Summary
Reprogramming of differentiated cells into induced pluripotent stem cells has been recently achieved in vivo in mice. Telomeres are essential for chromosomal stability and determine organismal life span as well as cancer growth. Here, we study whether tissue dedifferentiation induced by in vivo reprogramming involves changes at telomeres. We find telomerase-dependent telomere elongation in the reprogrammed areas. Notably, we found highly upregulated expression of the TRF1 telomere protein in the reprogrammed areas, which was independent of telomere length. Moreover, TRF1 inhibition reduced in vivo reprogramming efficiency. Importantly, we extend the finding of TRF1 upregulation to pathological tissue dedifferentiation associated with neoplasias, in particular during pancreatic acinar-to-ductal metaplasia, a process that involves transdifferentiation of adult acinar cells into ductal-like cells due to K-Ras oncogene expression. These findings place telomeres as important players in cellular plasticity both during in vivo reprogramming and in pathological conditions associated with increased plasticity, such as cancer.
Keywords: in vivo reprogramming, telomeres, stem cells, TRF1, tumorigenesis, cellular plasticity, cancer, transdifferentiation, ADM, regeneration
Highlights
-
•
Telomeres are elongated during in vivo reprogramming in a telomerase-dependent manner
-
•
Telomeric protein TRF1 is highly overexpressed during in vivo reprogramming
-
•
TRF1 is upregulated during acinar-to-ductal transdifferentiation
Blasco and colleagues study whether tissue dedifferentiation induced by in vivo reprogramming involves changes at telomeres. They show a telomerase-dependent telomere elongation and upregulated expression of TRF1 telomere protein in the in vivo reprogrammed areas. Furthermore, they demonstrate TRF1 upregulation in pathological tissue dedifferentiation during pancreatic acinar-to-ductal metaplasia. These findings place telomeres as important players in processes involving cellular plasticity.
Introduction
Reprogramming into full pluripotency has been achieved in vivo in the context of mouse tissues (Abad et al., 2013). Thus, induction of the reprogramming factors in transgenic mice (so-called reprogrammable mice) results in reprogramming events marked by the expression of the pluripotency factor NANOG in multiple organs, tissue dedifferentiation, and teratoma formation. Therefore, these mice could be useful for a deeper understanding of the molecular mechanisms that govern tissue dedifferentiation in vivo. Interestingly, mammalian cell reprogramming can also occur spontaneously during regeneration after injury or damage conditions (Yanger et al., 2013). Differentiated cells can be converted in vivo into another cell type and also into functional multipotent stem-like cells (Tata et al., 2013). This capacity of somatic cells to dedifferentiate into stem-like cells in vivo may have a pivotal role in physiological tissue regeneration or during tumorigenesis.
Telomeres are repetitive DNA structures at the chromosome ends that safeguard them from DNA repair and degradation activities. This protective function requires a proper telomere length and the binding of a six-protein complex known as shelterin, including TRF1, which is essential for telomere protection (Martinez and Blasco, 2010). Telomeres are elongated by telomerase (Greider and Blackburn, 1985), a reverse transcriptase present during early embryonic development and in adult stem cell compartments (Flores et al., 2005, Flores et al., 2006, Liu et al., 2007). However, telomerase activity in adult stem cells can only partially counterbalance the telomere shortening associated with cell division, so telomeres shorten in all cell types with aging (Flores et al., 2006). Telomere shortening in adult stem cells causes severe impairment of the ability of stem cells to regenerate tissues (Flores et al., 2005). Thus, pathological conditions that require acquisition of cellular immortality, such as many cancer types, show high telomerase activity to allow indefinite proliferation (Martinez and Blasco, 2011).
Interestingly, short telomeres in adult differentiated cells can regrow to the long telomeres of embryonic stem cells during in vitro reprogramming to induced pluripotent stem cells (iPSCs) (Marion et al., 2009b, Varela et al., 2011). Telomerase expression and telomere elongation are essential for the efficient generation and pluripotency of the iPSCs, as both the reprogramming efficiency and pluripotency features of these cells are impaired in telomerase-deficient cells with short telomeres (Marion et al., 2009a, Marion et al., 2009b). The shelterin component TRF1 is highly upregulated during iPSC generation, which in turn is essential both for initiation and maintenance of pluripotency in iPSCs (Schneider et al., 2013). Finally, in vitro reprogramming induces epigenetic changes at the telomeric chromatin, including decreased trimethylation of H3K9 and H4K20, indicative of a more “open” chromatin at telomeres (Marion et al., 2009b). These findings suggest substantial changes at telomeres during in vitro iPSC generation. However, whether changes at telomeres occur in association with tissue dedifferentiation induced by in vivo reprogramming or other pathological processes, such as cancer, remains unknown to date.
In view of the importance of telomere biology in tissue regeneration, aging, and cancer, here we study telomere changes during tissue dedifferentiation induced by reprogramming in vivo. We find that in vivo reprogrammed areas present longer telomeres and increased expression of the telomerase Terc RNA component than non-reprogrammed tissue, and this telomere elongation is telomerase dependent as it is abolished in Terc-deficient mice. In vivo reprogrammed cells highly overexpress TRF1 in a manner that coincides with OCT4 expression. Chemical inhibition of TRF1 decreases in vivo reprogramming, suggesting an important role of TRF1 upregulation for tissue reprogramming. These telomere-related changes are accompanied by drastic chromatin changes, including loss of histone trimethylation marks.
Finally, we extend these findings to pathological tissue dedifferentiation during cancer development. We found elevated TRF1 expression during pancreatic acinar-to-ductal metaplasia (ADM), which involves transdifferentiation of adult acinar cells into ductal-like cells as a result of K-Ras oncogene expression, which can subsequently progress to malignant adenocarcinoma. Telomeres were also elongated in a percentage of the lesions, in a manner uncoupled from TRF1 expression, also mimicking telomere changes during tissue dedifferentiation induced by reprogramming in vivo.
Our results uncover key molecular events at telomeres that occur during the dedifferentiation of adult cells induced by in vivo reprogramming or early stages of tumorigenesis. The understanding of the molecular events associated with dedifferentiation will open new roads to a better understanding of cellular plasticity, the control of regeneration of tissues in vivo, and pathological conditions such as cancer.
Results
Telomere Elongation during Reprogramming-Induced Tissue Dedifferentiation
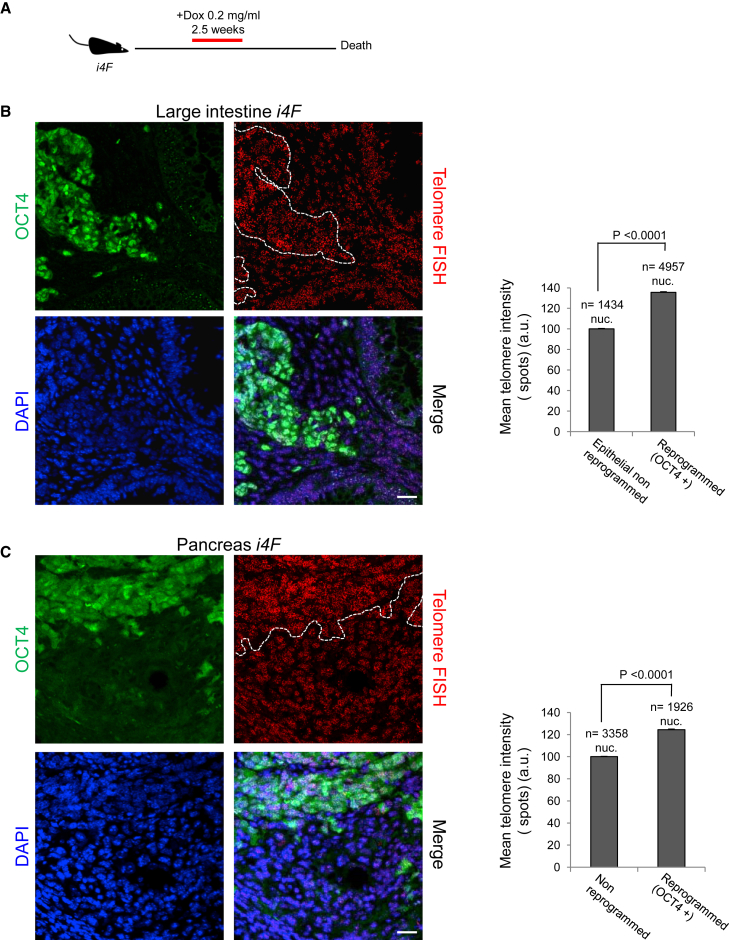
To induce tissue dedifferentiation and reprogramming in vivo, we used the reprogrammable mouse model (i4F mice), which carries a doxycycline-inducible cassette encoding the four reprogramming factors (OCT4, SOX2, KLF4, and cMYC) (Abad et al., 2013). We treated i4F mice for 2.5 weeks with a low doxycycline dose (0.2 mg/mL), followed by doxycycline withdrawal (Figure 1A). After a variable period of time, treated i4F mice succumb to the presence of teratomas in various tissues, indicating reprogramming into full pluripotency in vivo (Abad et al., 2013). In addition to teratomas, i4F mice show dedifferentiation and aberrant reprogrammed structures and masses containing undifferentiated dysplastic cells, some of them expressing the pluripotency marker NANOG (Figures S1A and S1B), indicating again full reprogramming in vivo.
Figure 1.
Telomeres Elongate during In Vivo Reprogramming
(A) Schematic representation of the induction of in vivo reprogramming in i4F mice.
(B and C) Left: representative immuno-FISH images showing OCT4 (green) and telomeres (red) in (B) the large intestine and (C) the pancreas of reprogrammable mice after induction of in vivo reprogramming. White dotted lines mark the reprogrammed areas. Scale bars, 25 μm. Right: quantification of telomere signal in in vivo reprogrammed cells and the corresponding non-reprogrammed control cells. Error bars denote SE. Statistical analysis by Student's t test. n, number of nuclei. Number of mice analyzed = 3.
See also Figures S1, S4, and S5.
Next, we studied whether in vivo reprogramming resulted in telomere lengthening. This would demonstrate that telomere elongation is possible in adult tissues by reprogramming-induced dedifferentiation. To address this exciting hypothesis, we combined quantitative telomere fluorescence in situ hybridization (Q-FISH) to measure the telomere length of individual cells, with OCT4 immunofluorescence to mark reprogrammed cells in the large intestine and the pancreas of reprogrammable i4F mice. Strikingly, we found significantly longer telomeres in the cells of the reprogrammed areas compared with the cells of the non-reprogrammed areas within the same tissue (Figures 1B and 1C). These results demonstrate telomere elongation in adult tissues upon dedifferentiation induced by in vivo reprogramming.
Telomere Elongation during Reprogramming-Induced Tissue Dedifferentiation Is Primarily Mediated by Telomerase
Next, we studied whether telomerase was upregulated during in vivo reprogramming and whether it was responsible for the observed telomere elongation. To do so, we generated a telomerase-deficient i4F reprogrammable mouse by crossing the i4F mouse to telomerase-deficient Terc−/− mice (Blasco et al., 1997). First-generation (G1) telomerase-deficient i4F mice (i4F G1Terc−/−) were treated for 2.5 weeks with low doxycycline (0.2 mg/mL) followed by doxycycline withdrawal to induce in vivo reprogramming. Interestingly, i4F G1Terc−/− mice also developed teratomas, with the three embryonic germ layers indicative of full in vivo reprogramming. We could also detect early reprogramming events by the presence of NANOG-positive cells within tissue structures (Figures S1C and S1D), demonstrating that tissue dedifferentiation induced by in vivo reprogramming can be achieved in the absence of telomerase activity.
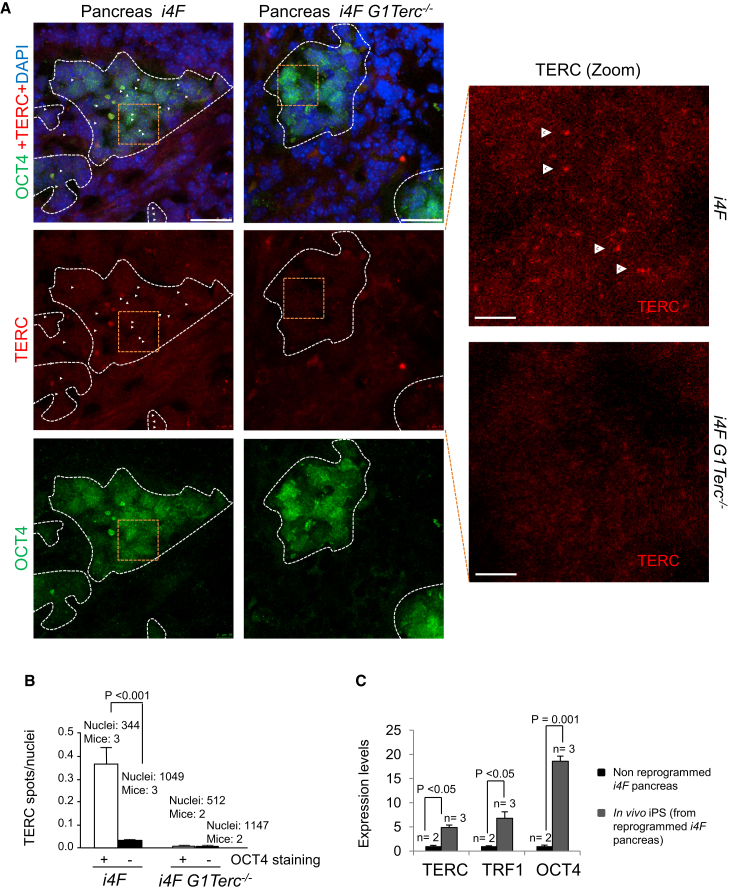
To determine whether telomerase expression was increased during in vivo reprogramming, we performed an in situ RNA-FISH using a probe specific for the Terc telomerase RNA gene (Figure S2), followed by immunofluorescence against OCT4 to detect the reprogrammed areas within the pancreas from both i4F wild-type and i4F G1Terc−/− reprogrammable mice. We found that the number of bright TERC spots was significantly increased in the OCT4-positive reprogrammed areas in the pancreas of i4F wild-type mice (Figures 2A and 2B), indicating that the telomerase RNA component is upregulated during in vivo reprogramming. As negative control, we failed to detect TERC RNA foci in the OCT4-positive reprogrammed tissues from the reprogrammed i4F G1Terc−/− controls (Figures 2A and 2B), validating the specificity of the assay. To further confirm upregulation of Terc RNA during in vivo reprogramming, we isolated in vivo iPSCs from pancreas of reprogrammed i4F mice. qRT-PCR analysis showed an elevated expression of Terc as well as OCT4 mRNAs in in vivo iPSCs of pancreatic origin when compared with normal non-reprogrammed pancreas (Figure 2C), thus confirming that the telomerase Terc RNA component is upregulated during in vivo reprogramming.
Figure 2.
TERC Expression Increases during In Vivo Reprogramming
(A) Representative images corresponding to TERC detection by RNA-FISH (red) followed by immunostaining to detect OCT4 (green) in the pancreas of induced reprogrammable mice of the indicated genotypes. Arrowheads indicate the presence of TERC foci. Scale bars, 25 μm. Zoomed areas (orange boxes) of TERC detection are shown on the right. Scale bars, 50 μm. White dashed lines contain OCT4-positive regions.
(B) Number of TERC spots per nuclei. Expression of TERC is increased in the reprogrammed areas. Error bars denote SE. Statistical analysis by Student's t test. Number of mice analyzed is indicated.
(C) Expression of TERC, TRF1, and OCT4 in non-reprogrammed pancreas compared with in vivo iPSCs of pancreatic origin as measured by qRT-PCR. Expression of these genes is strongly upregulated during the acquisition of pluripotency in vivo. Error bars denote SE. Statistical analysis by Student's t test. n, number of independent pancreas or in vivo iPSC clones obtained from independent reprogrammed pancreas, respectively.
See also Figures S1 and S2.
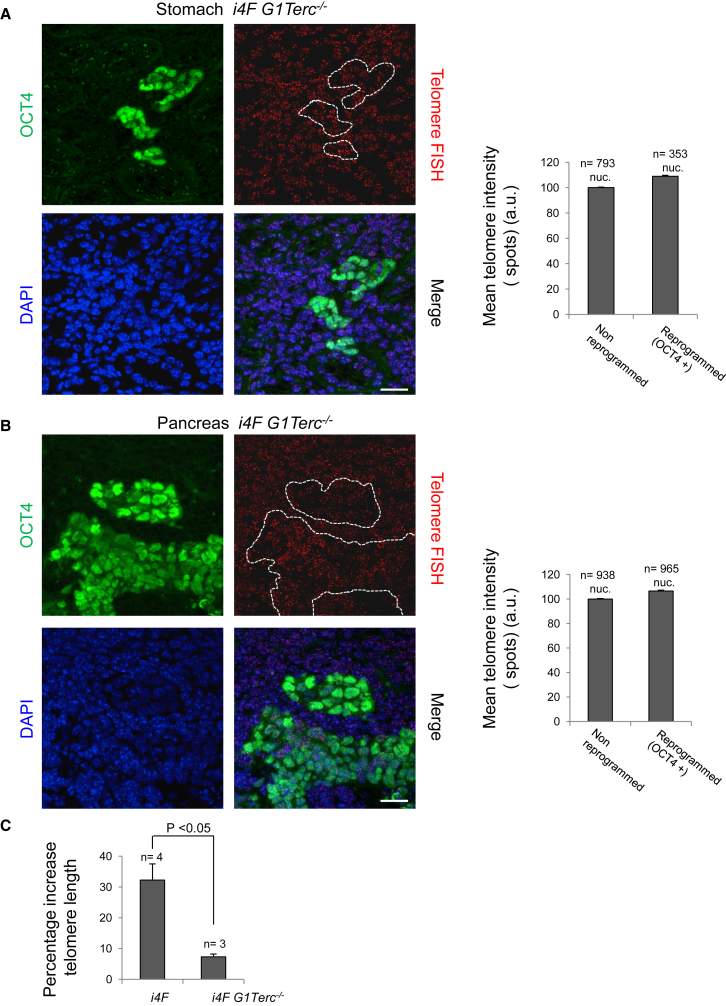
Next, we studied whether telomerase was necessary for the telomere elongation associated with in vivo reprogramming (Figures 1B and 1C). To this end, we performed an immuno-telomere Q-FISH with OCT4 antibodies to determine telomere length in both the OCT4-positive (reprogrammed areas) and OCT4-negative (normal non-reprogrammed areas from the same tissue) stomach (Figure 3A) and pancreas (Figure 3B) from i4F G1 Terc−/− mice. Importantly, we found that telomere lengthening in reprogrammed cells from the telomerase-deficient mice is severely impaired in the absence of a functional telomerase enzyme (compare Figures 3A and 3B with Figures 1B and 1C). Indeed, upon comparison of the average telomere elongation in i4F versus i4F G1Terc−/− reprogrammable mice in several tissues, we found that average telomere length was increased by 30% in the i4F wild-type reprogrammed areas, while this decreased to less than 10% in the case of the telomerase-deficient tissues (Figure 3C), suggesting that telomerase is the primary activity responsible for telomere elongation during in vivo reprogramming.
Figure 3.
Telomere Elongation during In Vivo Reprogramming Depends on Telomerase Activity
(A and B) Left: representative immuno-FISH images showing OCT4 (green) and telomeres (red) in (A) the stomach and (B) the pancreas of i4F G1Terc−/− reprogrammable mice after induction of in vivo reprogramming. White dashed lines mark the reprogrammed areas. Scale bars, 25 μm. Right: quantification of telomere signal in in vivo reprogrammed cells and the corresponding non-reprogrammed control cells. Telomere lengthening during in vivo reprogramming is abolished in the absence of telomerase activity. Error bars denote SE. n, number of nuclei. Number of mice analyzed = 3.
(C) Comparison of telomere lengthening during in vivo reprogramming in i4F and i4F G1Terc−/− reprogrammable mice. Note that most of the telomere elongation observed during in vivo reprogramming is dependent on telomerase activity. Error bars denote SE. Statistical analysis by Student's t test. n, number of tissues from independent mice analyzed.
See also Figure S1.
Upregulation of the Telomere Protection Factor TRF1 during Reprogramming-Induced Tissue Dedifferentiation
Next, we determined the levels of the TRF1 telomere-binding protein in the reprogrammed tissues. TRF1 levels are very high in embryonic stem cells, in vitro-generated iPSCs and in adult stem cell compartments (Schneider et al., 2013), where it is required for the induction and maintenance of pluripotency in iPSCs (Schneider et al., 2013) and for tissue homeostasis (Beier et al., 2012, Martinez et al., 2009, Povedano et al., 2015, Schneider et al., 2013), respectively.
To this end, we performed double immunofluorescence against TRF1 and NANOG, a marker of reprogrammed cells, in both large intestine and pancreas from i4F mice where we previously confirmed the presence of reprogramming events by NANOG immunohistochemistry (Figures S1A and S1B). We first studied the large intestine of i4F mice and found a significant increase in TRF1 levels in the reprogrammed masses compared with the non-reprogrammed areas (Figure S3A). We extended these findings to the pancreas of induced reprogrammable mice (Figure S3B). Interestingly, in both cases we found that high TRF1 expression in the reprogrammed masses is not restricted to NANOG-positive cells (Figures S3A and S3B).
The presence of cells within the reprogrammed areas showing high TRF1 expression but no expression of NANOG prompted us to determine their differentiation status. We performed a triple immunofluorescence against NANOG, TRF1, and the epithelial marker cytokeratin 19 (CK19) in the large intestine of i4F mice. We observed that NANOG-negative cells showing high TRF1 levels had fully lost the expression of CK19 (Figure S3C), demonstrating that they had indeed dedifferentiated and lost their epithelial cell identity. These findings suggest that high TRF1 expression in dedifferentiated cells persists independently of NANOG expression.
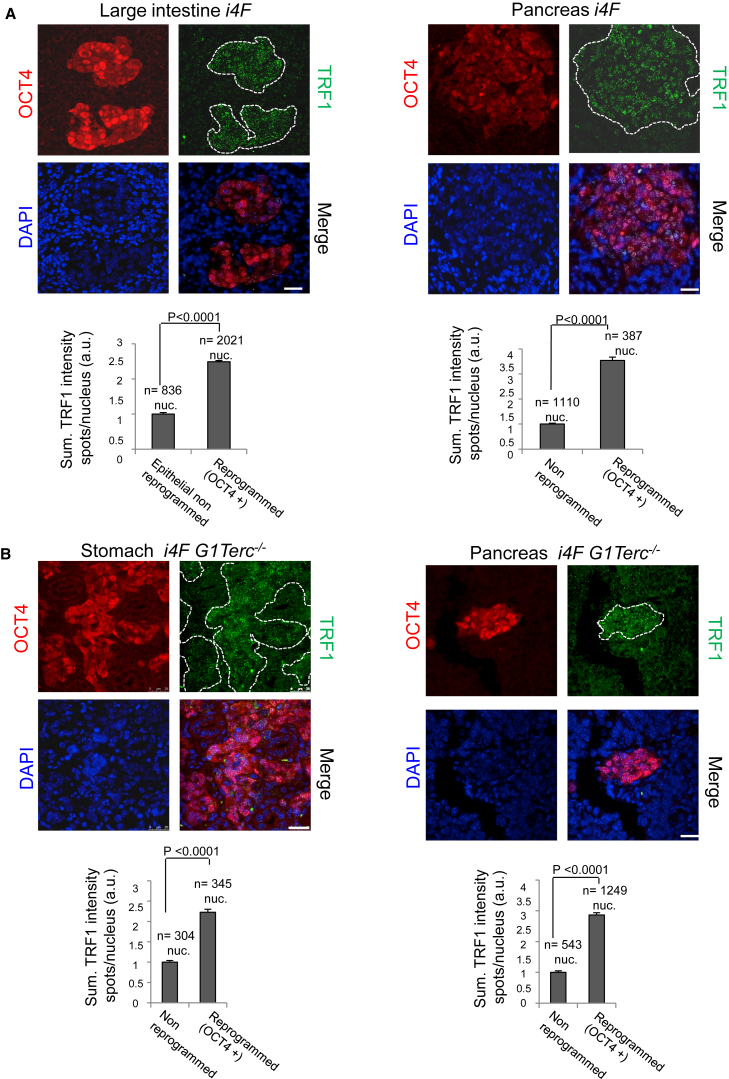
In this regard, pluripotency factor OCT4 has been shown to bind the Trf1 promoter and to upregulate Trf1 expression during in vitro iPSC generation (Schneider et al., 2013). Thus, we next addressed whether cells showing high TRF1 levels within the in vivo reprogrammed areas also showed high OCT4 expression. To do so, we performed double immunofluorescence against TRF1 and OCT4 in the large intestine and pancreas of induced reprogrammable i4F mice. We observed that OCT4 is highly expressed in the majority of the cells within the reprogrammed area, and that this expression fully coincides with high TRF1 levels (Figure 4A), suggesting that OCT4 expression is associated with TRF1 expression during in vivo reprogramming.
Figure 4.
Increased TRF1 Expression during In Vivo Reprogramming Correlates with OCT4 Expression and Is Uncoupled from Telomere Length
(A) Upper: representative images of double immunofluorescence against OCT4 (red) and TRF1 (green) proteins in (left) the large intestine and (right) the pancreas of reprogrammable mice after induction of in vivo reprogramming. A strong correlation between the presence of OCT4 and high TRF1 expression can be observed. Lower: quantification of TRF1 expression in in vivo reprogrammed cells and the corresponding non-reprogrammed control cells. Scale bars, 25 μm. Error bars denote SE. Statistical analysis by Student's t test. n, number of nuclei. Number of mice analyzed = 3.
(B) Upper: representative images of double immunofluorescence against OCT4 (red) and TRF1 (green) proteins in (left) the stomach and (right) the pancreas of i4F G1Terc−/− reprogrammable mice after induction of in vivo reprogramming. High TRF1 levels are observed in the in vivo reprogrammed areas even in the absence of a functional telomerase enzyme. Lower: quantification of TRF1 expression in in vivo reprogrammed cells and the corresponding non-reprogrammed control cells.
White dashed lines mark the reprogrammed area. Scale bars, 25 μm. Error bars denote SE. Statistical analysis by Student's t test. n, number of nuclei. Number of mice analyzed = 3.
See also Figures S1 and S3.
Finally, we confirmed increased TRF1 expression during in vivo reprogramming in in vivo iPSCs isolated from reprogrammed pancreas. qRT-PCR analysis showed an elevated expression of TRF1 (Figure 2C), thus confirming that TRF1 expression is upregulated during in vivo reprogramming.
Upregulation of TRF1 Expression during In Vivo Reprogramming-Induced Tissue Dedifferentiation Is Independent of Telomere Elongation and Is Important for Reprogramming
In vitro generation of iPSCs deficient for TPP1, a component of shelterin essential for telomere elongation, fail to elongate telomeres but still show highly upregulated TRF1 protein levels (Schneider et al., 2013). To address whether TRF1 upregulation is coupled to telomere elongation during in vivo reprogramming, we performed double immunofluorescence against TRF1 and OCT4 in the stomach and pancreas (Figure 4B) of induced telomerase-deficient i4F G1Terc−/− reprogrammable mice in which telomere elongation associated with reprogramming is abolished (Figures 3A and 3B). We observed that, even in the absence of telomerase, TRF1 levels were highly upregulated in the reprogrammed cells, thus demonstrating that increased TRF1 levels are uncoupled from telomere elongation during reprogramming in vivo. These findings reinforce the notion that TRF1 upregulation is one of the key events associated with cellular reprogramming and not a mere consequence of telomere elongation during this process.
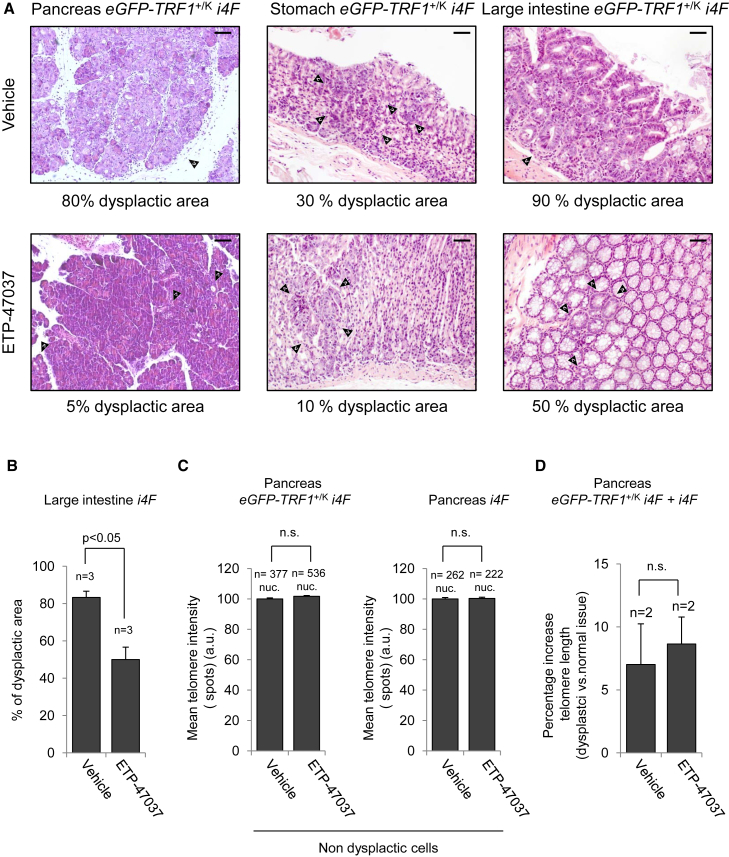
We recently demonstrated that blocking TRF1 by chemical (small molecules) and genetic means is an efficient way to block the initiation and progression of K-Ras-induced lung adenocarcinomas (Garcia-Beccaria et al., 2015). Thus, we wondered whether chemical inhibition of TRF1 would be sufficient to decrease in vivo reprogramming. We previously described that oral administration of these inhibitors for weeks was not leading to decreased mouse viability or loss of homeostasis of mouse tissues (Garcia-Beccaria et al., 2015). To this end, we treated i4F and eGFP-TRF1+/KI i4F (Schneider et al., 2013) reprogrammable mice for 1 week with high doxycycline (1 mg/mL) in the presence or absence of TRF1 inhibitor ETP-47037 administered orally, after which the mice were euthanized for analysis of reprogramming. As expected, we observed multifocal dysplasia in several organs, such as pancreas, stomach, and large intestine, in the absence of the inhibitor (Figure 5A, upper panel). Interestingly, the percentage of the area of the tissues showing dysplasia (as readout for reprogramming) was clearly reduced in mice treated with TRF1 inhibitor (Figure 5A, lower panel). We found that in reprogrammable mice induced under these conditions (1 week with high doxycycline), the large intestine presented the higher percentages of dysplastic areas, which were significantly reduced in mice treated with TRF1 inhibitor (Figure 5B). These findings suggest that blocking TRF1 may be impairing in vivo reprogramming. We recently showed that both genetic and chemical TRF1 downregulation did not induce changes in telomere length in mouse tumors (Garcia-Beccaria et al., 2015). To address whether chemical inhibition of TRF1 affects telomere length in the reprogrammed tissues, we performed an immuno-telomere Q-FISH with c-MYC antibodies (as a marker for dysplastic areas) to determine telomere length in both the dysplastic and non-dysplastic cells from the pancreas of both i4F and eGFP-TRF1+/KI i4F induced in the presence or absence of the TRF1 inhibitor ETP-47037 (Figures 5C and 5D). We found that chemical inhibition of TRF1 during in vivo reprogramming did not affect the telomere length of normal non-dysplastic cells of the pancreas (Figure 5C). Interestingly, we observed that dysplastic cells already show (1 week after induction) a moderate increase of telomere length when compared with normal cells of the same tissue (Figure 5D), and that chemical inhibition of TRF1 during the induction of in vivo reprogramming does not alter this telomere lengthening in the reprogrammed areas (Figure 5D). Altogether these results confirm that chemical TRF1 inhibition does not change telomere length (Garcia-Beccaria et al., 2015) and demonstrate that the decreased reprogramming efficiency observed by chemical inhibition of TRF1 during the induction of in vivo reprogramming is not due to the presence of shorter telomeres.
Figure 5.
In Vivo Treatment with TRF1 Inhibitor ETP-47037 Reduces Dysplasia during In Vivo Reprogramming without Affecting Telomere Length
(A) Representative images of pancreas, stomach, and large intestine from eGFP-TRF1+/KIi4F mice in vivo treated with vehicle (top) or ETP-47037 (bottom) compound during induction of in vivo reprogramming for 7 days with high doxycycline. Arrowheads point to foci of dysplasia. Treatment with TRF1 inhibitor reduces the dysplasia associated with in vivo reprogramming. Scale bars, 100 μm.
(B) Quantification of the percentage of dysplastic area in the large intestine of i4F mice treated with vehicle or TRF1 inhibitor ETP-47037. Note the strong reduction in the affected area in mice treated with ETP-47037. Error bars denote SE. Statistical analysis by Student's t test. n, number of mice analyzed in each group.
(C) Quantification of telomere length in normal non-dysplastic cells of pancreas from induced eGFP-TRF1+/KIi4F mice and i4F mice. Inhibition of TRF1 does not alter telomere length in normal tissue. Error bars denote SE. Statistical analysis by Student's t test. n, number of nuclei. Number of mice analyzed in each group = 2. n.s., not significant.
(D) Quantification of telomere elongation of dysplastic cells, compared with non-dysplastic cells from the same tissue, in mice treated with vehicle or TRF1 inhibitor ETP-47037. Inhibition of TRF1 does not change the telomere length increase of the dysplastic cells. Error bars denote SE. Statistical analysis by Student's t test. n, number of mice analyzed in each group. n.s., not significant.
In Vivo Reprogramming Involves Drastic Changes in Heterochromatin Architecture
In the course of these experiments we repetitively observed that the nucleus of the in vivo reprogrammed cells showed a weaker DAPI staining than the non-reprogrammed cells in the same tissue (Figures 1B, 1C, 3A, 3B, 4A, 4B, and S3A–S3C). Since DAPI-intense staining is associated with heterochromatin regions, this could be indicative of a less compacted chromatin. Indeed, differentiated cells present densely packed constitutive heterochromatin, while embryonic stem cells (ESCs) and iPSCs show a dispersed chromatin, suggesting that dispersed chromatin may be a hallmark of pluripotency (Fussner et al., 2010). In fact, chromatin undergoes large-scale epigenetic resetting during in vitro reprogramming, leading to a more open chromatin structure (Meshorer et al., 2006), which may be fundamental for the acquisition of pluripotency. Indeed, a decrease in both H3K9me3 and H4K20me3 heterochromatin marks has been described both in iPSCs and ESCs, which also affects the telomeric chromatin (Marion et al., 2009b, Varela et al., 2011). As a more relaxed telomeric chromatin has been described to favor telomere elongation (Blasco, 2007a, Marion et al., 2011), a less compacted heterochromatin during in vivo reprogramming could facilitate the observed telomere elongation in reprogrammed cells (Figures 1B and 1C).
Thus, here we studied whether in vivo reprogrammed areas show decreased histone heterochromatic marks. To do so, we performed double immunofluorescence against OCT4 and the heterochromatic marks H4K20me3 and H3K9me3 (Figures S4A and S4B) in the large intestine of induced i4F reprogrammable mice. We observed a drastic reduction in both marks in the dedifferentiated cells when compared with non-reprogrammed control cells from the same tissue (Figures S4A and S4B), thus confirming that reprogramming in vivo involves profound changes in heterochromatin structure.
In mouse interphase nuclei, centromeric constitutive heterochromatin is detected as DAPI-dense large spots marked by H3K9me3 and H4K20me3 (Guenatri et al., 2004). Interestingly, both ESCs (Meshorer et al., 2006) and iPSCs show decreased centromeric heterochromatin marks (Fussner et al., 2010). Thus, we studied whether centromeric heterochromatin was altered during tissue dedifferentiation upon in vivo reprogramming. To this end, we performed an immuno-FISH using an antibody against OCT4 and a probe specific for major satellite repeats (Figure S4C). Major satellite regions in differentiated cells were observed as dense spots, which correspond to DAPI-intense areas (Figure S4C). Remarkably, dedifferentiated OCT4-positive cells showed a drastic disruption of the centromeric clusters (Figure S4C, zoomed area), and present a less compacted pattern of heterochromatin, consistent with a more open chromatin structure that is characteristic of pluripotent cells.
In Vivo Reprogramming Results in Decreased Global Expression of the SA1 Cohesin
In addition to its role in sister chromatid cohesion, cohesin is an important player in the organization of interphase chromatin (Kagey et al., 2010). Somatic cells have two versions of cohesin with three common subunits RAD21/SCC1, SMC1, and SMC3, and either SA1 or SA1 subunits (Sumara et al., 2000). SA1 mediates telomere cohesion and contributes to cohesion along chromosome arms (Remeseiro et al., 2012). The telomeric proteins TIN2 and TRF1 associate in human cells specifically with cohesin SA1, which plays an important role in efficient telomere replication (Remeseiro et al., 2012). SA1 is also a major player in the control of architectural organization of chromatin loops that leads to alteration of the transcriptional profile (Kagey et al., 2010). In view of the upregulated TRF1 levels in the in vivo reprogrammed areas, we studied whether tissue dedifferentiation also leads to changes in SA1 expression and/or localization. To this end, we performed triple immunofluorescence against OCT4, TRF1, and SA1 in the pancreas of i4F reprogrammable mice. We observed that in vivo reprogrammed areas show decreased expression of SA1 protein coincidental with increased TRF1 expression (Figure S5), suggesting that the telomere changes associated to tissue dedifferentiation are also accompanied by lower expression of SA1 protein, which in turn can alter chromatin architecture and transcriptional control of gene expression.
Taken together, these data indicate drastic chromatin reorganization processes during tissue dedifferentiation upon reprogramming in vivo that could facilitate telomere elongation. This poses the interesting question of whether these changes may also be of relevance in other processes associated with cellular plasticity and dedifferentiation, such as tissue regeneration and cancer.
Telomere Changes during In Vivo Dedifferentiation Associated with Early Neoplastic Lesions
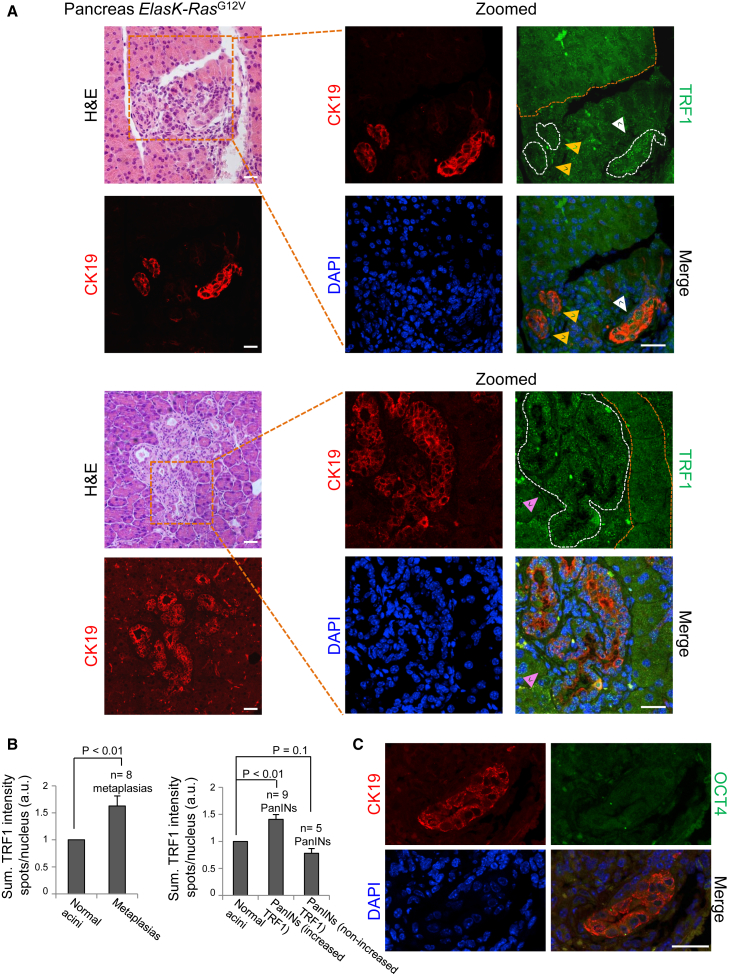
As loss of a defined differentiated state is emerging as a critical early step during cancer initiation (Puri et al., 2015, Roy and Hebrok, 2015), we studied whether telomeric events associated with cellular change identity, in particular TRF1 overexpression and telomere elongation, were present in plastic processes associated with the onset of cancer. To do so, we used a mouse model for pancreatic ductal adenocarcinoma (PDAC) (Guerra et al., 2007). Loss of cellular identity is a critical step in the initiation of PDAC (Puri et al., 2015). PDAC originates from a process known as ADM that involves transdifferentiation of adult acinar cells into ductal-like cells as a result of K-Ras oncogene expression, which can subsequently progress to malignant adenocarcinoma through a series of histopathological lesions known as pancreatic intraepithelial neoplasias (PanINs) (Maitra and Hruban, 2008). Mice K-Ras+/LSLG12Vgeo;Elas-tTA/tetO-Cre (ElasK-RasG12V), in which expression of their K-RasG12V oncogenic allele is turned on during late embryonic development in acinar cells, develop ADM and PanIN lesions that immunostain for CK19, confirming their ductal nature (Guerra et al., 2007). Thus, these mice provide an excellent model for the study of TRF1 changes during the cell fate changes that take place during cancer initiation. To determine whether TRF1 levels were altered during acinar-to-ductal transdifferentiation, we performed double immunofluorescence against TRF1 and CK19 (as a marker for ductal-like nature) in sections of pancreas from ElasK-RasG12V mice in which ADM and/or PanINs had been localized by histopathological analysis (Figure 6A, left). We found a significant increase in TRF1 levels in all the ADMs when compared with the normal acinar cells (orange dashed lines) from the same tissue (Figure 6A, white arrowhead, and Figure 6B, left). Interestingly, in the case of the PanINs, which constitute more advanced lesions, 64% of the PanINs analyzed showed increased TRF1 expression when compared with the normal acinar cells (orange dashed lines), similar to that of the metaplasias (Figure 6A, pink arrowhead and Figure 6B, right), while 36% of them did not (Figure 6A, yellow arrowheads and Figure 6B, right). These results confirm that TRF1 expression is increased during the early events of the acinar-to-ductal transdifferentiation associated to the onset of pancreatic cancer, and suggests that TRF1 overexpression could be a common feature of the initiation steps of different plastic processes.
Figure 6.
Increased TRF1 Expression in Acinar-to-Ductal Metaplasias and PanINs of ElasK-RasG12V Mice
(A) Left: representative images of pancreatic lesions (metaplasias and PanINs) of ElasK-RasG12V mice. Serial sections of pancreas from ElasK-RasG12V mice were stained with H&E and immunostained with cytokeratin 19 (CK19). Right: zoomed panel of the selected areas showing double immunofluorescence against CK19 (red) and TRF1 (green). White dashed lines mark pancreatic lesions. Orange dashed lines mark normal acinar cells used as control. Elevated TRF1 protein expression when compared with normal acinar cells can be detected in all the metaplasias analyzed (white arrowhead) and in most of the PanINs (pink arrowhead). A fraction of the PanINs analyzed did not show increased TRF1 levels (yellow arrowheads). Scale bars, 25 μm.
(B) Quantification of TRF1 expression in acinar-to-ductal metaplasias and PanINs and their corresponding normal acinar control cells. TRF1 levels are significantly elevated in metaplasias (left) and in a fraction of the PanINs analyzed (right). Error bars denote SE. Statistical analysis by Student's t test. n, number of pancreatic lesions analyzed. Number of mice analyzed = 4.
(C) Double immunofluorescence against CK19 and OCT4 in a pancreatic lesion expressing elevated levels of TRF1 (see A). OCT4 expression is not detected in the dedifferentiated acinar cells. Scale bars, 25 μm.
See also Figure S6.
Along the ADM dedifferentiation process, acinar cells increase expression of factors expressed in progenitor cells present during pancreatic development (Puri et al., 2015), supporting the idea that adult acinar cells retain the potential to revert to progenitor-like cells with multipotent capacities. We wondered whether ductal-like cells showing elevated TRF1 levels would also express the pluripotency marker OCT4, whose expression corresponds to high TRF1 levels in reprogramming-induced dedifferentiation (Figures 4A and 4B). To this end, we performed double immunofluorescence against CK19 and OCT4 in serial sections from the pancreas of ElasK-RasG12V mice analyzed in Figures 6A and 6B. We observed that OCT4 could not be detected in the pancreatic lesions (Figure 6C), even in those where high TRF1 expression was shown before (see Figure 6A), suggesting that other transcription factors regulate TRF1 expression during the acinar-to-ductal dedifferentiation process.
Finally, we addressed whether increased TRF1 expression in pancreatic lesions was uncoupled from telomere elongation. To this end, we performed Q-FISH of the different lesions. We found that 28% of the lesions showed clear telomere elongation compared with the normal tissue, while in the rest we observed either the same telomere length or shorter telomeres than the surrounding tissue (Figures S6A and S6B). It is interesting to note that some lesions with increased TRF1 expression presented shorter telomeres while others were associated with longer telomeres (Figure S6A). Thus, TRF1 upregulation in early stages of pancreatic cancer is uncoupled from telomere elongation. Interestingly, although a majority of the lesions showed shorter telomeres than the normal surrounding tissue, possibly as the consequence of telomere shortening associated with oncogene-induced cellular proliferation, the remaining 28% showed clear telomere elongation, anticipating the known telomerase upregulation associated with tumorigenesis (Martinez and Blasco, 2011).
Discussion
Here we describe dramatic changes at telomeres during dedifferentiation of mouse adult tissues by induction of in vivo reprogramming, including upregulation of telomerase RNA expression, telomerase-dependent telomere elongation, and upregulation of the TRF1 telomere protein. In addition, we describe dramatic changes in the global structure of chromatin, including decreased heterochromatic marks and decreased expression of the SA1 telomeric cohesin. As increased cellular plasticity and dedifferentiation are also proposed to be important during tissue regeneration as well as in pathological conditions such as cancer, our findings suggest that similar changes at telomeres and global chromatin could also underlie these processes. In support of this notion, we extend the finding of TRF1 upregulation to tissue dedifferentiation during early neoplastic lesions, in a mouse model of pancreatic cancer induced by expression of oncogenic K-Ras. Interestingly, we find increased expression of TRF1 uncoupled from telomere elongation during the acinar-to-ductal transdifferentiation, a process that leads to the initiation of PDAC. Moreover, although a majority of the lesions showed shorter telomeres than the normal surrounding tissue, the remaining 28% showed telomere elongation, in a manner similar to that of in vivo reprogramming-induced dedifferentiation, anticipating the known telomerase upregulation associated with tumorigenesis (Martinez and Blasco, 2011). Thus, here we show analogous changes at telomeres associated with dedifferentiation induced by either in vivo reprogramming or initiation of cancer.
Remarkably, the changes described suggest that in vivo reprogramming-induced dedifferentiation renders a change at telomeres toward a more “juvenile” state, which in turn resembles that of ESCs and adult stem cells (see Figure 7). In this regard, we have previously described telomerase-mediated telomere elongation during induction of skin and hair regeneration processes, which is essential to maintain skin homeostasis (Flores et al., 2005, Flores et al., 2008). In addition, we have previously shown that TRF1 is highly upregulated in mouse adult stem cell compartments, being also essential for tissue renewal and maintenance of tissue homeostasis in skin, gut, bone marrow, and lung (Beier et al., 2012, Martinez et al., 2009, Povedano et al., 2015, Schneider et al., 2013), thus highlighting the similarities between processes associated with normal tissue regeneration and in vivo reprogramming. While TRF1 is highly upregulated in ESCs and iPSCs and this upregulation seems to be linked to OCT4 (Schneider et al., 2013), the mechanisms by which TRF1 is upregulated in adult stem cell compartments are completely unknown. In particular, OCT4 has been shown to bind the Trf1 promoter of both in vitro cultured ESCs and iPSCs, and to be sufficient to induce TRF1 expression in differentiated cells (Schneider et al., 2013). Indeed, TRF1 upregulation coincides with OCT4 expression during the generation of iPSCs in vitro (Schneider et al., 2013) as well as during in vivo reprogramming (this article). This poses the interesting question of whether OCT4 may also be responsible for maintaining high TRF1 expression levels and adult stem cell functionality in the context of normal physiology. Finally, the fact that TRF1 is highly upregulated during in vivo reprogramming of adult tissues and during the transdifferentiation of acinar cells to ductal-like cells opens the intriguing possibility that TRF1 upregulation may be a common event in the early steps of different processes associated with induction of tissue plasticity and cell fate change, such as tissue repair or tumorigenesis. In the case of early stages of tumorigenesis, however, we demonstrated here that TRF1 upregulation occurs in the absence of OCT4 expression, opening the interesting possibility that alternative mechanisms and factors could be involved in activation of TRF1 expression in different processes involving cellular plasticity.
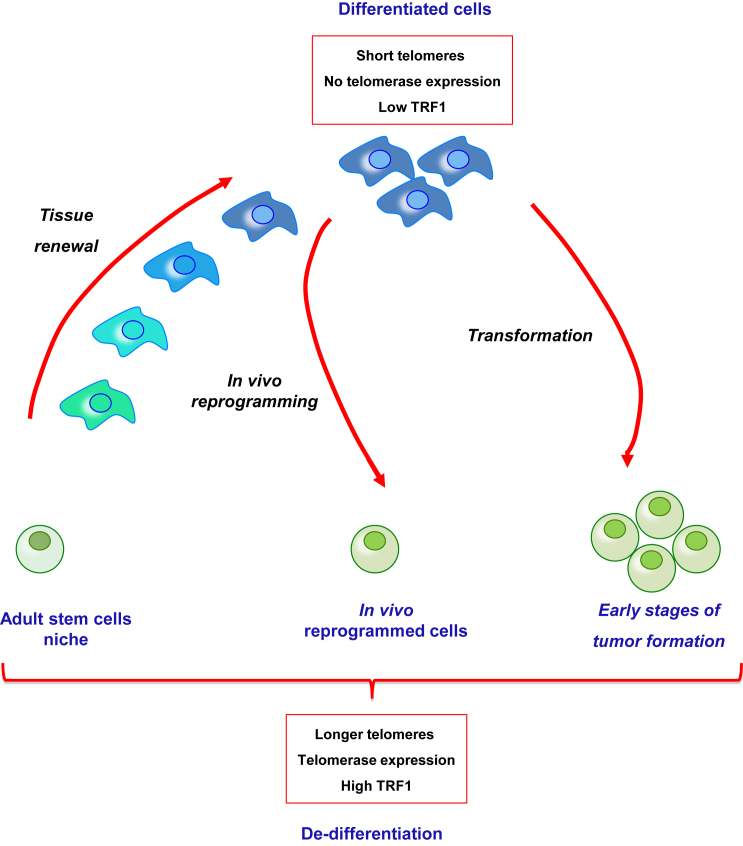
Figure 7.
Model for the Role of In Vivo Reprogramming in Tissue Repair and Cancer Initiation
Adult stem cells within a tissue show the longest telomeres, telomerase activity, and high levels of TRF1 protein. As they mobilize for tissue renewal, telomeres shorten, telomerase activity is lost, and TRF1 expression diminishes. In vivo reprogramming reverses these features generating dedifferentiated cells with longer telomeres, telomerase expression, and higher levels of TRF1. The similarities of the in vivo reprogrammed cells to the adult stem cells suggest the possibility that reprogrammed cells could have a role in tissue regeneration. Other cellular plastic processes, such as dedifferentiation associated with initiation of tumorigenesis, also show increased levels of TRF1 protein and elongation of telomeres, indicating a general role of these telomeric events in processes associated with cell fate change.
In vivo reprogramming-induced tissue dedifferentiation resulted in upregulation of telomerase expression and telomerase-dependent telomere elongation. Again, this is in analogy with the fact that stem cell compartments have increased telomerase activity and longer telomeres compared with the more differentiated compartments (Flores et al., 2008, Schneider et al., 2013) (Figure 7). Indeed, maintaining a sufficient telomere reserve is essential for proper adult stem cell mobilization and tissue renewal (Armanios and Blackburn, 2012, Blasco, 2007b, Flores et al., 2005). Again, the mechanisms by which telomerase expression and telomere length are regulated in these stem cell compartments are currently unknown. It has been shown that OCT4 binds to the TERC locus in human iPSCs as well as in murine ESCs (Agarwal et al., 2010). These results support the interesting possibility that OCT4 could be involved in upregulation of TERC during in vivo reprogramming. In this regard, telomerase upregulation and telomerase-dependent telomere elongation during in vivo reprogramming may shed light on the mechanism that regulates telomerase activity and telomere length in adult stem cell compartments.
We found here that in vivo reprogramming and the subsequent dedifferentiation of adult cell types involves drastic changes in heterochromatin architecture leading to a more open chromatin, which has been associated with acquisition of pluripotency during in vitro reprogramming (Meshorer et al., 2006). We find this fact particularly interesting, as a more relaxed telomeric chromatin has been described to favor telomere elongation, also in the context of in vitro generation of iPSCs (Blasco, 2007a, Marion et al., 2011). In particular, we described a dramatic decrease in the H3K9me3 and H4K20me3 marks at telomeric chromatin during in vitro generation of iPSCs (Marion et al., 2009b). Thus it is tempting to speculate that the telomerase-mediated telomere elongation that we observe during in vivo reprogramming may be facilitated by the global loss in heterochromatic marks, which have been previously described as repressors of telomere elongation (Blasco, 2007a, Marion et al., 2011).
Upregulation of the TRF1 protein and telomere elongation in in vivo reprogrammed areas associates with a weaker detection of cohesion component SA1 protein. SA1 is involved in telomere cohesion and replication, and has been detected bound to TRF1 protein in human cells (Remeseiro et al., 2012). Moreover, SA1 is emerging as a major player in chromatin folding and subsequent regulation of global transcription. Therefore, it is tempting to speculate that SA1 protein constitutes a bridge between the observed changes in telomeres during in vivo reprogramming and the global changes in chromatin structure and transcriptional regulation associated with acquisition of pluripotency.
In summary, the results presented here provide mechanistic insights involving important telomere changes in the reprogramming of cell fate in vivo. We have demonstrated that pathological cellular processes involving in vivo dedifferentiation and change of cell fate, such as tumorigenesis, show telomere changes resembling those of in vivo reprogramming. These findings also allow for a deeper understanding of the role of telomeres in other processes characterized by tissue dedifferentiation and increased plasticity, such as tissue regeneration or tumorigenesis.
Experimental Procedures
Animal Procedures
Animal experimentation at the Spanish National Cancer Research Center (CNIO) was performed according to protocols approved by the CNIO-ISCIII Ethics Committee for Research and Animal Welfare (CEIyBA). Doxycycline was administered at 0.2 mg/mL in the drinking water supplemented with 7.5% sucrose. Reprogramming experiments were performed with mice of both sexes and from 2 to 10 months of age. For in vivo reprogramming in the presence of the TRF1 inhibitor ETP-47037 (Garcia-Beccaria et al., 2015), eGFP-TRF1+/KI i4F and i4F reprogrammable mice were daily dosed orally with 75 mg/kg ETP-47037 formulated in 10% N-methylpyrrolidone and 90% polyethylene glycol 300 (or just vehicle as controls) for 7 days, while doxycycline was administered at 1 mg/mL in the drinking water supplemented with 7.5% sucrose. Pancreases were obtained from K-Ras+/LSLG12Vgeo;Elas-tTA/tetO-Cre mice from 6 weeks to 1 year old.
Immunofluorescence
Tissue samples were fixed in 10% formalin, paraffin embedded, and cut in 2.5-μm sections, then mounted in Superfrostplus portaobjects. Immunofluorescence was performed on deparaffinized samples processed for antigen retrieval and permeabilized with 0.5% Triton X-100 in PBS for 2 hr at room temperature. Samples were blocked in PBS with 4% BSA for 1 hr and incubated overnight at 4°C with the corresponding primary antibody. Antibodies used were OCT4 (Abcam, ab19857), NANOG (Cell Signaling Technology, 8822), rat CK19 (CNIO Monoclonal Antibodies Core Unit, AM-TROMA III), rabbit polyclonal antibody against TRF1 (raised in our laboratory against full-length mouse TRF1 protein), rat antibody against TRF1 (raised in our laboratory against full-length mouse TRF1 protein), H4K20me3 (Upstate, 07-749), H3K9me3 (Upstate, 07-442), and rabbit polyclonal sera against SA1 (kindly provided by Dr. A. Losada). Slides were washed for 3 × 10 min with PBS with 0.1% Tween 20 and incubated with the corresponding secondary antibody for 1 hr at room temperature. Secondary antibodies used were Alexa Fluor 488 anti-rabbit and Alexa Fluor 647 anti-rat. Samples were washed for 3 × 10 min with PBS with 0.1% Tween 20 and incubated for 2 hr at room temperature with the second primary antibody, directly labeled with the Zenon Alexa Fluor 555 Rabbit IgG Labeling Kit (ThermoFisher Scientific, Z-25305) following the manufacturer's recommendations. Samples were washed for 3 × 10 min with PBS with 0.1% Tween 20 and fixed for 15 min in 4% paraformaldehyde in PBS. Nuclei were counterstained in a 4-μg/mL DAPI/PBS solution before mounting with Vectashield.
Immunofluorescence Followed by FISH
Immunofluorescence was performed as described above. Primary antibodies used were OCT4 antibody (Abcam, ab19857), rabbit c-Myc antibody (Abcam, ab32072), or rat CK19 (CNIO Monoclonal Antibodies Core Unit, AM-TROMA III). After immunofluorescence, slides were fixed for 20 min in 4% paraformaldehyde in PBS. Quantitative FISH was performed as described previously (Gonzalo et al., 2006) with some modifications: samples were not treated with pepsin and were subjected directly to dehydration steps, formamide concentration during incubation with the probe and washes was reduced to 50%, and incubation of the sample with the probe was reduced to 30 min. Telomere or centromere (major satellite) PNA probes labeled with CY3 (Panagene) were used. Nuclei were counterstained in a 4-μg/mL DAPI/PBS solution before mounting with Vectashield.
Author Contributions
M.A.B. secured funding, conceived the project, designed experiments, and wrote the manuscript. R.M.M. designed experiments, performed most of the experiments, and wrote the manuscript. I.L.de.S. performed RNA-FISH experiments. C.G., L.M., and M.A. provided tissue samples and technical advice. B.G. aided with mice procedures and immunofluorescence. D.M. helped with confocal microscopy and performed the Definiens analysis. M.S. contributed with intellectual input.
Acknowledgments
We are indebted to R. Serrano for expert mouse colony management and the Comparative Pathology Unit at CNIO for technical assistance. We are grateful to Dr. Ana Losada for providing us with SA1 antibody. We thank Ana Carolina Moises da Silva for her assistance in setting-up RNA-FISH experiments. Work in the laboratory of M.A.B. is funded by the Spanish Ministry of Economy and Competiveness (PLAN RETOS SAF2013-45111-R) and by the Fundación Botín.
Published: February 2, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.01.001.
Supplemental Information
References
- Abad M., Mosteiro L., Pantoja C., Canamero M., Rayon T., Ors I., Grana O., Megias D., Dominguez O., Martinez D. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- Agarwal S., Loh Y.H., McLoughlin E.M., Huang J., Park I.H., Miller J.D., Huo H., Okuka M., Dos Reis R.M., Loewer S. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M., Blackburn E.H. The telomere syndromes. Nat. Rev. Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier F., Foronda M., Martinez P., Blasco M.A. Conditional TRF1 knockout in the hematopoietic compartment leads to bone marrow failure and recapitulates clinical features of dyskeratosis congenita. Blood. 2012;120:2990–3000. doi: 10.1182/blood-2012-03-418038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- Blasco M.A. Telomere length, stem cells and aging. Nat. Chem. Biol. 2007;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Lee H.W., Hande M.P., Samper E., Lansdorp P.M., DePinho R.A., Greider C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Flores I., Cayuela M.L., Blasco M.A. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- Flores I., Benetti R., Blasco M.A. Telomerase regulation and stem cell behaviour. Curr. Opin. Cell Biol. 2006;18:254–260. doi: 10.1016/j.ceb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Flores I., Canela A., Vera E., Tejera A., Cotsarelis G., Blasco M.A. The longest telomeres: a general signature of adult stem cell compartments. Genes Dev. 2008;22:654–667. doi: 10.1101/gad.451008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussner E., Ahmed K., Dehghani H., Strauss M., Bazett-Jones D.P. Changes in chromatin fiber density as a marker for pluripotency. Cold Spring Harb. Symp. Quant. Biol. 2010;75:245–249. doi: 10.1101/sqb.2010.75.012. [DOI] [PubMed] [Google Scholar]
- Garcia-Beccaria M., Martinez P., Mendez-Pertuz M., Martinez S., Blanco-Aparicio C., Canamero M., Mulero F., Ambrogio C., Flores J.M., Megias D. Therapeutic inhibition of TRF1 impairs the growth of p53-deficient K-RasG12V-induced lung cancer by induction of telomeric DNA damage. EMBO Mol. Med. 2015;7:930–949. doi: 10.15252/emmm.201404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S., Jaco I., Fraga M.F., Chen T., Li E., Esteller M., Blasco M.A. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Guenatri M., Bailly D., Maison C., Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C., Schuhmacher A.J., Canamero M., Grippo P.J., Verdaguer L., Perez-Gallego L., Dubus P., Sandgren E.P., Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Bailey S.M., Okuka M., Munoz P., Li C., Zhou L., Wu C., Czerwiec E., Sandler L., Seyfang A. Telomere lengthening early in development. Nat. Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- Maitra A., Hruban R.H. Pancreatic cancer. Annu. Rev. Pathol. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion R.M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion R.M., Strati K., Li H., Tejera A., Schoeftner S., Ortega S., Serrano M., Blasco M.A. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Marion R.M., Schotta G., Ortega S., Blasco M.A. Suv4-20h abrogation enhances telomere elongation during reprogramming and confers a higher tumorigenic potential to iPS cells. PLoS One. 2011;6:e25680. doi: 10.1371/journal.pone.0025680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P., Blasco M.A. Role of shelterin in cancer and aging. Aging Cell. 2010;9:653–666. doi: 10.1111/j.1474-9726.2010.00596.x. [DOI] [PubMed] [Google Scholar]
- Martinez P., Blasco M.A. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- Martinez P., Thanasoula M., Munoz P., Liao C., Tejera A., McNees C., Flores J.M., Fernandez-Capetillo O., Tarsounas M., Blasco M.A. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povedano J.M., Martinez P., Flores J.M., Mulero F., Blasco M.A. Mice with pulmonary fibrosis driven by telomere dysfunction. Cell Rep. 2015;12:286–299. doi: 10.1016/j.celrep.2015.06.028. [DOI] [PubMed] [Google Scholar]
- Puri S., Folias A.E., Hebrok M. Plasticity and dedifferentiation within the pancreas: development, homeostasis, and disease. Cell Stem Cell. 2015;16:18–31. doi: 10.1016/j.stem.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S., Cuadrado A., Carretero M., Martinez P., Drosopoulos W.C., Canamero M., Schildkraut C.L., Blasco M.A., Losada A. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N., Hebrok M. Regulation of cellular identity in cancer. Dev. Cell. 2015;35:674–684. doi: 10.1016/j.devcel.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R.P., Garrobo I., Foronda M., Palacios J.A., Marion R.M., Flores I., Ortega S., Blasco M.A. TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nat. Commun. 2013;4:1946. doi: 10.1038/ncomms2946. [DOI] [PubMed] [Google Scholar]
- Sumara I., Vorlaufer E., Gieffers C., Peters B.H., Peters J.M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata P.R., Mou H., Pardo-Saganta A., Zhao R., Prabhu M., Law B.M., Vinarsky V., Cho J.L., Breton S., Sahay A. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela E., Schneider R.P., Ortega S., Blasco M.A. Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc. Natl. Acad. Sci. USA. 2011;108:15207–15212. doi: 10.1073/pnas.1105414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K., Zong Y., Maggs L.R., Shapira S.N., Maddipati R., Aiello N.M., Thung S.N., Wells R.G., Greenbaum L.E., Stanger B.Z. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.