBUTIONS
YHS, FBW, RC, and SCR designed the experiments. FBW, SCR, RC, XLS, and YSZ performed the experiments, and WZ and TLJ collected the clinical data. FBW, RC, SCR, CZ, XG, YHS, CLX, and JGH analyzed the data. YHS, FBW, RC, and SCR drafted and revised the manuscript. All authors read and approved the final version of the manuscript.
COMPETING INTERESTS
The authors declared no competing interests.
ACKNOWLEDGMENTS
This work was supported by the Chinese Prostate Cancer Consortium, Program for Changjiang Scholars and the Innovative Research Team in University of the Ministry of Education of China (No. IRT1111, Ying-Hao Sun) and the National Basic Research Program of China (2012CB518300, 2012CB518306, Ying-Hao Sun).
Supplementary information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Center MM, Jemal A, Lortet-Tieulent J, Ward E, Ferlay J, et al. International variation in prostate cancer incidence and mortality rates. Eur Urol. 2012;61:1079–92. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 3.He JC. Chinese Cancer Registry Annual Report 2012. Beijing: China Military Medical Science Press; 2013. [Google Scholar]
- 4.Zhao P, Chen W. Chinese Cancer Registry Annual Report 2012. Beijing: China Military Medical Science Press; 2013. [Google Scholar]
- 5.Kimura T. East meets West: ethnic differences in prostate cancer epidemiology between East Asians and Caucasians. Chin J Cancer. 2012;31:421–9. doi: 10.5732/cjc.011.10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djavan B, Zlotta A, Remzi M, Ghawidel K, Basharkhah A, et al. Optimal predictors of prostate cancer on repeat prostate biopsy: a prospective study of 1,051 men. J Urol. 2000;163:1144–8. [PubMed] [Google Scholar]
- 7.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or=4.0 ng per milliliter. N Engl J Med. 2004;350:2239–46. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 8.Stephan C, Ralla B, Jung K. Prostate-specific antigen and other serum and urine markers in prostate cancer. Biochim Biophys Acta. 2014;1846:99–112. doi: 10.1016/j.bbcan.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Haese A, de la Taille A, van Poppel H, Marberger M, Stenzl A, et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol. 2008;54:1081–8. doi: 10.1016/j.eururo.2008.06.071. [DOI] [PubMed] [Google Scholar]
- 10.Bradley LA, Palomaki GE, Gutman S, Samson D, Aronson N. Comparative effectiveness review: prostate cancer antigen 3 testing for the diagnosis and management of prostate cancer. J Urol. 2013;190:389–98. doi: 10.1016/j.juro.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Ochiai A, Okihara K, Kamoi K, Oikawa T, Shimazui T, et al. Clinical utility of the prostate cancer gene 3 (PCA3) urine assay in Japanese men undergoing prostate biopsy. BJU Int. 2013;111:928–33. doi: 10.1111/j.1464-410X.2012.11683.x. [DOI] [PubMed] [Google Scholar]
- 12.Ochiai A, Okihara K, Kamoi K, Iwata T, Kawauchi A, et al. Prostate cancer gene 3 urine assay for prostate cancer in Japanese men undergoing prostate biopsy. Int J Urol: official J Jpn Urol Assoc. 2011;18:200–5. doi: 10.1111/j.1442-2042.2010.02711.x. [DOI] [PubMed] [Google Scholar]
- 13.Shen M, Chen W, Yu K, Chen Z, Zhou W, et al. The diagnostic value of PCA3 gene-based analysis of urine sediments after digital rectal examination for prostate cancer in a Chinese population. Exp Mol Pathol. 2011;90:97–100. doi: 10.1016/j.yexmp.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Ren S, Peng Z, Mao JH, Yu Y, Yin C, et al. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012;22:806–21. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–9. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–9. [PubMed] [Google Scholar]
- 17.Hessels D, Klein Gunnewiek JM, van Oort I, Karthaus HF, van Leenders GJ, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8–15. doi: 10.1016/s0302-2838(03)00201-x. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt U, Fuessel S, Koch R, Baretton GB, Lohse A, et al. Quantitative multi-gene expression profiling of primary prostate cancer. Prostate. 2006;66:1521–34. doi: 10.1002/pros.20490. [DOI] [PubMed] [Google Scholar]
- 19.Vaananen RM, Rissanen M, Kauko O, Junnila S, Vaisanen V, et al. Quantitative real-time RT-PCR assay for PCA3. Clin Biochem. 2008;41:103–8. doi: 10.1016/j.clinbiochem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Auprich M, Bjartell A, Chun FK, de la Taille A, Freedland SJ, et al. Contemporary role of prostate cancer antigen 3 in the management of prostate cancer. Eur Urol. 2011;60:1045–54. doi: 10.1016/j.eururo.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Groskopf J, Aubin SM, Deras IL, Blase A, Bodrug S, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52:1089–95. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 22.Roobol MJ. Contemporary role of prostate cancer gene 3 in the management of prostate cancer. Curr Opin Urol. 2011;21:225–9. doi: 10.1097/MOU.0b013e328344939c. [DOI] [PubMed] [Google Scholar]
- 23.Vickers AJ, Cronin AM, Roobol MJ, Hugosson J, Jones JS, et al. The relationship between prostate-specific antigen and prostate cancer risk: the prostate biopsy collaborative group. Clin Cancer Res. 2010;16:4374–81. doi: 10.1158/1078-0432.CCR-10-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao Z, Shen M, Zheng Y, Mao X, Chen Z, et al. PCA3 gene expression in prostate cancer tissue in a Chinese population: quantification by real-time FQ-RT-PCR based on exon 3 of PCA3. Exp Mol Pathol. 2010;89:58–62. doi: 10.1016/j.yexmp.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Ng CF, Yeung R, Chiu PK, Lam NY, Chow J, et al. The role of urine prostate cancer antigen 3 mRNA levels in the diagnosis of prostate cancer among Hong Kong Chinese patients. Hong Kong Med J=Xianggang Yi Xue Za Zhi/Hong Kong Acad Med. 2012;18:459–65. [PubMed] [Google Scholar]
- 26.Lazzeri M, Haese A, de la Taille A, Palou Redorta J, McNicholas T, et al. Serum isoform [-2] proPSA derivatives significantly improve prediction of prostate cancer at initial biopsy in a total PSA range of 2-10 ng/ml: a multicentric European study. Eur Urol. 2013;63:986–94. doi: 10.1016/j.eururo.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Lughezzani G, Lazzeri M, Haese A, McNicholas T, de la Taille A, et al. Multicenter European external validation of a prostate health index-based nomogram for predicting prostate cancer at extended biopsy. Eur Urol. 2014;66:906–12. doi: 10.1016/j.eururo.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Na R, Ye D, Liu F, Chen H, Qi J, et al. Performance of serum prostate-specific antigen isoform [-2] proPSA (p2PSA) and the prostate health index (PHI) in a Chinese hospital-based biopsy population. Prostate. 2014;74:1569–75. doi: 10.1002/pros.22876. [DOI] [PubMed] [Google Scholar]
- 29.Filella X, Foj L, Mila M, Auge JM, Molina R, et al. PCA3 in the detection and management of early prostate cancer. Tumour Biol: j Int Soc Oncodev Biol Med. 2013;34:1337–47. doi: 10.1007/s13277-013-0739-6. [DOI] [PubMed] [Google Scholar]
- 30.Crawford ED, Rove KO, Trabulsi EJ, Qian J, Drewnowska KP, et al. Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: a prospective study of 1,962 cases. J Urol. 2012;188:1726–31. doi: 10.1016/j.juro.2012.07.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multivariate logistic analysis for predicting prostate cancer in patients with PSA levels of 4.0-10.0 ng/ml
Net benefit and reduction in avoidable biopsies for the base model and PCA3-based model compared to the “treat all” strategy to biopsy every patient for different threshold probabilities in the same range for patients with a PSA of 4.0-10.0 ng/ml
Number of total and HGPCa missed and reduction in biopsies according to threshold probability in the range of 15–40% for the base model and PCA3.based model for patients with a PSA of 4.0–10.0 ng/ml