Graphical abstract
Highlights
-
•
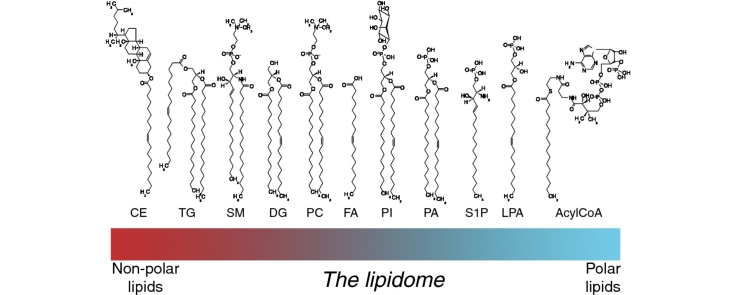
The extreme diversity of the lipidome poses a significant analytical challenge.
-
•
A variety of mass spectrometry-based approaches exist for the analysis of lipids.
-
•
Combining extraction and separation procedures maximizes coverage of the lipidome.
Abstract
The lipidome comprises a large array of molecules with diverse physicochemical properties. Lipids are structural components of cells, act as a source of energy, and function as signaling mediators. Alterations in lipid metabolism are involved in the onset and progression of a variety of diseases, including metabolic syndrome and cancer. Because of this, interest in lipidomics, the comprehensive characterization of the lipidome by mass spectrometry, has intensified in recent years. However, obtaining a truly complete overview of all lipids in a sample has remained very challenging due to their enormous structural diversity. Here, we provide an overview of the collection of analytical approaches used to study various lipid classes, emphasizing innovations in sample preparation and liquid chromatography–mass spectrometry (LC–MS). Additionally, we provide practical suggestions for increasing the coverage of the lipidome.
Current Opinion in Biotechnology 2017, 43:127–133
This review comes from a themed issue on Analytical biotechnology
Edited by Jurre J Kamphorst and Ian A Lewis
For a complete overview see the Issue and the Editorial
Available online 1st December 2016
http://dx.doi.org/10.1016/j.copbio.2016.11.008
0958-1669/© 2016 The Author(s). Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
Lipids are low molecular weight biomolecules characterized by their high hydrophobicity. They are involved in nearly all major aspects of cell biology. For instance, triglycerides store energy in the form of fatty acids, phospholipids form cellular membranes, and various lipid classes initiate or transduce signaling events: lysophosphatidic acid stimulates proliferation and migration [1], and specific phosphatidylinositol lipid species transduce insulin signaling [2]. Because of their intricate involvement in many physiological processes, it is not surprising that lipids play important roles in a variety of diseases such as cancer, cardiovascular disorders, neurodegenerative diseases, obesity and diabetes [3, 4, 5, 6, 7].
Particularly due to their involvement in pathological processes, there is a strong interest in investigating the variety of lipids present in samples and their functional roles in disease. It has recently been estimated that mammalian cells contain 10 000 individual lipid species [8]. As much as 50% of these remain without assigned functions [9••]. Therefore, many novel structures with potential medical relevance are left to be discovered. This is perhaps best illustrated by the recent discovery of branched fatty acid esters of hydroxy fatty acids (FAHFAs), which were found to improve glucose tolerance and to stimulate insulin secretion in diabetic mice [10••].
Lipidomics has emerged as a key technology for investigating the metabolism and cellular functions of known lipids, as well as for discovering and characterizing novel lipid structures. Lipidomics can be defined as the comprehensive characterization of lipids in biological systems [11]. In recent years, there have been considerable advancements in various aspects of the lipidomics ‘pipeline’. For instance, the generation of lipid databases and the tools to cross-compare them with experimentally obtained lipidomics data has been an important development. Examples are LipidMaps, LipidBank, LipidHome, LipidBlast, and LipidSearch [12•, 13, 14, 15, 16]. The lipidomics field has particularly benefited from continued developments in mass spectrometry; the ever-increasing sensitivity, resolution, speed, and dynamic range of modern instruments allow researchers to probe the lipid composition in unprecedented detail. These developments in mass spectrometry have been exploited in different ways. For example, direct infusion or ‘shotgun’ lipidomics approaches introduce samples into the mass spectrometer without prior separation, instead relying on the resolution and dynamic range of modern instruments [17, 18]. This approach enables the rapid analysis of samples, but is unable to resolve isobaric species and may compromise the detection of lower abundant species due to ion suppression effects and insufficient dynamic range. Instead, although requiring more time, liquid chromatography-based separation followed by mass spectrometry (LC–MS) remains very popular as a way to increase lipidome coverage, separate isobaric species and maximize dynamic range [17, 19, 20]. The enormous potential of LC–MS in comprehensive lipidomic analysis is arguably best demonstrated by recent study exploring the lipid composition of platelets, where approximately 5600 unique lipids were detected [9••].
It is important to note that, despite exciting advances in mass spectrometry and bioinformatics, the degree to which the lipidome can truly be ‘covered’ comprehensively actually depends on the sample extraction and liquid chromatography separation. Due to the considerable chemical diversity of lipids, any single extraction (and likewise separation) procedure will invariably create a bias toward certain lipid species at the expense of others. We therefore argue that combining multiple extraction and separation procedures is essential to maximize coverage of both the more hydrophilic and hydrophobic lipid classes. To support our argument, we provide an overview of recently published literature on lipid extraction and LC-MS procedures, and suggest a practical approach for maximizing the coverage of the lipidome.
Lipid extraction
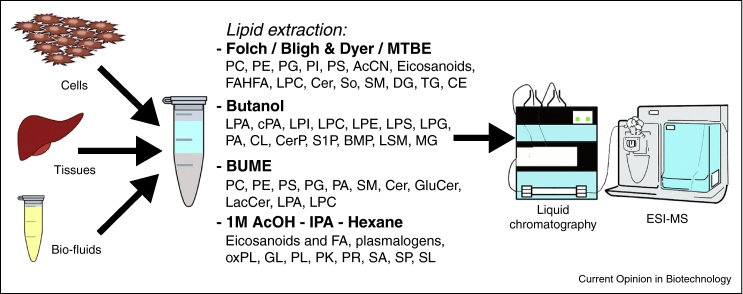
Lipidomic sample preparation protocols exploit the hydrophobic nature of lipids to extract them while eliminating other components of the biological matrix (i.e. proteins, sugars, inorganic salts) that could potentially interfere with the chromatographic separation and mass spectrometry analysis (Figure 1) [21, 22]. As will be discussed below, the most commonly used lipid extraction procedures involve chloroform, as originally described by Folch et al. [23], and Bligh and Dyer [24]. More recently, extraction procedures with less hazardous solvents have been introduced [17]. Finally, several protocols for the extraction of more hydrophilic lipid species have been published [25, 26, 27].
Figure 1.
Commonly used extraction procedures in lipidomics and the lipid classes they cover. Published methods include chloroform and methyl tert-butyl ether (MTBE) based extractions [23, 24, 28, 29, 30, 31, 32], butanol and butanol-methanol (BUME) extraction procedures [17, 25, 33, 34, 35], and an extraction procedure using acetic acid (AcOH) with isopropanol and hexane [9••]. (L)PC, (lyso)phosphatidylcholine; (L)PE, (lyso)phosphatidylethanolamie; (L)PG, (lyso)phosphatidylglycerol; (L)PI, (lyso)phosphatidylinositol; (L)PS, (lyso)phosphatidylserine; AcCN, acyl-carnitine; FAHFA, branched fatty acid esters of hydroxy fatty acid; Cer, ceramide; So, sphingosine; (L)SM, (lyso)sphingomyelin; DG, diglyceride; TG, triglyceride; CE, cholesterol ester; (L)PA, (lyso)phosphatidic acid; cPA, cyclic phosphatidic acid; CL, cardiolipin; CerP, ceramide-phosphate; S(1)P, sphingosine-1-phosphate; BMP, bis(monoglyceride)phosphate; MG, monoglyceride; GluCer, glucosyl-ceramide; LacCer, lactosyl-ceramide; FA, fatty acid; oxPL, oxidized phospholipids; GL, glycerides; PL, phospholipids; PK, polyketides; PR, prenols; SL, sphingolipids.
Chloroform-based lipid extraction
Variations on the lipid extraction protocol developed originally by Folch in 1957 remain very widely used to this day [23]. It efficiently extracts the abundant classes of lipids such as most phospholipids, glycerides, cholesterol and cholesterol-esters, sphingolipids, and waxes [28]. The original method is based on liquid-liquid partitioning using chloroform and methanol in a 2:1 (v/v) ratio [23]. A method modified to use less organic solvents and to be suitable for water-based samples was later described by Bligh and Dyer [24, 29]. Chloroform-based protocols directed toward specific lipid classes have also been published. For example, a method for extracting branched fatty acid esters of hydroxyl fatty acids (FAHFAs), a newly discovered class of endogenous mammalian lipids with antidiabetic and anti-inflammatory effects, uses a modified Bligh and Dyer extraction method from acidified serum samples [10••]. A downside of chloroform-based protocols is the inability to extract charged and more polar lipids like some of the lysophospholipids (LPA), phosphatidic acid lipids (PA), acylcarnitines, acyl-CoAs, and sphingosine phosphates [17].
MTBE-based lipid extraction
Recently, extraction procedures using methyl tert-butyl ether (MTBE) have gained popularity as a less toxic alternative to chloroform [30]. An added advantage of MTBE is that in contrast to chloroform, its density is lower than water. Thus, MTBE will separate to the upper phase during extraction of water-based samples, facilitating a convenient removal of the lipid extract [31]. Importantly, it was also shown that a modified MTBE method can provide near quantitative recovery of more polar lipids such as LPAs and PAs [32]. A downside of using MTBE, however, is the significant carry-over of water, causing the typical sample drying step to be lengthy, and the increased chance for ion suppression and adduct formation due to the co-extraction of salts and other metabolites [17, 21].
Butanol-based lipid extraction
An extraction procedure using butanol was originally used for the analysis of lysophospholipids and acylcarnitines [33]. Later, it was shown that butanol can additionally efficiently recover cardiolipins (CL), bis(monoacylglycero)phosphate (BMP), as well as their precursors phosphoglycerols (PGs) and phosphatidic acids (PAs) from heart tissues and primary human skin fibroblasts [25]. Because of this observation, this extraction procedure was modified for global lipid analysis [17]. In one procedure, mixing of a sample, first with butanol:methanol (3:1, v/v) leads to single extraction phase, or ‘BUME’ (BUtanol MEthanol) mixture. Thereafter, heptane:ethyl acetate (3:1, v/v) and 1% acetic acid are sequentially added to BUME mixture. The top organic layer of this extraction system contains most major lipid classes. This BUME method can be used as an alternative to MTBE with less salts and metabolites carrying over into the organic phase. Also, BUME extraction results in lipid recovery highly comparable to a modified Bligh-Dyer method [34, 35]. A consideration is, however, that co-extracted water in the organic phase may significantly increase the time needed for sample evaporation.
Other solvent systems for lipid extraction
A few studies describe lipid extraction procedures employing alternative solvent systems such as methanol/dichloromethane (DCM), isopropanol, or a combination of acetic acid solution/isopropanol/hexane [9••, 36•]. The rationale for using DCM is that it is less toxic than chloroform, and extraction efficiency of plasma lipids appears comparable to chloroform [36•]. In this paper, a strong case is made for isopropanol as a precipitation solvent for plasma lipid analysis. In a study investigating the platelet lipidome, use of a hexane/isopropanol protocol with addition of 1 M acetic acid was found to be able to extract ∼5600 different lipid species including fatty acids, glycerolipids, phospholipids, polyketides, prenol lipids, saccharolipids, sphingolipids and sterol lipids [9••].
Specialized lipid derivatization approaches for polar and low-abundant lipids
Many physiological processes in cells are regulated by low-abundant bioactive lipids, such as phosphoinositol phosphates, eicosanoids and prostaglandins [37, 38, 39, 40, 41]. Their analysis is easily compromised due to a combination of their low abundance, limited extraction efficiency in regularly used solvents, and susceptibility to ion suppression. Chemical derivatization of the polar head group of these lipids appears to be a promising approach to increase the stability, extraction efficiency, and ionization [40, 42, 43]. For example, it was demonstrated that phosphoinositol phosphates, mediators of insulin signaling and other signaling cascades, can be readily recovered, analyzed and quantified after methylation using a trimethylsilyl diazomethane solution [40]. Derivatization has also successfully been used for the analysis of fatty acids and eicosanoids [42, 43].
Mass spectrometry-based lipidomics
Recent innovations in separation and analysis science have boosted lipidomics applications. Most lipidomics methods using mass spectrometry can be subdivided in two distinct strategies; those that directly infuse lipid extracts into the mass spectrometer, also called ‘shotgun’ lipidomics, and those where lipid samples undergo chromatographic separation prior to analysis, i.e. LC–MS.
Direct infusion, or ‘shotgun’ lipidomics
Shotgun lipidomics is favored by many in the lipidomics community [44, 45, 46]. Here, lipid extracts are directly introduced into the mass spectrometer using electrospray ionization (ESI), often at the nano-scale [47, 48, 49]. This approach benefits significantly from high resolution instruments that have become increasingly available. One of the strong advantages of shotgun lipidomics is that sample analysis time can be on the order of minutes, making it amenable for high-throughput, routine analysis of major lipid species [21]. However, when the aim is to maximize coverage of the entire lipidome, several aspects need to be considered. First, the common occurrence of isobaric species complicates shotgun lipidomics approaches. For example, in our (LC–MS) lipidomic screens we frequently find phosphatidylcholine (PC) species that have the same exact mass as phosphatidylethanolamines (PEs). With LC–MS these species can be differentiated based on differences in retention time. This is not possible with shotgun lipidomics, and although fragmentation may provide a solution, its interpretation is not trivial [50]. Second, in-source fragmentation can easily confound lipid identification and quantification. From our own experience, lysophosphatidylcholines (LPCs) in negative mode can lose their choline moiety during ionization to become lysophosphatidic lipids (LPAs). Although this may only happen to a fraction of the LPCs, their massive over-abundance relative to LPAs can easily lead to significant over-estimation of LPA levels. Finally, as all lipids, both high and low abundant, are introduced into the mass spectrometer and analyzed simultaneously, ion-suppression and a limited dynamic range increase the likelihood of compounds in the lower concentration range being lost.
Liquid chromatography–mass spectrometry
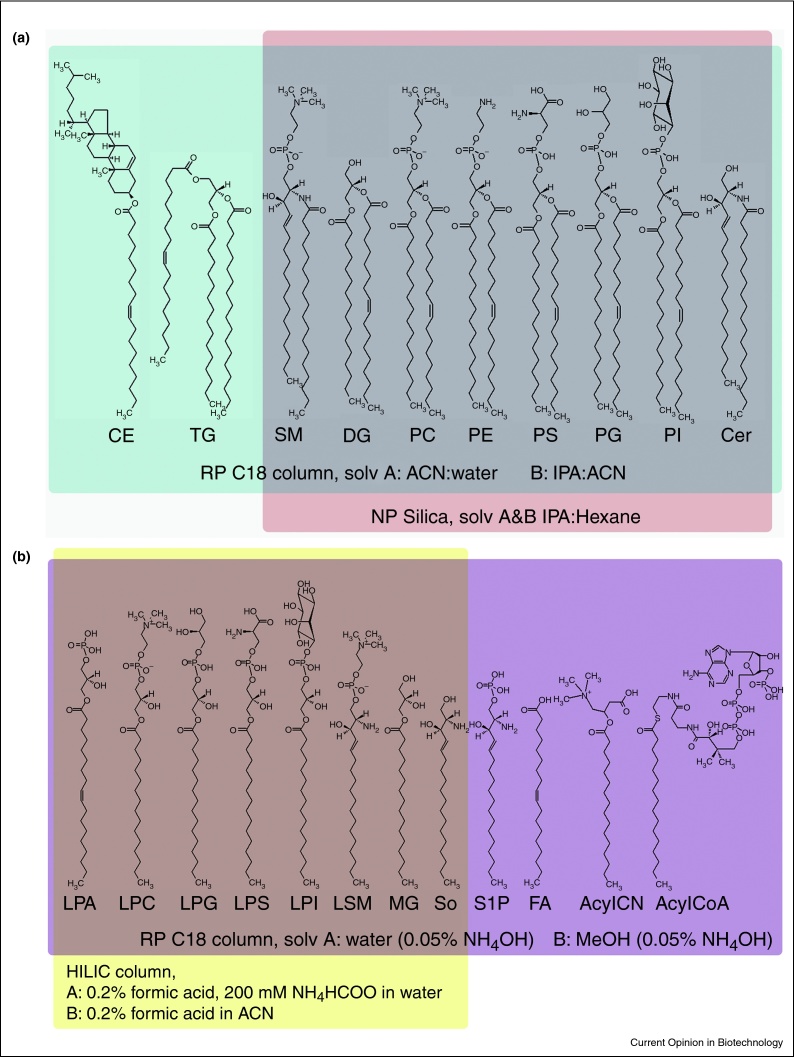
For the comprehensive characterization of the lipidome, liquid chromatography–mass spectrometry (LC–MS) remains the method of choice for many. At the cost of increased run times, LC separations reduce the complexity of the eluent introduced into the mass spectrometer. This reduces the risk for ion-suppression during electrospray ionization, and is less tasking for the mass spectrometer with respect to the dynamic range. As a result, low abundant (and often biologically relevant) species are more readily detected. Various modes of separation have been used for lipid analysis (Figure 2). For example, normal phase chromatography has been used in the past to separate phospholipid classes based on head group polarity [3, 51]. More recently, hydrophilic interaction liquid chromatography (HILIC) was used for the separation of lipid extracts [19, 52]. It is worth mentioning that HILIC-based separations appeared particularly suitable for separating lysophospholipid regioisomers (mono-acyl lipids with same fatty acid carbon length but a different double bond position) [19]. Reversed phase chromatography appears to be the most popular mode of separation. A very typical separation involves a C8/C18 column and a gradient of (mobile phase A) acetonitrile–water (60:40), and (mobile phase B) isopropanol–acetonitrile (90:10), with gradient run times commonly lasting 10–40 min depending on the flow rate used [53, 54]. This platform is very suitable for the analysis of many of the phospholipids species (PC, PE, PG, PI, PS), sphingomyelins, ceramides, cholesterol-esters, diglycerides, and triglycerides. Although lysophospholipids and phosphatidic acid (PA) lipids can also be detected with this setup, a more polar gradient, such as a methanol-water gradient, results in our experience in a better analytical performance, i.e. improved peak shape and ionization.
Figure 2.
Coverage of the (a) apolar, and (b) polar lipid classes by commonly used separation methods. RP, reversed phase; ACN, acetonitrile; IPA, isopropanol; NP, normal phase; HILIC, hydrophilic interaction liquid chromatography; CE, cholesterol ester; TG, triglyceride; (L)SM, (lyso)sphingomyelin; DG, diglyceride; (L)PC, (lyso)phosphatidylcholine; PE, phosphatidylethanolamine; (L)PS, (lyso)phosphatidylserine; (L)PG, (lyso)phosphatidylglycerol; (L)PI, (lyso)phosphatidylinositol; Cer, ceramide; MG, monoglyceride; So, sphingosine; S1P, sphingosine-1-phosphate; FA, fatty acid; AcylCN, acyl-carnitine.
Two recent studies in particular demonstrate the merit of LC–MS-based lipidomics for the detection and characterization of unknown lipids. In one study lipidomic analysis was performed on the adipose tissue of both wildtype mice and mice with adipose tissue-specific overexpression of the glucose transporter Glut4, which leads to increased glucose tolerance [10••]. Tissue lipids were extracted using a modified Bligh and Dyer protocol and subsequently analyzed in both positive (on a C5 column) and negative (on C18 column) mode. In both cases a 45 min gradient of (A) 95:5 v/v water:methanol and (B) 60:35:5 v/v isopropanol:methanol:water was used, with in negative mode additionally 0.1% ammonium hydroxide and in positive mode 0.1% formic acid plus 5 mM ammonium formate. The mass spectrometer used was an Agilent 6220 TOF instrument. Untargeted data analysis was then performed using XCMS [55•], resulting in the identification of a cluster of novel lipids that were elevated in the Glut4 overexpressing mice. Follow up work revealed that these molecules are branched fatty acid esters of hydroxyl fatty acids (FAHFAs), which were found to have strong antidiabetic and anti-inflammatory effects. In the second study a very comprehensive analysis was conducted to map the lipidome remodeling upon platelet activation [9••]. Here, extraction was performed with a 2:20:30 v/v 1 M acetic acid:isopropanol:hexane mixture. Samples were then subjected to two gradient setups, one a 55 min 50:50 v/v acetonitrile:water and 70:30 v/v isopropanol:acetonitrile gradient for the analysis of the ‘non-polar’ lipids (lipophilic phospholipids, neutral lipids), and the other a 30 min gradient based on 75:25 v/v water:acetonitrile and 60:40 methanol:acetonitrile, for the analysis of ‘polar’ species (eicosanoids, fatty acids). The analysis was done on an Orbitrap elite in full scan only, and for both gradients samples were run both in positive and negative mode and at two mass ranges (100–900 and 900–1800 m/z), totaling 8 runs per condition. In this tour-de-force profiling study, data was then analyzed with a program called Sieve and the resulting list of lipid features was interrogated against multiple databases (HMDB, Lipidhome, LipidMaps, and METLIN). This led to the detection of approximately 5600 unique lipid species, of which only 50% could be putatively identified. Of these 5600 lipids, 900 increased upon platelet activation by thrombin.
Conclusion and future perspectives
Cells, tissues, and bio-fluids contain many thousands of structurally diverse lipids. In one of the most comprehensive lipidomics screens thus far, only 50% of the ∼5600 detected lipids could be identified [9••]. While the numbers detected and percentages identified might differ from one biological system to the next, it is apparent that tremendous opportunities exist to discover new aspects of lipid biology with potential relevance to medicine. Lipidomics will continue to play a key role in seizing these opportunities.
We conclude that no single extraction and separation gradient modality sufficiently captures the full spectrum of chemical diversity that exists between lipid classes. Rather, a combination of sample extraction and separation procedures is required. For example, a good approach would be to subject samples to both a chloroform-based extraction in conjunction with acetonitrile–isopropanol reversed phase gradient, and a butanol extraction followed by water-methanol gradient on the same or a different reversed phase column. This covers the entire spectrum from very lipophilic lipid species (triglycerides, cholesterol-esters, etc.) to the relatively hydrophilic lipids (acyl-CoAs, LPAs, etc.). While this increases workload and commitment of mass spectrometry time, it is necessary when one's aim is to maximize coverage of the lipidome.
Continued developments in chromatography and mass spectrometry are driving ongoing improvements in analytical performance. Together with a well thought out sample preparation approach and optimized separation and mass spectrometer settings, we are guaranteed to detect and identify more lipid species. It will be important for the community to actively populate existing public databases with these findings. In short, we anticipate that the use of lipidomics will lead to exciting new discoveries in the field of lipid biology in the coming years.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
The Kamphorst lab is supported by funding from Cancer Research UK and the Rosetrees Trust. JJK is a Cancer Research UK Career Development Fellow (C50242/A17728). Owing to space constraints, we apologize for failing to cite many relevant articles.
References
- 1.Willier S., Butt E., Grunewald T.G.P. Lysophosphatidic acid (LPA) signalling in cell migration and cancer invasion: a focussed review and analysis of LPA receptor gene expression on the basis of more than 1700 cancer microarrays. Biol Cell. 2013;105:317–333. doi: 10.1111/boc.201300011. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B., Stephens L., Hawkins P. PI3K signalling: the path to discovery and understanding. Nat Rev Mol Cell Biol. 2012;13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 3.Shui G., Stebbins J.W., Lam B.D., Cheong W.F., Lam S.M., Gregoire F., Kusonoki J., Wenk M.R. Comparative plasma lipidome between human and cynomolgus monkey: are plasma polar lipids good biomarkers for diabetic monkeys? PLoS One. 2011;6:e19731. doi: 10.1371/journal.pone.0019731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopperton K.E., Duncan R.E., Bazinet R.P., Archer M.C. Fatty acid synthase plays a role in cancer metabolism beyond providing fatty acids for phospholipid synthesis or sustaining elevations in glycolytic activity. Exp Cell Res. 2014;320:302–310. doi: 10.1016/j.yexcr.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Fhaner C.J., Liu S., Ji H., Simpson R.J., Reid G.E. Comprehensive lipidome profiling of isogenic primary and metastatic colon adenocarcinoma cell lines. Anal Chem. 2012;84:8917–8926. doi: 10.1021/ac302154g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng H., Wang M., Li J.L., Cairns N.J., Han X. Specific changes of sulfatide levels in individuals with pre-clinical Alzheimer's disease: an early event in disease pathogenesis. J Neurochem. 2013;127:733–738. doi: 10.1111/jnc.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu B., Ackerman D., Sanchez D.J., Li B., Ochocki J.D., Grazioli A., Bobrovnikova-Marjon E., Diehl J.A., Keith B., Simon M.C. HIF2α-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. 2015;5:652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q., Wakelam M.J.O. Lipidomics in the analysis of malignancy. Adv Biol Regul. 2014;54:93–98. doi: 10.1016/j.jbior.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 9••.Slatter D.A., Aldrovandi M., O’Connor A., Allen S.M., Brasher C.J., Murphy R.C., Mecklemann S., Ravi S., Darley-Usmar V., O’Donnell V.B. Mapping the human platelet lipidome reveals cytosolic phospholipase A2 as a regulator of mitochondrial bioenergetics during activation. Cell Metab. 2016;23:930–944. doi: 10.1016/j.cmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; The most comprehensive lipidomics profiling study to date. Here, the authors combine multiple gradients, column chemistries, polarities, and mass scan ranges to cover as much of the lipidome as possible. Of the ∼5600 unique lipid species detected in lipid extracts of platelets, only 50% could be identified. A great reminder that a lot of novel lipid biochemistry remains to be characterized.
- 10••.Yore M.M., Syed I., Moraes-Vieira P.M., Zhang T., Herman M.A., Homan E.A., Patel R.T., Lee J., Chen S., Peroni O.D. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159:318–332. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the discovery of the previously unrecognized fatty acid esters of hydroxy fatty acids (FAHFAs) lipids. These lipids elicit a variety of favorable effects that counteract type 2 diabetes, including lowering ambient glycemia, improving glucose tolerance, and reducing adipose tissue inflammation. A great example of how untargeted lipidomics can lead to the discovery of novel lipids with important functions in physiology and disease.
- 11.Brügger B. Lipidomics: analysis of the lipid composition of cells and subcellular organelles by electrospray ionization mass spectrometry. Annu Rev Biochem. 2014;83:79–98. doi: 10.1146/annurev-biochem-060713-035324. [DOI] [PubMed] [Google Scholar]
- 12•.Fahy E., Subramaniam S., Murphy R.C., Nishijima M., Raetz C.R., Shimizu T., Spener F., van Meer G., Wakelam M.J., Dennis E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50 Suppl:S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]; Over the last decennium, the LIPID MAPS consortium has innovated the design of a novel lipid classification and nomenclature system, which has now been widely adopted by researchers worldwide. Today, the LIPID MAPS website is a very comprehensive online resource for lipidomics and lipid biology. The website provides access to a large, publicly available database of lipid structures and contains useful information as well as a variety of tools.
- 13.Watanabe K., Yasugi E., Oshima M. How to search the glycolipid data in “LIPID BANK for Web”, the newly developed lipid database in Japan. Trends Glycosci Glycotechnol. 2000;12:175–184. [Google Scholar]
- 14.Foster J.M., Moreno P., Fabregat A., Hermjakob H., Steinbeck C., Apweiler R., Wakelam M.J.O., Vizcaíno J.A. LipidHome: a database of theoretical lipids optimized for high throughput mass spectrometry lipidomics. PLoS One. 2013;8:e61951. doi: 10.1371/journal.pone.0061951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kind T., Liu K.H., Lee do Y., DeFelice B., Meissen J.K., Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taguchi R., Ishikawa M. Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine Lipid Search. J Chromatogr A. 2010;1217:4229–4239. doi: 10.1016/j.chroma.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 17.Wang M., Wang C., Han R.H., Han X. Novel advances in shotgun lipidomics for biology and medicine. Prog Lipid Res. 2016;61:83–108. doi: 10.1016/j.plipres.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papan C., Penkov S., Herzog R., Thiele C., Kurzchalia T., Shevchenko A. Systematic screening for novel lipids by shotgun lipidomics. Anal Chem. 2014;86:2703–2710. doi: 10.1021/ac404083u. [DOI] [PubMed] [Google Scholar]
- 19.Koistinen K.M., Suoniemi M., Simolin H., Ekroos K. Quantitative lysophospholipidomics in human plasma and skin by LC-MS/MS. Anal Bioanal Chem. 2015;407:5091–5099. doi: 10.1007/s00216-014-8453-9. [DOI] [PubMed] [Google Scholar]
- 20.Cajka T., Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal Chem. 2016;88:524–545. doi: 10.1021/acs.analchem.5b04491. [DOI] [PubMed] [Google Scholar]
- 21.Han X., editor. Lipidomics: Comprehensive Mass Spectrometry of Lipids. John Wiley & Sons, Inc.; Hoboken, New Jersey: 2016. [Google Scholar]
- 22.Zhao Y.Y., Wu S.P., Liu S., Zhang Y., Lin R.C. Ultra-performance liquid chromatography-mass spectrometry as a sensitive and powerful technology in lipidomic applications. Chem Biol Interact. 2014;220:181–192. doi: 10.1016/j.cbi.2014.06.029. [DOI] [PubMed] [Google Scholar]
- 23.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 24.Bligh E.G., Dyer W.J. A rapid method for total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 25.Scherer M., Schmitz G., Liebisch G. Simultaneous quantification of cardiolipin, bis(monoacylglycero)phosphate and their precursors by hydrophilic interaction LC–MS/MS including correction of isotopic overlap. Anal Chem. 2010;82:8794–8799. doi: 10.1021/ac1021826. [DOI] [PubMed] [Google Scholar]
- 26.Masoodi M., Eiden M., Koulman A., Spaner D., Volmer D.A. Comprehensive lipidomics analysis of bioactive lipids in complex regulatory networks. Anal Chem. 2010;82:8176–8185. doi: 10.1021/ac1015563. [DOI] [PubMed] [Google Scholar]
- 27.Popa I., Vlad C., Bodennec J., Portoukalian J. Recovery of gangliosides from aqueous solutions on styrene-divinylbenzene copolymer columns. J Lipid Res. 2002;43:1335–1340. [PubMed] [Google Scholar]
- 28.Lam S.M., Tong L., Reux B., Lear M.J., Wenk M.R., Shui G. Rapid and sensitive profiling of tear wax ester species using high performance liquid chromatography coupled with tandem mass spectrometry. J Chromatogr A. 2013;1308:166–171. doi: 10.1016/j.chroma.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Ogiso H., Suzuki T., Taguchi R. Development of a reverse-phase liquid chromatography electrospray ionization mass spectrometry method for lipidomics, improving detection of phosphatidic acid and phosphatidylserine. Anal Biochem. 2008;375:124–131. doi: 10.1016/j.ab.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 30.Jiang S., Li Y., Lin S., Yang H., Guan X., Zhou H., Luan T., Cai Z. Mass spectrometry-based lipidomics analysis using methyl tert-butyl ether extraction in human hepatocellular carcinoma tissues. Anal Methods. 2015;7:8466–8471. [Google Scholar]
- 31.Matyash V., Liebisch G., Kurzchalia T.V., Shevchenko A., Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byeon S.K., Lee J.Y., Moon M.H. Optimized extraction of phospholipids and lysophospholipids for nanoflow liquid chromatography-electrospray ionization-tandem mass spectrometry. Analyst. 2012;137:451–458. doi: 10.1039/c1an15920h. [DOI] [PubMed] [Google Scholar]
- 33.Morrison W.R., Tan S.L., Hargin K.D. Methods for the quantitative analysis of lipids in cereal grains and similar tissues. J Sci Food Agric. 1980;31:329–340. doi: 10.1002/jsfa.2740310402. [DOI] [PubMed] [Google Scholar]
- 34.Ametaj B.N., Bobe G., Lu Y., Young J.W., Beitz D.C. Effect of sample preparation, length of time, and sample size on quantification of total lipids from bovine liver. J Agric Food Chem. 2003;51:2105–2110. doi: 10.1021/jf0259011. [DOI] [PubMed] [Google Scholar]
- 35.Virot M., Tomao V., Colnagui G., Visinoni F., Chemat F. New microwave-integrated Soxhlet extraction. An advantageous tool for the extraction of lipids from food products. J Chromatogr A. 2007;1174:138–144. doi: 10.1016/j.chroma.2007.09.067. [DOI] [PubMed] [Google Scholar]
- 36•.Sarafian M.H., Gaudin M., Lewis M.R., Martin F.-P., Holmes E., Nicholson J.K., Dumas M.-E. Objective set of criteria for optimization of sample preparation procedures for ultra-high throughput untargeted blood plasma lipid profiling by ultra performance liquid chromatography−mass spectrometry. Anal Chem. 2014;86:5766–5774. doi: 10.1021/ac500317c. [DOI] [PubMed] [Google Scholar]; In this paper a rigorous comparison is made between a variety of protein precipitation and liquid-liquid extraction sample preparation procedures for the analysis of the plasma lipidome. The authors benchmarked the procedures against the following criteria: protein removal efficiency, selectivity, repeatability, and recovery efficiency. Based on this the authors recommend isopropanol-mediated protein precipitation as a simple and effective sample preparation strategy for the lipidomic analysis of plasma.
- 37.Kimura T., Jennings W., Epand R.M. Roles of specific lipid species in the cell and their molecular mechanism. Prog Lipid Res. 2016;62:75–92. doi: 10.1016/j.plipres.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Kuerschner L., Thiele C. Multiple bonds for the lipid interest. Biochim Biophys Acta - Mol Cell Biol Lipids. 2014;1841:1031–1037. doi: 10.1016/j.bbalip.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 39.Mulugeta S., Suzuki T., Hernandez N.T., Griesser M., Boeglin W.E., Schneider C. Identification and absolute configuration of dihydroxy-arachidonic acids formed by oxygenation of 5S-HETE by native and aspirin-acetylated COX-2. J Lipid Res. 2010;51:575–585. doi: 10.1194/jlr.M001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark J., Anderson K.E., Juvin V., Smith T.S., Karpe F., Wakelam M.J.O., Stephens L.R., Hawkins P.T. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat Methods. 2011;8:267–272. doi: 10.1038/nmeth.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hankin J.A., Wheelan P., Murphy R.C. Identification of novel metabolites of prostaglandin E2 formed by isolated rat hepatocytes. Arch Biochem Biophys. 1997;340:317–330. doi: 10.1006/abbi.1997.9921. [DOI] [PubMed] [Google Scholar]
- 42.Bollinger J.G., Thompson W., Lai Y., Oslund R.C., Hallstrand T.S., Sadilek M., Turecek F., Gelb M.H. Improved sensitivity mass spectrometric detection of eicosanoids by charge reversal derivatization. Anal Chem. 2010;82:6790–6796. doi: 10.1021/ac100720p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bollinger J.G., Rohan G., Sadilek M., Gelb M.H. LC/ESI-MS/MS detection of FAs by charge reversal derivatization with more than four orders of magnitude improvement in sensitivity. J Lipid Res. 2013;54:3523–3530. doi: 10.1194/jlr.D040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han X., Jiang X. A review of lipidomic technologies applicable to sphingolipidomics and their relevant applications. Eur J Lipid Sci Technol. 2009;111:39–52. doi: 10.1002/ejlt.200800117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han X., Yang K., Gross R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuhmann K., Almeida R., Baumert M., Herzog R., Bornstein S.R., Shevchenko A. Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J Mass Spectrom. 2012;47:96–104. doi: 10.1002/jms.2031. [DOI] [PubMed] [Google Scholar]
- 47.Haag M., Schmidt A., Sachsenheimer T., Brügger B. Quantification of signaling lipids by nano-electrospray ionization tandem mass spectrometry (Nano-ESI MS/MS) Metabolites. 2012;2:57–76. doi: 10.3390/metabo2010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan E., Reid G.E. Chemical derivatization and ultrahigh resolution and accurate mass spectrometry strategies for “Shotgun” lipidome analysis. Acc Chem Res. 2016;49:1596–1604. doi: 10.1021/acs.accounts.6b00030. [DOI] [PubMed] [Google Scholar]
- 49.Özbalci C., Sachsenheimer T., Brügger B. Membrane biogenesis. In: Rapaport D., Herrmann J.M., editors. Membrane Biogenesis. Methods and Protocols. Humana Press; 2013. pp. 3–20. [Google Scholar]
- 50.Schwudke D., Schuhmann K., Herzog R., Bornstein S.R., Shevchenko A. Shotgun lipidomics on high resolution mass spectrometers. Cold Spring Harb Perspect Biol. 2011;3:a004614. doi: 10.1101/cshperspect.a004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hermansson M., Uphoff A., Käkelä R., Somerharju P. Automated quantitative analysis of complex lipidomes by liquid chromatography/mass spectrometry. Anal Chem. 2005;77:2166–2175. doi: 10.1021/ac048489s. [DOI] [PubMed] [Google Scholar]
- 52.Scherer M., Leuthäuser-Jaschinski K., Ecker J., Schmitz G., Liebisch G. A rapid and quantitative LC-MS/MS method to profile sphingolipids. J Lipid Res. 2010;51:2001–2011. doi: 10.1194/jlr.D005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aicheler F., Li J., Hoene M., Lehmann R., Xu G., Kohlbacher O. Retention time prediction improves identification in nontargeted lipidomics approaches. Anal Chem. 2015;87:7698–7704. doi: 10.1021/acs.analchem.5b01139. [DOI] [PubMed] [Google Scholar]
- 54.Basit A., Pontis S., Piomelli D., Armirotti A. Ion mobility mass spectrometry enhances low-abundance species detection in untargeted lipidomics. Metabolomics. 2016;12:50. doi: 10.1007/s11306-016-0971-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Smith C.A., Want E.J., O’Maille G., Abagyan R., Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]; This paper describes the development of XCMS, a popular program for the untargeted analysis of metabolomics and lipidomics datasets. An easy to use online version of this program is now publicly available.