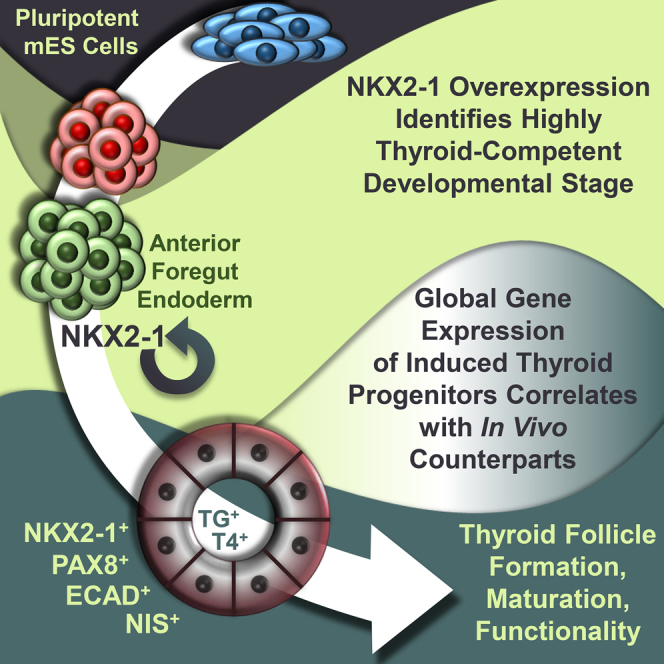
Summary
The clinical importance of anterior foregut endoderm (AFE) derivatives, such as thyrocytes, has led to intense research efforts for their derivation through directed differentiation of pluripotent stem cells (PSCs). Here, we identify transient overexpression of the transcription factor (TF) NKX2-1 as a powerful inductive signal for the robust derivation of thyrocyte-like cells from mouse PSC-derived AFE. This effect is highly developmental stage specific and dependent on FOXA2 expression levels and precise modulation of BMP and FGF signaling. The majority of the resulting cells express thyroid TFs (Nkx2-1, Pax8, Foxe1, Hhex) and thyroid hormone synthesis-related genes (Tg, Tpo, Nis, Iyd) at levels similar to adult mouse thyroid and give rise to functional follicle-like epithelial structures in Matrigel culture. Our findings demonstrate that NKX2-1 overexpression converts AFE to thyroid epithelium in a developmental time-sensitive manner and suggest a general methodology for manipulation of cell-fate decisions of developmental intermediates.
Keywords: mouse embryonic stem cell, pluripotent stem cell, directed differentiation, thyroid development, Nkx2-1, transcription factor, overexpression, progenitor specification, foregut, inducible
Graphical Abstract
Highlights
-
•
Transient Nkx2-1 overexpression robustly converts foregut endoderm to thyroid lineage
-
•
The effect is highly developmental stage specific and dependent on BMP/FGF signals
-
•
The progeny self-organizes to 3D structures reminiscent of follicular thyroid tissue
-
•
Mature cultures produce T4 hormone at levels comparable with mouse thyroid tissue
Ikonomou and colleagues found that pluripotent stem cell-derived anterior foregut endoderm can be robustly converted to thyroid progenitors through transient expression of the transcription factor Nkx2-1. This inductive effect is highly stage specific and relies on thyroid specification signals. The thyrocyte-like progeny self-organize into follicle structures in 3D culture and express mature thyroid proteins indicative of functionality (TG, NIS, T4).
Introduction
Since tissue progenitors, giving rise to all mature cell types within a given tissue, are essential intermediates in embryonic development, their in vitro derivation has important implications for the fields of pluripotent stem cell (PSC) biology and regenerative medicine. Significant advances in anterior foregut endoderm (AFE) progenitor biology in recent years (Ikonomou and Kotton, 2015) have led to derivation of AFE lung and thyroid progenitors (TPs) and their clinically relevant progeny from PSCs (Green et al., 2011, Huang et al., 2014, Kurmann et al., 2015, Longmire et al., 2012, Mou et al., 2012). However, with one notable exception (Huang et al., 2014), efficiencies of progenitor derivation have been relatively low (<40%).
Overexpression of transcription factors (TFs) is a well-established approach to manipulate cellular identities as it results in reconfiguration or emergence of core TF networks, with derivation of induced PSCs from somatic cells being the most prominent example (Takahashi and Yamanaka, 2006). Inducible TF expression in PSCs has been used to potentiate the derivation of specific lineages and facilitate mechanistic understanding of cell specification (Bondue et al., 2008, Mazzoni et al., 2013, Petros et al., 2013, Seguin et al., 2008). For example, Costagliola and coworkers (Antonica et al., 2012) used forced overexpression of the thyroid TFs NKX2-1/PAX8 in mouse embryonic stem cells (mESCs) to produce thyrocyte-like cells with high efficiency (∼60%) that formed in vitro follicular structures and rescued athyroid mice upon transplantation. However, studies that systematically investigate the mechanistic interplay between pulsed heterologous TF expression and developmental stages in directed differentiation of PSCs are lacking. To address this question, we used thyroid-directed differentiation (Kurmann et al., 2015) in combination with transient NKX2-1 overexpression as our model system.
Here, we report that transient expression of NKX2-1 functions as an inductive signal during thyroid-directed differentiation to convert AFE-stage cells to thyrocyte-like cells that self-organize to epithelial, follicle-like structures in 3D Matrigel culture. This thyroid conversion effect pertains only to a narrow developmental window of competence contingent on several parameters, including dual bone morphogenetic protein (BMP)/fibroblast growth factor (FGF) signaling and correct anterior patterning of definitive endoderm (DE). We employ emerging computational methods (linear algebra projections; Lang et al., 2014, Pusuluri et al., 2015) and genome-wide gene expression analysis by RNA-sequencing (RNA-seq) to demonstrate that the resulting cells are similar to mouse embryonic thyrocytes. Finally, we suggest using mathematical modeling that the induction effect is potentially governed by a time-dependent bistable switch.
Results
Efficient Temporal Control of Dox-Inducible Nkx2-1 Transgene Expression
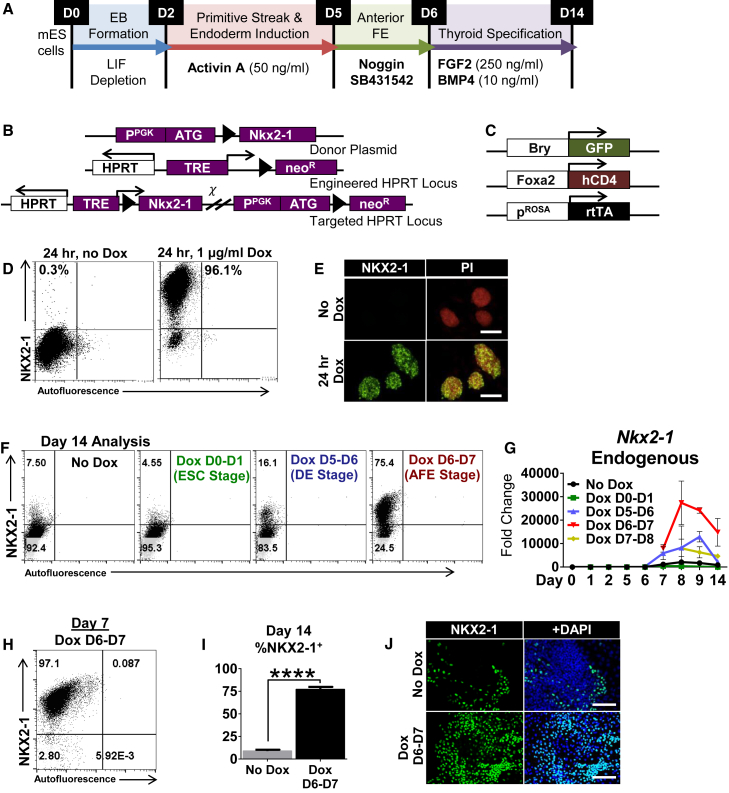
We hypothesized that transient, temporally regulated Nkx2-1 overexpression during directed differentiation of mESCs would lead to distinct differentiation outcomes due to differential competence of in vitro developmental stages. We created an mESC line (iNkx2-1) with a doxycycline (Dox)-inducible Nkx2-1 transgene (Figures 1B, 1C, S1A, and S1B) (Ting et al., 2005). This line was previously targeted with knockin reporters for Foxa2 (Foxa2hCD4) and T (BryGFP) (Gadue et al., 2006). The iNkx2-1 line displayed rapid on/off kinetics in response to Dox (Figures 1D, 1E, and S1C–S1E). Neither the transgene addition nor Dox affected pluripotency or standard directed differentiation (Figures S1F and S1G). This system allowed for efficient manipulation of NKX2-1 expression, resulting in robust induction of the Nkx2-1 transgene (>95% NKX2-1+ cells by flow cytometry) following 24-hr exposure to Dox (1 μg/ml).
Figure 1.
Stage-Specific Effect of Nkx2-1 Overexpression on Derivation of NKX2-1+ Progenitors
(A) Directed differentiation protocol for NKX2-1+ TPs.
(B) Integration schematic of the Nkx2-1 transgene into the HPRT locus.
(C) Schematic of the knockin reporters (BrachyuryGFP and Foxa2hCD4) and rtTA engineered into the iNkx2-1 line.
(D) Intracellular flow cytometry for NKX2-1 in undifferentiated cells with and without 24 hr of Dox treatment.
(E) Immunostaining of undifferentiated iNkx2-1 cells post-24-hr Dox; nuclear counterstain with propidium iodide (PI). Scale bars represent 100 μm.
(F) Intracellular NKX2-1 flow cytometry plots from D14 following 24-hr pulses of Dox added at indicated intermediate stages. Representative of three differentiations.
(G) Kinetics of endogenous Nkx2-1 expression by RT-qPCR following 24-hr staged pulses of Dox. Fold changes relative to undifferentiated cells, error bars represent SD (n = 3 wells from same differentiation). Representative of three independent experiments.
(H) Representative flow cytometry plot of NKX2-1 expression directly post-24-hr Dox.
(I) Averaged flow cytometry data (n = 12 independent experiments) comparing untreated (no Dox) and treated (Dox D6–D7) samples. Error bars represent SEM. ∗∗∗∗p < 0.0001.
(J) Representative immunostaining of Dox-induced and uninduced cultures at D14. Scale bars represent 100 μm.
See also Figure S1.
Stage-Specific Effect of Transient NKX2-1 Overexpression at the AFE Stage Results in Efficient Lineage Conversion
To test the temporal effects of Nkx2-1 overexpression on thyroid derivation, we employed our iNkx2-1 cell line in combination with our thyroid differentiation protocol (Kurmann et al., 2015) (Figure 1A), which recapitulates key stages of thyroid development. In short, DE is induced by Activin A (Kubo et al., 2004), followed by further patterning to AFE (anteriorization) (Green et al., 2011, Longmire et al., 2012) using brief dual BMP/transforming growth factor β (TGF-β) inhibition through Noggin/SB431542 (NS) treatment, and specification to NKX2-1+ TPs with BMP4 and FGF2 signaling. While this protocol is effective and reproducible, the yield is generally low (up to 25% NKX2-1+ cells) with a fraction (∼5%) coexpressing PAX8 (Kurmann et al., 2015), indicative of thyroid.
We first examined the effect of single 24-hr pulses of Dox-mediated transgene induction at biologically intermediate stages of differentiation. Parallel cultures received Dox at one of the following stages (D = day of differentiation): D0–D1 (exiting pluripotency); D5–D6 (DE stage); D6–D7 (AFE stage); or D7–D8 (early thyroid specification stage). In response to Dox, transgene activation resulted in >90%–95% NKX2-1+ cells within 24 hr (Figure 1H and data not shown) at all time points and subsequently extinguished within 2 days of Dox withdrawal (Figure S1I). Not all intermediate stages of development demonstrated a lasting response to the NKX2-1 transgene overexpression, and a substantial increase of resulting NKX2-1+ cells was highly restricted to a narrow window of induction at the AFE stage (Dox D6–D7) (Figures 1F and 1G). In addition, all other stages had low endogenous Nkx2-1 expression immediately post-Dox (Figure 1G), showing a relatively weak response to the transgene overexpression (Oguchi and Kimura, 1998).
The effect of Nkx2-1 overexpression at the AFE stage consistently resulted in a dramatic increase in the derivation of NKX2-1+ cells (Figures 1I and 1J), indicating a singular competent stage where NKX2-1 acts as a lineage specification signal.
AFE-Stage NKX2-1 Overexpression Leads to Efficient Thyroid Derivation
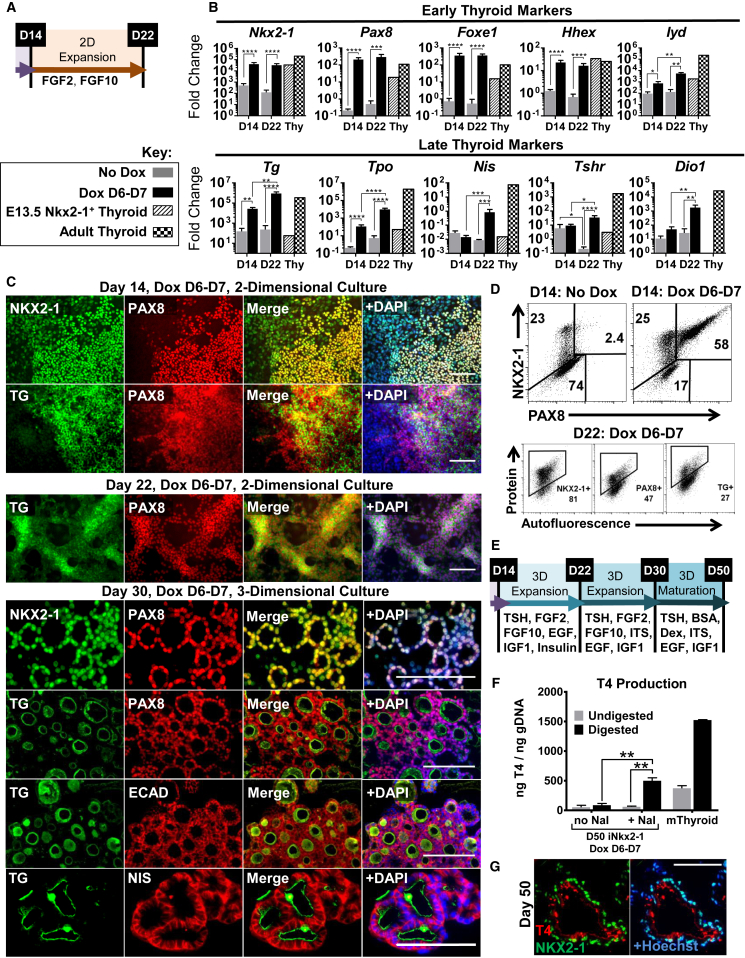
Next we asked: does stage-specific, transient Nkx2-1 overexpression potentiate the thyroid lineage or the derivation of non-thyroid NKX2-1+ lineages? Early (Pax8, Hhex, Foxe1) and more mature (Tg, Tpo, Nis, Tshr, Iyd, Dio1) thyroid markers showed distinct activation kinetics along the differentiation course (Figure S1H) and were upregulated at D14 and D22 in the Dox D6–D7 induced cultures at levels comparable with in vivo purified NKX2-1GFP+ E13.5 thyrocytes (Longmire et al., 2012) and adult mouse thyroid tissue (Figures 2A and 2B). In addition, we observed extensive coexpression of NKX2-1 and PAX8 (Figures 2C, upper panel, 2D, and S2E) with the increasing presence of TG to D22/30 (Figures 2B–2D and S2E).
Figure 2.
Nkx2-1 Overexpression at AFE Stage Results in Efficient Thyroid Differentiation
(A) Experimental schematic of the extended culture conditions in (B), (C) (top three panels), and (D).
(B) RT-qPCR for thyroid marker expression. Fold changes calculated relative to undifferentiated cells, error bars represent SEM (n = 5 independent experiments, n = 1 control).
(C) Immunostaining of induced cultures at D14, D22 (gelatin substratum), and D30 (Matrigel embedded). Scale bars represent 100 μm.
(D) Intracellular flow cytometry for populations at D14 (top), separate differentiation single stains at D22 (bottom).
(E) Schematic of the maturation culture conditions for (F) and (G).
(F) T4 ELISA from D50 (Dox D6–D7) cells ± 10 μM NaI from D40 to D50 (n = 3 wells from the same differentiation). Mouse thyroid tissue for reference (n = 2 tissue samples).
(G) D50 immunostaining for +NaI cultures corresponding with (F). Scale bar represents 100 μm.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. See also Figure S2.
To test the functionality of our thyrocyte-like cells, we cultured cells in 3D Matrigel to induce organoid formation (Kurmann et al., 2015, Martin et al., 1993), which promoted expansion and maturation of the thyroid-like cells (Figure S2G) and organization into follicle-like structures (Figure S2F). These structures expressed NKX2-1, PAX8, E-cadherin, TG secreted into the follicle lumen, and importantly basolateral NIS, indicating maturation (Figure 2C). Following culture to D50 in maturation media (Figures 2E and S2H), organoids exposed to iodide produced T4 (Figures 2F and 2G), indicating full functional capability.
Although NKX2-1 is also critical for the development of the lung and forebrain (Kimura et al., 1996, Xu et al., 2008), we found minimal evidence of derived forebrain or neuronal lineages (Figures S2C and S2D), lung lineages (Figures S2A and S2B), or ectopic expression of liver or foregut markers (Figure S2D and data not shown).
These results show that overexpression of NKX2-1 can be used as an inductive signal to efficiently and robustly specify thyrocyte-like cells from AFE resulting in the derivation of in vitro cells exhibiting characteristics typical of murine thyrocytes.
NKX2-1-Induced Thyroid Specification Is Dependent on Efficient Derivation of AFE
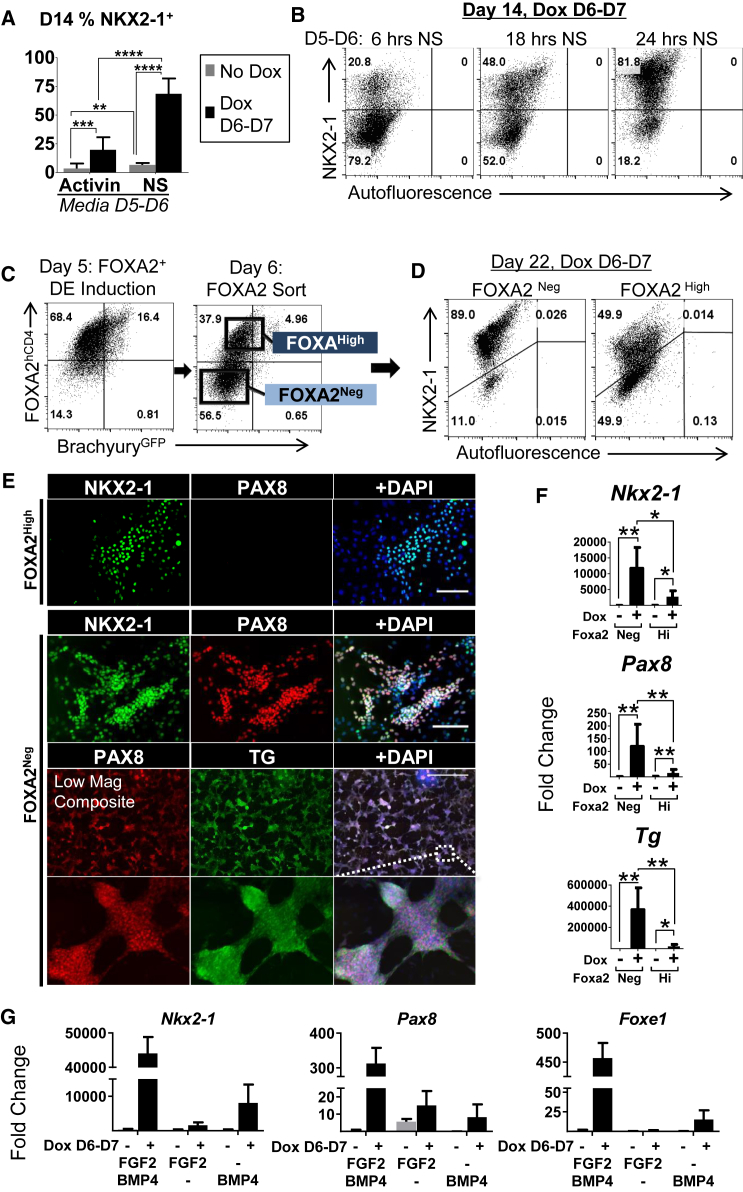
Next, we sought to define the parameters of the robust AFE response to transient heterologous NKX2-1 expression. Omission of NS-mediated anteriorization at the DE stage, previously shown to be essential for lung/thyroid specification (Longmire et al., 2012), significantly reduced the percentage of NKX2-1+ cells, both with the standard and induced protocols (Figure 3A). More so, we found that abbreviation of this stage resulted in a time-proportional reduction in the D14 NKX2-1+ cell percentage (Figure 3B), demonstrating that the inductive effect of NKX2-1 overexpression is dependent on efficient AFE patterning.
Figure 3.
Efficient Thyroid Specification Is Dependent on Precise Modulation of BMP/FGF Signaling and Derivation of the Thyroid-Competent FOXA2Neg AFE Population
(A) Intracellular D14 flow cytometry for NKX2-1 following D5–D6 NS or Activin treatment (n = 3 independent experiments).
(B) Intracellular D14 flow cytometry plots from varying duration of the anteriorization stage followed by 24 hr of Dox treatment.
(C) Flow cytometry sort schematic of D6 FOXA2+ AFE.
(D) Intracellular D22 flow cytometry from the sorted and induced populations shown in (C). Representative of three independent experiments.
(E) D22 immunostaining from FOXA2High and FOXA2Neg sorted populations. Top, scale bar represents 100 μm; bottom, scale bars represent 1,000 μm (composite image).
(F) D22 RT-qPCR data from the sorted populations shown in (C), with and without Dox D6–D7. Fold changes relative to undifferentiated cells, statistics from paired t tests, (n = 5 independent experiments).
(G) D14 RT-qPCR from induced and uninduced cells with specification factor variations. Fold changes relative to undifferentiated cells (n = 3 wells from same differentiation). Results are representative of three independent experiments.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. See also Figure S3.
Efficient Thyroid Conversion of FOXA2Neg AFE Subpopulation
Next, we wanted to determine if the AFE contained subpopulations with variations in thyroid competency. We interrogated FOXA2 due to its dynamic expression kinetics during in vivo and in vitro foregut endoderm derivation (Fagman et al., 2011, Gadue et al., 2006) (Figures S3A–S3C).
We sorted cells at the AFE stage based on FOXA2hCD4 expression (Figure 3C) and replated with or without Dox (D6–D7). The population induced by FOXA2Neg, relative to FOXA2High, resulted in higher numbers of NKX2-1+ and NKX2-1+PAX8+ cells (Figures 3D and 3E) and higher expression of thyroid markers (Figure 3F). In addition, no thyroid specification was observed from FOXA2low mesoderm (Figures S3D and S3E) treated with Dox and cultured in thyroid specification media (Figure S3F). These results indicate the FOXA2Neg population likely acquires this stage-specific thyroid competence through sequential progression from FOXA2+ DE to FOXA2Neg AFE.
Combinatorial FGF2 and BMP4 Signaling Is Required for Thyroid Specification and Conversion
FGF2 and BMP4 have been identified as necessary and sufficient signals for thyroid specification in several species, including mouse (Kurmann et al., 2015). Withdrawal of either factor resulted in a severe loss of NKX2-1+ thyroid specification and thyroid marker expression, as well as reduced cell expansion in both uninduced and induced conditions (Figures 3G, S3G, and S3H). Last, Dox-treated pre-endoderm-stage cells cultured in various media including TGF-β/BMP inhibition followed by BMP4/FGF2 stimulation did not show thyroid competence (data not shown).
Taken collectively, these data demonstrate that only appropriately patterned AFE in conjunction with essential signaling pathways can specify at high efficiency to TPs in response to NKX2-1 overexpression.
Genome-wide Analysis of NKX2-1-Induced Thyroid Specification Reveals Putative Regulators of AFE Thyroid Competence
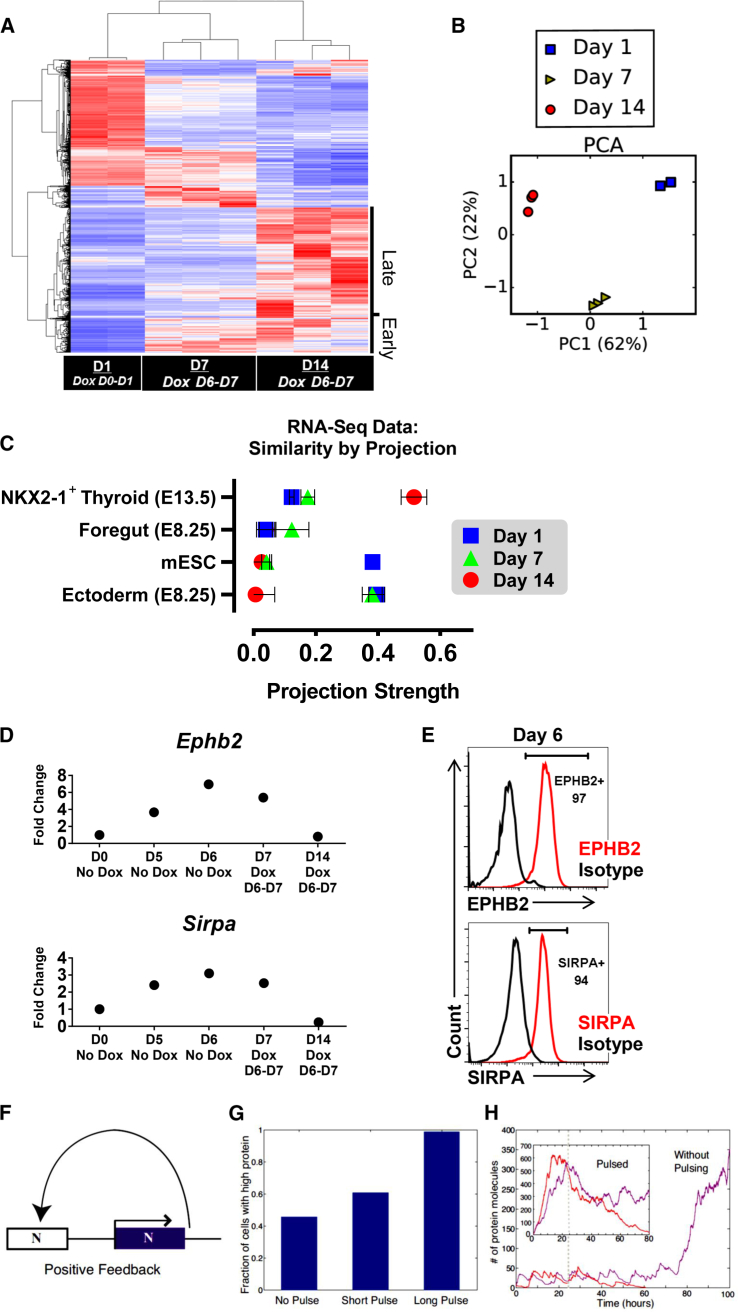
To gain further insights in the differential response to Nkx2-1 overexpression, we performed RNA-seq on three populations (data available in GEO: GSE92572): Dox-induced D1 cells (minimal competence), Dox-induced D7 cells (AFE stage, maximum competence) and D14 cells, Dox D6–D7 (NKX2-1-induced TPs). Unsupervised hierarchical clustering and principal component analysis (PCA) demonstrated distinct transcriptional profiles by population (Figures 4A and 4B). Gene ontology analysis showed strong segregation of processes relating to differentiating cells (Figure S4A) and expression profiles confirmed the relevance of active BMP and FGF signaling pathways on D7 and D14 populations for thyroid specification (Figure S4B). To investigate the similarity of our populations to known cell types, we employed a linear algebra method using projection scores to measure cell similarity based on global gene expression (Lang et al., 2014, Pusuluri et al., 2015). D14 cells projected strongly on purified E13.5 mouse Nkx2-1GFP+ thyrocytes, while D1 cells projected on mESCs, as expected (Figure 4C). Overall, these data suggest that transient NXK2-1 overexpression in the course of directed mESC thyroid differentiation does not fundamentally alter the identity of the derived TPs.
Figure 4.
RNA-Sequencing Data Reveal Thyroid Signature
(A) RNA-seq heatmap of the top 9,088 differentially expressed genes with highest variance (false discovery rate <0.05) among samples, clustered by samples (columns) and genes (rows). Late and early clusters are indicated.
(B) PCA plot of RNA-seq populations.
(C) Projection graph representing the degree of similarity (x axis, exact match = 1) between RNA-seq samples (dots) and reference gene expression datasets (y axis).
(D) RT-qPCR validation data of cell surface markers identified in the early cluster.
(E) Flow cytometry data corresponding with (D).
(F) Bistable model: a protein that cooperatively binds its own promoter leads to a positive feedback-based, bistable switch.
(G) Percentage of high-protein-expressing cells in Monte-Carlo simulations of the bistable switch shown in (F) for three pulse lengths of protein expression. Results were calculated using 1,000 simulations in each condition.
(H) Stochastic trajectories of protein number as a function of time in the absence of pulsing (main figure) and in the presence of a long pulse (inset). Colored lines show stochastic trajectories that successfully activate the Nkx2-1 feedback loop (purple lines) or fail to activate the feedback loop (red lines). The dashed line indicates the time the basal transcription rate was turned off in all simulations.
See also Figure S4.
From two clusters of interest (“early” and “late”) (Figures 4A and S4C), we identified potential transcriptional regulators and cell surface markers (CSMs) of thyroid fate (Tables S1 and S2). These lists contain genes with potentially novel roles in thyroid development, as well as genes previously implicated in foregut and early thyroid development both in vitro and in vivo (Fagman et al., 2011, Longmire et al., 2012, Millien et al., 2008). Of the early cluster CSMs, EPHB2 and SIRPA were identified to be highly expressed in DE (Figures 4D and 4E). Interestingly, Ephb2 expression in in vivo foregut endoderm was reported recently (Dravis and Henkemeyer, 2011). From the late cluster, reflective of a differentiated phenotype (Figure S4C), out of several CSMs correlating with the RNA-seq data, Col25a1 and Gabre were found to be enriched in both our induced and purified in vitro Nkx2-1mCherry+ TPs (Figures S4D and S4E).
The Stage-Specific Effect of NKX2-1 Overexpression May Be Governed by a Bistable Switch
The central observation underlying our work is that activation of an Nkx2-1 transgene at different stages during thyroid-directed differentiation results in a wide percentage range of endogenous NKX2-1+ TPs on D14 (Figure 1F). As NKX2-1 is known to bind its own promoter (Boggaram, 2009) and almost all cells are transgene NKX2-1+ post-Dox induction (Figure 1H), the behavior of the system implies the existence of an AFE-stage-specific positive feedback loop at the Nkx2-1 locus. This is further supported by the immediate activation of the endogenous Nkx2-1 expression at the Dox D6–D7 condition (Figure 1G).
Based on these data, we modeled the D6–D7 cell response to the Dox pulse as a bistable switch (Ferrell, 2012), where the two states are “on” and “off” expression of endogenous Nkx2-1 (Figures 4F–4H). Overall, this model could qualitatively reproduce our basic experimental observations, suggesting that a time-dependent bistable switch may underlie the effect of transgene overexpression on cell-fate decisions during directed differentiation of PSCs.
Discussion
PSC-based systems hold great promise for the mass production of transplantable, clinically relevant cell types and for in vitro modeling of complex disease states. Currently, a major roadblock to achieving these goals is the poor or variable differentiation efficiency of many differentiation protocols. This study used developmental-stage-specific overexpression of a single TF, NKX2-1, to (1) investigate how cell competence changes in a dynamic, developmentally relevant system and (2) improve the efficiency of TP specification.
Our results demonstrate that the inductive effect of NKX2-1 is restricted to a singular stage of competence contingent on several synergistic parameters (FOXA2 levels, duration of anteriorization, and BMP4/FGF2 signaling), implying that AFE-staged cells possess a unique epigenetic status allowing for robust conversion to thyroid. Respiratory lineages were not derived as indicated by insignificant numbers of SPC+ cells and the only presumed lung marker with high levels of expression (Sftpb) is expressed in both adult mouse (Figure S2A) and human thyroid (http://gtexportal.org/home/gene/SFTPB). We presume that the absence of Wnt agonists, an important signal for lung specification (Goss et al., 2009, Harris-Johnson et al., 2009, Huang et al., 2014), is the major reason for the paucity of respiratory lineages following NKX2-1 overexpression in our system.
Our study also achieved the practical goal of highly efficient thyroid differentiation. Our data indicate that the majority of the derived progenitors acquired a thyroid identity comparable with their in vivo counterparts, and importantly, their progeny gave rise to follicular-like structures in a 3D environment, expressed genes of thyroid hormone biosynthesis at levels comparable with adult thyroid, and produced high levels of T4 hormone. Overall, it appears that brief NKX2-1 exogenous expression during a well-defined window of maximum thyroid competence is sufficient to dramatically increase the yield and robustness of PSC thyroid specification and differentiation. Although a similar end-stage result has been previously reported through direct reprogramming of mESCs (Antonica et al., 2012), our approach delves into the mechanistic aspects of thyroid fate decisions and competence at a developmentally relevant stage. Further work is needed to define whether the thyrocyte-like cells produced downstream in our system are functionally equivalent to the reprogrammed cells and to the progeny of purified bona fide TPs (Kurmann et al., 2015).
This system can also be useful as a discovery tool in the absence of lineage-specific reporters. Analysis of the RNA-seq has identified both CSMs as well as potential regulators of thyroid fate in AFE and early TPs. Some of the TFs identified (Irx5, Hoxb8, Isl1) have been involved in mouse foregut and thyroid development (Millien et al., 2008, Westerlund et al., 2008), while others, such as PROX1, have been implicated in human thyroid disease (Ishii et al., 2016). Future PSC-based and in vivo studies will unravel the thyroid-related function of select TFs during thyroid specification and development.
Our theoretical model proposes a bistable positive feedback loop as the underlying mechanism to describe the endogenous Nkx2-1 locus activation at the AFE stage leading to subsequent stabilization of the core TF network, establishing thyroid identity. Future work will focus on experimental validation of the model and investigate the possibility that bistable switches controlling bifurcation dynamics in cell-fate decisions (Loh et al., 2014) can lead to the development of highly efficient and robust protocols of general applicability.
Experimental Procedures
All mouse work was approved by the Institutional Animal Care and Use Committee of Boston University School of Medicine.
Generation and Characterization of Inducible Line
The Ainv15 mESC line (Kyba et al., 2002) was a kind gift from Dr. Paul Gadue. As previously described (Ting et al., 2005), this line was engineered with a constitutively expressed rtTA from the Rosa26 locus and a promoter with tetO sites upstream of a loxP site at the HPRT locus. The Nkx2-1 transgene was inserted via co-electroporation of the plox-Nkx2-1 and pSalk-Cre plasmids. Colonies with restored neomycin resistance were screened for efficient Dox-mediated Nkx2-1 induction.
mESC-Directed Differentiation
Directed differentiation was performed in serum-free media as previously described (Kurmann et al., 2015, Longmire et al., 2012) (see Supplemental Experimental Procedures for detailed description).
Statistical Analysis
Error bars in graphs represent SD or SEM as indicated in the figure legends. Biological sample replicates (N) are also indicated in the legends. Statistically significant differences between conditions (ΔCt values used for RT-qPCR calculations) were determined using two-tailed unpaired Student t tests or as specified in figure legends. Significance is represented as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See Supplemental Experimental Procedures for additional methods.
Author Contributions
L.I. and D.N.K. conceived the work and D.N.K. provided critical intellectual input. L.I. and K.D. designed experiments and wrote the manuscript. S.C. and R.S. performed experiments. A.L. performed additional RNA-seq computational analysis and generated projection scores. L.Z. performed and S.M. supervised the RNA-seq computational analysis. J.C. performed the T4 ELISA with supervision from A.N.H. T.W. characterized the iNkx2-1 cell line. L.K. engineered and characterized the plox-Nkx2-1 vector. M.K. performed gene set enrichment pathway analysis. P.M. generated the bistability model and wrote the manuscript. All authors reviewed, edited, and approved the manuscript.
Acknowledgments
The targeted Ainv15 mESC line with was a kind gift from Paul Gadue (University of Pennsylvania). We would like to thank Brian R. Tilton and Patrick Autissier from the BU Flow Cytometry Core Facility as well as Michael T. Kirber from the BU Cellular Imaging Core for technical assistance. We thank Nicholas Skvir for help with visualization of RNA-seq data and Matthew Lawton for his help with immunostaining and technical assistance. K.D. was supported by a CTSI TL1 grant TL1TR001410, J.C. and A.N.H. by NIH DK105029, L.I. by NIH R01 HL111574, D.N.K. by NIH R01 HL095993, NIH R01 HL122442, and NIH DK105029, and P.M. by a Simons Investigator in MMLS (Simons Foundation).
Published: February 2, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.12.024.
Supplemental Information
References
- Antonica F., Kasprzyk D.F., Opitz R., Iacovino M., Liao X.H., Dumitrescu A.M., Refetoff S., Peremans K., Manto M., Kyba M. Generation of functional thyroid from embryonic stem cells. Nature. 2012;491:66–71. doi: 10.1038/nature11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggaram V. Thyroid transcription factor-1 (TTF-1/Nkx2.1/TITF1) gene regulation in the lung. Clin. Sci. (Lond) 2009;116:27–35. doi: 10.1042/CS20080068. [DOI] [PubMed] [Google Scholar]
- Bondue A., Lapouge G., Paulissen C., Semeraro C., Lacovino M., Kyba M., Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Dravis C., Henkemeyer M. Ephrin-B reverse signaling controls septation events at the embryonic midline through separate tyrosine phosphorylation-independent signaling avenues. Dev. Biol. 2011;355:138–151. doi: 10.1016/j.ydbio.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagman H., Amendola E., Parrillo L., Zoppoli P., Marotta P., Scarfò M., De Luca P., de Carvalho D.P., Ceccarelli M., De Felice M. Gene expression profiling at early organogenesis reveals both common and diverse mechanisms in foregut patterning. Dev. Biol. 2011;359:163–175. doi: 10.1016/j.ydbio.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J.E. Bistability, bifurcations, and Waddington's epigenetic landscape. Curr. Biol. 2012;22:R458–R466. doi: 10.1016/j.cub.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P., Huber T.L., Paddison P.J., Keller G.M. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss A.M., Tian Y., Tsukiyama T., Cohen E.D., Zhou D., Lu M.M., Yamaguchi T.P., Morrisey E.E. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev. Cell. 2009;17:290–298. doi: 10.1016/j.devcel.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.D., Chen A., Nostro M.C., d'Souza S.L., Schaniel C., Lemischka I.R., Gouon-Evans V., Keller G., Snoeck H.W. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 2011;29:267–272. doi: 10.1038/nbt.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Johnson K.S., Domyan E.T., Vezina C.M., Sun X. beta-Catenin promotes respiratory progenitor identity in mouse foregut. Proc. Natl. Acad. Sci. USA. 2009;106:16287–16292. doi: 10.1073/pnas.0902274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.X.L., Islam M.N., O'Neill J., Hu Z., Yang Y.G., Chen Y.W., Mumau M., Green M.D., Vunjak-Novakovic G., Bhattacharya J. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomou L., Kotton D.N. Derivation of endodermal progenitors from pluripotent stem cells. J. Cell Physiol. 2015;230:246–258. doi: 10.1002/jcp.24771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii J., Yazawa T., Chiba T., Shishido-Hara Y., Arimasu Y., Sato H., Kamma H. PROX1 promotes secretory granule formation in medullary thyroid cancer cells. Endocrinology. 2016;157:1289–1298. doi: 10.1210/en.2015-1973. [DOI] [PubMed] [Google Scholar]
- Kimura S., Hara Y., Pineau T., FernandezSalguero P., Fox C.H., Ward J.M., Gonzalez F.J. The T/ebp null mouse thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- Kubo A., Shinozaki K., Shannon J.M., Kouskoff V., Kennedy M., Woo S., Fehling H.J., Keller G. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- Kurmann A.A., Serra M., Hawkins F., Rankin S.A., Mori M., Astapova I., Ullas S., Lin S., Bilodeau M., Rossant J. Regeneration of thyroid function by transplantation of differentiated pluripotent stem cells. Cell Stem Cell. 2015;17:527–542. doi: 10.1016/j.stem.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyba M., Perlingeiro R.C.R., Daley G.Q. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- Lang A.H., Li H., Collins J.J., Mehta P. Epigenetic landscapes explain partially reprogrammed cells and identify key reprogramming genes. PLoS Comput. Biol. 2014;10:e1003734. doi: 10.1371/journal.pcbi.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.M., Ang L.T., Zhang J.Y., Kumar V., Ang J., Auyeong J.Q., Lee K.L., Choo S.H., Lim C.Y.Y., Nichane M. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14:237–252. doi: 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire T.A., Ikonomou L., Hawkins F., Christodoulou C., Cao Y.X., Jean J.C., Kwok L.W., Mou H.M., Rajagopal J., Shen S.S. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell. 2012;10:398–411. doi: 10.1016/j.stem.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Valentine M., Unger P., Lichtenstein C., Schwartz A.E., Friedman E.W., Shultz L.D., Davies T.F. Preservation of functioning human thyroid organoids in the scid mouse. 1. System characterization. J. Clin. Endocrinol. Metab. 1993;77:305–310. doi: 10.1210/jcem.77.2.8345031. [DOI] [PubMed] [Google Scholar]
- Mazzoni E.O., Mahony S., Closser M., Morrison C.A., Nedelec S., Williams D.J., An D.S., Gifford D.K., Wichterle H. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 2013;16:1219–1286. doi: 10.1038/nn.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millien G., Beane J., Lenburg M., Tsao P.-N., Lü J., Spira A., Ramirez M.I. Characterization of the mid-foregut transcriptome identifies genes regulated during lung bud induction. Gene Expr. Patterns. 2008;8:124–139. doi: 10.1016/j.modgep.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H.M., Zhao R., Sherwood R., Ahfeldt T., Lapey A., Wain J., Sicilian L., Izvolsky K., Musunuru K., Cowan C. Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell Stem Cell. 2012;10:385–397. doi: 10.1016/j.stem.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguchi H., Kimura S. Multiple transcripts encoded by the thyroid-specific enhancer-binding protein (T/EBP) thyroid-specific transcription factor-1 (TTF-1) gene: evidence of autoregulation. Endocrinology. 1998;139:1999–2006. doi: 10.1210/endo.139.4.5933. [DOI] [PubMed] [Google Scholar]
- Petros T.J., Maurer C.W., Anderson S.A. Enhanced derivation of mouse ESC-derived cortical interneurons by expression of Nkx2.1. Stem Cell Res. 2013;11:647–656. doi: 10.1016/j.scr.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusuluri, S.T., Lang, A.H., Mehta, P., and Castillo, H.E. (2015). Cellular reprogramming dynamics follow a simple one-dimensional reaction coordinate. arXiv 1505.03889 [q-bio.MN] [DOI] [PMC free article] [PubMed]
- Seguin C.A., Draper J.S., Nagy A., Rossant J. Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell. 2008;3:182–195. doi: 10.1016/j.stem.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Ting D.T., Kyba M., Daley G.Q. Inducible transgene expression in mouse stem cells. Methods Mol. Med. 2005;105:23–46. doi: 10.1385/1-59259-826-9:023. [DOI] [PubMed] [Google Scholar]
- Westerlund J., Andersson L., Carlsson T., Zoppoli P., Fagman H., Nilsson M. Expression of Islet1 in thyroid development related to budding, migration, and fusion of primordia. Dev. Dyn. 2008;237:3820–3829. doi: 10.1002/dvdy.21772. [DOI] [PubMed] [Google Scholar]
- Xu Q., Tam M., Anderson S.A. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.