SUMMARY
Bacterial chromosomes initiate replication at a fixed time in the cell cycle, whereas there is generally no particular time for plasmid replication initiation or chromosomal replication initiation from integrated plasmids. In bacteria with divided genomes, the replication system of one of the chromosomes typically resembles that of bacteria with undivided genomes, whereas the remaining chromosomes have plasmid-like replication systems. For example, in Vibrio cholerae, a bacterium with two chromosomes (chromosome 1 [Chr1] and Chr2), the Chr1 system resembles that of the Escherichia coli chromosome, and the Chr2 system resembles that of iteron-based plasmids. However, Chr2 still initiates replication at a fixed time in the cell cycle and thus offers an opportunity to understand the molecular basis for the difference between random and cell cycle-regulated modes of replication. Here we review studies of replication control in Chr2 and compare it to those of plasmids and chromosomes. We argue that although the Chr2 control mechanisms in many ways are reminiscent of those of plasmids, they also appear to combine more regulatory features than are found on a typical plasmid, including some that are more typical of chromosomes. One of the regulatory mechanisms is especially novel, the coordinated timing of replication initiation of Chr1 and Chr2, providing the first example of communication between chromosomes for replication initiation.
KEYWORDS: cell cycle control of replication timing, plasmid replication, Vibrio cholerae
INTRODUCTION
Failures to control the frequency or timing of DNA replication can cause slowed growth and cell death or result in genome instability and uncontrolled cell proliferation. The underlying control systems are therefore among the most important in biology, from both basic science and clinical perspectives, and have been extensively studied in all three domains of life: bacteria, archaea, and eukarya (1). In bacteria, there is typically a single chromosome, and initiation is triggered at a single origin of replication. However, about 10% of sequenced bacteria have their genomes divided into two or more chromosomes (2, 3). This raises several questions regarding the genesis of such divided genomes, the mechanisms that ensure their stability, and the extent to which the different chromosomes communicate with each other. Here we discuss these questions in the context of the human pathogen Vibrio cholerae, where the genome is divided into two chromosomes: chromosome 1 (Chr1) and Chr2. This is the only bacterium to date with a divided genome where the relationship between replication and the cell cycle has been studied in detail and is thus the most likely bacterium to provide initial answers to these questions.
V. cholerae Chr1 has similarity to the well-studied chromosome of Escherichia coli and is controlled by the replication initiator protein DnaA, as is the norm in bacteria (4). The region responsible for the replication initiation of Chr2 resembles those of large, low-copy-number E. coli plasmids, such as F and P1 (5). These plasmids have an array of repeated sequences (iterons) that bind the plasmid-specific initiator protein for both initiation and control of initiation. The two chromosomes of V. cholerae are also distinguished by the essential genes that they carry. As in other bacteria with divided genomes, the majority of the essential genes are present in one of the chromosomes (the largest [Chr1 in the present context]), and a very limited number (and those are variable) are present on other chromosomes (6). These characteristics give rise to the general view that bacteria with divided genomes become so by the recombinational transfer of a few essential genes to plasmids. Since such bacteria belong to several different phyla, divided genomes appear to have arisen on more than one occasion during bacterial evolution. (Note that what exactly distinguishes a chromosome from a plasmid is still a matter of debate [6, 7], but for the purposes of this review, we consider a chromosome to be a natural self-replicating DNA that carries essential genes.)
Although plasmids are common in bacteria, and different plasmids often cohabit the same bacterial cell (8), plasmid-like replicons are not normally found to drive chromosomal replication. Chromosomes differ from plasmids in the timing of their replication in relation to the cell cycle. Chromosome replication initiates at a particular cellular state (such as accumulation of cell mass or reaching a certain cell cycle stage), while plasmid replication does not. Wherever studied, plasmid replication in different cells of a culture can occur at any point in the cell cycle. The high copy numbers and small size of plasmids relative to chromosomes presumably remove the risk that replication late in the cell cycle will retard or disrupt cell division (9–11) or that the cell will divide before replication occurs. For chromosomes, it appears that their large size necessitates controlling the timing of replication initiation for the timely completion of the cell cycle. However, integrated plasmids are in some circumstances able to ensure chromosome replication and cell viability in the absence of the normal replication system (integrative suppression) (12), at the price of discoordination with the cell cycle, leading to long interdivision times, cell shape abnormality, and slow growth (13, 14). One question that we focus on here is how the natural plasmid-like replicons of secondary chromosomes behave, i.e., whether they overcome the lack of timing of the original plasmids and link replication to the cell cycle.
It was clear early on that unlike that of plasmids, replication initiation of V. cholerae Chr2 is indeed cell cycle regulated (15). Since the control of replication is reasonably well understood for the E. coli chromosome and iteron-based plasmids, a comparative study of these replication systems with that of Chr2 appears relevant to address how the timing of an important biological process switched from being random to being cell cycle regulated. Here, we discuss Chr2 replication in terms of the principles of control learned from studying the replication of the E. coli chromosome and plasmids. We find that the replication control of Chr2 is significantly more complex than that of related plasmids, more comparable to chromosomes than to plasmids, even though the molecular mechanisms are more like those of plasmids. We also discuss the coordination of Chr2 replication with the cell cycle.
NEGATIVE-FEEDBACK CONTROL OF REPLICATION INITIATION
The mechanism of replication initiation was proposed initially in the replicon model in the early 1960s (16, 17). The proposal was elegant in its simplicity and specified that self-replicating units of DNA (replicons), such as plasmids and chromosomes, would depend upon two basic elements: the replicator, later called the origin of replication, and a structural gene for a protein, called the initiator, which binds the origin. The specificity of the initiator-origin interactions prevents replication initiation from ectopic sites in the molecule. Pritchard et al. later pointed out that the replicon model and a few other subsequent models were inadequate because they lacked features that could adjust for fluctuations in initiation times (18). Such fluctuations are expected for all chemical reactions in single cells, e.g., due to the probabilistic nature of initiator synthesis, and are particularly important for self-replicating systems where fluctuations may not be corrected at all without active control. Some of the variation in timing could come from fluctuations in the abundances of an initiator, but even without such fluctuations, heterogeneity in the timing of individual events is unavoidable because chemical events are fundamentally probabilistic.
To meet this concern, Pritchard et al. proposed that replicons should have a negative regulator of replication—an inhibitor—whose concentration determines the replication initiation probability. A round of initiation is permitted when the inhibitor concentration drops below a critical concentration (the inhibitor dilution model), and premature rereplication is prevented due to a rise in the inhibitor concentration due to a higher gene dosage postreplication or more dramatically, e.g., via a burst of transcription of the inhibitor gene triggered by replication. This forms a negative-feedback loop that solves many problems of self-replication. For example, a self-replicating system without some sort of feedback control could never set the replication frequency to exactly once per cell cycle, even on average, and even the smallest offset would accumulate over time and lead to either losses or high numbers of replicons. A nonfeedback system that was perfectly tuned to one condition would also over- or underreplicate under another condition, whereas feedback can adjust automatically to ensure a single replication per cell cycle on average. Finally, because all chemical reactions are probabilistic, even a hypothetical nonfeedback self-replicating system that on average replicated exactly once per cell cycle would eventually accumulate extreme random fluctuations in copy numbers, for two reasons. First, even if there were no fluctuations during the cell cycle, a cell with 10 plasmid copies at cell division that accidentally placed 4 into one cell and 6 into the other would, without feedback, create one lineage that replicated from 4 to 8 copies and another that replicated from 6 to 12 copies, until new errors occurred, creating further variation between lineages. Second, keeping a constant replication probability per time unit would additionally cause a large random variation in the number of replication events per cell cycle; specifically, the number of events would be Poisson distributed in any given time window. The requirement of an inhibitor-based control system thus seems obligatory to stably maintain a replicon copy number (Fig. 1). Indeed, to date, no replicon has been found without its own inhibitor and negative feedback, and subsequent research on plasmid replication control has largely been an exploration of mechanisms of negative regulation. Many regulatory mechanisms have been discovered and characterized. Here we describe those relevant to iteron-based plasmids and their host (mainly E. coli) chromosomes.
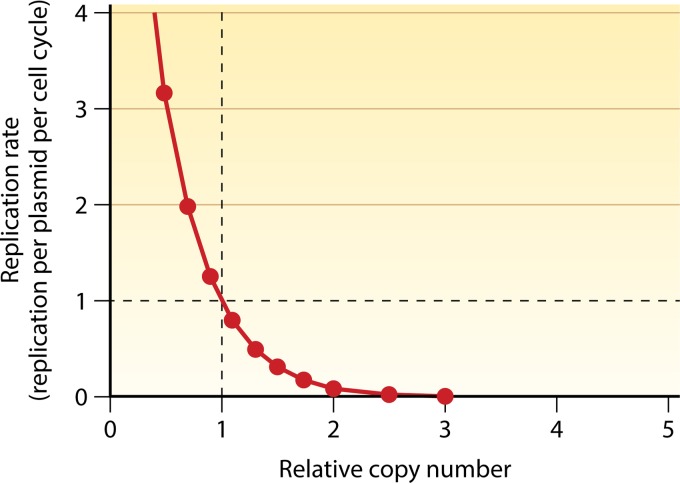
FIG 1.
Self-regulating plasmid copy number control system based on negative feedback. A plasmid copy number of 1 in abscissa refers to the average copy number of a plasmid per cell in a steady-state culture. During exponential growth, the copy number is maintained by replicating every plasmid on average once per cell cycle (the coordinates 1, 1). The control system realizes this by actively sensing deviations from the average copy number in either direction and adjusts the replication initiation rate accordingly. At a reduced copy number, the replication rate increases to >1, and at an increased copy number, the replication rate decreases to <1 (negative-feedback control). Since plasmids control replication by sensing their own copy number, the control system is “self-regulating.” (Adapted from reference 97 with permission of the publisher [copyright Elsevier {1984}].)
COPY NUMBER CONTROL OF ITERON-BASED PLASMIDS BY NEGATIVE FEEDBACK
Plasmids such as P1, F, RK2, R6K, pSC101, and pPS10 are well-characterized members of the iteron-based replicon family (Fig. 2) (5). Iterons are the initiator binding sites in these plasmids. Although essential for initiation, iterons also serve as inhibitors, particularly when their number increases upon replication, because the sister origins compete for the initiator, making it a limiting factor for initiation (“initiator titration”) (19–21). However, this mechanism may appear more straightforward than it is. If plasmids were able to keep a constant total level of the initiator, then titration would immediately create a negative-feedback loop by spreading out the initiators over more sites upon replication. However, when the plasmid replicates, the gene dosage for the initiator also increases, and thus, more of the initiator is made as well. Thus, on its own, titration would not necessarily be sufficient for control, and indeed, several additional mechanisms are found to control initiator synthesis and activity.
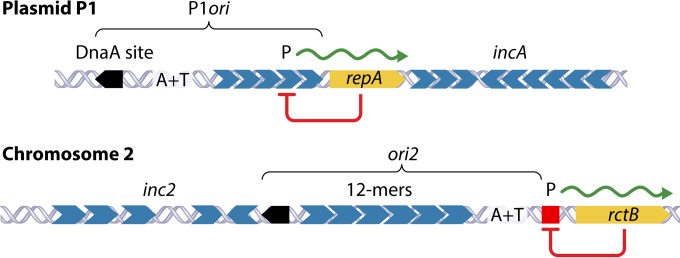
FIG 2.
Maps showing features common in the origin regions of plasmid P1 and chromosome 2 (Chr2) of V. cholerae. Both regions have a DnaA binding site (black pentagon), an AT-rich region (A+T), multiple initiator binding sites (gray hexagons) (called iterons in plasmids and 12-mers in Chr2), and an autorepressed initiator gene (repA and rctB, respectively). Autorepression is achieved by RepA binding to iterons in P1 and RctB binding to a 29-mer (red square), which is unrelated to 12-mers in sequence. P indicates promoters. The minimal regions that suffice as origins are marked as P1ori and ori2. The iterons and 12-mers outside P1ori and ori2 (called incA and inc2) reduce origin activity, although in Chr2, the activity of inc2 12-mers reverses in the presence of another regulator (39-mer) (shown in Fig. 7).
Initiator synthesis is controlled by transcriptional autorepression. The initiator promoters are sometimes buried within the array of origin iterons, and initiator binding to origin iterons also serves to repress the promoter (21). More commonly, initiator promoters have adjacent inverted repeats of half-iterons, which serve as dedicated operators of transcription of the initiator gene. Transcriptional autorepression is often thought of as a simple negative-feedback mechanism that dampens changes in the initiator concentration due to, e.g., gene dosage. However, again, the control mechanism sounds more straightforward than it is: once initiator molecules are titrated away on other plasmid iterons, reducing the cytoplasmic concentration, it also relieves transcriptional autorepression. In fact, these kinds of systems can create an uncontrolled rise in plasmid copy numbers (“runaway replication”) (Fig. 3) despite the combination of autocontrol of initiator production and subsequent titration (5). Another mechanism used to dampen the increase in initiator monomer levels upon plasmid replication is initiator dimerization. This can serve a role similar to that of transcriptional autoregulation. Specifically, if the dimer concentration is much higher than the monomer concentration, the dimer concentration will be approximately the same as the total concentration. Because the rate of dimerization depends on the square of the monomer concentration, the monomer concentration will in turn stay roughly proportional to the square root of the total. Thus, if an increase in the plasmid level creates a proportional increase in the total level of the initiator, the monomer form of the initiator would thus follow in proportion to the square root of the increase in plasmid copy number. For example, a 9-fold increase in the total initiator concentration would produce only a 3-fold increase in the monomer concentration, dampening the self-replicative tendency. This significant but imperfect (or partial) dampening effect is quantitatively similar to what is expected for transcriptional autoregulation without cooperativity in the repression mechanism (Fig. 3) (5), and the combination of the two mechanisms can in principle create much more dampened responses in the monomeric form of the initiator. However, in the context of iteron-based control, this mechanism suffers the same problem as the one described above: as new iterons titrate away initiators, the monomer-dimer balance will reequilibrate to compensate for the titrated molecules.
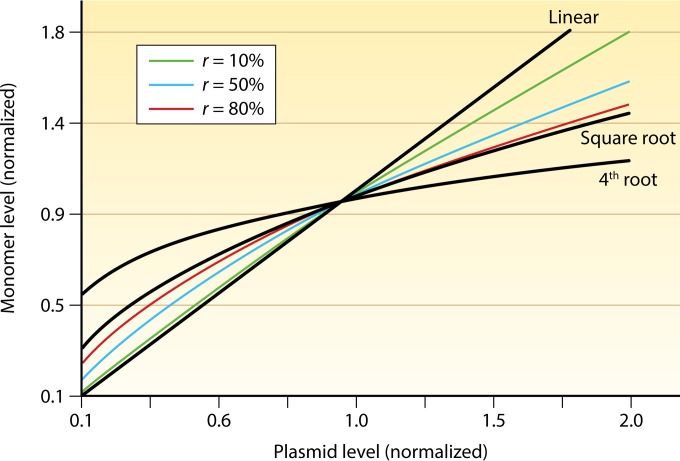
FIG 3.
Steady-state levels of the initiator monomer at different plasmid concentrations. The concentration of the initiator increases linearly with increasing plasmid copy numbers when it is produced constitutively. The increase is dampened when synthesis is controlled by transcriptional autorepression (in a monomer-only system) or by dimerization of the initiator (in a monomer-dimer-equilibrating system, without involving autorepression). The degree of dampening depends on r, the fraction of promoters repressed or the fraction of the initiator present as a dimer. r is zero for the linear line (no autorepression or dimerization). The square-root and the 4th-root plots are when r approaches 1. The square-root plot is obtained when autorepression or dimerization is present alone, and the 4th-root plot is obtained when both are present. Note that even in the most damped case, the initiator monomer level increases upon an increase of the plasmid copy number, which would lead to uncontrolled (“runaway”) replication if monomers are limiting for replication initiation. (Reprinted from reference 98 with permission [copyright {2005} National Academy of Sciences, U.S.A.].)
Thus, the seemingly corrective mechanisms of titration, autorepression, and dimerization fail to ensure that the probability of replication initiation decreases after a round of replication initiation. Indeed, there is another key mechanism to restrict initiation, referred to as “handcuffing.” In handcuffing, sister origins are inactivated by coupling them through initiator bridges that require the participation of dimeric initiators (Fig. 4) (22, 23). This bridging is believed to cause steric hindrance to the origin activity. In principle, this could create a stable system on its own, since at higher plasmid concentrations, each plasmid copy would be more likely to be handcuffed, ensuring less initiation of replication per copy when there are many copies per cell. This mechanism also takes advantage of the dynamics of initiator dimers: when a plasmid replicates, initiator synthesis increases, which increases the concentration of dimer initiators. Replication thus means both that each plasmid copy has more copies with which to form handcuffing pairs and that each pair can form more easily due to the presence of more dimer initiator proteins. This lowers the replication probability following a round of replication and could also provide a cooperative response, i.e., a sharp downshift in replication when the desired copy number is reached.
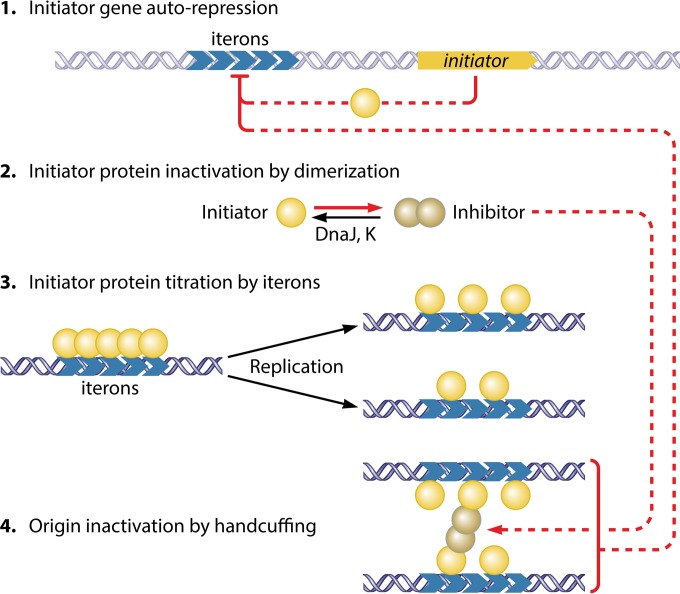
FIG 4.
Control of replication in iteron-carrying plasmid P1. The controls target the initiator as well as the iterons. The initiator is controlled negatively by three mechanisms: repression of transcription, inactivation by dimerization, and limiting availability by titration (steps 1 to 3). In P1, initiator binding to the origin also causes repression of the initiator promoter (autorepression), with the promoter being buried within the iteron array. The handcuffing reaction described below (step 4) possibly strengthens transcriptional repression. The initiator dimerizes, but the dimers do not bind iterons. Upon initiation, iteron numbers increase, which titrate the limiting amount of monomers, preventing the saturation of the iteron arrays, and without saturation, origins cannot be activated. Moreover, the nascent origins are paired via initiator bridges (handcuffing). The dimers, although inactive in iteron binding, participate in handcuffing and thereby serve as inhibitors. The dimers and the iterons are the two inhibitors in this system, and they cooperate in shutting down origins following replication initiation. Both of their concentrations are reduced upon cell growth. Moreover, the chaperones (DnaJ and DnaK) actively convert dimers to monomers. The increase in levels of monomers over dimers reduces handcuffing and helps to saturate the origin with monomers, allowing replication initiation. The inhibitory interactions are in red. For clarity, the iterons outside the origin (incA iterons) are not shown. They serve inhibitory roles only by increasing titration and handcuffing with the origin iterons. (Adapted from reference 5 with permission of the publisher.)
In view of the presence of a mechanism like handcuffing, it is possible that initiator titration also plays a feedback role. The key issue is that transcriptional autorepression (and dimerization) typically leads to a damped increase in cytoplasmic levels in response to a higher gene dosage, while what is needed is a damped increase in the total levels so that titration can lead to lower cytoplasmic levels. However, if the initiators bound to iterons can contribute to transcriptional autorepression via handcuffing (Fig. 4) (24), this mechanism could regulate total levels and not just the cytoplasmic fraction. Such a mechanism would allow initiator titration to play a real role in feedback control (23).
In summary, both iterons and initiator dimers satisfy the requirement to be inhibitors set forth by Pritchard et al.; that is, their concentration must increase following replication (18). Experimentally, evidence for the inhibitory role of iterons has always been clear-cut, as increasing their numbers increasingly inhibited replication (19). The inhibitory role of dimers became evident only when their importance in mediating handcuffing was realized (25–28). The inhibitory role of initiator dimers was not obvious earlier because the induced supply of excess initiators increased the concentrations of both monomers and dimers, i.e., both initiator and inhibitor concentrations, which partly neutralized the dominance of one over the other (Fig. 5). It now appears that the primary mechanism of inhibition is by dimer-mediated handcuffing of iterons, the probability of which increases because of increased dimer concentrations due to increases in plasmid copy numbers. We note that, as speculated by Pritchard et al., in plasmid P1, new initiators are synthesized in a burst when replication initiates (18, 29). (In Bacillus subtilis, the autorepressed initiator gene dnaA is also induced at the time of replication initiation [30].) An elongating replication fork most likely dislodges the bound initiators from the promoter and thereby activates transcription (“replication-induced transcription”) (29). This causes a more sudden increase in the dimer-to-monomer (inhibitor-to-initiator) ratio.
FIG 5.
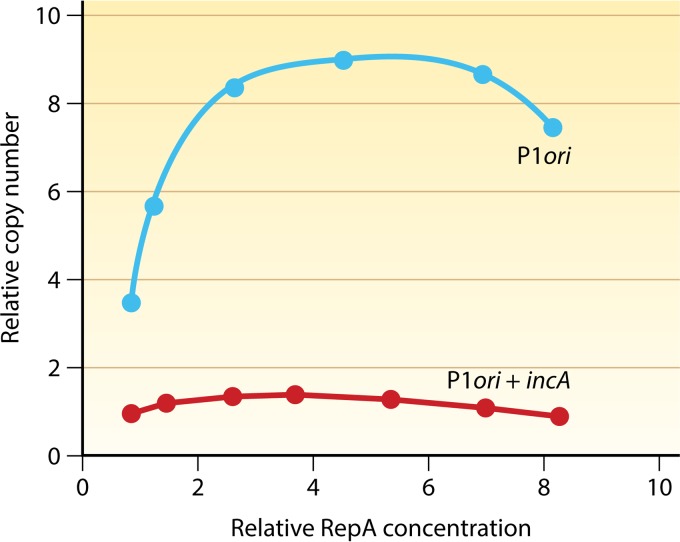
Response of P1 plasmid copy numbers to increasing initiator (RepA) concentrations. P1 plasmids carried either the minimal origin or the origin with the incA iterons. RepA was supplied in trans from an inducible promoter, and its levels and plasmid copy numbers are relative to the values seen in cells carrying the wild-type P1 plasmid. Both curves show that the copy number reaches a plateau and then decreases with further increases in RepA levels. Replication is more inhibited with further increases in RepA levels (data not shown). Inhibition is interpreted to be due to the prevalence of dimers over monomers at higher RepA levels.
How does replication resume? Apparently, replication resumes by inhibitor dilution (20). Cell growth dilutes both iterons and dimers, which can have a cooperative effect in reducing handcuffing.
CONTROL OF CHROMOSOMAL REPLICATION IN E. COLI
In bacteria, chromosomal replication is often regulated by the binding of the initiator DnaA to multiple sites within and outside the origin (31). As in plasmids, multiple mechanisms regulate the initiator, including transcriptional autorepression of the dnaA gene, titration of DnaA, and inactivation of DnaA (31, 32). Additionally, analogous to handcuffing, nascent sister origins are targeted for inactivation not by pairing but by binding of a protein, a process that is called sequestration. The inhibitory mechanisms are further detailed below.
Autorepression.
The dnaA autorepression circuit is more complex than that found in plasmids, as there are two promoters and a multitude of operators to which DnaA binds (33–35). Additionally, the promoter region is under methylation control, as discussed below for sequestration (36).
Titration.
DnaA has 10 or more binding sites within the origin and around 300 additional sites distributed throughout the chromosome (37, 38). In plasmids, titration sites are also found outside origins but in numbers comparable to those found within the origin (Fig. 2). The role of titration appears to be stronger in chromosomal replication than in plasmid replication.
Inactivation.
DnaA binds to ATP or ADP, with the ATP-bound form (DnaA-ATP) being the species required for initiation. The intrinsic ATPase activity of DnaA is weak and is not required for replication initiation. This activity is stimulated by two mechanisms, the major one being RIDA (regulatory inactivation of DnaA). In RIDA, DnaA-ATP bound to titration sites throughout the chromosome is converted to DnaA-ADP when encountered by the replication fork (31). This requires the participation of DNA-loaded clamps (DNA polymerase processivity factor DnaN) and Hda, a homolog of DnaA. A second mechanism also catalyzes the conversion of DnaA-ATP to DnaA-ADP by making use of a cluster of DnaA binding sites in a locus called datA (39).
Sequestration.
The replication origin of E. coli is unusually rich in Dam methylation sites. Due to semiconservative replication, these sites become hemimethylated, allowing the specific binding of a protein, SeqA (40). The nascent sister origins are thus made largely inaccessible to other proteins (“sequestration”) until the sites become fully methylated, which takes about one-third of the cell doubling time. The dnaA promoter region is also sequestered due to nearby Dam methylation sites, preventing new DnaA synthesis following replication initiation. This, together with titration and an expanding cell volume, lowers the DnaA concentration sufficiently so that when the origins are released from sequestration, the DnaA concentration would not be enough to trigger initiation in the remainder of the cell cycle.
There are also mechanisms that promote replication initiation by increasing the concentration of DnaA-ATP. The de novo-synthesized DnaA is likely to be largely ATP bound, with the ATP concentration being 10 times higher than that of ADP in the cell (41). Additionally, there are two clusters of DnaA binding sites, DARS1 and DARS2 (DnaA-reactivating sequence), involved in nucleotide exchange that convert DnaA-ADP to DnaA-ATP (42, 43). Reactivation to DnaA-ATP is also accomplished by acidic phospholipids (44, 45). Although the dynamics of these processes are largely unknown, their effect results in maximizing the DnaA-ATP/DnaA-ADP ratio just before initiation (46).
In sum, the controls for chromosomal replication are more elaborate than the ones operating on plasmid replication. How the chromosomal inhibitory mechanisms provide negative feedback has been extensively discussed elsewhere (31, 32) and is briefly summarized below, where the mechanisms are compared with those operating on plasmids.
COMMON GROUND IN REPLICATION CONTROL OF ITERON-BASED PLASMIDS AND THE E. COLI CHROMOSOME
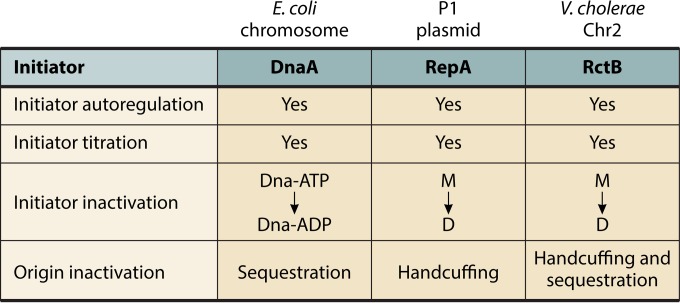
In E. coli, the chromosome and iteron-based plasmids control their replication independently, employing replicon-specific initiators and inhibitors. The basic principles of control, however, have many similarities. Both control systems are centered on the initiator and the origin, the two major players in initiation. In both systems, initiator synthesis is controlled by transcriptional autorepression, initiator availability by titration, and initiator activity by dimerization (in plasmids) and binding of cofactors (ATP versus ADP) (in the chromosome) (Fig. 6). The origin is targeted by plasmid-plasmid handcuffing in one case and by SeqA-mediated sequestration in another. In E. coli, the regulatory mechanisms with negative-feedback features are autorepression, titration, RIDA, and sequestration. These mechanisms become effective or more pronounced following replication initiation (32).
FIG 6.
Common principles that control P1 plasmid, E. coli chromosome, and V. cholerae Chr2 replication. The targets of control are the initiator and the origin. The initiators are controlled by autorepression, titration, and inactivation, and the origins are controlled by sequestration and handcuffing.
In both iteron-based plasmids and the E. coli chromosome, a striking feature of the replication systems is the degree to which the different modes of control appear to be interdependent. In plasmids, the balance between the monomeric and dimeric forms of the initiator, which appears to be at the heart of control, depends on how much of the initiator is synthesized through the autorepression circuit. As discussed above, the increase in the dimer concentration upon plasmid replication is counteracted to some extent by autorepression. Initiator titration is in turn counteracted by both dimerization and autorepression, unless handcuffing ensures that the titrated initiators in their bound state can participate in control (24, 47). In fact, the same control-defective mutants that show increased replication initiation (the copy-up phenotype) can be defective in all three processes of autorepression, dimerization, and handcuffing, as would be expected from their interdependence (48). Such tightly interwoven systems are particularly challenging to analyze, and the relative importance of the different control steps is not known, although the handcuffing interaction is considered necessary.
The interdependence of control mechanisms is also apparent in chromosomal replication. The functioning of RIDA, datA, and DARS is contingent upon DnaA titration, the effectiveness of which depends on the initiator supply, which is in turn controlled by autorepression and sequestration. Sequestration of the dnaA promoter is controlled by Dam methylase, and DnaA in turn controls the expression of the dam methylase gene (49). As in plasmids, it is difficult to single out any one step as being more important than the others, but the balance between DnaA-ATP and DnaA-ADP appears to be at the heart of this control. Not only do multiple regulators directly control the balance (RIDA, datA, DARS, and acidic phospholipids), but the DnaA-ATP/DnaA-ADP ratio also peaks before initiation (46). In sum, although the mechanisms are not always the same in details, many principles of negative-feedback control apply to both plasmid and E. coli chromosomal replication, and in both cases, the balance between initiators and inhibitors appears to determine the initiation probability (5, 50).
CONTROL OF V. CHOLERAE CHROMOSOME 2 REPLICATION
To discuss how far the above-described control principles apply to a plasmid-like replicon in charge of a chromosome, we now take up the case of V. cholerae Chr2. Chr2 appears to have retained essentially all the features of replication control present in iteron-based plasmids. The origin of Chr2 contains an array of repeats (12-mers) for initiator binding and an adjacent autorepressed initiator gene, rctB (Fig. 2) (51, 52). The DNA binding of the initiator RctB is activated by chaperones, as is the case for plasmid initiators (48, 53, 54). RctB can dimerize and can bridge the 12-mers (55). Several additional inhibitory features are also found (Fig. 7). (i) Although monomers are the active initiators for Chr2, RctB binds to 12-mers in both monomer and dimer forms, and chaperones activate the binding of both initiator forms (53, 56). In plasmids, dimers generally do not bind iterons, and chaperones reduce dimerization of the initiator (48, 53, 54). In Chr2, the dimers appear to serve as direct competitors of monomers for 12-mer binding, providing an additional inhibitory mechanism (53). (ii) The Chr2 origin has Dam methylation sites at a density as high as that in the E. coli origin. The methylation sites are invariably present in each of the 12-mers, and methylation of the 12-mers is required for efficient RctB binding (57). Dam is essential for V. cholerae because of its role in RctB binding to 12-mers, which is essential for Chr2 replication. (Dam-minus cells can survive only when Chr2 is passively maintained by integration into Chr1 [58].) Methylation also allows the nascent hemimethylated origin to be under SeqA control, as is the case in E. coli. (iii) RctB also binds to a second kind of site, the 39-mers. There are three such sites in the origin, and they are the primary inhibitors of Chr2 replication (55). Chr2 thus uses two kinds of sites, 12-mers to activate replication and 39-mers to inhibit it, as opposed to plasmids, where iterons play both roles. The 39-mers can handcuff with 12-mers of ori2 via RctB bridges (Fig. 7), and this appears to be the mechanism of inhibition, although it remains to be firmly established. RctB binding to the origin has been reported to be inhibited in the presence of ATP (not ADP), although the initiator lacks any known ATP binding motif (59).
FIG 7.
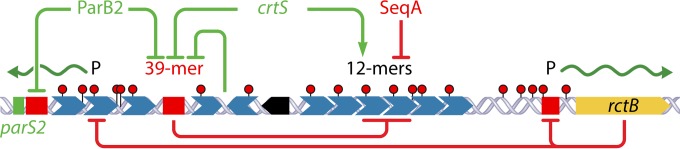
Features of V. cholerae Chr2 replication control in addition to those present in plasmid P1. The added features that inhibit replication are marked in red, and the ones that promote replication are in green. Other symbols are the same as those described in the legend of Fig. 2. In Chr2, initiation requires initiator (RctB) binding to the array of 12-mers, but this requires the sites to be methylated (red dots). The methylation requirement also subjects the nascent hemimethylated sister origins to be inhibited by SeqA. In addition to 12-mers, RctB binds to a second kind of site, 39-mers (red boxes). The 39-mers interact with 12-mers within ori2 and inhibit the origin strongly. Several activities reduce RctB binding to 39-mers and thereby reduce their inhibitory activity. The interaction of 39-mers with 12-mers within ori2 is competed (reduced) by 12-mers outside ori2. The leftmost 39-mer is transcribed from a promoter that overlaps 12-mers. When the 12-mers are not bound, the promoter is active, and transcription across the 39-mer interferes with RctB binding to that site and thus decreases its inhibitory activity (60). The same 39-mer is also restrained by ParB2 spreading from a nearby centromeric site, parS2 (61). The centrally located 39-mer is bound directly by ParB2, which competes with RctB binding. There is a site in Chr1, crtS, that remodels RctB such that its binding to 39-mers is reduced and its binding to 12-mers is increased, both of which favor initiation (Fig. 8). The rightmost red box (a truncated 39-mer) serves as the operator for rctB autorepression (99).
There are several mechanisms to stimulate initiation. The majority of them do so by reducing the inhibitory activity of 39-mers. (i) The 39-mer at the left end of the origin (Fig. 7) is transcribed, which interferes with RctB binding to that 39-mer, and without binding, the site cannot serve as an inhibitor (60). (ii) The handcuffing of 39-mers with 12-mers of ori2 inhibits replication, but the effect is the opposite when handcuffing occurs with 12-mers of inc2 (outside ori2) (Fig. 2) (55). The 12-mers of inc2 are believed to serve as competitors of 12-mers of ori2 for handcuffing with 39-mers, which reduces the inhibitory activity of the 39-mers on ori2. (iii) ParB2, a protein that binds to Chr2-specific centromeres for the segregation of Chr2, also binds to 39-mers by two mechanisms: by spreading into the leftmost 39-mer from a nearby centromeric site, parS2, and by directly binding to the central 39-mer (Fig. 7) (61). ParB2 thus serves as a competitor of RctB for binding to 39-mers. (iv) There is a novel site in Chr1 that can enhance Chr2 replication (62). This site remodels RctB in a way that increases its affinity for 12-mers and reduces its affinity for 39-mers. Since 39-mer binding inhibits and 12-mer binding promotes replication, remodeling biases binding toward the initiation mode. The location of this site in Chr1, called crtS (Chr2 replication-triggering site), is such that it would replicate just before the time of Chr2 replication initiation (Fig. 8) (63). The requirement that crtS be duplicated prior to the replication of Chr2 has been demonstrated. Duplication affords a mechanism to coordinate the replication of Chr2 with that of Chr1. Indeed, increasing or decreasing the distance of the site from the Chr1 origin changes the timing of Chr2 replication correspondingly.
FIG 8.
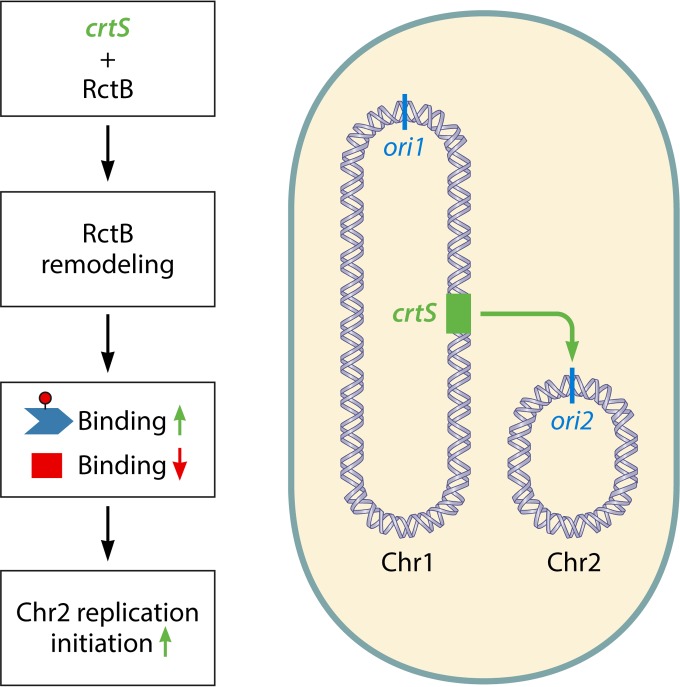
Coordination of replication between the two chromosomes of V. cholerae. A novel site in Chr1 (crtS [marked in green]) binds and remodels RctB such that its binding to 12-mers increases and its binding to 39-mers decreases (62). Since 12-mer binding helps to activate initiation and 39-mer binding helps to inhibit it, the changes in both kinds of binding by crtS are favorable to Chr2 replication. The position of the site (around 3 o'clock) is such that a replication fork would travel across it shortly before the time of Chr2 replication initiation. The timing of Chr2 replication is thus made dependent on the position of the crtS site (63).
The Chr2 replication system thus appears to have evolved from a plasmid-like system by acquiring three types of additional controls that (i) modify initiator-origin interactions by allowing initiator dimer binding to 12-mers as an additional inhibitory mechanism, by separating initiator binding sites involved in promoting (12-mer) and in inhibiting (39-mer) initiation, and by introducing a methylation requirement for 12-mer binding and thus making the origin sequesterable; (ii) involve a Par protein in the regulation of replication; and (iii) allow coordination of replication with that of a site in Chr1. The presence of these additional controls can help explain how inhibitory activities could dominate postinitiation and how they are overcome to allow initiation in the next cell cycle. Since the 12-mers are hemimethylated and are bound by SeqA following initiation, this would prevent RctB binding to 12-mers. The 39-mers that are devoid of methylation sites remain competent in RctB binding and available for their inhibitory activities. The initiator promoter is also likely to be repressed at this stage, being controlled by a 39-mer, which would dampen the increase in new initiator synthesis. The 39-mers could also serve as titrating sites to decrease the availability of RctB for initiating replication. When the 12-mers become fully methylated and competent in RctB binding, they can restrain the 39-mers by DNA looping (Fig. 7). The 39-mers will also be restrained by ParB2, and one of them will be restrained by transcription across it. The timing of these events in the cell cycle is not yet clear. Finally, the activation of crtS by replication also appears to reduce 39-mer binding and promote 12-mer binding, both of which would favor initiation (64). A noteworthy feature of crtS is that its activation leads to the firing of an ori2 once in a cell cycle, which conforms to one of the basic requirements of cell cycle control of replication (63).
In E. coli, a miniplasmid that carries the entire Chr2 origin replicates at a copy number comparable to that of the E. coli chromosome, whereas if the miniplasmid carries only the minimal origin, ori2 (Fig. 2), the copy number can increase at least 10-fold (55, 56, 59, 64). ori2 thus appears to have a considerable capacity for overinitiation. Even then, the ori2-containing plasmids are not maintained in cells if a 39-mer is added to it, indicating that the 39-mers have excess inhibitory capacity. Chr2 thus has both strong positive and negative regulatory capacities and several mechanisms to fine-tune these activities, particularly to restrain the inhibitory activity of the 39-mers.
In rich medium, V. cholerae, like E. coli, grows rapidly, and under these conditions, the cell generation time decreases, but the replication elongation rate does not increase correspondingly (65). Consequently, depending on the chromosomal length, the time to complete a round of replication can significantly exceed the generation time. This challenge is handled by initiating DNA replication on a partially duplicated chromosome, causing overlapping rounds of replication (65, 66). The number of overlapping rounds required depends on the chromosomal length. For example, at a certain growth rate, while E. coli (4.6 Mb) requires two overlapping rounds of replication, V. cholerae Chr1 (3.0 Mb) requires one, and Chr2 (1.1 Mb) requires none (67). Under these conditions, only Chr2 duplicates in a time that is less than the generation time. Dividing the genome thus has reduced the requirement for overlapping replication cycles. Other than the chromosomal length and generation time, no dedicated genes for controlling replication specifically during rapid growth have been identified.
DIFFERENCES BETWEEN RANDOM AND CELL CYCLE-REGULATED REPLICATION
Chr2 is the first bacterial example of a chromosomal replicon not regulated by DnaA, meaning that an increased DnaA concentration does not increase the Chr2 copy number (68). However, DnaA is a factor required for the replication of Chr2, as it is in the replication of plasmids such as P1 and F (51, 69). Except for crtS, the Chr2-specific regulatory elements (12-mers, 39-mers, and RctB) are encoded in Chr2 (55, 68, 70). Nonetheless, Chr2 replication control has many features that are either similar or analogous to those that control the replication of the E. coli chromosome and V. cholerae Chr1. Like these chromosomes, Chr2 has a high density of methylation sites in the origin and uses SeqA to inactivate the origin for a significant part of the cell cycle. This appears to be the key feature for replication once per cell cycle as well as for replication at a defined time in the cell cycle (57, 63). The 39-mers can be likened to the datA locus, as they both bind their cognate initiators for inhibitory purposes. Similarly, the crtS site in Chr1 can be likened to DARS, as they both serve to promote replication. Like SeqA, the Chr1 site is a determinant of the timing of Chr2 replication.
The replication of Chr2 also involves a Vibrio-specific element, the crtS site (63). This site appears to have evolved out of the requirement to coordinate the replication of the two chromosomes, a necessity irrelevant for bacteria with undivided genomes. Since the replication of Chr1 is cell cycle regulated apparently similarly to the E. coli chromosome, an economical mechanism to regulate Chr2 replication in the cell cycle could have been to add to its plasmid-like features the dependence on the crtS site. The presence of many additional features in Chr2 compared to plasmids suggests that these features make the intrinsic Chr2 replication control system efficient enough that an external signal from Chr1 can be effective in coordinating the timing of replication of the two chromosomes.
The elongation phase that is related to chromosomal length is not utilized in Chr2 replication control as much as it is used in E. coli replication, perhaps because the time of elongation for Chr2 is short compared to the time between initiation events. Chr2 does not have as many titration sites outside the origin as those present in the E. coli chromosome (62). The origin-centric replication control of Chr2 is reminiscent of its origin from a plasmid.
In sum, although Chr2 replication control is similar in many ways to that of its counterpart in plasmids, it is also considerably more complex, and the number and variety of regulators are similar to those that control chromosomal replication in E. coli and other bacteria (described below). In fact, although the core control processes seem to have come from plasmids, the Chr2 regulators also include E. coli-like and V. cholerae-specific control elements. In E. coli, inactivation of almost any of the regulators disrupts the timing of replication initiation, which could compromise the fitness of the mutants in competitive environments (43). The fitness cost of mutating the Chr2 regulators is not known. While Chr2 exhibits some of the features seen in cell cycle-coordinated chromosomes (primarily dependence on SeqA and the datA analog 39-mers) (Fig. 6), they are not sufficient for cell cycle coupling of initiation without the additional participation of a novel regulator, the crtS site. This site also appears to be required for Chr2 replication initiation and not merely for controlling the timing of initiation (63).
SPECIES-SPECIFIC CONTROL
As discussed above, the regulators of Chr2 include a species-specific element, the crtS site. Although bacterial DNA replication control mechanisms are based primarily on DnaA, its availability and activity are controlled rather differently in different bacteria by using a variety of species-specific elements. Below we illustrate the existence of such elements in B. subtilis and Caulobacter crescentus, the two bacteria other than E. coli and V. cholerae for which replication control has been extensively studied. The species-specific elements appear to be used for the adaptation of these bacteria to their complex lifestyles.
During vegetative growth, B. subtilis replication initiation is controlled by Soj (71). Soj is a homolog of the highly conserved segregation protein ParA. Soj monomers inhibit replication by inhibiting DnaA oligomerization (72–74). Upon dimerization, which requires binding to ATP, Soj activates replication, possibly by promoting DnaA oligomerization. (Likewise, ParA1 promotes the replication of V. cholerae Chr1 by interacting with DnaA, although V. cholerae is distantly related to B. subtilis [75].) In addition to the Soj monomer, B. subtilis uses another inhibitor, YabA, which interacts with DnaA and could also be preventing the polymerization of DnaA, similarly to Soj monomers (76). An additional role of YabA could be to sequester DnaA to within the elongating replisome, thus depriving the origin of the initiator (77). YabA interacts with the clamp of the replisome, and in this sense, it is analogous to the Hda protein of the E. coli RIDA system. It is not known whether YabA can convert DnaA-ATP to DnaA-ADP; nonetheless, it appears to be the primary inhibitor of replication in B. subtilis. Another noteworthy difference from E. coli is the absence of dam and seqA genes in B. subtilis (SeqA is found in only a subset of Gram-negative bacteria [78]). Thus, although DnaA is regulated by autorepression and titration by sites outside the origin, as in E. coli (79, 80), several other regulators in B. subtilis are quite distinct, some of which are present because of the option to sporulate. DNA replication initiation is inhibited when the cells enter sporulation. This is achieved through a sporulation-specific transcription factor, Spo0A, which also binds to the replication origin (81, 82). This interferes with DnaA binding to the origin and, consequently, origin opening. Replication initiation is further inhibited by another protein, SirA (sporulation inhibitor of replication A), which is induced by Spo0A. SirA targets the DnaA protein directly and displaces it from the origin (83, 84). These controls ensure that unnecessary replication is not initiated when cells are proceeding to dormancy.
C. crescentus is another bacterium with a complex lifestyle, as it differentiates into two cell types, swarmer and stalked, in each cell cycle. A novel inhibitor, CtrA, is used to control replication initiation in a cell type-specific fashion (85). Phosphorylated CtrA binds to multiple sites in the origin and serves as a competitor of DnaA binding (86). The negative feedback following replication initiation seems to come from decreasing DnaA activity and availability and increasing CtrA activity and availability (87, 88). DnaA activity is decreased primarily by Hda, as in E. coli. Availability is decreased because the efficiency of the dnaA promoter decreases when it becomes hemimethylated following replication initiation. DnaA synthesis is further repressed by GcrA, a potent cell cycle regulator that accumulates in replicating stalked cells, and by an antisense noncoding RNA posttranscriptionally (89). Moreover, DnaA is proteolyzed in a timely manner with the help of a ClpP protease and possibly the Lon protease (88, 90). Hda, in addition to controlling DnaA activity, helps to accelerate DnaA turnover (91). CtrA availability increases following replication initiation because the ctrA promoter is activated upon hemimethylation and by GcrA, opposite from what happens to the dnaA promoter. In C. crescentus, in addition to cell cycle (temporal) control, there is also spatial control. The bacterium replicates DNA only in stalked cells because of the differential presence of CtrA in the two cell types. In stalked cells, CtrA is dephosphorylated (rendering it inactive in DNA binding) and is proteolyzed, allowing replication initiation. At the end of the cell cycle, CtrA is maximally synthesized in predivisional cells but is preferentially found in the swarmer cell compartment because proteolysis is restricted in the stalked-cell compartment. From this limited description, it should be clear that changes in the balance between initiators and inhibitors (DnaA and CtrA) are also critical for replication control in C. crescentus. The timing of replication initiation is perhaps best understood in this system because of a clear understanding of the temporal control of cell cycle-regulated genes (92).
Some of the other examples of species-specific replication regulators include AdpA, which helps to coordinate replication in Streptomyces coelicolor with the developmental program of the bacterium (93). In Mycobacterium tuberculosis and Helicobacter pylori, the signal transduction proteins (response regulators) MtrA and HP1021, respectively, control replication by interacting with the origin and influencing DnaA binding (94, 95). Like CtrA, the role of MtrA in the cell cycle regulation of replication has been determined.
CONCLUDING REMARKS
Here we have reviewed some features of the replication control system of Chr2 that apparently allow plasmid-like random timing of replication initiation to become nonrandom in the cell cycle, the norm in chromosomal replication. Many of these features make sense in that they are mechanistically or conceptually similar to the mechanisms used by other chromosomes. However, it is still unclear why plasmid-like mechanisms would need these additional features to ensure nonrandom timing, when timing in fact could also be produced by the most basic replication control systems. Specifically, consider the simplest version of inhibitor dilution mechanisms with a cooperative inhibition mechanism and where random effects in the control system are minor, i.e., where attempts to initiate replication are successful only when the inhibitor concentration is below a fixed threshold concentration. Under conditions where the average copy number is so low that plasmids typically change from a single copy in the beginning of the cell cycle to two copies at the end of the cell cycle, which can be achieved by tuning rate constants such as inhibitor expression, dilution of all components due to cell growth eventually causes the inhibitor concentration to drop below the replication threshold for some critical cell size. Cells will then replicate the plasmid close to the time when cells reach the critical size, and because the increase in gene dosage quickly leads to an increase in the inhibitor concentration, further replication events are blocked. Cell size of course does not correlate perfectly with the time in the cell cycle, but even for chromosomes, it is the cell size and not time that is believed to correlate with the initiation of replication (96).
Such perfect threshold mechanisms would also create timed initiations at higher average plasmid copy numbers but then at multiple specific cell sizes. For example, if a plasmid on average doubles from 10 to 20 copies during the cell cycle and uses a sharp threshold mechanism, an increase from, e.g., 12 to 13 would block further replication attempts only briefly after each replication and soon permit another replication. Because of measurement errors, or slight differences between cells, 10 pulses of replication would likely appear “smeared,” as if there were no cell cycle pattern at all.
Thus, sharp replication control mechanisms that sense the exact concentration and thus the cell volume for a given copy number would likely appear random at high average copy numbers and cell cycle specific at very low copy numbers. Most so-called low-copy-number plasmids are indeed present in multiple copies per cell under most growth conditions, and when copy numbers of 1 to 2 are reported in the literature, they are typically not in units of plasmid copies per cell but in plasmid copies per chromosome equivalent under growth conditions where there are multiple copies of the chromosomal reference gene per cell. Even though cell cycle-specific patterns are generally not observed for existing plasmids, it is thus possible that their control systems could be capable of such feats if the copy numbers were simply lowered further. This raises the question of why secondary chromosomes could not merely tweak the parameters of preexisting plasmid control systems in order to achieve timing.
One possible explanation is that systems with sharp thresholds, i.e., where a small fold change in the inhibitor concentration cooperatively produces a large fold change in the initiation frequency, would be too sensitive to other system imperfections. For example, as demonstrated by the mathematical study that started control theory as a field, Maxwell's 1867 study of steam engine governors, negative-feedback loops that respond too strongly to small changes can more easily be destabilized by delays. For plasmids, there is an elongation delay between committing to replication and producing additional inhibitor molecules, and because the inhibitors have some finite half-life, the inhibitor concentration cannot adjust immediately to changes in plasmid copy numbers. The same is true for fluctuations that arise inherently in the control system because of the probabilistic nature of all chemical reactions. Highly cooperative mechanisms can respond too sensitively to such fluctuations, thereby randomizing the plasmid level. Thus, a sharply cooperative control mechanism may not be enough to reliably create cell cycle-specific replication given other constraints on control, explaining why chromosomes use additional control.
That said, the core principles of control are not that different from plasmids to chromosomes, as we discuss above in the section on common grounds (Fig. 6). The additional regulators that are found for chromosomes primarily control the initiator, e.g., either its origin binding (using competitors such as SeqA, SirA, CtrA, MtrA, and HP1021), its synthesis (GcrA in C. crescentus and replication-induced transcription in B. subtilis), its availability by sequestration (YabA in B. subtilis) and proteolysis (ClpP in C. crescentus), or its activity by oligomerization (Soj and YabA in B. subtilis). Thus, the core features of control still remain centered on the initiator and the origin.
Studies in plasmids and chromosomes have made it abundantly clear that without inhibitors, replication cannot be controlled, as argued by Pritchard et al. A corollary to their proposal that the inhibitor concentration must rise after replication initiation has also gained considerable support, as the initiator-to-inhibitor ratio (monomer to dimer, DnaA-ATP to DnaA-ADP, and DnaA to CtrA) flips following replication initiation (50). Replication switch-off would be more rapid if both the initiator and inhibitor are changed but in opposite directions, as is the case in E. coli and C. crescentus and as has been argued in the case of plasmid P1. The crtS site in Chr1 promotes initiator binding that favors initiation but decreases binding that inhibits initiation, thus causing a synergistic effect on initiation.
A distinguishing feature of chromosomal replication is the use of the bulk of the chromosomal DNA for regulatory purposes, as replication-elongation-mediated controls are the most common ones in bacteria, including replication-induced transcription, sequestration by SeqA and YabA, increased titration due to a duplication of DnaA binding sites, and DNA-loaded clamp dependence for RIDA (31, 32). The coordination of replication timing of Chr2 with that of Chr1 apparently also depends on the passage of the replication fork over the crtS site (Fig. 8). Plasmids control their replication at the stage of initiation, most likely due to their brief elongation phase.
Replication control has been studied in only a few bacteria. It appears that control mechanisms can vary considerably among different bacteria: one obvious reason appears to be for the accommodation of species-specific requirements. Identification of these species-specific regulators is likely to reveal the common control principles more clearly. On the contrary, new control principles are likely to emerge from the study of bacteria with exotic lifestyles. If V. cholerae is any guide, the study of communication among chromosomes in bacteria with divided genomes is likely to reveal new checkpoint control mechanisms (63).
ACKNOWLEDGMENTS
We thank Dave Lane, Manolo Espinosa, Anders Løbner-Olesen, Heath Murray, Justine Collier, Ramon Diaz, Peter Barth, Peter Young, Martin Marinus, and Michael Yarmolinsky for many thoughtful comments and Jemima Barrowman and Tara Filsuf for help with editing.
This research was supported (in part) by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, and a National Institutes of Health grant (GM081563) to J.P.
We have no conflict of interest to declare.
Biographies

Revathy Ramachandran is a Postdoctoral Fellow in the Laboratory of Dr. Dhruba Chattoraj at the National Cancer Institute, NIH. She received her Ph.D. in Biology from the Virginia Polytechnic Institute and State University, Blacksburg, VA, in 2014, where she studied primarily the quorum-sensing regulon in Pantoea stewartii subsp. stewartii by transcriptomic and proteomic analyses. At the NIH, she is studying chromosome maintenance in a bacterium with a divided genome, Vibrio cholerae. In particular, she is addressing the mechanism of communication between the two chromosomes of V. cholerae.

Jyoti Jha is a Research Fellow in the Laboratory of Dr. Dhruba Chattoraj at the National Cancer Institute, NIH. He did his postgraduate studies at the Indian Institute of Technology, Kharagpur, India, and received his Ph.D. in Biotechnology from the University of Kolkata, India, in 2008. As a graduate student, he developed technologies for genetic engineering in plants and studied primarily fatty acid biosynthesis. At the NIH, he is studying cell cycle regulation of chromosomal DNA replication focusing on the structure-function analysis of a unique replication initiator protein specific to chromosome 2 of V. cholerae.

Johan Paulsson is a professor of Systems Biology at Harvard University. He received his Ph.D. from Uppsala University, Sweden, in 2000, focusing on mathematical models of stochastic processes in cells, particularly plasmid copy number control. He was then a Lewis-Thomas Fellow for his postdoc at Princeton University, for mathematicians and physicists going into biology. In 2003, he joined the Department of Applied Mathematics and Theoretical Physics at the University of Cambridge, United Kingdom, but moved to his current department in 2005 to set up an experimental group focused on quantifying properties of individual microbial cells.

Dhruba Chattoraj is a Senior Investigator at the National Cancer Institute, NIH. He received his Ph.D. in Biophysics from of the University of Calcutta, India, in 1970. After postdoctoral studies at the University of Wisconsin, Madison; the Institute of Molecular Biology, University of Oregon; and the Karolinska Institutet, Sweden, Dr. Chattoraj joined the NIH in 1978 and is currently the Chief of the Section on DNA Replication Control in the Laboratory of Biochemistry and Molecular Biology, NCI. His interest has been on the maintenance of plasmids and chromosomes in bacteria. His current interest is on the maintenance of chromosomes in bacteria with divided genomes, specifically on replication and segregation of chromosomes in the cell cycle of Vibrio cholerae.
This review is dedicated to the memory of Robert Pritchard for his seminal contributions in the field of DNA replication control.
REFERENCES
- 1.O'Donnell M, Langston L, Stillman B. 2013. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol 5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egan ES, Fogel MA, Waldor MK. 2005. Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol Microbiol 56:1129–1138. doi: 10.1111/j.1365-2958.2005.04622.x. [DOI] [PubMed] [Google Scholar]
- 3.Jha JK, Baek JH, Venkova-Canova T, Chattoraj DK. 2012. Chromosome dynamics in multichromosome bacteria. Biochim Biophys Acta 1819:826–829. doi: 10.1016/j.bbagrm.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackiewicz P, Zakrzewska-Czerwinska J, Zawilak A, Dudek MR, Cebrat S. 2004. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res 32:3781–3791. doi: 10.1093/nar/gkh699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulsson J, Chattoraj DK. 2006. Origin inactivation in bacterial DNA replication control. Mol Microbiol 61:9–15. doi: 10.1111/j.1365-2958.2006.05229.x. [DOI] [PubMed] [Google Scholar]
- 6.Harrison PW, Lower RP, Kim NK, Young JP. 2010. Introducing the bacterial ‘chromid’: not a chromosome, not a plasmid. Trends Microbiol 18:141–148. doi: 10.1016/j.tim.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Anda M, Ohtsubo Y, Okubo T, Sugawara M, Nagata Y, Tsuda M, Minamisawa K, Mitsui H. 2015. Bacterial clade with the ribosomal RNA operon on a small plasmid rather than the chromosome. Proc Natl Acad Sci U S A 112:14343–14347. doi: 10.1073/pnas.1514326112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macrina FL, Kopecko DJ, Jones KR, Ayers DJ, McCowen SM. 1978. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid 1:417–420. doi: 10.1016/0147-619X(78)90056-2. [DOI] [PubMed] [Google Scholar]
- 9.Botello E, Nordstrom K. 1998. Effects of chromosome underreplication on cell division in Escherichia coli. J Bacteriol 180:6364–6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cambridge J, Blinkova A, Magnan D, Bates D, Walker JR. 2014. A replication-inhibited un-segregated nucleoid at mid-cell blocks Z-ring formation and cell division independently of SOS and the SlmA nucleoid occlusion protein in Escherichia coli. J Bacteriol 196:36–49. doi: 10.1128/JB.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernard R, Marquis KA, Rudner DZ. 2010. Nucleoid occlusion prevents cell division during replication fork arrest in Bacillus subtilis. Mol Microbiol 78:866–882. doi: 10.1111/j.1365-2958.2010.07369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura Y, Caro L, Berg CM, Hirota Y. 1971. Chromosome replication in Escherichia coli. IV. Control of chromosome replication and cell division by an integrated episome. J Mol Biol 55:441–456. [DOI] [PubMed] [Google Scholar]
- 13.Koppes L, Nordstrom K. 1986. Insertion of an R1 plasmid into the origin of replication of the E. coli chromosome: random timing of replication of the hybrid chromosome. Cell 44:117–124. doi: 10.1016/0092-8674(86)90490-3. [DOI] [PubMed] [Google Scholar]
- 14.Eliasson A, Bernander R, Nordstrom K. 1996. Random initiation of replication of plasmids P1 and F (oriS) when integrated into the Escherichia coli chromosome. Mol Microbiol 20:1025–1032. doi: 10.1111/j.1365-2958.1996.tb02543.x. [DOI] [PubMed] [Google Scholar]
- 15.Egan ES, Lobner-Olesen A, Waldor MK. 2004. Synchronous replication initiation of the two Vibrio cholerae chromosomes. Curr Biol 14:R501–R502. doi: 10.1016/j.cub.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 16.Jacob F. 1993. The replicon: thirty years later. Cold Spring Harb Symp Quant Biol 58:383–387. doi: 10.1101/SQB.1993.058.01.045. [DOI] [PubMed] [Google Scholar]
- 17.Jacob F, Brenner S, Cuzin F. 1964. On the regulation of DNA replication in bacteria. Cold Spring Harb Symp Quant Biol 28:329–348. [Google Scholar]
- 18.Pritchard RH, Barth PT, Collins J. 1969. Control of DNA synthesis in bacteria. Symp Soc Gen Microbiol 19:263–297. [Google Scholar]
- 19.Tolun A, Helinski DR. 1981. Direct repeats of the F plasmid incC region express F incompatibility. Cell 24:687–694. doi: 10.1016/0092-8674(81)90095-7. [DOI] [PubMed] [Google Scholar]
- 20.Tsutsui H, Fujiyama A, Murotsu T, Matsubara K. 1983. Role of nine repeating sequences of the mini-F genome for expression of F-specific incompatibility phenotype and copy number control. J Bacteriol 155:337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chattoraj D, Cordes K, Abeles A. 1984. Plasmid P1 replication: negative control by repeated DNA sequences. Proc Natl Acad Sci U S A 81:6456–6460. doi: 10.1073/pnas.81.20.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pal SK, Chattoraj DK. 1988. P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J Bacteriol 170:3554–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McEachern MJ, Bott MA, Tooker PA, Helinski DR. 1989. Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc Natl Acad Sci U S A 86:7942–7946. doi: 10.1073/pnas.86.20.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chattoraj DK, Mason RJ, Wickner SH. 1988. Mini-P1 plasmid replication: the autoregulation-sequestration paradox. Cell 52:551–557. doi: 10.1016/0092-8674(88)90468-0. [DOI] [PubMed] [Google Scholar]
- 25.Toukdarian AE, Helinski DR. 1998. TrfA dimers play a role in copy-number control of RK2 replication. Gene 223:205–211. doi: 10.1016/S0378-1119(98)00370-9. [DOI] [PubMed] [Google Scholar]
- 26.Das N, Chattoraj DK. 2004. Origin pairing (‘handcuffing') and unpairing in the control of P1 plasmid replication. Mol Microbiol 54:836–849. doi: 10.1111/j.1365-2958.2004.04322.x. [DOI] [PubMed] [Google Scholar]
- 27.Kunnimalaiyaan S, Inman RB, Rakowski SA, Filutowicz M. 2005. Role of pi dimers in coupling (“handcuffing”) of plasmid R6K's gamma ori iterons. J Bacteriol 187:3779–3785. doi: 10.1128/JB.187.11.3779-3785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zzaman S, Bastia D. 2005. Oligomeric initiator protein-mediated DNA looping negatively regulates plasmid replication in vitro by preventing origin melting. Mol Cell 20:833–843. doi: 10.1016/j.molcel.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 29.Mukhopadhyay S, Chattoraj DK. 2000. Replication-induced transcription of an autorepressed gene: the replication initiator gene of plasmid P1. Proc Natl Acad Sci U S A 97:7142–7147. doi: 10.1073/pnas.130189497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogasawara N, Moriya S, von Meyenburg K, Hansen FG, Yoshikawa H. 1985. Conservation of genes and their organization in the chromosomal replication origin region of Bacillus subtilis and Escherichia coli. EMBO J 4:3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skarstad K, Katayama T. 2013. Regulating DNA replication in bacteria. Cold Spring Harb Perspect Biol 5:a012922. doi: 10.1101/cshperspect.a012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. 2010. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol 8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 33.Atlung T, Clausen ES, Hansen FG. 1985. Autoregulation of the dnaA gene of Escherichia coli K12. Mol Gen Genet 200:442–450. doi: 10.1007/BF00425729. [DOI] [PubMed] [Google Scholar]
- 34.Braun RE, O'Day K, Wright A. 1985. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell 40:159–169. doi: 10.1016/0092-8674(85)90319-8. [DOI] [PubMed] [Google Scholar]
- 35.Leonard AC, Grimwade JE. 5 January 2010. Initiation of DNA replication. EcoSal Plus 2010 doi: 10.1128/ecosalplus.4.4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell JL, Kleckner N. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967–979. doi: 10.1016/0092-8674(90)90271-F. [DOI] [PubMed] [Google Scholar]
- 37.Hansen FG, Christensen BB, Atlung T. 1991. The initiator titration model: computer simulation of chromosome and minichromosome control. Res Microbiol 142:161–167. doi: 10.1016/0923-2508(91)90025-6. [DOI] [PubMed] [Google Scholar]
- 38.Leonard AC, Grimwade JE. 2015. The orisome: structure and function. Front Microbiol 6:545. doi: 10.3389/fmicb.2015.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasho K, Katayama T. 2013. DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc Natl Acad Sci U S A 110:936–941. doi: 10.1073/pnas.1212070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu M, Campbell JL, Boye E, Kleckner N. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 41.Bochner BR, Ames BN. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem 257:9759–9769. [PubMed] [Google Scholar]
- 42.Kasho K, Fujimitsu K, Matoba T, Oshima T, Katayama T. 2014. Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res 42:13134–13149. doi: 10.1093/nar/gku1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frimodt-Moller J, Charbon G, Krogfelt KA, Lobner-Olesen A. 2015. Control regions for chromosome replication are conserved with respect to sequence and location among Escherichia coli strains. Front Microbiol 6:1011. doi: 10.3389/fmicb.2015.01011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crooke E, Castuma CE, Kornberg A. 1992. The chromosome origin of Escherichia coli stabilizes DnaA protein during rejuvenation by phospholipids. J Biol Chem 267:16779–16782. [PubMed] [Google Scholar]
- 45.Xia W, Dowhan W. 1995. In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc Natl Acad Sci U S A 92:783–787. doi: 10.1073/pnas.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. 1999. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J 18:6642–6652. doi: 10.1093/emboj/18.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cournac A, Plumbridge J. 2013. DNA looping in prokaryotes: experimental and theoretical approaches. J Bacteriol 195:1109–1119. doi: 10.1128/JB.02038-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dibbens JA, Muraiso KM, Chattoraj DK. 1997. Chaperone-mediated reduction of RepA dimerization is associated with RepA conformational change. Mol Microbiol 25:185–195. [DOI] [PubMed] [Google Scholar]
- 49.Jonczyk P, Hines R, Smith DW. 1989. The Escherichia coli dam gene is expressed as a distal gene of a new operon. Mol Gen Genet 217:85–96. doi: 10.1007/BF00330946. [DOI] [PubMed] [Google Scholar]
- 50.Donachie WD, Blakely GW. 2003. Coupling the initiation of chromosome replication to cell size in Escherichia coli. Curr Opin Microbiol 6:146–150. doi: 10.1016/S1369-5274(03)00026-2. [DOI] [PubMed] [Google Scholar]
- 51.Egan ES, Waldor MK. 2003. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114:521–530. doi: 10.1016/S0092-8674(03)00611-1. [DOI] [PubMed] [Google Scholar]
- 52.Val ME, Soler-Bistue A, Bland MJ, Mazel D. 2014. Management of multipartite genomes: the Vibrio cholerae model. Curr Opin Microbiol 22:120–126. doi: 10.1016/j.mib.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Jha JK, Demarre G, Venkova-Canova T, Chattoraj DK. 2012. Replication regulation of Vibrio cholerae chromosome II involves initiator binding to the origin both as monomer and as dimer. Nucleic Acids Res 40:6026–6038. doi: 10.1093/nar/gks260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickner S, Hoskins J, McKenney K. 1991. Monomerization of RepA dimers by heat shock proteins activates binding to DNA replication origin. Proc Natl Acad Sci U S A 88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Venkova-Canova T, Chattoraj DK. 2011. Transition from a plasmid to a chromosomal mode of replication entails additional regulators. Proc Natl Acad Sci U S A 108:6199–6204. doi: 10.1073/pnas.1013244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koch B, Ma X, Lobner-Olesen A. 2012. rctB mutations that increase copy number of Vibrio cholerae oriCII in Escherichia coli. Plasmid 68:159–169. doi: 10.1016/j.plasmid.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 57.Demarre G, Chattoraj DK. 2010. DNA adenine methylation is required to replicate both Vibrio cholerae chromosomes once per cell cycle. PLoS Genet 6:e1000939. doi: 10.1371/journal.pgen.1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Val ME, Kennedy SP, Soler-Bistue AJ, Barbe V, Bouchier C, Ducos-Galand M, Skovgaard O, Mazel D. 2014. Fuse or die: how to survive the loss of Dam in Vibrio cholerae. Mol Microbiol 91:665–678. doi: 10.1111/mmi.12483. [DOI] [PubMed] [Google Scholar]
- 59.Duigou S, Yamaichi Y, Waldor MK. 2008. ATP negatively regulates the initiator protein of Vibrio cholerae chromosome II replication. Proc Natl Acad Sci U S A 105:10577–10582. doi: 10.1073/pnas.0803904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venkova-Canova T, Srivastava P, Chattoraj DK. 2006. Transcriptional inactivation of a regulatory site for replication of Vibrio cholerae chromosome II. Proc Natl Acad Sci U S A 103:12051–12056. doi: 10.1073/pnas.0605120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venkova-Canova T, Baek JH, Fitzgerald PC, Blokesch M, Chattoraj DK. 2013. Evidence for two different regulatory mechanisms linking replication and segregation of Vibrio cholerae chromosome II. PLoS Genet 9:e1003579. doi: 10.1371/journal.pgen.1003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baek JH, Chattoraj DK. 2014. Chromosome I controls chromosome II replication in Vibrio cholerae. PLoS Genet 10:e1004184. doi: 10.1371/journal.pgen.1004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Val ME, Marbouty M, de Lemos Martins F, Kennedy SP, Kemble H, Bland MJ, Possoz C, Koszul R, Skovgaard O, Mazel D. 2016. A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae. Sci Adv 2:e1501914. doi: 10.1126/sciadv.1501914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Messerschmidt SJ, Kemter FS, Schindler D, Waldminghaus T. 2015. Synthetic secondary chromosomes in Escherichia coli based on the replication origin of chromosome II in Vibrio cholerae. Biotechnol J 10:302–314. doi: 10.1002/biot.201400031. [DOI] [PubMed] [Google Scholar]
- 65.Cooper S, Helmstetter CE. 1968. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol 31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 66.Skarstad K, Boye E, Steen HB. 1986. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J 5:1711–1717. (Erratum, 5:3074.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srivastava P, Chattoraj DK. 2007. Selective chromosome amplification in Vibrio cholerae. Mol Microbiol 66:1016–1028. doi: 10.1111/j.1365-2958.2007.05973.x. [DOI] [PubMed] [Google Scholar]
- 68.Duigou S, Knudsen KG, Skovgaard O, Egan ES, Lobner-Olesen A, Waldor MK. 2006. Independent control of replication initiation of the two Vibrio cholerae chromosomes by DnaA and RctB. J Bacteriol 188:6419–6424. doi: 10.1128/JB.00565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen EB, Yarmolinsky MB. 1986. Host participation in plasmid maintenance: dependence upon dnaA of replicons derived from P1 and F. Proc Natl Acad Sci U S A 83:4423–4427. doi: 10.1073/pnas.83.12.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pal D, Venkova-Canova T, Srivastava P, Chattoraj DK. 2005. Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II. J Bacteriol 187:7167–7175. doi: 10.1128/JB.187.21.7167-7175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ogura Y, Ogasawara N, Harry EJ, Moriya S. 2003. Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J Bacteriol 185:6316–6324. doi: 10.1128/JB.185.21.6316-6324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scholefield G, Whiting R, Errington J, Murray H. 2011. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol Microbiol 79:1089–1100. doi: 10.1111/j.1365-2958.2010.07507.x. [DOI] [PubMed] [Google Scholar]
- 73.Scholefield G, Errington J, Murray H. 2012. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J 31:1542–1555. doi: 10.1038/emboj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murray H, Errington J. 2008. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 75.Kadoya R, Baek JH, Sarker A, Chattoraj DK. 2011. Participation of chromosome segregation protein ParAI of Vibrio cholerae in chromosome replication. J Bacteriol 193:1504–1514. doi: 10.1128/JB.01067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scholefield G, Murray H. 2013. YabA and DnaD inhibit helix assembly of the DNA replication initiation protein DnaA. Mol Microbiol 90:147–159. doi: 10.1111/mmi.12353. [DOI] [PubMed] [Google Scholar]
- 77.Soufo CD, Soufo HJ, Noirot-Gros MF, Steindorf A, Noirot P, Graumann PL. 2008. Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev Cell 15:935–941. doi: 10.1016/j.devcel.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 78.Brezellec P, Hoebeke M, Hiet MS, Pasek S, Ferat JL. 2006. DomainSieve: a protein domain-based screen that led to the identification of dam-associated genes with potential link to DNA maintenance. Bioinformatics 22:1935–1941. doi: 10.1093/bioinformatics/btl336. [DOI] [PubMed] [Google Scholar]
- 79.Breier AM, Grossman AD. 2009. Dynamic association of the replication initiator and transcription factor DnaA with the Bacillus subtilis chromosome during replication stress. J Bacteriol 191:486–493. doi: 10.1128/JB.01294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okumura H, Yoshimura M, Ueki M, Oshima T, Ogasawara N, Ishikawa S. 2012. Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic Acids Res 40:220–234. doi: 10.1093/nar/gkr716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Castilla-Llorente V, Munoz-Espin D, Villar L, Salas M, Meijer WJ. 2006. Spo0A, the key transcriptional regulator for entrance into sporulation, is an inhibitor of DNA replication. EMBO J 25:3890–3899. doi: 10.1038/sj.emboj.7601266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boonstra M, de Jong IG, Scholefield G, Murray H, Kuipers OP, Veening JW. 2013. Spo0A regulates chromosome copy number during sporulation by directly binding to the origin of replication in Bacillus subtilis. Mol Microbiol 87:925–938. doi: 10.1111/mmi.12141. [DOI] [PubMed] [Google Scholar]
- 83.Wagner JK, Marquis KA, Rudner DZ. 2009. SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol Microbiol 73:963–974. doi: 10.1111/j.1365-2958.2009.06825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jameson KH, Rostami N, Fogg MJ, Turkenburg JP, Grahl A, Murray H, Wilkinson AJ. 2014. Structure and interactions of the Bacillus subtilis sporulation inhibitor of DNA replication, SirA, with domain I of DnaA. Mol Microbiol 93:975–991. doi: 10.1111/mmi.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barnett MJ, Hung DY, Reisenauer A, Shapiro L, Long SR. 2001. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J Bacteriol 183:3204–3210. doi: 10.1128/JB.183.10.3204-3210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Collier J. 2012. Regulation of chromosomal replication in Caulobacter crescentus. Plasmid 67:76–87. doi: 10.1016/j.plasmid.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 87.Collier J, Shapiro L. 2009. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J Bacteriol 191:5706–5716. doi: 10.1128/JB.00525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jonas K, Liu J, Chien P, Laub MT. 2013. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154:623–636. doi: 10.1016/j.cell.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou B, Schrader JM, Kalogeraki VS, Abeliuk E, Dinh CB, Pham JQ, Cui ZZ, Dill DL, McAdams HH, Shapiro L. 2015. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet 11:e1004831. doi: 10.1371/journal.pgen.1004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gorbatyuk B, Marczynski GT. 2005. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol 55:1233–1245. [DOI] [PubMed] [Google Scholar]
- 91.Wargachuk R, Marczynski GT. 2015. The Caulobacter crescentus homolog of DnaA (HdaA) also regulates the proteolysis of the replication initiator protein DnaA. J Bacteriol 197:3521–3532. doi: 10.1128/JB.00460-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Collier J, McAdams HH, Shapiro L. 2007. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci U S A 104:17111–17116. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wolanski M, Jakimowicz D, Zakrzewska-Czerwinska J. 2014. Fifty years after the replicon hypothesis: cell-specific master regulators as new players in chromosome replication control. J Bacteriol 196:2901–2911. doi: 10.1128/JB.01706-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Purushotham G, Sarva KB, Blaszczyk E, Rajagopalan M, Madiraju MV. 2015. Mycobacterium tuberculosis oriC sequestration by MtrA response regulator. Mol Microbiol 98:586–604. doi: 10.1111/mmi.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Donczew R, Makowski L, Jaworski P, Bezulska M, Nowaczyk M, Zakrzewska-Czerwinska J, Zawilak-Pawlik A. 2015. The atypical response regulator HP1021 controls formation of the Helicobacter pylori replication initiation complex. Mol Microbiol 95:297–312. doi: 10.1111/mmi.12866. [DOI] [PubMed] [Google Scholar]
- 96.Hill NS, Kadoya R, Chattoraj DK, Levin PA. 2012. Cell size and the initiation of DNA replication in bacteria. PLoS Genet 8:e1002549. doi: 10.1371/journal.pgen.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nordstrom K. 2006. Plasmid R1—replication and its control. Plasmid 55:1–26. doi: 10.1016/j.plasmid.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 98.Das N, Valjavec-Gratian M, Basuray AN, Fekete RA, Papp PP, Paulsson J, Chattoraj DK. 2005. Multiple homeostatic mechanisms in the control of P1 plasmid replication. Proc Natl Acad Sci U S A 102:2856–2861. doi: 10.1073/pnas.0409790102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Venkova-Canova T, Saha A, Chattoraj DK. 2012. A 29-mer site regulates transcription of the initiator gene as well as function of the replication origin of Vibrio cholerae chromosome II. Plasmid 67:102–110. doi: 10.1016/j.plasmid.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]