Abstract
Azacitidine (AZA) and decitabine (DAC) are cytidine azanucleoside analogs with clinical activity in myelodysplastic syndromes (MDS) and potential activity in solid tumors. To better understand the mechanism of action of these drugs, we examined the effects of AZA and DAC in a panel of non-small cell lung cancer (NSCLC) cell lines. Of 5 NSCLC lines tested in a cell viability assay, all were sensitive to AZA (EC50 of 1.8–10.5 µM), while only H1299 cells were equally sensitive to DAC (EC50 of 5.1 µM). In the relatively DAC-insensitive cell line A549, both AZA and DAC caused DNA methyltransferase I depletion and DNA hypomethylation; however, only AZA significantly induced markers of DNA damage and apoptosis, suggesting that mechanisms in addition to, or other than, DNA hypomethylation are important for AZA-induced cell death. Cell cycle analysis indicated that AZA induced an accumulation of cells in sub-G1 phase, whereas DAC mainly caused an increase of cells in G2/M. Gene expression analysis of AZA- and DAC-treated cells revealed strikingly different profiles, with many genes distinctly regulated by each drug. In summary, while both AZA and DAC caused DNA hypomethylation, distinct effects were demonstrated on regulation of gene expression, cell cycle, DNA damage, and apoptosis.
Keywords: apoptosis, azacitidine, decitabine, gene expression, non-small cell lung cancer
Introduction
Azacitidine (AZA) (5-azacytidine, Vidaza®; Celgene Corporation, Summit, NJ) and decitabine (DAC) (2′-deoxy-5-azacytidine, Dacogen®; Eisai Inc., Woodcliff Lake, NJ) are used clinically for the treatment of myelodysplastic syndromes (MDS), a heterogeneous group of bone marrow stem cell disorders.1,2 Both AZA and DAC are cytidine nucleoside analogs that become incorporated into newly synthesized DNA, where they bind DNA methyltransferases (DNMTs) in an irreversible, covalent manner.3,4 The sequestration of DNMTs prevents maintenance of the methylation state of DNA, leading to DNA hypomethylation.5,6 As a consequence, genes previously silenced by DNA hypermethylation can be re-expressed upon treating cancer cell lines with these DNMT inhibitors.7,8 Re-expression of aberrantly methylated genes involved in normal cell cycle control, differentiation, and apoptotic pathways is believed to contribute to the anticancer effects of these drugs.9
Clinical activities of AZA and DAC are best established in the hematological malignancies MDS and acute myeloid leukemia (AML), cancers with a high frequency of aberrantly methylated genes.10 Aberrant DNA methylation of genes involved in DNA repair, cell adhesion, cell cycle, and cell death has also been reported in multiple types of solid cancers, including colon, stomach, breast, ovary, kidney, and lung.11 For example, in non-small cell lung cancer (NSCLC), hypermethylation of tumor suppressor genes RAS association domain family 1A (RASSF1A), adenomatous polyposis coli (APC), fragile histidine triad (FHIT), and p16INK4A has been associated with poor survival.12–15 Clinical trials investigating the use of AZA and DAC in solid tumors have been reported, although response rates were poor. In a Phase I study of DAC in patients with cancers involving the lungs, esophagus, and pleura, no objective responses were observed.16 Similar outcomes were obtained with DAC in patients with other forms of solid tumors.17 In a Phase II trial of AZA in patients with solid tumors, the responses were minimal and transient.18 The clinical response rate was also low for the combination of AZA and phenylbutyrate in patients with refractory solid tumors.19
A better understanding of the mechanistic activities of AZA and DAC will provide insights into rational use of these agents as therapies for solid tumor patients, including potential uses as combination therapies, adjuvant therapies, and maintenance therapies. Here, we directly compared the in vitro effects of AZA and DAC on cell viability, DNMT1 protein levels, DNA methylation, DNA damage, apoptosis, cell cycle, and gene expression in NSCLC cell lines. Although AZA and DAC caused similar effects on DNA-mediated markers such as DNMT1 depletion and DNA methylation, the drugs showed very different effects on cell viability, DNA damage, apoptosis, cell cycle, and gene expression.
Results
AZA and DAC have differential effects on NSCLC cell viability
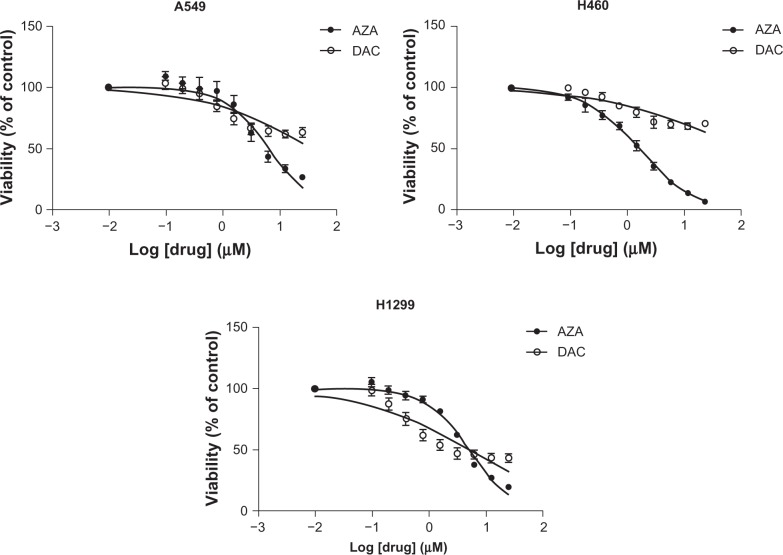
AZA and DAC were compared in a panel of 5 NSCLC cell lines (A549, H1975, H460, H23, and H1299) for their effects on cell viability (Figure 1 and Supporting Information Figure 1). AZA reduced cell viability by at least 75% at high concentrations, with EC50 values of 1.8–10.5 µM (Table 1). In contrast, DAC did not reduce cell viability more than 55%, and EC50 values were not reached in 4 (A549, H1975, H460, and H23) of the 5 NSCLC cell lines tested. In H1299 cells, DAC EC50 values were calculated; however, the 95% confidence intervals for the EC50 values were poor (data not shown). The EC50 values for AZA and DAC are similar to those reported for drugs commonly used in NSCLC, including gemcitabine, cisplatin, and carboplatin.20–22 The distinct dose-response curves and EC50 values indicate differential sensitivities of these NSCLC cell lines to AZA and DAC.
Figure 1.
AZA and DAC differentially affect cell viability in a panel of NSCLC cell lines. Viability of A549, H460, and H1299 cells was assessed after 72 hours of treatment with AZA or DAC (0–25 µM). Error bars represent the standard error of mean of 3 independent experiments, with triplicate wells per experiment.
Abbreviations: AZA, azacitidine; DAC, decitabine; NSCLC, non-small cell lung cancer.
Table 1.
EC50 values for AZA and DAC on NSCLC cell viability
| AZA EC50 ± SEM (μM) | DAC EC50 ± SEM (μM) | |
|---|---|---|
| A549 | 6.3 ± 1.1 | >25 |
| H1975 | 8.6 ± 2.9 | >25 |
| H460 | 1.8 ± 0.3 | >25 |
| H23 | 10.5 ± 1.8 | >25 |
| H1299 | 5.1 ± 0.2 | 5.9 ± 2.1 |
Note: EC50 values were calculated from 3 independent experiments using Graphpad Prism software.
Abbreviations: AZA, azacitidine; DAC, decitabine; NSCLC, non-small cell lung cancer.
AZA and DAC cause DNMT1 depletion and DNA hypomethylation
To determine whether the differential sensitivities of NSCLC cell lines to AZA versus DAC in cell viability assays reflected differences in the incorporation of each drug into DNA, DNMT1 protein depletion and DNA hypomethylation were evaluated as indirect measures of drug incorporation into DNA. When A549 and H1299 cells were treated with AZA or DAC for 20 hours, DNMT1 protein levels were reduced (Figure 2). Dose-dependent decreases in DNMT1 protein were observed with AZA, while near-maximal reduction of DNMT1 protein was observed at the lowest concentration (0.05 µM) of DAC. In A549 cells, DNMT1 depletion caused by 5 µM AZA was not as much as that caused by 0.5 or 1 µM AZA, possibly as a consequence of cell growth inhibition at the higher AZA concentration.23 Reduced DNMT1 levels were detected as early as 4 hours after drug treatment (Supporting Information Figure 2). Similar results were obtained in the H460 and H23 cell lines (data not shown).
Figure 2.
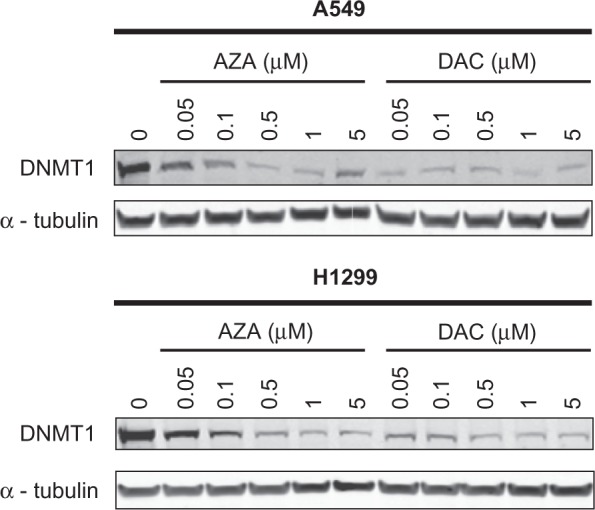
AZA and DAC cause DNMT1 depletion in NSCLC cell lines. A549 and H1299 cells were treated with AZA or DAC (0–5 µM) for 20 hours and DNMT1 protein was detected by Western blotting of cell extracts. Alpha-tubulin was used as a loading control.
Abbreviations: AZA, azacitidine; DAC, decitabine; DNMT1, DNA methyltransferase 1; NSCLC, non-small cell lung cancer.
We next determined whether AZA and DAC caused DNA hypomethylation by examining the methylat ion status of LINE-1 elements in A549 and H1299 cells treated for 48 hours (Figure 3) or 72 hours (Supporting Information Figure 3). Both AZA and DAC decreased LINE-1 methylation; however, DAC was 3- to 10-fold more potent. Peak hypomethylation was observed at 0.3–1.0 µM AZA and 0.1 µM DAC. LINE-1 methylation was unaffected at the highest DAC concentration tested, possibly as a consequence of cell growth inhibition.23 DAC modulated the DNA-mediated markers (DNMT1 depletion and DNA hypomethylation) in both cell lines, suggesting that the relative insensitivity to DAC in cell viability assays cannot be attributed to a lack of drug uptake, phosphorylation, and DNA incorporation. These findings rule out dysfunctional deoxycytidine kinase, the rate-limiting kinase in the phosphorylation of DAC, as a possible mechanism of relative DAC-insensitivity,24 and suggest that mechanisms in addition to DNA incorporation are responsible for the greater sensitivity of NSCLC cell viability to AZA.
Figure 3.
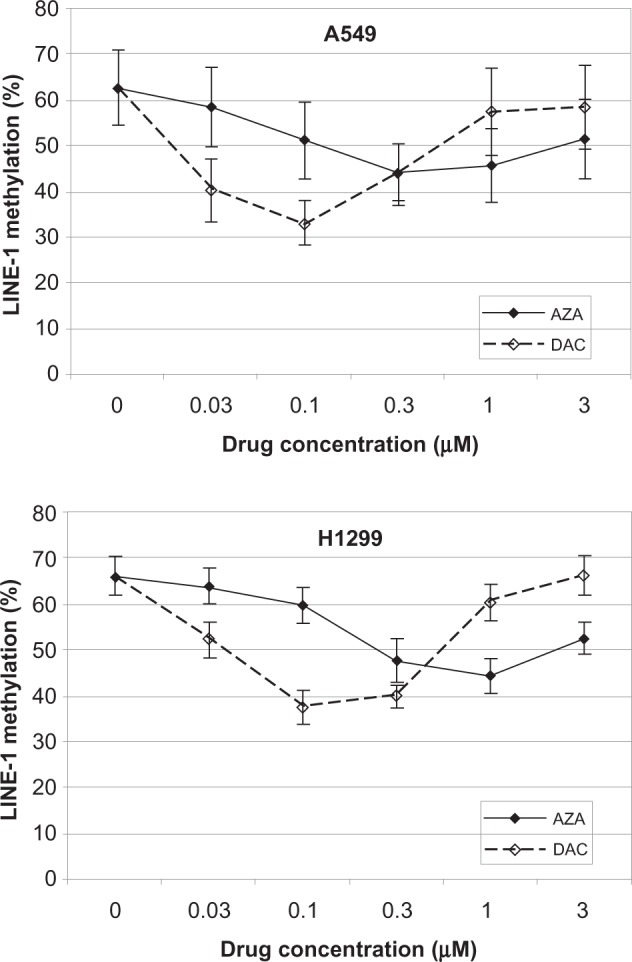
AZA and DAC reduce DNA methylation in A549 and H1299 cells. LINE-1 DNA methylation was assessed in A549 and H1299 cells after 48 hours of treatment with AZA or DAC (0–3 µM). Percentage LINE-1 methylation represents the average percentage methylation of 4 CpG sites in duplicate samples, with error bars representing the standard deviation.
Abbreviations: AZA, azacitidine; DAC, decitabine.
AZA, but not DAC, robustly induces markers of DNA damage and apoptosis
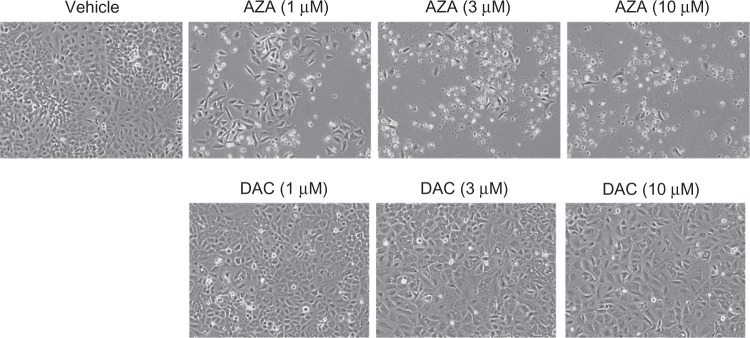
Phase contrast images of A549 cell cultures after 3 days of drug treatment showed reduced cell numbers and increased debris in AZA-treated cell cultures, but healthy-looking cells in DAC-treated cultures (Figure 4). These findings confirmed results of the cell viability assays (Figure 1). To examine the mechanism(s) of drug-induced cell death, A549 and H1299 NSCLC cell lines were treated with AZA or DAC for 24 or 48 hours, and markers of double-strand DNA (dsDNA) damage (histone-H2AX(ser139) phosphorylation) and apoptosis (PARP cleavage) were evaluated by Western blot (Figure 5 and data not shown). AZA dose-dependently induced histone-H2AX(ser139) phosphorylation and PARP cleavage in A549 cells. Similar results were observed in the H460 cell line (data not shown). There was relatively high basal phosphorylation of histone-H2AX(ser139) in H1299 cells, which was further increased by 10 µM AZA. High concentrations of AZA also induced PARP cleavage in H1299 cells. In A549 and H1299 cells, DNMT1 protein was completely depleted by DAC treatment; however, neither histone-H2AX(ser139) phosphorylation nor PARP cleavage were induced.
Figure 4.
AZA-treated A549 cultures show reduced cell numbers. A549 cells, seeded in 6-well plates, were treated with vehicle or 1, 3, and 10 µM AZA or DAC for 72 hours. The CoolSNAP ES2 CCD camera (Photometrics) was used to take phase-contrast images of cells under the Plan Fluor 10X objective (Nikon) on the Eclipse Ti-S microscope (Nikon).
Abbreviations: AZA, azacitidine; DAC, decitabine.
Figure 5.
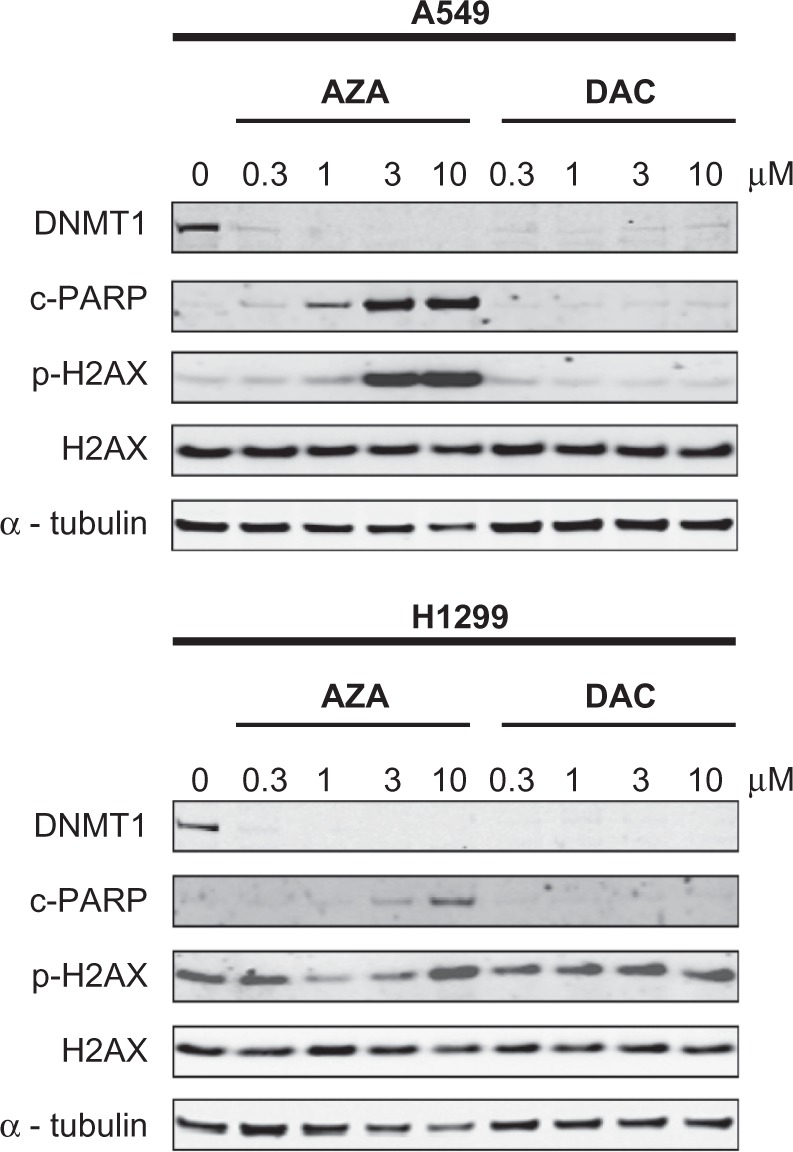
AZA, but not DAC, induces markers of DNA damage and apoptosis in NSCLC cell lines. A549 and H1299 cells were treated with AZA or DAC (0–10 µM) for 48 hours and Western blotting of cell extracts was used to detect DNMT1, cleaved-PARP, phospho-histone-H2AX(ser139), and total histone-H2AX. alpha-tubulin was used as a loading control.
Abbreviations: AZA, azacitidine; DAC, decitabine; DNMT1, DNA methyltransferase 1; NSCLC, non-small cell lung cancer.
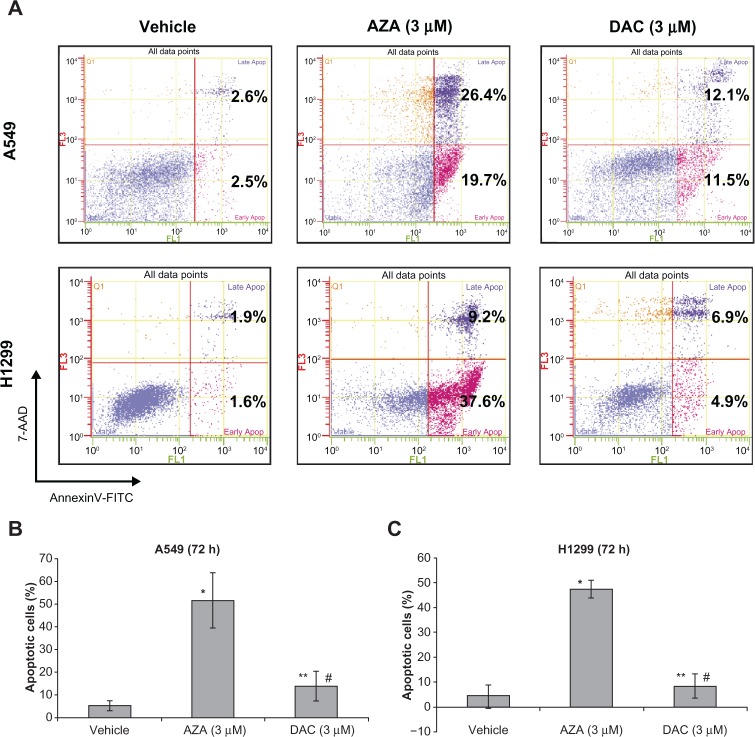
As AZA induced PARP cleavage, we further examined early-apoptotic (AnnexinV-FITC+ and 7-AAD−) and late-apoptotic (AnnexinV-FITC+ and 7-AAD+) cell populations by flow cytometry in A549 and H1299 cells treated with AZA (3 µM) or DAC (3 µM) for 72 hours (Figure 6). AZA (3 µM) treatment of A549 and H1299 cells caused a significant increase in the early- and late-apoptotic populations (Figures 6B and 6C). DAC did not significantly cause an increase in these populations. These results demonstrated that AZA, but not DAC, induced dsDNA damage and apoptosis in NSCLC cell lines.
Figure 6.
AZA, but not DAC, strongly induces apoptosis in NSCLC cell lines. A549 and H1299 cells were treated with AZA or DAC (3 µM) for 72 hours, and staining for AnnexinV-FITC (x-axis) and 7-AAD (y-axis) was detected by flow cytometry. A) The percentages of early apoptotic cells and late apoptotic cells are represented in the lower right and upper right quadrants, respectively. Representative data of 4 independent experiments are shown. B) Percentage (mean ± SD; n = 4) of apoptotic (early and late) cells with AZA or DAC treatment of A549 cells. *P < 0.001 versus “vehicle”. **P = 0.328 versus “vehicle”. #P < 0.001 versus “AZA (3 µM)”. C) Percentage (mean ± SD; n = 4) of apoptotic (early and late) cells with AZA or DAC treatment of H1299 cells. *P < 0.001 versus “vehicle”. **P = 0.442 versus “vehicle”. #P < 0.001 versus “AZA (3 µM)”.
Abbreviations: AZA, azacitidine; DAC, decitabine; NSCLC, non-small cell lung cancer.
DAC-treated H1299 cells show delayed DNA damage response
AZA and DAC appear to be incorporated into DNA of NSCLC cell lines, as both drugs induced DNMT1 depletion (Figure 2) and DNA hypomethylation (Figure 3). It was therefore surprising that 48-hour treatment with DAC did not induce dsDNA damage (histone-H2AX(ser139) phosphorylation) in A549 and H1299 cells (Figure 5). To better define the DNA damage response of NSCLC cell lines treated with AZA and DAC, we treated NSCLC cell lines with the drugs for an extended period of time. A549 and H1299 cells were treated with AZA or DAC for 6 days and lysates were collected on days 3 and 6 (Figure 7). At the 3-day time point in both cell lines, the results were similar to those at the 24- and 48-hour time points; AZA, but not DAC, induced histone-H2AX(ser139) phosphorylation and PARP cleavage. In A549 cells, even after 6 days of daily treatment with DAC, there was no induction of histone-H2AX(ser139) phosphorylation and PARP cleavage (Figure 7). The EC50 values for AZA and DAC were 4.4 µM and 2.5 µM, respectively, for A549 cells after 6 days of treatment (Supporting Information Table 1). Although the calculated EC50 value for DAC was lower than that of AZA, DAC did not reduce cell viability more than 75%, while AZA almost completely inhibited cell viability (Supporting Information Figure 4). In H1299 cells, substantial histone-H2AX(ser139) phosphorylation, without much effect on PARP cleavage, was observed after 6 days of DAC treatment (Figure 7). Consistent with these results, phase contrast images of H1299 cells treated with DAC for a prolonged period did not show many cells undergoing apoptosis. Rather, prolonged treatment of H1299 cells resulted in fewer cells that are enlarged (data not shown). These results suggest that DAC may have a delayed effect on inducing DNA damage in NSCLC cell lines.
Figure 7.
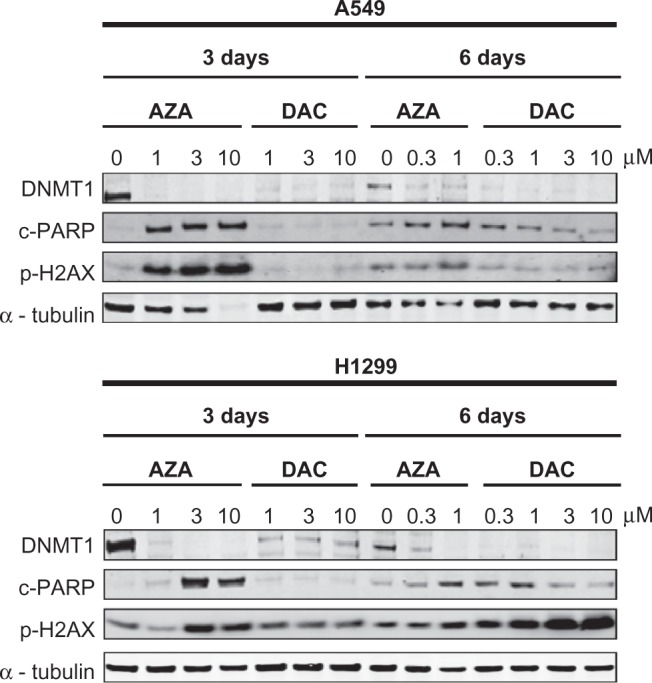
DAC-treated H1299 cells show delayed DNA damage response. A549 and H1299 cells were treated with AZA or DAC (0–10 µM) for 3 and 6 days, and Western blotting of cell extracts was used to detect DNMT1, cleaved-PARP, and phospho-histone-H2AX(ser139). Alpha-tubulin was used as a loading control.
Abbreviations: AZA, azacitidine; DAC, decitabine; DNMT1, DNA methyl-transferase 1.
AZA and DAC differentially affect the cell cycle
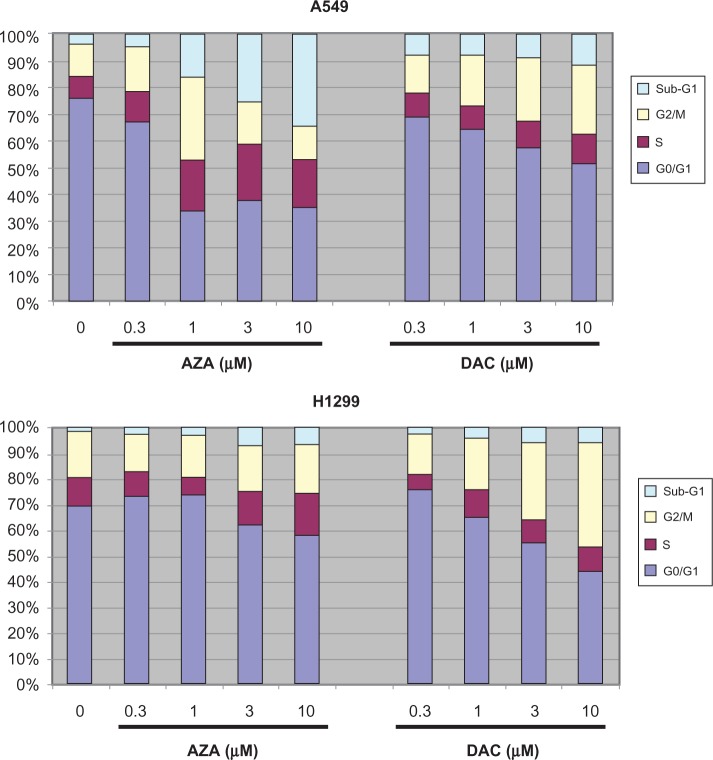
The effects of AZA and DAC on cell cycle distribution were evaluated in A549 and H1299 cells treated for 72 hours (Figure 8). AZA dose-dependently increased the sub-G1 population in A549 cells, consistent with the induction of apoptosis (Figures 4–6). AZA also caused a minor increase in the sub-G1 population in H1299 cells (Figure 8), consistent with the induction of early-, rather than late-, apoptotic cell population at this time point (Figure 6). DAC also caused a minor increase in the sub-G1 population in these cell lines; however, the more prominent effect of DAC was an increase in the G2/M population.
Figure 8.
AZA increases the sub-G1 population of cells, while DAC increases the G2/M population. A549 and H1299 cells were stained with NIM-DAPI after 72 hours of treatment with AZA or DAC at 0, 0.3, 1, 3, and 10 µM. The percentage of cells in sub-G1, G2/M, S, and G0/G1 was quantified by flow cytometry. Representative data of 3 independent experiments are shown.
Abbreviations: AZA, azacitidine; DAC, decitabine.
AZA and DAC modulate expression of different sets of genes
Although both AZA and DAC caused DNMT1 depletion and DNA hypomethylation in NSCLC cell lines, the drugs had very different effects on cell viability, DNA damage, apoptosis, and cell cycle. To better understand the molecular pathways regulated by each drug, A549 and H1299 cells were treated with a dose range (0.3–3.0 µM) of AZA or DAC for 48 hours, and effects on gene expression were assessed by microarray analysis. The total number of genes regulated by AZA or DAC, and the overlap of regulated genes, are presented in Table 2. At the lower drug concentration (0.3 µM), AZA and DAC modulated few genes, with DAC modulating 4- to 20-fold more genes than AZA. At the higher drug concentrations (1 and 3 µM), many more genes were modulated, with AZA typically modulating 2- to 5-fold more genes than DAC. Interestingly, the number of genes modulated in common between the 2 drugs was low (6%–22%). For example, in A549 cells, AZA (3 µM) and DAC (3 µM) commonly upregulated 66 genes, while AZA uniquely upregulated 636 genes and DAC uniquely upregulated 413 genes (Table 2).
Table 2.
Number of genes regulated by AZA and/or DAC
| Cell line | [Drug] (μM) | Upregulated genes
|
Downregulated genes
|
||||
|---|---|---|---|---|---|---|---|
| AZA-specific genes | Genes in common | DAC-specific genes | AZA-specific genes | Genes in common | DAC-specific genes | ||
| A549 | 0.3 | 16 | 17 | 139 | 14 | 11 | 55 |
| 1.0 | 279 | 45 | 261 | 273 | 30 | 111 | |
| 3.0 | 636 | 66 | 413 | 560 | 55 | 239 | |
| H1299 | 0.3 | 10 | 55 | 214 | 33 | 45 | 121 |
| 1.0 | 435 | 135 | 238 | 393 | 107 | 170 | |
| 3.0 | 1368 | 173 | 303 | 991 | 153 | 257 | |
Notes: A549 and H1299 cells were treated with AZA or DAC (0–3.0 µM) for 48 hours, and RNA was isolated for evaluation of gene expression using Affymetrix human U133A 2.0 gene chipset. The table shows the number of genes regulated by AZA and DAC at different drug concentrations. Duplicate samples of each were averaged and compared with untreated samples. A fold change of ≥1.7 in gene expression was considered as regulated.
Abbreviations: AZA, azacitidine; DAC, decitabine.
Functional groupings of the modulated genes were determined using Gene Ontology classifications in NextBio. Different biogroups were regulated by each drug. The top 200 biogroups most significantly regulated by each drug (at 3 µM) are shown in Supporting Information Tables 2–5. In H1299 cells, AZA treatment caused a general downregulation of genes within the “cell cycle”, “metabolic process”, and “biosynthetic process” biogroups. DAC treatment of H1299 cells caused a general upregulation of genes within the “cell differentiation” biogroup. In A549 cells, AZA treatment caused downregulation of genes involved in extracellular matrix, while DAC treatment caused downregulation of genes involved in cell cycle. Aside from the regulation of genes related to extracellular matrix, these results are similar to the gene expression data from AML cell lines treated with AZA and DAC.25 Interestingly, AZA treatment of A549 and H1299 cells caused a general upregulation of genes within the “response to DNA damage stimulus” and “DNA repair” biogroups (Figure 9, Supporting Information Tables 2 and 4). These results are consistent with the induction of the dsDNA damage marker (histone-H2AX(ser139) phosphorylation) by AZA in these cells (Figure 5). On the contrary, DAC treatment caused a general downregulation of genes within these bio-groups in A549 cells (Figure 9, Supporting Information Table 3), and DAC did not significantly modulate these biogroups in H1299 cells (Supporting Information Table 5). Collectively, these results indicate that AZA and DAC regulate different cellular pathways.
Figure 9.
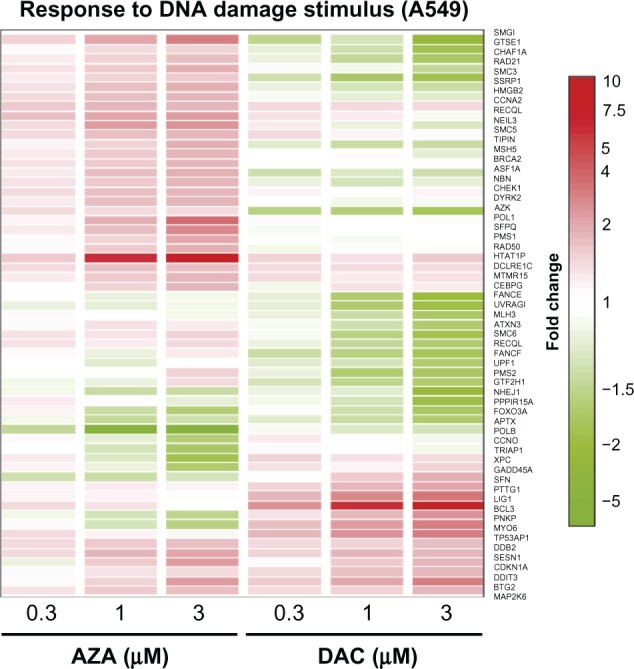
AZA upregulates, while DAC downregulates, genes important in the response to DNA damage stimulus. Gene expression profiling was performed in A549 cells after 48 hours of treatment with AZA or DAC at 0, 0.3, 1, and 3 µM. NextBio (http://www.nextbio.com/) was used to identify regulated Gene Ontology biogroups from lists of regulated genes. The genes displayed represent all genes within the “response to DNA damage stimulus” biogroup that were modulated 1.7-fold or greater by AZA or DAC.
Abbreviations: AZA, azacitidine; DAC, decitabine.
Discussion
In this study, we revealed differential effects of AZA and DAC on cell viability in a panel of NSCLC cell lines, with AZA inducing greater cellular toxicity and markers of apoptosis (PARP cleavage and AnnexinV staining) in comparison to DAC. Furthermore, AZA induced phosphorylation of histone-H2AX(ser139), a marker of dsDNA damage, while DAC had no, or delayed, effect on this endpoint. The striking differences in the response of NSCLC cell lines to these structurally similar cytidine nucleoside analogs further support emerging evidence that the common perception of these agents as mechanistically interchangeable DNA hypomethylating agents should be reconsidered.25,26
Other recent publications also provide data which differentiate AZA from DAC. For example, an in vitro study evaluating the response of a panel of human cancer cell lines to AZA and DAC showed no correlation in the EC50 values of the drugs.24 Another study comparing AZA and DAC activity in the Kasumi-1 AML cell line showed that these drugs had distinct and largely non-overlapping effects on gene expression profiles.26 We have recently demonstrated that AZA and DAC have different effects on cell viability, protein synthesis, cell cycle, and gene expression in AML cell lines.25 Similar to the findings in AML cell lines,25,26 we now demonstrate notable differences between AZA and DAC effects on NSCLC cell lines.
Despite the differences in the activities of AZA and DAC on cytotoxicity and induction of dsDNA damage, both AZA and DAC were active in modulating the DNA-mediated markers of DNMT1 protein depletion and LINE-1 hypomethylation. While DNA methylation undeniably contributes to cancer development and progression,27 it is not clear that the anticancer effects of cytidine azanucleoside analogs are solely driven by their DNA hypomethylating activity. Findings from several clinical studies suggest that DNA hypomethylation may not correlate with clinical response. For example, a study found that DNMT depletion caused by DAC treatment did not necessarily result in clinical response.28 Another clinical trial demonstrated that DAC-induced LINE-1 hypomethylation tended to be greater in patients who did not respond to therapy than in patients who did respond.29 Stresemann et al showed that a subset of patients who responded to AZA treatment did not display detectable DNA hypomethylation.30 These results suggest that mechanisms in addition to, or other than, DNA hypomethylation may be critical for the anticancer effects of these drugs.
DAC’s potent activity on DNA-mediated markers (DNMT1 depletion and DNA hypomethylation) demonstrates that the lack of cytotoxic activity with DAC was not due to a lack of cellular uptake, drug phosphorylation, and DNA incorporation. It is unclear why DAC does not induce dsDNA damage, despite depleting DNMT1 protein and hypomethylating DNA in the NSCLC cell lines tested. The lack of DAC effects on dsDNA damage and on cytotoxicity is consistent with mounting evidence suggesting that DNA damage may be important for the antitumor effects observed with nucleoside analogs.31–34 Published data surrounding DAC-induced DNA damage are mixed. In HeLa and HCT116 cells, DAC induced histone-H2AX(ser139) phosphorylation in a DNMT1-dependent and ataxia-telangiectasia-mutated (ATM)-dependent manner;34 however, other researchers found that DAC induced DNA single-strand breaks, but not DNA double-strand breaks (DSBs).35–37 Our results suggest that AZA induces DSBs in NSCLC cell lines, coincident with its induction of apopto-sis (Figure 5). DAC did not induce as much DSBs and cell death as AZA in A549 cells. Thus, DSBs may correlate with tumor cell death. Dose and schedule will influence mechanism of action, so the potential for cumulative effects of each drug given at low doses or extended schedules should be tested. Furthermore, potential activities of AZA and DAC on cancer stem cell viability and/or differentiation were not tested here.
In summary, we found that AZA and DAC differentially affected the viability of NSCLC cell lines. While AZA and DAC similarly caused DNMT1 depletion and DNA hypomethylation, the drugs differed in their effects on DNA damage, apoptosis, cell cycle, and gene expression. Perhaps a key difference is that AZA can be incorporated into both RNA and DNA, while DAC is only incorporated into DNA.25,38–41 The functional consequences of AZA incorporation into RNA can include (1) alterations in the synthesis and processing of various species of RNA, (2) inhibition of transcription, and (3) disruption of protein synthesis.25,38,42–45 The in vitro anticancer activity of AZA in NSCLC models warrants its evaluation in the clinic. It will be important to consider the multiple mechanisms of AZA activity when selecting therapies for use in combination.
Materials and methods
Cell culture and drug treatments
NSCLC cell lines (H460, H1299, A549, and H1975) were purchased from American Type Culture Collection (ATCC, Manassas, VA). The H23 NSCLC cell line was obtained from the National Cancer Institute (NCI) (Bethesda, MD). Cell lines were cultured in their respective media, as recommended by ATCC and NCI. AZA was manufactured at Aptuit (Greenwich, CT) for Celgene Corporation, while DAC was purchased from Sigma-Aldrich (St Louis, MO). In all experiments, cells were seeded 24 hours before drug treatment and incubated at 37°C and 5% CO2. For cell viability assays, H460, H1299, A549, H23, and H1975 cells were seeded in triplicate at 1 × 103, 1 × 103, 1 × 103, 4 × 103, and 4 × 103 cells per well, respectively, in 96-well plates using 200 µL of medium per well. As the half-lives of AZA and DAC in cell culture are short (∼8–12 hours) (data not shown), fresh drug was added every 24 hours by replacing medium with drug-containing medium. For all other assays, cells were seeded at 0.6–1.2 × 105 cells per well, in 6-well plates, using 4 mL of medium per well, with fresh drug added directly to the medium every 24 hours. At this seeding density, cells are 30%–40% confluent at the start of drug treatments. The concentrations of AZA and DAC used in these experiments are similar to the maximum concentrations (Cmax) achieved in human plasma at clinically used dosages and schedules of administration (3–11 µM AZA and 0.3–1.6 µM DAC).28,46,47
Cell viability
Cell viability was assessed 72 hours after the initial drug treatment, using the CyQUANT assay (Life Technologies Corporation, Carlsbad, CA). Fluorescence was measured with a spectrophotometer (Molecular Devices, Sunnyvale, CA), and EC50 values were calculated from three independent experiments using Prism version 5.01 (GraphPad Software, Inc., La Jolla, CA).
Western analysis
For Western analyses of protein levels, cells were washed with phosphate-buffered saline (PBS) and lysed with radio immuno precipitation assay (RIPA) buffer (Cell Signaling Technology, Inc., Danvers, MA) supplemented with 350 mM NaCl and 0.1% sodium dodecyl sulfate (SDS). Cell lysates were sonicated with two 5-second bursts under low amplitude (20%) using the Digital Sonic Dismembrator (ThermoFisher Scientific, Inc., Waltham, MA). Proteins were separated on 4%–12% Bis-Tris NuPAGE gels (Life Technologies Corporation) and transferred to nitrocellulose membranes. DNMT1, phospho-histone-H2AX(ser139), total histone-H2AX, cleaved-PARP, and alpha-tubulin were detected using the Li-Cor Odyssey imaging system (Li-Cor Biotechnology, Lincoln, NE), following incubation with the appropriate primary and secondary antibodies. The phospho-histone-H2AX(ser139) and cleaved-PARP antibodies were obtained from Cell Signaling Technology, Inc. The total histone-H2AX (C-20) antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The alpha-tubulin and DNMT1 antibodies were purchased from EMD Chemicals, Inc. (Gibbstown, NJ) and Abcam, Inc. (Cambridge, MA), respectively. The goat anti-Rabbit IRDye 680, goat anti-Mouse IRDye 800CW, and donkey anti-Goat IRDye 800CW secondary antibodies were obtained from Li-Cor Biotechnology.
DNA methylation analysis
Genomic DNA was purified from cells using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions. DNA yield was quantitated with a NanoDrop 8000 spectrophotometer (ThermoFisher Scientific, Inc.). Genomic DNA (1 µg/sample) was submitted to EpigenDx (Worcester, MA) for bisulfite conversion and pyrosequencing of LINE-1 elements. Briefly, 1 µg of DNA was bisulfite treated using the Zymo DNA Methylation Kit (Zymo Research, Orange, CA) and eluted in 10 µL volume. DNA eluate (1 µL) was used for polymerase chain reaction (PCR) with biotinylated primers to the LINE-1 locus, converting the PCR product to single-stranded DNA templates. PCR products (each 10 µL) were sequenced by the Pyrosequencing PSQ96 HS System (Biotage AB), following the manufacturer’s instructions (Biotage, Kungsgatan, Sweden). The methylation status of each locus was analyzed individually as a T/C SNP using QCpG software (Biotage). Percentage LINE-1 methylation represents the average percentage methylation of 4 CpG sites in duplicate samples. EpigenDx provided 3 controls for the LINE-1 methylation assay: (1) low methylated DNA control, which is human genomic DNA that has been chemically and enzymatically treated to remove the methyl groups; (2) high methylated DNA control, which is human genomic DNA that has been methylated in vitro; and (3) 50/50 mix control, which is an equal mixture of the low methylated DNA and high methylated DNA controls. The percentages of LINE-1 methylation for the low methylated DNA control, the 50/50 mix control, and the high methylated DNA control were 25.8 ± 8.1, 56.2 ± 4.6, and 86.3 ± 6.5, respectively (data not shown).
Flow cytometry
For cell cycle distribution, cells were stained with the NIM-DAPI reagent (Beckman Coulter, Inc., Fullerton, CA). For measurement of early- and late-apoptotic cell populations, cells were stained with AnnexinV-FITC and 7-AAD reagents (Beckman Coulter, Inc.). Samples were processed according to manufacturer’s instructions and analyzed on a Cell Lab Quanta MPL flow cytometer (Beckman Coulter, Inc.). The effects of treatment were compared using one-way ANOVA, followed by single step method for adjusting P-values in multiple testing with the bioconductor package multcomp.48
Gene expression analysis
Cells were lysed using the TRIzol reagent (Life Technologies Corporation), and total RNA was isolated using the miRNeasy kit (Qiagen). Double-stranded cDNA and biotin-labeled cRNA were synthesized using 100 ng of total RNA with Ambion’s MessageAmp Premier RNA Amplification Kit (ABI, Foster City, CA). Biotin-labeled cRNA (10 µg) was fragmented and hybridized to each human U133A 2.0 genechip (Affymetrix, Santa Clara, CA). The GC-RMA algorithm was used for normalization, and all analyses were done using GeneSpring 7.3 (Agilent, Santa Clara, CA). Averaged signals from biological duplicate samples were used to determine fold change (treated versus untreated), with an absolute fold change of ≥1.7 defining regulated genes. NextBio (http://www.nextbio.com/) was used to identify regulated gene ontology biogroups from lists of regulated genes. The top 200 biogroups are those with the lowest P-values calculated within NextBio.
Supporting information figures and tables
Viability of H23 and H1975 cells was assessed after 72 hours of treatment with AZA or DAC (0–25 µM). Error bars represent the standard error of mean of three independent experiments, with triplicate wells per experiment.
AZA and DAC cause DNMT1 depletion in NSCLC cell lines. A549 and H1299 cells were treated with AZA or DAC (5 µM) for 4, 8, or 16 hours and DNMT1 protein was detected by Western blotting of cell extracts. Alpha-tubulin was used as a loading control.
AZA and DAC reduce DNA methylation in A549 and H1299 cells. LINE-1 DNA methylation was assessed in A549 and H1299 cells after 72 hours of treatment with AZA or DAC (0–3 µM). Percentage LINE-1 methylation represents the average percentage methylation of 4 CpG sites in duplicate samples, with error bars representing the standard deviation.
Viability of A549, H460, and H1299 cells was assessed after 6 days of treatment with AZA or DAC (0–25 µM).
Table S1.
EC50 values for AZA and DAC on NSCLC cell viability (6 days)
| AZA EC50 (μM) | DAC EC50 (μM) | |
|---|---|---|
| A549 | 4.4 | 2.5 |
| H460 | 2.2 | 4.4 |
| H1299 | 4.1 | 0.5 |
Abbreviations: AZA, azacitidine; DAC, decitabine; NSCLC, non-small cell lung cancer.
Table S2.
Top 200 biogroups modulated by azacitidine (AZA) in A549 cells
| A549 cells treated with 3 μM AZA (48 hours)
| ||
|---|---|---|
| Biogroup name | Direction | P value |
| Proteinaceous extracellular matrix | down | 3.40E-18 |
| Extracellular matrix | down | 4.70E-18 |
| Transcription | up | 2.50E-16 |
| Extracellular matrix structural constituent | down | 4.90E-16 |
| Glycosaminoglycan binding | down | 7.30E-15 |
| Polysaccharide binding | down | 9.60E-15 |
| Pattern binding | down | 2.80E-14 |
| Lipid biosynthetic process | down | 3.60E-14 |
| Fibrillar collagen | down | 1.60E-13 |
| Calcium ion binding | down | 2.20E-13 |
| Fibrinogen complex | down | 7.20E-13 |
| Humoral immune response | down | 8.90E-12 |
| Protein binding, bridging | down | 9.00E-12 |
| Collagen | down | 1.70E-11 |
| Response to wounding | down | 6.00E-11 |
| Response to external stimulus | down | 1.50E-10 |
| Platelet activation | down | 2.20E-10 |
| Ligase activity, forming aminoacyl-tRNA and related compounds | up | 3.30E-10 |
| Ligase activity, forming carbon-oxygen bonds | up | 3.30E-10 |
| Response to nutrient | up | 3.70E-10 |
| Carbohydrate binding | down | 3.80E-10 |
| Basement membrane | down | 1.00E-09 |
| Lipid metabolic process | down | 2.20E-09 |
| Response to nutrient levels | up | 2.30E-09 |
| Steroid biosynthetic process | down | 2.30E-09 |
| Collagen binding | down | 2.40E-09 |
| Response to extracellular stimulus | up | 3.70E-09 |
| Inflammatory response | down | 7.80E-09 |
| Nucleoplasm | up | 8.70E-09 |
| Acute inflammatory response | down | 1.30E-08 |
| Response to stress | down | 1.50E-08 |
| Blood pressure regulation | down | 1.50E-08 |
| RNA binding | up | 1.60E-08 |
| Cell motility | down | 3.40E-08 |
| Localization of cell | down | 3.40E-08 |
| Epithelial cell differentiation | down | 4.70E-08 |
| tRNA binding | up | 4.80E-08 |
| Steroid metabolic process | down | 7.20E-08 |
| Endoplasmic reticulum | down | 8.00E-08 |
| Translation | up | 1.30E-07 |
| Fatty acid biosynthetic process | down | 1.90E-07 |
| Parturition | down | 2.50E-07 |
| Sterol metabolic process | down | 2.50E-07 |
| Blood coagulation | down | 2.90E-07 |
| Coagulation | down | 3.10E-07 |
| Humoral immune response mediated by circulating immunoglobulin | down | 3.60E-07 |
| Hemostasis | down | 3.60E-07 |
| Organic acid biosynthetic process | down | 4.40E-07 |
| Regulation of body fluids | down | 6.70E-07 |
| Wound healing | down | 6.90E-07 |
| ER-Golgi intermediate compartment | down | 8.00E-07 |
| Actin binding | down | 9.70E-07 |
| Anion transport | down | 1.10E-06 |
| Extracellular structure organization and biogenesis | down | 1.10E-06 |
| Transaminase activity | up | 1.10E-06 |
| Complement activation | down | 1.20E-06 |
| Extracellular matrix organization and biogenesis | down | 1.20E-06 |
| Calmodulin binding | down | 1.90E-06 |
| Circulation | down | 1.90E-06 |
| Female pregnancy | down | 2.50E-06 |
| Cellular homeostasis | down | 2.60E-06 |
| Morphogenesis of an epithelium | down | 2.70E-06 |
| Cell proliferation | down | 3.10E-06 |
| Alkene metabolic process | down | 3.10E-06 |
| Ribosome biogenesis and assembly | up | 3.20E-06 |
| Complement activation, classical pathway | down | 3.50E-06 |
| Ribonucleoprotein complex biogenesis and assembly | up | 4.90E-06 |
| Sodium:potassium-exchanging ATPase complex | down | 5.20E-06 |
| Transferase activity, transferring nitrogenous groups | up | 5.80E-06 |
| Cell activation | down | 6.10E-06 |
| Endoplasmic reticulum lumen | down | 6.60E-06 |
| Fatty acid metabolic process | down | 6.90E-06 |
| Vesicular fraction | down | 8.20E-06 |
| Cellular ion homeostasis | down | 1.10E-05 |
| Cellular chemical homeostasis | down | 1.10E-05 |
| Positive regulation of immune system process | down | 1.10E-05 |
| Positive regulation of immune response | down | 1.10E-05 |
| Phosphoinositide binding | down | 1.40E-05 |
| Activation of immune response | down | 1.50E-05 |
| Positive regulation of multicellular organismal process | down | 1.60E-05 |
| Cofactor transporter activity | up | 1.60E-05 |
| Soluble fraction | up | 1.70E-05 |
| Enzyme inhibitor activity | down | 1.70E-05 |
| Development of primary sexual characteristics | up | 1.80E-05 |
| NAD binding | down | 1.90E-05 |
| Amine biosynthetic process | up | 1.90E-05 |
| Cytoskeleton | down | 2.20E-05 |
| Lymphocyte mediated immunity | down | 2.40E-05 |
| Receptor binding | down | 2.50E-05 |
| Transcription corepressor activity | up | 2.50E-05 |
| Response to DNA damage stimulus | up | 2.60E-05 |
| Cartilage development | down | 2.70E-05 |
| SNARE complex | up | 2.80E-05 |
| Gastrulation | up | 3.20E-05 |
| mRNA transport | up | 3.50E-05 |
| Epidermis development | down | 3.80E-05 |
| Cell migration | down | 4.20E-05 |
| Immune effector process | down | 4.20E-05 |
| Response to hypoxia | down | 4.30E-05 |
| Leukocyte mediated immunity | down | 4.50E-05 |
| Adaptive immune response | down | 4.90E-05 |
| Adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | down | 4.90E-05 |
| Endopeptidase inhibitor activity | down | 5.00E-05 |
| Protease inhibitor activity | down | 5.00E-05 |
| Oxidoreductase activity, acting on heme group of donors | down | 5.30E-05 |
| Oxidoreductase activity, acting on heme group of donors, oxygen as acceptor | down | 5.30E-05 |
| Cytochrome-c oxidase activity | down | 5.30E-05 |
| Heme-copper terminal oxidase activity | down | 5.30E-05 |
| Germ cell migration | up | 6.00E-05 |
| Coenzyme binding | down | 6.20E-05 |
| Regulation of translation | up | 6.90E-05 |
| Cytokine biosynthetic process | up | 7.30E-05 |
| Neurotransmitter:sodium symporter activity | up | 7.40E-05 |
| Ectoderm development | down | 7.50E-05 |
| Establishment of RNA localization | up | 8.00E-05 |
| RNA transport | up | 8.00E-05 |
| Nucleic acid transport | up | 8.00E-05 |
| Transcription factor binding | up | 8.30E-05 |
| Regulation of immune response | down | 8.50E-05 |
| Regulation of immune system process | down | 8.50E-05 |
| Ligase activity | up | 8.60E-05 |
| RNA localization | up | 9.10E-05 |
| Neurotransmitter transporter activity | up | 9.30E-05 |
| RNA export from nucleus | up | 9.80E-05 |
| Phospholipid binding | down | 9.90E-05 |
| Cell cycle | up | 0.0001 |
| Cytosol | down | 0.0001 |
| Cytoskeletal protein binding | down | 0.0001 |
| Response to endogenous stimulus | up | 0.0001 |
| Gonad development | up | 0.0001 |
| Nucleobase, nucleoside, nucleotide and nucleic acid transport | up | 0.0001 |
| Nucleolus | up | 0.0001 |
| Regulation of cytokine biosynthetic process | up | 0.0001 |
| Rhythmic process | up | 0.0001 |
| Reproductive structure development | up | 0.0002 |
| Mitochondrion organization and biogenesis | up | 0.0002 |
| Structural constituent of cytoskeleton | down | 0.0002 |
| Sex differentiation | up | 0.0002 |
| Transcription repressor activity | up | 0.0002 |
| Peroxidase activity | down | 0.0002 |
| Oxidoreductase activity, acting on peroxide as acceptor | down | 0.0002 |
| Laminin-1 complex | down | 0.0002 |
| Transcription cofactor activity | up | 0.0002 |
| Female sex differentiation | up | 0.0002 |
| Development of primary female sexual characteristics | up | 0.0002 |
| Oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | down | 0.0002 |
| Nitrogen compound biosynthetic process | up | 0.0002 |
| Cell structure disassembly during apoptosis | up | 0.0003 |
| Amino acid transport | up | 0.0003 |
| Acyl-CoA binding | down | 0.0003 |
| Response to dsRNA | up | 0.0003 |
| Neuron development | down | 0.0003 |
| Integrator complex | up | 0.0003 |
| Immune response | down | 0.0003 |
| Protein dimerization activity | up | 0.0004 |
| Laminin complex | down | 0.0004 |
| Cofactor binding | down | 0.0004 |
| Germ-line sex determination | down | 0.0004 |
| Intramolecular oxidoreductase activity | down | 0.0004 |
| DNA repair | up | 0.0004 |
| Cell soma | down | 0.0004 |
| Cellular morphogenesis during differentiation | down | 0.0004 |
| RNA polymerase II transcription factor activity | up | 0.0005 |
| Regulation of epithelial cell proliferation | down | 0.0005 |
| Regulation of biosynthetic process | up | 0.0005 |
| UDP-glycosyltransferase activity | up | 0.0005 |
| Pyridoxal phosphate binding | up | 0.0005 |
| Lipid binding | down | 0.0005 |
| Positive regulation of programmed cell death | up | 0.0005 |
| Helicase activity | up | 0.0006 |
| Cell redox homeostasis | down | 0.0006 |
| Cell death | up | 0.0006 |
| Death | up | 0.0006 |
| Epithelial cell proliferation | down | 0.0006 |
| Mesenchymal cell development | down | 0.0006 |
| Ovulation | up | 0.0006 |
| Positive regulation of locomotion | down | 0.0006 |
| Positive regulation of cell motility | down | 0.0006 |
| DNA catabolic process | up | 0.0006 |
| Cell differentiation | down | 0.0006 |
| Basal lamina | down | 0.0007 |
| Insulin-like growth factor binding mesenchymal cell differentiation | Down down |
0.0007 0.0007 |
| Sequestering of metal ion | down | 0.0007 |
| Neurotransmitter transport | up | 0.0007 |
| Specific RNA polymerase II transcription factor activity | up | 0.0007 |
| Intramolecular oxidoreductase activity, transposing C=C bonds | down | 0.0007 |
| Cellular component disassembly | up | 0.0008 |
| Heme binding | down | 0.0008 |
| Tetrapyrrole binding | down | 0.0008 |
| Presynaptic active zone | up | 0.0008 |
| Amine transport | up | 0.0009 |
| Sequestering of calcium ion | down | 0.0009 |
| Cell recognition | down | 0.0009 |
| Endoplasmic reticulum part | down | 0.0009 |
| Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | down | 0.001 |
| Myosin binding | down | 0.001 |
| Lyase activity | up | 0.001 |
| Transferase activity, transferring hexosyl groups | down | 0.001 |
| Neuron differentiation | down | 0.001 |
Notes: Functional groupings of the modulated genes were determined using Gene Ontology classifications in NextBio. The top 200 biogroups most significantly regulated by AZA (at 3 µM) are shown.
Table S3.
Top 196 biogroups modulated by decitabine (DAC) in A549 cells
| A549 cells treated with 3 μM DAC (48 hours)
| ||
|---|---|---|
| Biogroup name | Direction | P value |
| Mitosis | down | 2.00E-09 |
| Cell cycle | down | 2.90E-09 |
| Cell division | down | 5.70E-08 |
| Transferase activity, transferring sulfur-containing groups | up | 5.80E-08 |
| Meiosis | down | 1.50E-07 |
| Meiotic cell cycle | down | 1.70E-07 |
| Response to DNA damage stimulus | down | 1.80E-07 |
| Male gamete generation | up | 6.10E-07 |
| Response to endogenous stimulus | down | 7.60E-07 |
| Chromosome segregation | down | 2.10E-06 |
| Aromatic compound metabolic process | up | 2.70E-06 |
| Phenol metabolic process | up | 2.70E-06 |
| Structural constituent of cytoskeleton | up | 5.10E-06 |
| Sister chromatid cohesion | down | 5.90E-06 |
| Cellular lipid catabolic process | up | 6.70E-06 |
| DNA repair | down | 7.20E-06 |
| Alkali metal ion binding | up | 9.00E-06 |
| Regulation of neurotransmitter levels | up | 1.10E-05 |
| DNA damage response, signal transduction | down | 1.30E-05 |
| Cofactor transporter activity | up | 1.60E-05 |
| Sulfotransferase activity | up | 1.70E-05 |
| Intermediate filament | up | 3.20E-05 |
| Neurotransmitter:sodium symporter activity | up | 3.40E-05 |
| Chromatin assembly | down | 3.90E-05 |
| Neurotransmitter transporter activity | up | 4.60E-05 |
| Cytokinesis | down | 5.10E-05 |
| Chromosome | down | 5.30E-05 |
| Mitotic spindle organization and biogenesis | down | 5.30E-05 |
| Negative regulation of enzyme activity | up | 5.60E-05 |
| Establishment of mitotic spindle localization | down | 6.50E-05 |
| Establishment of spindle localization | down | 6.50E-05 |
| Spindle localization | down | 6.50E-05 |
| Retinol binding | up | 6.50E-05 |
| Microtubule organizing center part | down | 7.60E-05 |
| Mitotic sister chromatid segregation | down | 8.30E-05 |
| Alcohol metabolic process | up | 8.60E-05 |
| Sister chromatid segregation | down | 9.30E-05 |
| Catabolic process | up | 0.0001 |
| Soluble fraction | up | 0.0001 |
| Retinal binding | up | 0.0001 |
| Positive regulation of programmed cell death | up | 0.0001 |
| Steroid biosynthetic process | up | 0.0002 |
| Response to stress | down | 0.0002 |
| Gamma-tubulin complex | down | 0.0002 |
| Mitotic chromosome condensation | down | 0.0002 |
| Transporter activity | up | 0.0002 |
| Phosphopyruvate hydratase complex | up | 0.0002 |
| Amino acid derivative metabolic process | up | 0.0003 |
| Vitamin binding | up | 0.0003 |
| Lipid catabolic process | up | 0.0003 |
| Nuclear chromosome | down | 0.0003 |
| Retinoid binding | up | 0.0003 |
| Isoprenoid binding | up | 0.0003 |
| Homologous chromosome segregation | down | 0.0003 |
| Meiotic chromosome segregation | down | 0.0003 |
| Meiotic spindle organization and biogenesis | down | 0.0003 |
| Cell differentiation | up | 0.0003 |
| NADP binding | down | 0.0004 |
| Steroid metabolic process | up | 0.0004 |
| Lipid raft | up | 0.0004 |
| Cohesin complex | down | 0.0004 |
| Meiosis I | down | 0.0004 |
| Sodium:potassium-exchanging ATPase complex | up | 0.0004 |
| Negative regulation of cell proliferation | up | 0.0004 |
| Actin binding | down | 0.0005 |
| Nuclear matrix | down | 0.0005 |
| Cytoskeletal protein binding | down | 0.0005 |
| Protein kinase inhibitor activity | up | 0.0005 |
| Cell proliferation | up | 0.0006 |
| Cytoskeleton | down | 0.0006 |
| Cytoskeleton organization and biogenesis | down | 0.0006 |
| Fat cell differentiation | down | 0.0006 |
| Hormone metabolic process | up | 0.0006 |
| Positive regulation of progression through cell cycle | down | 0.0006 |
| Kinase inhibitor activity | up | 0.0006 |
| Oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | down | 0.0007 |
| Neurotransmitter transport | up | 0.0007 |
| Membrane invagination | down | 0.0008 |
| Endocytosis | down | 0.0008 |
| Amide metabolic process | up | 0.0008 |
| Spindle | down | 0.0008 |
| Ion transport | up | 0.0009 |
| Blastocyst growth | down | 0.0009 |
| Interleukin binding | down | 0.0009 |
| RNA export from nucleus | down | 0.0009 |
| Tubulin binding | down | 0.0009 |
| Epidermis development | up | 0.0009 |
| Neurotransmitter metabolic process | up | 0.0011 |
| Translation activator activity | up | 0.0011 |
| Spindle pole | down | 0.0011 |
| Synaptic transmission | up | 0.0012 |
| Intracellular cyclic nucleotide activated cation channel complex | up | 0.0012 |
| Biogenic amine metabolic process | up | 0.0012 |
| Cell fate determination | up | 0.0013 |
| Oxidoreductase activity, acting on iron-sulfur proteins as donors | up | 0.0013 |
| Ion channel activity | up | 0.0013 |
| Lipoprotein binding | down | 0.0014 |
| Positive regulation of neurogenesis | down | 0.0014 |
| Cytosol | up | 0.0018 |
| Microtubule organizing center | down | 0.002 |
| Microtubule | down | 0.002 |
| Glutathione peroxidase activity | up | 0.0021 |
| Odontogenesis | down | 0.0022 |
| Passive transmembrane transporter activity | up | 0.0022 |
| Transmission of nerve impulse | up | 0.0023 |
| Oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor | down | 0.0024 |
| Dynein binding | down | 0.0024 |
| Humoral immune response | down | 0.0024 |
| Ectoderm development | up | 0.0025 |
| Arginine metabolic process | up | 0.0025 |
| Myosin binding | down | 0.0025 |
| Lipid biosynthetic process | up | 0.0026 |
| Muscle contraction | up | 0.0027 |
| Mitochondrion organization and biogenesis | up | 0.0027 |
| Fat-soluble vitamin metabolic process | up | 0.0028 |
| Female gamete generation | down | 0.0028 |
| Urea cycle intermediate metabolic process | up | 0.0029 |
| Inclusion body | down | 0.0029 |
| Folic acid transporter activity | down | 0.0029 |
| Protein heterodimerization activity | down | 0.003 |
| Angiogenesis | up | 0.003 |
| Replication fork | down | 0.0031 |
| Nucleoside metabolic process | down | 0.0031 |
| Regulation of axonogenesis | down | 0.0032 |
| Anatomical structure formation | up | 0.0033 |
| Protein kinase regulator activity | up | 0.0034 |
| Lipid metabolic process | up | 0.0037 |
| Glycoprotein binding | up | 0.0037 |
| Pyridoxal phosphate binding | up | 0.0037 |
| Blood vessel morphogenesis | up | 0.004 |
| Carbohydrate metabolic process | up | 0.0041 |
| Tissue regeneration | up | 0.0041 |
| Regeneration | up | 0.0041 |
| Germ cell development | up | 0.0042 |
| Growth factor binding | down | 0.0042 |
| Peptide transporter activity | up | 0.0043 |
| Nitrogen compound biosynthetic process | up | 0.0044 |
| Cytokine binding | down | 0.0044 |
| Nitric oxide metabolic process | up | 0.0047 |
| Nitric oxide biosynthetic process | up | 0.0047 |
| Centrosome | down | 0.0047 |
| Embryonic morphogenesis | down | 0.0048 |
| Regulation of neurogenesis | down | 0.0048 |
| Oxidoreductase activity, acting on the aldehyde or oxo group of donors | up | 0.0048 |
| Cytokinesis during cell cycle | down | 0.0049 |
| Cell–cell signaling | up | 0.005 |
| Calmodulin binding | up | 0.005 |
| Structure-specific DNA binding | down | 0.0051 |
| Oxidoreductase activity, acting on the CH–CH group of donors | down | 0.0051 |
| Peroxidase activity | up | 0.0051 |
| Oxidoreductase activity, acting on peroxide as acceptor | up | 0.0051 |
| Microfibril | up | 0.0052 |
| Protein–DNA complex assembly | down | 0.0052 |
| Vasculature development | up | 0.0054 |
| Excretion | up | 0.0055 |
| mRNA transport | down | 0.0056 |
| Identical protein binding | up | 0.0056 |
| Vitamin transporter activity | down | 0.0057 |
| Response to organic cyclic substance | up | 0.0059 |
| Response to alkaloid | up | 0.0059 |
| Kinase regulator activity | up | 0.006 |
| Chromatin | down | 0.006 |
| Electron carrier activity | up | 0.0061 |
| Vitamin biosynthetic process | down | 0.0062 |
| RNA transport | down | 0.0067 |
| Nucleic acid transport | down | 0.0067 |
| Establishment of RNA localization | down | 0.0067 |
| Protein domain specific binding | up | 0.0068 |
| Homophilic cell adhesion | down | 0.0068 |
| RNA localization | down | 0.0069 |
| Hormone biosynthetic process | up | 0.007 |
| Protein dimerization activity | down | 0.0071 |
| RNA binding | down | 0.0073 |
| Blastocyst development | down | 0.0074 |
| Cyclin binding | up | 0.0075 |
| Nucleobase, nucleoside, nucleotide and nucleic acid transport | down | 0.0077 |
| Cartilage development | down | 0.0077 |
| Folic acid binding | down | 0.0079 |
| Positive regulation of developmental process | down | 0.0081 |
| Chordate embryonic development | down | 0.0082 |
| NAD binding | up | 0.0082 |
| Cofactor binding | up | 0.0082 |
| Vesicle docking during exocytosis | up | 0.0084 |
| Developmental maturation | up | 0.0085 |
| Hydrolase activity, acting on carbon–nitrogen (but not peptide) bonds, in cyclic amidines | up | 0.0086 |
| Lysosome | up | 0.0086 |
| Embryonic digit morphogenesis | down | 0.0086 |
| DNA helicase activity | down | 0.0089 |
| Axon guidance | down | 0.0091 |
| Membrane docking | up | 0.0093 |
| Vesicle docking | up | 0.0093 |
| Voltage-gated sodium channel complex | down | 0.0093 |
| mRNA binding | up | 0.0094 |
| Establishment of organelle localization | down | 0.0096 |
| Vitamin metabolic process | up | 0.0096 |
| Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | down | 0.0099 |
Notes: Functional groupings of the modulated genes were determined using Gene Ontology classifications in NextBio. The top 196 biogroups most significantly regulated by DAC (at 3 µM) are shown.
Table S4.
Top 200 biogroups modulated by azacitidine (AZA) in H1299 cells
| H1299 cells treated with 3 μM AZA (48 hours)
| ||
|---|---|---|
| Biogroup name | Direction | P value |
| Transcription | up | 1.90E-25 |
| Cell cycle | down | 7.60E-25 |
| Mitosis | down | 8.00E-24 |
| Cell division | down | 1.00E-22 |
| Cytoskeleton | down | 5.60E-14 |
| Microtubule | down | 1.30E-13 |
| Spindle | down | 1.80E-13 |
| Mitochondrion | down | 1.80E-12 |
| Sterol metabolic process | down | 1.40E-11 |
| Chromosome | down | 1.20E-10 |
| Alcohol metabolic process | down | 2.20E-10 |
| Ligase activity | up | 2.40E-10 |
| Lipid biosynthetic process | down | 2.70E-10 |
| Steroid biosynthetic process | down | 3.00E-10 |
| Mitotic sister chromatid segregation | down | 1.90E-09 |
| Steroid metabolic process | down | 2.50E-09 |
| Sister chromatid segregation | down | 2.90E-09 |
| Endoplasmic reticulum | down | 3.60E-09 |
| Envelope | down | 4.70E-09 |
| Lipid metabolic process | down | 2.60E-08 |
| Response to nutrient | down | 7.00E-08 |
| Collagen binding | down | 1.60E-07 |
| Centrosome | down | 1.70E-07 |
| Wound healing | down | 1.90E-07 |
| Response to nutrient levels | down | 2.00E-07 |
| Intramolecular oxidoreductase activity | down | 2.50E-07 |
| Response to extracellular stimulus | down | 2.70E-07 |
| Mitochondrial membrane | down | 2.80E-07 |
| Microtubule organizing center | down | 2.80E-07 |
| Acid–amino acid ligase activity | up | 3.10E-07 |
| Cell proliferation | down | 3.60E-07 |
| Blood coagulation | down | 3.70E-07 |
| Establishment of chromosome localization | down | 4.40E-07 |
| Coagulation | down | 4.90E-07 |
| Chromosome segregation | down | 5.00E-07 |
| Nitrogen compound catabolic process | down | 5.20E-07 |
| Kinase binding | up | 5.30E-07 |
| Beta-catenin binding | down | 5.50E-07 |
| Nucleoplasm | up | 6.10E-07 |
| Enzyme inhibitor activity | down | 8.80E-07 |
| Alcohol catabolic process | down | 9.80E-07 |
| Hemostasis | down | 1.10E-06 |
| Transcription cofactor activity | up | 1.10E-06 |
| Transcription repressor activity | up | 1.20E-06 |
| Midbody | down | 1.30E-06 |
| Ligase activity, forming carbon-nitrogen bonds | up | 1.70E-06 |
| Establishment of organelle localization | down | 2.00E-06 |
| Germ-line sex determination | down | 2.00E-06 |
| Oligosaccharyl transferase complex | down | 2.10E-06 |
| Response to external stimulus | down | 2.20E-06 |
| Oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor | down | 2.20E-06 |
| Amine catabolic process | down | 2.20E-06 |
| Cytoskeleton organization and biogenesis | down | 2.20E-06 |
| Cofactor binding | down | 2.30E-06 |
| Coenzyme binding | down | 2.70E-06 |
| Transcription corepressor activity | up | 3.00E-06 |
| Cell differentiation | up | 3.20E-06 |
| mRNA binding | down | 3.30E-06 |
| Meiotic chromosome segregation | down | 3.60E-06 |
| Homologous chromosome segregation | down | 3.60E-06 |
| Nuclear envelope-endoplasmic reticulum network | down | 3.90E-06 |
| Oxidoreductase activity, acting on the CH–CH group of donors, NAD or NADP as acceptor | down | 5.00E-06 |
| Intramolecular oxidoreductase activity, transposing C=C bonds | down | 5.30E-06 |
| Mitotic chromosome condensation | down | 5.50E-06 |
| Endoplasmic reticulum part | down | 5.50E-06 |
| Transcription factor binding | up | 5.60E-06 |
| Organic acid transport | up | 5.70E-06 |
| Carboxylic acid transport | up | 5.70E-06 |
| Acyl-CoA binding | down | 5.70E-06 |
| DNA-directed RNA polymerase II, holoenzyme | up | 5.80E-06 |
| Interphase of mitotic cell cycle | down | 6.00E-06 |
| Primary sex determination | down | 6.00E-06 |
| Cell-matrix adhesion | down | 6.20E-06 |
| Hormone activity | down | 6.30E-06 |
| Organic acid transmembrane transporter activity | up | 6.30E-06 |
| Mitochondrial inner membrane | down | 7.30E-06 |
| Cell-substrate adhesion | down | 7.60E-06 |
| Transcription activator activity | up | 7.70E-06 |
| Mitotic spindle organization and biogenesis | down | 7.90E-06 |
| Lyase activity | down | 8.10E-06 |
| Sterol transport | down | 8.70E-06 |
| Arginine metabolic process | down | 8.70E-06 |
| Chromatin assembly | down | 1.00E-05 |
| Nitrogen compound biosynthetic process | down | 1.10E-05 |
| RNA polymerase II transcription factor activity | up | 1.10E-05 |
| Transaminase activity | up | 1.20E-05 |
| Meiotic spindle organization and biogenesis | down | 1.30E-05 |
| Organelle localization | down | 1.40E-05 |
| Isomerase activity | down | 1.40E-05 |
| Interphase | down | 1.40E-05 |
| Urea cycle intermediate metabolic process | down | 1.50E-05 |
| Fatty acid biosynthetic process | down | 1.70E-05 |
| Receptor binding | down | 1.70E-05 |
| Regulation of body fluids | down | 1.80E-05 |
| Condensin complex | down | 1.80E-05 |
| Regulation of coagulation | down | 1.90E-05 |
| Nuclear envelope | down | 2.20E-05 |
| Caveola | down | 2.30E-05 |
| Organic acid biosynthetic process | down | 2.30E-05 |
| Meiotic cell cycle | down | 2.30E-05 |
| Amino acid transport | up | 2.50E-05 |
| NADP binding | down | 2.80E-05 |
| Protein dimerization activity | up | 2.90E-05 |
| Spindle pole | down | 3.40E-05 |
| Ubiquitin–protein ligase activity | up | 3.50E-05 |
| Transferase activity, transferring nitrogenous groups | up | 3.60E-05 |
| Nucleoside metabolic process | down | 3.80E-05 |
| Structural constituent of cytoskeleton | down | 3.90E-05 |
| Carbohydrate catabolic process | down | 4.10E-05 |
| Endoplasmic reticulum membrane | down | 4.30E-05 |
| Nucleosome | down | 4.40E-05 |
| Nucleotide catabolic process | down | 4.60E-05 |
| Cellular chemical homeostasis | down | 4.70E-05 |
| Cellular ion homeostasis | down | 4.70E-05 |
| Cytokinesis | down | 4.90E-05 |
| Muscle cell differentiation | up | 5.00E-05 |
| Myeloid cell differentiation | up | 5.20E-05 |
| Catabolic process | down | 5.20E-05 |
| Oxygen and reactive oxygen species metabolic process | down | 5.20E-05 |
| Chromatin | down | 5.30E-05 |
| Epidermis development | down | 5.40E-05 |
| Oxidoreductase activity, acting on the CH–NH group of donors | down | 5.60E-05 |
| SNARE complex | up | 5.60E-05 |
| Ligase activity, forming carbon–oxygen bonds | up | 5.70E-05 |
| Ligase activity, forming aminoacyl–tRNA and related compounds | up | 5.70E-05 |
| Soluble fraction | up | 5.80E-05 |
| Endopeptidase inhibitor activity | down | 5.90E-05 |
| Protease inhibitor activity | down | 5.90E-05 |
| Small protein conjugating enzyme activity | up | 6.00E-05 |
| AP-type membrane coat adaptor complex | down | 6.40E-05 |
| Cell–cell signaling | down | 6.70E-05 |
| Amine transport | up | 6.80E-05 |
| Response to DNA damage stimulus | up | 7.20E-05 |
| Male sex determination | down | 7.20E-05 |
| Cell–cell adhesion | down | 8.00E-05 |
| Protein heterodimerization activity | down | 8.00E-05 |
| Enzyme binding | up | 8.40E-05 |
| Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | down | 9.10E-05 |
| Ectoderm development | down | 9.40E-05 |
| Positive regulation of progression through cell cycle | down | 0.0001 |
| One-carbon compound metabolic process | down | 0.0001 |
| Heterogeneous nuclear ribonucleoprotein complex | down | 0.0001 |
| Response to stress | down | 0.0001 |
| Response to endogenous stimulus | up | 0.0001 |
| Cell death | up | 0.0001 |
| Death | up | 0.0001 |
| Meiosis | down | 0.0001 |
| Dioxygenase activity | up | 0.0001 |
| Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen | up | 0.0001 |
| Isoprenoid biosynthetic process | down | 0.0001 |
| Carbon–carbon lyase activity | down | 0.0001 |
| Lipid digestion | down | 0.0001 |
| Nitric oxide metabolic process | down | 0.0001 |
| Nitric oxide biosynthetic process | down | 0.0001 |
| Cellular homeostasis | down | 0.0001 |
| Positive regulation of locomotion | down | 0.0001 |
| Positive regulation of cell motility | down | 0.0001 |
| Sulfur compound biosynthetic process | up | 0.0001 |
| Proton-transporting ATP synthase complex, catalytic core F(1) | down | 0.0001 |
| Mitochondrial proton-transporting ATP synthase complex | down | 0.0002 |
| Muscle development | up | 0.0002 |
| Integrator complex | up | 0.0002 |
| Caspase inhibitor activity | down | 0.0002 |
| Fatty acid binding | down | 0.0002 |
| Isoprenoid metabolic process | down | 0.0002 |
| Blood vessel morphogenesis | up | 0.0002 |
| Vasculogenesis | up | 0.0002 |
| Kinetochore | down | 0.0002 |
| Low-density lipoprotein binding | down | 0.0002 |
| Cell structure disassembly during apoptosis | up | 0.0002 |
| Endoplasmic reticulum lumen | down | 0.0002 |
| Negative regulation of multicellular organismal process | down | 0.0002 |
| Intestinal absorption | down | 0.0002 |
| Organelle outer membrane | down | 0.0002 |
| Proton-transporting two-sector ATPase complex, catalytic domain | down | 0.0002 |
| Regulation of transforming growth factor beta receptor signaling pathway | down | 0.0002 |
| Clathrin adaptor complex | down | 0.0002 |
| Fatty acid metabolic process | down | 0.0003 |
| Replication fork | down | 0.0003 |
| Lipoprotein binding | down | 0.0003 |
| Insemination | up | 0.0003 |
| Behavior | down | 0.0003 |
| Leukocyte differentiation | up | 0.0003 |
| Single-stranded RNA binding | down | 0.0003 |
| Histone acetyltransferase complex | up | 0.0003 |
| Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen | up | 0.0003 |
| Hemopoiesis | up | 0.0003 |
| Outer kinetochore of condensed | down | 0.0003 |
| chromosome | ||
| Response to virus | up | 0.0003 |
| Vasculature development | up | 0.0003 |
| DNA-directed RNA polymerase complex | up | 0.0003 |
| Perinuclear region of cytoplasm | down | 0.0003 |
| Sterol binding | down | 0.0003 |
| Generation of precursor metabolites and energy | down | 0.0003 |
| Copulation | up | 0.0004 |
| Spindle localization | down | 0.0004 |
| Establishment of mitotic spindle localization | down | 0.0004 |
| Establishment of spindle localization | down | 0.0004 |
| Neural crest cell development | down | 0.0004 |
| GTPase inhibitor activity | down | 0.0004 |
Notes: Functional groupings of the modulated genes were determined using Gene Ontology classifications in NextBio. The top 200 biogroups most significantly regulated by AZA (at 3 µM) are shown.
Table S5.
Top 200 biogroups modulated by decitabine (DAC) in H1299 cells
| H1299 cells treated with 3 μM DAC (48 hours)
| |||
|---|---|---|---|
| Biogroup name | Direction | P value | |
| Cofactor binding | down | 4.70E-12 | |
| Lipid metabolic process | down | 5.10E-12 | |
| Cell differentiation | up | 2.50E-08 | |
| Coenzyme binding | down | 3.70E-08 | |
| Transcription | up | 1.00E-07 | |
| Inner ear development | up | 2.20E-07 | |
| Cell fate determination | up | 2.50E-07 | |
| Fatty acid metabolic process | down | 2.90E-07 | |
| Collagen binding | up | 4.90E-07 | |
| Oxidoreductase activity, acting on the CH–OH group of donors, NAD or NADP as acceptor | down | 1.20E-06 | |
| Enzyme inhibitor activity | up | 1.30E-06 | |
| Nucleosome | up | 1.30E-06 | |
| Aldehyde metabolic process | down | 1.40E-06 | |
| Sensory organ development | up | 2.30E-06 | |
| Oxidoreductase activity, acting on CH–OH group of donors | down | 2.50E-06 | |
| Response to external stimulus | up | 2.80E-06 | |
| Hormone biosynthetic process | up | 3.00E-06 | |
| CoA–ligase activity | down | 4.00E-06 | |
| Insulin-like growth factor binding | up | 4.20E-06 | |
| Response to stress | up | 4.40E-06 | |
| Mitochondrion | down | 5.20E-06 | |
| Acid–thiol ligase activity | down | 6.30E-06 | |
| Proteinaceous extracellular matrix | up | 7.40E-06 | |
| Transcription repressor activity | up | 7.90E-06 | |
| Extracellular matrix | up | 8.60E-06 | |
| Muscle cell differentiation | up | 9.40E-06 | |
| Response to wounding | up | 9.50E-06 | |
| Alcohol metabolic process | down | 9.90E-06 | |
| Enzyme regulator activity | up | 9.90E-06 | |
| Neurotransmitter metabolic process | up | 1.00E-05 | |
| Muscle fiber development | up | 1.10E-05 | |
| Skeletal muscle fiber development | up | 1.10E-05 | |
| Peroxisome | down | 1.10E-05 | |
| Microbody | down | 1.10E-05 | |
| Ligase activity, forming carbon-sulfur bonds | down | 1.40E-05 | |
| Regulation of epidermis development | up | 2.00E-05 | |
| Cell fate commitment | up | 2.20E-05 | |
| Hormone metabolic process | up | 2.20E-05 | |
| Sterol metabolic process | down | 2.30E-05 | |
| Inflammatory response | up | 2.30E-05 | |
| Oxidoreductase activity, acting on the CH–CH group of donors, NAD or NADP as acceptor | down | 2.60E-05 | |
| Death | up | 3.20E-05 | |
| Cell death | up | 3.20E-05 | |
| Steroid metabolic process | down | 3.50E-05 | |
| Lipid biosynthetic process | down | 3.80E-05 | |
| Lyase activity | down | 4.10E-05 | |
| Glycosaminoglycan binding | up | 4.50E-05 | |
| Polysaccharide binding | up | 5.30E-05 | |
| Pyridoxal phosphate binding | down | 5.50E-05 | |
| Muscle development | up | 6.70E-05 | |
| Nitrogen compound biosynthetic process | down | 6.90E-05 | |
| Protease inhibitor activity | up | 7.60E-05 | |
| Endopeptidase inhibitor activity | up | 7.60E-05 | |
| Phosphatase activator activity | up | 8.00E-05 | |
| Phenol metabolic process | up | 8.40E-05 | |
| Epidermis development | up | 8.50E-05 | |
| Regulation of neurotransmitter levels | up | 9.20E-05 | |
| Pattern binding | up | 9.60E-05 | |
| Cofactor catabolic process | down | 9.80E-05 | |
| Positive regulation of developmental process | up | 1.00E-04 | |
| Vitamin binding | down | 0.0001 | |
| Oxidoreductase activity, acting on the CH–CH group of donors | down | 0.0001 | |
| Amino acid derivative metabolic process | up | 0.0001 | |
| Chromatin assembly | up | 0.0001 | |
| Protein–DNA complex assembly | up | 0.0001 | |
| Amino acid derivative biosynthetic process | up | 0.0001 | |
| Positive regulation of cell differentiation | up | 0.0001 | |
| Acute inflammatory response | up | 0.0001 | |
| Dopamine metabolic process | up | 0.0001 | |
| Growth factor binding | up | 0.0002 | |
| Endothelial cell development | down | 0.0002 | |
| Transcription corepressor activity | up | 0.0002 | |
| Keratinocyte differentiation | up | 0.0002 | |
| Ectoderm development | up | 0.0002 | |
| Cellular respiration | down | 0.0002 | |
| RNA polymerase II transcription elongation factor activity | up | 0.0002 | |
| Angiogenesis | up | 0.0003 | |
| Calcium-dependent phospholipid binding | down | 0.0003 | |
| Suckling behavior | down | 0.0003 | |
| Oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | down | 0.0003 | |
| Germ-line sex determination | down | 0.0003 | |
| RNA polymerase II transcription factor activity | up | 0.0003 | |
| Protein kinase inhibitor activity | up | 0.0004 | |
| Translation activator activity | up | 0.0004 | |
| Regulation of Notch signaling pathway | up | 0.0004 | |
| Fatty acid biosynthetic process | down | 0.0004 | |
| Kinase inhibitor activity | up | 0.0004 | |
| Regulation of cell differentiation | up | 0.0004 | |
| ER-Golgi intermediate compartment | up | 0.0005 | |
| UDP-glycosyltransferase activity | down | 0.0005 | |
| Cell maturation | up | 0.0005 | |
| Cell surface | down | 0.0005 | |
| Inner ear receptor cell fate commitment | up | 0.0005 | |
| Organic acid biosynthetic process | down | 0.0005 | |
| Negative regulation of signal transduction | up | 0.0006 | |
| Hydro–Lyase activity | down | 0.0006 | |
| Epidermal cell differentiation | up | 0.0007 | |
| Inner ear morphogenesis | up | 0.0007 | |
| Regulation of cell growth | up | 0.0007 | |
| Response to bacterium | up | 0.0007 | |
| Blood vessel morphogenesis | up | 0.0007 | |
| Carbohydrate metabolic process | down | 0.0007 | |
| Lipoprotein binding | down | 0.0007 | |
| Multicellular organismal movement | down | 0.0008 | |
| Aromatic compound metabolic process | up | 0.0008 | |
| Amine biosynthetic process | down | 0.0008 | |
| tRNA binding | down | 0.0008 | |
| Cell migration | up | 0.0008 | |
| Transcription elongation factor complex | up | 0.0009 | |
| Glutathione peroxidase activity | up | 0.0009 | |
| Oxidoreductase activity, acting on the CH–NH group of donors, NAD or NADP as acceptor | down | 0.0009 | |
| Primary sex determination | down | 0.0009 | |
| Negative regulation of cell differentiation | up | 0.001 | |
| Defense response to bacterium | up | 0.001 | |
| Nuclear envelope | down | 0.001 | |
| Ear morphogenesis | up | 0.0011 | |
| Ligase activity, forming carbon–oxygen bonds | down | 0.0011 | |
| Ligase activity, forming aminoacyl–tRNA and related compounds | down | 0.0011 | |
| Generation of precursor metabolites and energy | down | 0.0011 | |
| Developmental maturation | up | 0.0011 | |
| Peripheral nervous system development | down | 0.0011 | |
| Extracellular matrix structural constituent | up | 0.0012 | |
| Oxidoreductase activity, acting on the aldehyde or oxo group of donors | down | 0.0012 | |
| Biogenic amine metabolic process | up | 0.0012 | |
| Epidermis morphogenesis | up | 0.0013 | |
| Endothelial cell differentiation | down | 0.0013 | |
| Endoplasmic reticulum | down | 0.0013 | |
| Carbon–oxygen lyase activity | down | 0.0013 | |
| Germ cell development | up | 0.0014 | |
| Cell proliferation | down | 0.0015 | |
| Peroxidase activity | up | 0.0015 | |
| Oxidoreductase activity, acting on peroxide as acceptor | up | 0.0015 | |
| Specific RNA polymerase II transcription factor activity | up | 0.0015 | |
| Positive regulation of programmed cell death | up | 0.0016 | |
| Response to nutrient levels | down | 0.0016 | |
| Myeloid cell differentiation | up | 0.0016 | |
| Lung development | up | 0.0017 | |
| Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | down | 0.0017 | |
| Vasculature development | up | 0.0017 | |
| Respiratory tube development | up | 0.0018 | |
| Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen | down | 0.0018 | |
| Dioxygenase activity | down | 0.0018 | |
| Negative regulation of enzyme activity | up | 0.0019 | |
| Odontogenesis | up | 0.0019 | |
| Positive regulation of locomotion | down | 0.0019 | |
| Positive regulation of cell motility | down | 0.0019 | |
| Cell cycle | up | 0.0019 | |
| Anatomical structure formation | up | 0.002 | |
| Cell growth | up | 0.0021 | |
| Regulation of cell size | up | 0.0021 | |
| NAD binding | down | 0.0021 | |
| Steroid biosynthetic process | down | 0.0022 | |
| Response to extracellular stimulus | down | 0.0022 | |
| Endocytosis | down | 0.0023 | |
| Membrane invagination | down | 0.0023 | |
| Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen | down | 0.0025 | |
| Musculoskeletal movement | down | 0.0025 | |
| Brain development | up | 0.0026 | |
| Immune system development | up | 0.0027 | |
| Negative regulation of cell growth | down | 0.0028 | |
| Negative regulation of cell size | down | 0.0028 | |
| Envelope | down | 0.0029 | |
| Negative regulation of developmental process | up | 0.0029 | |
| Chromatin | up | 0.003 | |
| mRNA binding | up | 0.0032 | |
| Antioxidant activity | up | 0.0033 | |
| Selenium binding | up | 0.0035 | |
| Receptor signaling protein serine/threonine kinase activity | down | 0.0036 | |
| Outer membrane-bounded periplasmic space | down | 0.0036 | |
| Cell envelope | down | 0.0036 | |
| Axon | up | 0.0036 | |
| Transferase activity, transferring hexosyl groups | down | 0.0038 | |
| Energy derivation by oxidation of organic compounds | down | 0.0038 | |
| Response to reactive oxygen species | up | 0.0038 | |
| Meiosis | up | 0.0038 | |
| Meiotic cell cycle | up | 0.0038 | |
| Enzyme activator activity | up | 0.0039 | |
| Hemopoiesis | up | 0.0043 | |
| Positive regulation of biosynthetic process | up | 0.0043 | |
| Regulation of transforming growth factor beta receptor signaling pathway | down | 0.0043 | |
| FAD binding | down | 0.0044 | |
| Negative regulation of growth | down | 0.0045 | |
| Defense response to Gram-positive bacterium | up | 0.0048 | |
| Sensory perception of light stimulus | down | 0.0048 | |
| Catabolic process | down | 0.0049 | |
| Hemopoietic or lymphoid organ development | up | 0.005 | |
| Positive regulation of protein metabolic process | up | 0.0051 | |
| Intermediate filament | up | 0.0051 | |
| Ras GTPase activator activity | down | 0.0052 | |
| Response to nutrient | down | 0.0053 | |
| Protein kinase regulator activity | up | 0.0053 | |
| Carbohydrate binding | up | 0.0054 | |
| Calcium ion binding | down | 0.0055 | |
| Oxidoreductase activity, acting on sulfur group of donors | down | 0.0056 | |
| Response to hypoxia | down | 0.0057 | |
| Transferase activity, transferring aldehyde or ketonic groups | up | 0.0058 | |
| Positive regulation of multicellular organismal process | up | 0.0059 | |
| Xenobiotic transporter activity | up | 0.0063 | |
| Xenobiotic-transporting ATPase activity | up | 0.0063 | |
| Phospholipase inhibitor activity | up | 0.0064 | |
Notes: Functional groupings of the modulated genes were determined using Gene Ontology classifications in NextBio. The top 200 biogroups most significantly regulated by DAC (at 3 µM) are shown.
Acknowledgments
We would like to acknowledge Sharon Aukerman, Victoria Sung, and Sharianne Louie for critical review of the manuscript. We thank Gina Fusaro and Marianna Shafarenko for their assistance with manuscript editing. We also thank Xiaoyue Zhao for statistical analyses. Vidaza is a marketed product with azacitidine as the active pharmaceutical ingredient. Dacogen is a marketed product with decitabine as the active pharmaceutical ingredient.
Footnotes
Disclosure
ANN, PWH, NR, AL-M, HB, CH, and KJM are employees of Celgene and as such own stock in the company.
References
- 1.Silverman LR, Demakos EP, Peterson BL, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20(10):2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 2.Kantarjian H, Issa JP, Rosenfeld CS, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106(8):1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 3.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci U S A. 1984;81(22):6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol Cancer Res. 2004;2(1):62–72. [PubMed] [Google Scholar]
- 5.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 6.Haaf T. The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and function: implications for methylation-associated cellular processes. Pharmacol Ther. 1995;65(1):19–46. doi: 10.1016/0163-7258(94)00053-6. [DOI] [PubMed] [Google Scholar]
- 7.Chuang JC, Yoo CB, Kwan JM, et al. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol Cancer Ther. 2005;4(10):1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- 8.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66(5):2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 9.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5(2):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M. Profiling aberrant DNA methylation in hematologic neoplasms: a view from the tip of the iceberg. Clin Immunol. 2003;109(1):80–88. doi: 10.1016/s1521-6616(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 11.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21(35):5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 12.Burbee DG, Forgacs E, Zöchbauer-Müller S, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93(9):691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brabender J, Usadel H, Danenberg KD, et al. Adenomatous polyposis coli gene promoter hypermethylation in non-small cell lung cancer is associated with survival. Oncogene. 2001;20(27):3528–3532. doi: 10.1038/sj.onc.1204455. [DOI] [PubMed] [Google Scholar]
- 14.Kim DH, Nelson HH, Wiencke JK, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61(8):3419–3424. [PubMed] [Google Scholar]
- 15.Maruyama R, Sugio K, Yoshino I, Maehara Y, Gazdar AF. Hypermethylation of FHIT as a prognostic marker in nonsmall cell lung carcinoma. Cancer. 2004;100(7):1472–1477. doi: 10.1002/cncr.20144. [DOI] [PubMed] [Google Scholar]
- 16.Schrump DS, Fischette MR, Nguyen DM, et al. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res. 2006;12(19):5777–5785. doi: 10.1158/1078-0432.CCR-06-0669. [DOI] [PubMed] [Google Scholar]
- 17.Samlowski WE, Leachman SA, Wade M, et al. Evaluation of a 7-day continuous intravenous infusion of decitabine: inhibition of promoter-specific and global genomic DNA methylation. J Clin Oncol. 2005;23(17):3897–3905. doi: 10.1200/JCO.2005.06.118. [DOI] [PubMed] [Google Scholar]
- 18.Weiss AJ, Metter GE, Nealon TF, et al. Phase II study of 5-azacytidine in solid tumors. Cancer Treat Rep. 1977;61(1):55–58. [PubMed] [Google Scholar]
- 19.Lin J, Gilbert J, Rudek MA, et al. A phase I dose-finding study of 5-azacytidine in combination with sodium phenylbutyrate in patients with refractory solid tumors. Clin Cancer Res. 2009;15(19):6241–6249. doi: 10.1158/1078-0432.CCR-09-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small cell lung cancer. J Clin Oncol. 2006;24(29):4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 21.Oguri T, Achiwa H, Sato S, et al. The determinants of sensitivity and acquired resistance to gemcitabine differ in non-small cell lung cancer: a role of ABCC5 in gemcitabine sensitivity. Mol Cancer Ther. 2006;5(7):1800–1806. doi: 10.1158/1535-7163.MCT-06-0025. [DOI] [PubMed] [Google Scholar]
- 22.Voortman J, Checinska A, Giaccone G, Rodriguez JA, Kruyt FA. Bortezomib, but not cisplatin, induces mitochondria-dependent apoptosis accompanied by up-regulation of noxa in the non-small cell lung cancer cell line NCI-H460. Mol Cancer Ther. 2007;6(3):1046–1053. doi: 10.1158/1535-7163.MCT-06-0577. [DOI] [PubMed] [Google Scholar]
- 23.Qin T, Youssef EM, Jelinek J, et al. Effect of cytarabine and decitabine in combination in human leukemic cell lines. Clin Cancer Res. 2007;13(14):4225–4232. doi: 10.1158/1078-0432.CCR-06-2762. [DOI] [PubMed] [Google Scholar]
- 24.Qin T, Jelinek J, Si J, Shu J, Issa JP. Mechanisms of resistance to 5-aza-2′-deoxycytidine in human cancer cell lines. Blood. 2009;113(3):659–667. doi: 10.1182/blood-2008-02-140038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollenbach PW, Nguyen AN, Brady H, et al. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS One. 2010;5(2):e9001. doi: 10.1371/journal.pone.0009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flotho C, Claus R, Batz C, et al. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23(6):1019–1028. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
- 27.Sadikovic B, Al-Romaih K, Squire JA, Zielenska M. Cause and consequences of genetic and epigenetic alterations in human cancer. Curr Genomics. 2008;9(6):394–408. doi: 10.2174/138920208785699580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blum W, Klisovic RB, Hackanson B, et al. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J Clin Oncol. 2007;25(25):3884–3891. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]
- 29.Oki Y, Kantarjian HM, Gharibyan V, et al. Phase II study of low-dose decitabine in combination with imatinib mesylate in patients with accelerated or myeloid blastic phase of chronic myelogenous leukemia. Cancer. 2007;109(5):899–906. doi: 10.1002/cncr.22470. [DOI] [PubMed] [Google Scholar]
- 30.Stresemann C, Bokelmann I, Mahlknecht U, Lyko F. Azacytidine causes complex DNA methylation responses in myeloid leukemia. Mol Cancer Ther. 2008;7(9):2998–3005. doi: 10.1158/1535-7163.MCT-08-0411. [DOI] [PubMed] [Google Scholar]
- 31.Karpf AR, Moore BC, Ririe TO, Jones DA. Activation of the p53 DNA damage response pathway after inhibition of DNA methyltransferase by 5-aza-2′-deoxycytidine. Mol Pharmacol. 2001;59(4):751–757. [PubMed] [Google Scholar]
- 32.Kiziltepe T, Hideshima T, Catley L, et al. 5-Azacytidine, a DNA methyltransferase inhibitor, induces ATR-mediated DNA double-strand break responses, apoptosis, and synergistic cytotoxicity with doxorubicin and bortezomib against multiple myeloma cells. Mol Cancer Ther. 2007;6(6):1718–1727. doi: 10.1158/1535-7163.MCT-07-0010. [DOI] [PubMed] [Google Scholar]
- 33.Jiemjit A, Fandy TE, Carraway H, et al. p21(WAF1/CIP1) induction by 5-azacytosine nucleosides requires DNA damage. Oncogene. 2008;27(25):3615–3623. doi: 10.1038/sj.onc.1211018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palii SS, van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28(2):752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Incalci M, Covey JM, Zaharko DS, Kohn KW. DNA alkali-labile sites induced by incorporation of 5-aza-2′-deoxycytidine into DNA of mouse leukemia L1210 cells. Cancer Res. 1985;45(7):3197–3202. [PubMed] [Google Scholar]
- 36.Covey JM, D’Incalci M, Tilchen EJ, Zaharko DS, Kohn KW. Differences in DNA damage produced by incorporation of 5-aza-2′-deoxycytidine or 5,6-dihydro-5-azacytidine into DNA of mammalian cells. Cancer Res. 1986;46(11):5511–5517. [PubMed] [Google Scholar]
- 37.Chai G, Li L, Zhou W, et al. HDAC inhibitors act with 5-aza-2′-deoxycytidine to inhibit cell proliferation by suppressing removal of incorporated abases in lung cancer cells. PLoS One. 2008;3(6):e2445. doi: 10.1371/journal.pone.0002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li LH, Olin EJ, Buskirk HH, Reineke LM. Cytotoxicity and mode of action of 5-azacytidine on L1210 leukemia. Cancer Res. 1970;30(11):2760–2769. [PubMed] [Google Scholar]
- 39.Veselý J, Cihák A. Incorporation of a potent antileukemic agent, 5-aza-2′-deoxycytidine, into DNA of cells from leukemic mice. Cancer Res. 1977;37(10):3684–3689. [PubMed] [Google Scholar]
- 40.Bouchard J, Momparler RL. Incorporation of 5-Aza-2′-deoxycytidine-5′-triphosphate into DNA. Interactions with mammalian DNA polymerase alpha and DNA methylase. Mol Pharmacol. 1983;24(1):109–114. [PubMed] [Google Scholar]
- 41.Glazer RI, Knode MC. 1-beta-D-arabinosyl-5-azacytosine. Cytocidal activity and effects on the synthesis and methylation of DNA in human colon carcinoma cells. Mol Pharmacol. 1984;26(2):381–387. [PubMed] [Google Scholar]
- 42.Cihák A. Biological effects of 5-azacytidine in eukaryotes. Oncology. 1974;30(5):405–422. doi: 10.1159/000224981. [DOI] [PubMed] [Google Scholar]
- 43.Lu LJ, Randerath K. Effects of 5-azacytidine on transfer RNA methyltransferases. Cancer Res. 1979;39(3):940–949. [PubMed] [Google Scholar]
- 44.Cohen MB, Glazer RI. Cytotoxicity and the inhibition of ribosomal RNA processing in human colon carcinoma cells. Mol Pharmacol. 1985;27(2):308–313. [PubMed] [Google Scholar]
- 45.Glover AB, Leyland-Jones B. Biochemistry of azacitidine: a review. Cancer Treat Rep. 1987;71(10):959–964. [PubMed] [Google Scholar]
- 46.Marcucci G, Silverman L, Eller M, Lintz L, Beach CL. Bioavailability of azacitidine subcutaneous versus intravenous in patients with the myelodysplastic syndromes. J Clin Pharmacol. 2005;45(5):597–602. doi: 10.1177/0091270004271947. [DOI] [PubMed] [Google Scholar]
- 47.Cashen AF, Shah AK, Todt L, Fisher N, DiPersio J. Pharmacokinetics of decitabine administered as a 3-h infusion to patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) Cancer Chemother Pharmacol. 2008;61(5):759–766. doi: 10.1007/s00280-007-0531-7. [DOI] [PubMed] [Google Scholar]
- 48.Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50(3):346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Viability of H23 and H1975 cells was assessed after 72 hours of treatment with AZA or DAC (0–25 µM). Error bars represent the standard error of mean of three independent experiments, with triplicate wells per experiment.
AZA and DAC cause DNMT1 depletion in NSCLC cell lines. A549 and H1299 cells were treated with AZA or DAC (5 µM) for 4, 8, or 16 hours and DNMT1 protein was detected by Western blotting of cell extracts. Alpha-tubulin was used as a loading control.
AZA and DAC reduce DNA methylation in A549 and H1299 cells. LINE-1 DNA methylation was assessed in A549 and H1299 cells after 72 hours of treatment with AZA or DAC (0–3 µM). Percentage LINE-1 methylation represents the average percentage methylation of 4 CpG sites in duplicate samples, with error bars representing the standard deviation.
Viability of A549, H460, and H1299 cells was assessed after 6 days of treatment with AZA or DAC (0–25 µM).
Table S1.
EC50 values for AZA and DAC on NSCLC cell viability (6 days)
| AZA EC50 (μM) | DAC EC50 (μM) | |
|---|---|---|
| A549 | 4.4 | 2.5 |
| H460 | 2.2 | 4.4 |
| H1299 | 4.1 | 0.5 |
Abbreviations: AZA, azacitidine; DAC, decitabine; NSCLC, non-small cell lung cancer.
Table S2.
Top 200 biogroups modulated by azacitidine (AZA) in A549 cells
| A549 cells treated with 3 μM AZA (48 hours)
| ||
|---|---|---|
| Biogroup name | Direction | P value |
| Proteinaceous extracellular matrix | down | 3.40E-18 |
| Extracellular matrix | down | 4.70E-18 |
| Transcription | up | 2.50E-16 |
| Extracellular matrix structural constituent | down | 4.90E-16 |
| Glycosaminoglycan binding | down | 7.30E-15 |
| Polysaccharide binding | down | 9.60E-15 |
| Pattern binding | down | 2.80E-14 |
| Lipid biosynthetic process | down | 3.60E-14 |
| Fibrillar collagen | down | 1.60E-13 |
| Calcium ion binding | down | 2.20E-13 |
| Fibrinogen complex | down | 7.20E-13 |
| Humoral immune response | down | 8.90E-12 |
| Protein binding, bridging | down | 9.00E-12 |
| Collagen | down | 1.70E-11 |
| Response to wounding | down | 6.00E-11 |
| Response to external stimulus | down | 1.50E-10 |
| Platelet activation | down | 2.20E-10 |
| Ligase activity, forming aminoacyl-tRNA and related compounds | up | 3.30E-10 |
| Ligase activity, forming carbon-oxygen bonds | up | 3.30E-10 |
| Response to nutrient | up | 3.70E-10 |
| Carbohydrate binding | down | 3.80E-10 |
| Basement membrane | down | 1.00E-09 |
| Lipid metabolic process | down | 2.20E-09 |
| Response to nutrient levels | up | 2.30E-09 |
| Steroid biosynthetic process | down | 2.30E-09 |
| Collagen binding | down | 2.40E-09 |
| Response to extracellular stimulus | up | 3.70E-09 |
| Inflammatory response | down | 7.80E-09 |
| Nucleoplasm | up | 8.70E-09 |
| Acute inflammatory response | down | 1.30E-08 |
| Response to stress | down | 1.50E-08 |
| Blood pressure regulation | down | 1.50E-08 |
| RNA binding | up | 1.60E-08 |
| Cell motility | down | 3.40E-08 |
| Localization of cell | down | 3.40E-08 |
| Epithelial cell differentiation | down | 4.70E-08 |
| tRNA binding | up | 4.80E-08 |
| Steroid metabolic process | down | 7.20E-08 |
| Endoplasmic reticulum | down | 8.00E-08 |
| Translation | up | 1.30E-07 |
| Fatty acid biosynthetic process | down | 1.90E-07 |
| Parturition | down | 2.50E-07 |
| Sterol metabolic process | down | 2.50E-07 |
| Blood coagulation | down | 2.90E-07 |
| Coagulation | down | 3.10E-07 |
| Humoral immune response mediated by circulating immunoglobulin | down | 3.60E-07 |
| Hemostasis | down | 3.60E-07 |
| Organic acid biosynthetic process | down | 4.40E-07 |
| Regulation of body fluids | down | 6.70E-07 |
| Wound healing | down | 6.90E-07 |
| ER-Golgi intermediate compartment | down | 8.00E-07 |
| Actin binding | down | 9.70E-07 |
| Anion transport | down | 1.10E-06 |
| Extracellular structure organization and biogenesis | down | 1.10E-06 |
| Transaminase activity | up | 1.10E-06 |
| Complement activation | down | 1.20E-06 |
| Extracellular matrix organization and biogenesis | down | 1.20E-06 |
| Calmodulin binding | down | 1.90E-06 |
| Circulation | down | 1.90E-06 |
| Female pregnancy | down | 2.50E-06 |
| Cellular homeostasis | down | 2.60E-06 |
| Morphogenesis of an epithelium | down | 2.70E-06 |
| Cell proliferation | down | 3.10E-06 |
| Alkene metabolic process | down | 3.10E-06 |
| Ribosome biogenesis and assembly | up | 3.20E-06 |
| Complement activation, classical pathway | down | 3.50E-06 |
| Ribonucleoprotein complex biogenesis and assembly | up | 4.90E-06 |
| Sodium:potassium-exchanging ATPase complex | down | 5.20E-06 |
| Transferase activity, transferring nitrogenous groups | up | 5.80E-06 |
| Cell activation | down | 6.10E-06 |
| Endoplasmic reticulum lumen | down | 6.60E-06 |
| Fatty acid metabolic process | down | 6.90E-06 |
| Vesicular fraction | down | 8.20E-06 |
| Cellular ion homeostasis | down | 1.10E-05 |
| Cellular chemical homeostasis | down | 1.10E-05 |
| Positive regulation of immune system process | down | 1.10E-05 |
| Positive regulation of immune response | down | 1.10E-05 |
| Phosphoinositide binding | down | 1.40E-05 |
| Activation of immune response | down | 1.50E-05 |
| Positive regulation of multicellular organismal process | down | 1.60E-05 |
| Cofactor transporter activity | up | 1.60E-05 |
| Soluble fraction | up | 1.70E-05 |
| Enzyme inhibitor activity | down | 1.70E-05 |
| Development of primary sexual characteristics | up | 1.80E-05 |
| NAD binding | down | 1.90E-05 |
| Amine biosynthetic process | up | 1.90E-05 |
| Cytoskeleton | down | 2.20E-05 |
| Lymphocyte mediated immunity | down | 2.40E-05 |
| Receptor binding | down | 2.50E-05 |
| Transcription corepressor activity | up | 2.50E-05 |
| Response to DNA damage stimulus | up | 2.60E-05 |
| Cartilage development | down | 2.70E-05 |
| SNARE complex | up | 2.80E-05 |
| Gastrulation | up | 3.20E-05 |
| mRNA transport | up | 3.50E-05 |
| Epidermis development | down | 3.80E-05 |
| Cell migration | down | 4.20E-05 |
| Immune effector process | down | 4.20E-05 |
| Response to hypoxia | down | 4.30E-05 |
| Leukocyte mediated immunity | down | 4.50E-05 |
| Adaptive immune response | down | 4.90E-05 |
| Adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | down | 4.90E-05 |
| Endopeptidase inhibitor activity | down | 5.00E-05 |
| Protease inhibitor activity | down | 5.00E-05 |
| Oxidoreductase activity, acting on heme group of donors | down | 5.30E-05 |
| Oxidoreductase activity, acting on heme group of donors, oxygen as acceptor | down | 5.30E-05 |
| Cytochrome-c oxidase activity | down | 5.30E-05 |
| Heme-copper terminal oxidase activity | down | 5.30E-05 |
| Germ cell migration | up | 6.00E-05 |
| Coenzyme binding | down | 6.20E-05 |
| Regulation of translation | up | 6.90E-05 |
| Cytokine biosynthetic process | up | 7.30E-05 |
| Neurotransmitter:sodium symporter activity | up | 7.40E-05 |
| Ectoderm development | down | 7.50E-05 |
| Establishment of RNA localization | up | 8.00E-05 |
| RNA transport | up | 8.00E-05 |
| Nucleic acid transport | up | 8.00E-05 |
| Transcription factor binding | up | 8.30E-05 |
| Regulation of immune response | down | 8.50E-05 |
| Regulation of immune system process | down | 8.50E-05 |
| Ligase activity | up | 8.60E-05 |
| RNA localization | up | 9.10E-05 |
| Neurotransmitter transporter activity | up | 9.30E-05 |
| RNA export from nucleus | up | 9.80E-05 |
| Phospholipid binding | down | 9.90E-05 |
| Cell cycle | up | 0.0001 |
| Cytosol | down | 0.0001 |
| Cytoskeletal protein binding | down | 0.0001 |
| Response to endogenous stimulus | up | 0.0001 |
| Gonad development | up | 0.0001 |
| Nucleobase, nucleoside, nucleotide and nucleic acid transport | up | 0.0001 |
| Nucleolus | up | 0.0001 |
| Regulation of cytokine biosynthetic process | up | 0.0001 |
| Rhythmic process | up | 0.0001 |
| Reproductive structure development | up | 0.0002 |
| Mitochondrion organization and biogenesis | up | 0.0002 |
| Structural constituent of cytoskeleton | down | 0.0002 |
| Sex differentiation | up | 0.0002 |
| Transcription repressor activity | up | 0.0002 |
| Peroxidase activity | down | 0.0002 |
| Oxidoreductase activity, acting on peroxide as acceptor | down | 0.0002 |
| Laminin-1 complex | down | 0.0002 |
| Transcription cofactor activity | up | 0.0002 |
| Female sex differentiation | up | 0.0002 |
| Development of primary female sexual characteristics | up | 0.0002 |
| Oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | down | 0.0002 |
| Nitrogen compound biosynthetic process | up | 0.0002 |
| Cell structure disassembly during apoptosis | up | 0.0003 |
| Amino acid transport | up | 0.0003 |
| Acyl-CoA binding | down | 0.0003 |
| Response to dsRNA | up | 0.0003 |
| Neuron development | down | 0.0003 |
| Integrator complex | up | 0.0003 |
| Immune response | down | 0.0003 |
| Protein dimerization activity | up | 0.0004 |
| Laminin complex | down | 0.0004 |
| Cofactor binding | down | 0.0004 |
| Germ-line sex determination | down | 0.0004 |
| Intramolecular oxidoreductase activity | down | 0.0004 |
| DNA repair | up | 0.0004 |
| Cell soma | down | 0.0004 |
| Cellular morphogenesis during differentiation | down | 0.0004 |
| RNA polymerase II transcription factor activity | up | 0.0005 |
| Regulation of epithelial cell proliferation | down | 0.0005 |
| Regulation of biosynthetic process | up | 0.0005 |
| UDP-glycosyltransferase activity | up | 0.0005 |
| Pyridoxal phosphate binding | up | 0.0005 |
| Lipid binding | down | 0.0005 |
| Positive regulation of programmed cell death | up | 0.0005 |
| Helicase activity | up | 0.0006 |
| Cell redox homeostasis | down | 0.0006 |
| Cell death | up | 0.0006 |
| Death | up | 0.0006 |
| Epithelial cell proliferation | down | 0.0006 |
| Mesenchymal cell development | down | 0.0006 |
| Ovulation | up | 0.0006 |
| Positive regulation of locomotion | down | 0.0006 |
| Positive regulation of cell motility | down | 0.0006 |
| DNA catabolic process | up | 0.0006 |
| Cell differentiation | down | 0.0006 |
| Basal lamina | down | 0.0007 |
| Insulin-like growth factor binding mesenchymal cell differentiation | Down down |
0.0007 0.0007 |
| Sequestering of metal ion | down | 0.0007 |
| Neurotransmitter transport | up | 0.0007 |
| Specific RNA polymerase II transcription factor activity | up | 0.0007 |
| Intramolecular oxidoreductase activity, transposing C=C bonds | down | 0.0007 |
| Cellular component disassembly | up | 0.0008 |
| Heme binding | down | 0.0008 |
| Tetrapyrrole binding | down | 0.0008 |
| Presynaptic active zone | up | 0.0008 |
| Amine transport | up | 0.0009 |
| Sequestering of calcium ion | down | 0.0009 |
| Cell recognition | down | 0.0009 |
| Endoplasmic reticulum part | down | 0.0009 |
| Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | down | 0.001 |
| Myosin binding | down | 0.001 |
| Lyase activity | up | 0.001 |
| Transferase activity, transferring hexosyl groups | down | 0.001 |
| Neuron differentiation | down | 0.001 |
Notes: Functional groupings of the modulated genes were determined using Gene Ontology classifications in NextBio. The top 200 biogroups most significantly regulated by AZA (at 3 µM) are shown.
Table S3.
Top 196 biogroups modulated by decitabine (DAC) in A549 cells
| A549 cells treated with 3 μM DAC (48 hours)
| ||
|---|---|---|
| Biogroup name | Direction | P value |
| Mitosis | down | 2.00E-09 |
| Cell cycle | down | 2.90E-09 |
| Cell division | down | 5.70E-08 |
| Transferase activity, transferring sulfur-containing groups | up | 5.80E-08 |
| Meiosis | down | 1.50E-07 |
| Meiotic cell cycle | down | 1.70E-07 |
| Response to DNA damage stimulus | down | 1.80E-07 |
| Male gamete generation | up | 6.10E-07 |
| Response to endogenous stimulus | down | 7.60E-07 |
| Chromosome segregation | down | 2.10E-06 |
| Aromatic compound metabolic process | up | 2.70E-06 |
| Phenol metabolic process | up | 2.70E-06 |
| Structural constituent of cytoskeleton | up | 5.10E-06 |
| Sister chromatid cohesion | down | 5.90E-06 |
| Cellular lipid catabolic process | up | 6.70E-06 |
| DNA repair | down | 7.20E-06 |
| Alkali metal ion binding | up | 9.00E-06 |
| Regulation of neurotransmitter levels | up | 1.10E-05 |
| DNA damage response, signal transduction | down | 1.30E-05 |
| Cofactor transporter activity | up | 1.60E-05 |
| Sulfotransferase activity | up | 1.70E-05 |
| Intermediate filament | up | 3.20E-05 |
| Neurotransmitter:sodium symporter activity | up | 3.40E-05 |
| Chromatin assembly | down | 3.90E-05 |
| Neurotransmitter transporter activity | up | 4.60E-05 |
| Cytokinesis | down | 5.10E-05 |
| Chromosome | down | 5.30E-05 |
| Mitotic spindle organization and biogenesis | down | 5.30E-05 |
| Negative regulation of enzyme activity | up | 5.60E-05 |
| Establishment of mitotic spindle localization | down | 6.50E-05 |
| Establishment of spindle localization | down | 6.50E-05 |
| Spindle localization | down | 6.50E-05 |
| Retinol binding | up | 6.50E-05 |
| Microtubule organizing center part | down | 7.60E-05 |
| Mitotic sister chromatid segregation | down | 8.30E-05 |
| Alcohol metabolic process | up | 8.60E-05 |
| Sister chromatid segregation | down | 9.30E-05 |
| Catabolic process | up | 0.0001 |
| Soluble fraction | up | 0.0001 |
| Retinal binding | up | 0.0001 |
| Positive regulation of programmed cell death | up | 0.0001 |
| Steroid biosynthetic process | up | 0.0002 |
| Response to stress | down | 0.0002 |
| Gamma-tubulin complex | down | 0.0002 |
| Mitotic chromosome condensation | down | 0.0002 |
| Transporter activity | up | 0.0002 |
| Phosphopyruvate hydratase complex | up | 0.0002 |
| Amino acid derivative metabolic process | up | 0.0003 |
| Vitamin binding | up | 0.0003 |
| Lipid catabolic process | up | 0.0003 |
| Nuclear chromosome | down | 0.0003 |
| Retinoid binding | up | 0.0003 |
| Isoprenoid binding | up | 0.0003 |
| Homologous chromosome segregation | down | 0.0003 |
| Meiotic chromosome segregation | down | 0.0003 |
| Meiotic spindle organization and biogenesis | down | 0.0003 |
| Cell differentiation | up | 0.0003 |
| NADP binding | down | 0.0004 |
| Steroid metabolic process | up | 0.0004 |
| Lipid raft | up | 0.0004 |
| Cohesin complex | down | 0.0004 |
| Meiosis I | down | 0.0004 |
| Sodium:potassium-exchanging ATPase complex | up | 0.0004 |
| Negative regulation of cell proliferation | up | 0.0004 |
| Actin binding | down | 0.0005 |
| Nuclear matrix | down | 0.0005 |
| Cytoskeletal protein binding | down | 0.0005 |
| Protein kinase inhibitor activity | up | 0.0005 |
| Cell proliferation | up | 0.0006 |
| Cytoskeleton | down | 0.0006 |
| Cytoskeleton organization and biogenesis | down | 0.0006 |
| Fat cell differentiation | down | 0.0006 |
| Hormone metabolic process | up | 0.0006 |
| Positive regulation of progression through cell cycle | down | 0.0006 |
| Kinase inhibitor activity | up | 0.0006 |
| Oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | down | 0.0007 |
| Neurotransmitter transport | up | 0.0007 |
| Membrane invagination | down | 0.0008 |
| Endocytosis | down | 0.0008 |
| Amide metabolic process | up | 0.0008 |
| Spindle | down | 0.0008 |
| Ion transport | up | 0.0009 |
| Blastocyst growth | down | 0.0009 |
| Interleukin binding | down | 0.0009 |
| RNA export from nucleus | down | 0.0009 |
| Tubulin binding | down | 0.0009 |
| Epidermis development | up | 0.0009 |
| Neurotransmitter metabolic process | up | 0.0011 |
| Translation activator activity | up | 0.0011 |
| Spindle pole | down | 0.0011 |
| Synaptic transmission | up | 0.0012 |
| Intracellular cyclic nucleotide activated cation channel complex | up | 0.0012 |
| Biogenic amine metabolic process | up | 0.0012 |
| Cell fate determination | up | 0.0013 |
| Oxidoreductase activity, acting on iron-sulfur proteins as donors | up | 0.0013 |
| Ion channel activity | up | 0.0013 |
| Lipoprotein binding | down | 0.0014 |
| Positive regulation of neurogenesis | down | 0.0014 |
| Cytosol | up | 0.0018 |
| Microtubule organizing center | down | 0.002 |
| Microtubule | down | 0.002 |
| Glutathione peroxidase activity | up | 0.0021 |
| Odontogenesis | down | 0.0022 |
| Passive transmembrane transporter activity | up | 0.0022 |
| Transmission of nerve impulse | up | 0.0023 |
| Oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor | down | 0.0024 |
| Dynein binding | down | 0.0024 |
| Humoral immune response | down | 0.0024 |
| Ectoderm development | up | 0.0025 |
| Arginine metabolic process | up | 0.0025 |
| Myosin binding | down | 0.0025 |
| Lipid biosynthetic process | up | 0.0026 |
| Muscle contraction | up | 0.0027 |
| Mitochondrion organization and biogenesis | up | 0.0027 |
| Fat-soluble vitamin metabolic process | up | 0.0028 |
| Female gamete generation | down | 0.0028 |
| Urea cycle intermediate metabolic process | up | 0.0029 |
| Inclusion body | down | 0.0029 |
| Folic acid transporter activity | down | 0.0029 |
| Protein heterodimerization activity | down | 0.003 |
| Angiogenesis | up | 0.003 |
| Replication fork | down | 0.0031 |
| Nucleoside metabolic process | down | 0.0031 |
| Regulation of axonogenesis | down | 0.0032 |
| Anatomical structure formation | up | 0.0033 |
| Protein kinase regulator activity | up | 0.0034 |
| Lipid metabolic process | up | 0.0037 |
| Glycoprotein binding | up | 0.0037 |
| Pyridoxal phosphate binding | up | 0.0037 |
| Blood vessel morphogenesis | up | 0.004 |
| Carbohydrate metabolic process | up | 0.0041 |
| Tissue regeneration | up | 0.0041 |
| Regeneration | up | 0.0041 |
| Germ cell development | up | 0.0042 |
| Growth factor binding | down | 0.0042 |
| Peptide transporter activity | up | 0.0043 |
| Nitrogen compound biosynthetic process | up | 0.0044 |
| Cytokine binding | down | 0.0044 |
| Nitric oxide metabolic process | up | 0.0047 |
| Nitric oxide biosynthetic process | up | 0.0047 |
| Centrosome | down | 0.0047 |
| Embryonic morphogenesis | down | 0.0048 |
| Regulation of neurogenesis | down | 0.0048 |
| Oxidoreductase activity, acting on the aldehyde or oxo group of donors | up | 0.0048 |
| Cytokinesis during cell cycle | down | 0.0049 |
| Cell–cell signaling | up | 0.005 |
| Calmodulin binding | up | 0.005 |
| Structure-specific DNA binding | down | 0.0051 |
| Oxidoreductase activity, acting on the CH–CH group of donors | down | 0.0051 |
| Peroxidase activity | up | 0.0051 |
| Oxidoreductase activity, acting on peroxide as acceptor | up | 0.0051 |
| Microfibril | up | 0.0052 |
| Protein–DNA complex assembly | down | 0.0052 |
| Vasculature development | up | 0.0054 |
| Excretion | up | 0.0055 |
| mRNA transport | down | 0.0056 |
| Identical protein binding | up | 0.0056 |
| Vitamin transporter activity | down | 0.0057 |
| Response to organic cyclic substance | up | 0.0059 |
| Response to alkaloid | up | 0.0059 |
| Kinase regulator activity | up | 0.006 |
| Chromatin | down | 0.006 |
| Electron carrier activity | up | 0.0061 |
| Vitamin biosynthetic process | down | 0.0062 |
| RNA transport | down | 0.0067 |
| Nucleic acid transport | down | 0.0067 |
| Establishment of RNA localization | down | 0.0067 |
| Protein domain specific binding | up | 0.0068 |
| Homophilic cell adhesion | down | 0.0068 |
| RNA localization | down | 0.0069 |
| Hormone biosynthetic process | up | 0.007 |
| Protein dimerization activity | down | 0.0071 |
| RNA binding | down | 0.0073 |
| Blastocyst development | down | 0.0074 |
| Cyclin binding | up | 0.0075 |
| Nucleobase, nucleoside, nucleotide and nucleic acid transport | down | 0.0077 |
| Cartilage development | down | 0.0077 |
| Folic acid binding | down | 0.0079 |
| Positive regulation of developmental process | down | 0.0081 |
| Chordate embryonic development | down | 0.0082 |
| NAD binding | up | 0.0082 |
| Cofactor binding | up | 0.0082 |
| Vesicle docking during exocytosis | up | 0.0084 |
| Developmental maturation | up | 0.0085 |
| Hydrolase activity, acting on carbon–nitrogen (but not peptide) bonds, in cyclic amidines | up | 0.0086 |
| Lysosome | up | 0.0086 |
| Embryonic digit morphogenesis | down | 0.0086 |
| DNA helicase activity | down | 0.0089 |
| Axon guidance | down | 0.0091 |
| Membrane docking | up | 0.0093 |
| Vesicle docking | up | 0.0093 |
| Voltage-gated sodium channel complex | down | 0.0093 |
| mRNA binding | up | 0.0094 |
| Establishment of organelle localization | down | 0.0096 |
| Vitamin metabolic process | up | 0.0096 |
| Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | down | 0.0099 |
Notes: Functional groupings of the modulated genes were determined using Gene Ontology classifications in NextBio. The top 196 biogroups most significantly regulated by DAC (at 3 µM) are shown.
Table S4.
Top 200 biogroups modulated by azacitidine (AZA) in H1299 cells
| H1299 cells treated with 3 μM AZA (48 hours)
| ||
|---|---|---|
| Biogroup name | Direction | P value |
| Transcription | up | 1.90E-25 |
| Cell cycle | down | 7.60E-25 |
| Mitosis | down | 8.00E-24 |
| Cell division | down | 1.00E-22 |
| Cytoskeleton | down | 5.60E-14 |
| Microtubule | down | 1.30E-13 |
| Spindle | down | 1.80E-13 |
| Mitochondrion | down | 1.80E-12 |
| Sterol metabolic process | down | 1.40E-11 |
| Chromosome | down | 1.20E-10 |
| Alcohol metabolic process | down | 2.20E-10 |
| Ligase activity | up | 2.40E-10 |
| Lipid biosynthetic process | down | 2.70E-10 |
| Steroid biosynthetic process | down | 3.00E-10 |
| Mitotic sister chromatid segregation | down | 1.90E-09 |
| Steroid metabolic process | down | 2.50E-09 |
| Sister chromatid segregation | down | 2.90E-09 |
| Endoplasmic reticulum | down | 3.60E-09 |
| Envelope | down | 4.70E-09 |
| Lipid metabolic process | down | 2.60E-08 |
| Response to nutrient | down | 7.00E-08 |
| Collagen binding | down | 1.60E-07 |
| Centrosome | down | 1.70E-07 |
| Wound healing | down | 1.90E-07 |
| Response to nutrient levels | down | 2.00E-07 |
| Intramolecular oxidoreductase activity | down | 2.50E-07 |
| Response to extracellular stimulus | down | 2.70E-07 |
| Mitochondrial membrane | down | 2.80E-07 |
| Microtubule organizing center | down | 2.80E-07 |
| Acid–amino acid ligase activity | up | 3.10E-07 |
| Cell proliferation | down | 3.60E-07 |
| Blood coagulation | down | 3.70E-07 |
| Establishment of chromosome localization | down | 4.40E-07 |
| Coagulation | down | 4.90E-07 |
| Chromosome segregation | down | 5.00E-07 |
| Nitrogen compound catabolic process | down | 5.20E-07 |
| Kinase binding | up | 5.30E-07 |
| Beta-catenin binding | down | 5.50E-07 |
| Nucleoplasm | up | 6.10E-07 |
| Enzyme inhibitor activity | down | 8.80E-07 |
| Alcohol catabolic process | down | 9.80E-07 |
| Hemostasis | down | 1.10E-06 |
| Transcription cofactor activity | up | 1.10E-06 |
| Transcription repressor activity | up | 1.20E-06 |
| Midbody | down | 1.30E-06 |
| Ligase activity, forming carbon-nitrogen bonds | up | 1.70E-06 |
| Establishment of organelle localization | down | 2.00E-06 |
| Germ-line sex determination | down | 2.00E-06 |
| Oligosaccharyl transferase complex | down | 2.10E-06 |
| Response to external stimulus | down | 2.20E-06 |
| Oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor | down | 2.20E-06 |
| Amine catabolic process | down | 2.20E-06 |
| Cytoskeleton organization and biogenesis | down | 2.20E-06 |
| Cofactor binding | down | 2.30E-06 |
| Coenzyme binding | down | 2.70E-06 |
| Transcription corepressor activity | up | 3.00E-06 |
| Cell differentiation | up | 3.20E-06 |
| mRNA binding | down | 3.30E-06 |
| Meiotic chromosome segregation | down | 3.60E-06 |
| Homologous chromosome segregation | down | 3.60E-06 |
| Nuclear envelope-endoplasmic reticulum network | down | 3.90E-06 |
| Oxidoreductase activity, acting on the CH–CH group of donors, NAD or NADP as acceptor | down | 5.00E-06 |
| Intramolecular oxidoreductase activity, transposing C=C bonds | down | 5.30E-06 |
| Mitotic chromosome condensation | down | 5.50E-06 |
| Endoplasmic reticulum part | down | 5.50E-06 |
| Transcription factor binding | up | 5.60E-06 |
| Organic acid transport | up | 5.70E-06 |
| Carboxylic acid transport | up | 5.70E-06 |
| Acyl-CoA binding | down | 5.70E-06 |
| DNA-directed RNA polymerase II, holoenzyme | up | 5.80E-06 |
| Interphase of mitotic cell cycle | down | 6.00E-06 |
| Primary sex determination | down | 6.00E-06 |
| Cell-matrix adhesion | down | 6.20E-06 |
| Hormone activity | down | 6.30E-06 |
| Organic acid transmembrane transporter activity | up | 6.30E-06 |
| Mitochondrial inner membrane | down | 7.30E-06 |
| Cell-substrate adhesion | down | 7.60E-06 |
| Transcription activator activity | up | 7.70E-06 |
| Mitotic spindle organization and biogenesis | down | 7.90E-06 |
| Lyase activity | down | 8.10E-06 |
| Sterol transport | down | 8.70E-06 |
| Arginine metabolic process | down | 8.70E-06 |
| Chromatin assembly | down | 1.00E-05 |
| Nitrogen compound biosynthetic process | down | 1.10E-05 |
| RNA polymerase II transcription factor activity | up | 1.10E-05 |
| Transaminase activity | up | 1.20E-05 |
| Meiotic spindle organization and biogenesis | down | 1.30E-05 |
| Organelle localization | down | 1.40E-05 |
| Isomerase activity | down | 1.40E-05 |
| Interphase | down | 1.40E-05 |
| Urea cycle intermediate metabolic process | down | 1.50E-05 |
| Fatty acid biosynthetic process | down | 1.70E-05 |
| Receptor binding | down | 1.70E-05 |
| Regulation of body fluids | down | 1.80E-05 |
| Condensin complex | down | 1.80E-05 |
| Regulation of coagulation | down | 1.90E-05 |
| Nuclear envelope | down | 2.20E-05 |
| Caveola | down | 2.30E-05 |
| Organic acid biosynthetic process | down | 2.30E-05 |
| Meiotic cell cycle | down | 2.30E-05 |
| Amino acid transport | up | 2.50E-05 |
| NADP binding | down | 2.80E-05 |
| Protein dimerization activity | up | 2.90E-05 |
| Spindle pole | down | 3.40E-05 |
| Ubiquitin–protein ligase activity | up | 3.50E-05 |
| Transferase activity, transferring nitrogenous groups | up | 3.60E-05 |
| Nucleoside metabolic process | down | 3.80E-05 |
| Structural constituent of cytoskeleton | down | 3.90E-05 |
| Carbohydrate catabolic process | down | 4.10E-05 |
| Endoplasmic reticulum membrane | down | 4.30E-05 |
| Nucleosome | down | 4.40E-05 |
| Nucleotide catabolic process | down | 4.60E-05 |
| Cellular chemical homeostasis | down | 4.70E-05 |
| Cellular ion homeostasis | down | 4.70E-05 |
| Cytokinesis | down | 4.90E-05 |
| Muscle cell differentiation | up | 5.00E-05 |
| Myeloid cell differentiation | up | 5.20E-05 |
| Catabolic process | down | 5.20E-05 |
| Oxygen and reactive oxygen species metabolic process | down | 5.20E-05 |
| Chromatin | down | 5.30E-05 |
| Epidermis development | down | 5.40E-05 |
| Oxidoreductase activity, acting on the CH–NH group of donors | down | 5.60E-05 |
| SNARE complex | up | 5.60E-05 |
| Ligase activity, forming carbon–oxygen bonds | up | 5.70E-05 |
| Ligase activity, forming aminoacyl–tRNA and related compounds | up | 5.70E-05 |
| Soluble fraction | up | 5.80E-05 |
| Endopeptidase inhibitor activity | down | 5.90E-05 |
| Protease inhibitor activity | down | 5.90E-05 |
| Small protein conjugating enzyme activity | up | 6.00E-05 |
| AP-type membrane coat adaptor complex | down | 6.40E-05 |
| Cell–cell signaling | down | 6.70E-05 |
| Amine transport | up | 6.80E-05 |
| Response to DNA damage stimulus | up | 7.20E-05 |
| Male sex determination | down | 7.20E-05 |
| Cell–cell adhesion | down | 8.00E-05 |
| Protein heterodimerization activity | down | 8.00E-05 |
| Enzyme binding | up | 8.40E-05 |
| Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | down | 9.10E-05 |
| Ectoderm development | down | 9.40E-05 |
| Positive regulation of progression through cell cycle | down | 0.0001 |
| One-carbon compound metabolic process | down | 0.0001 |
| Heterogeneous nuclear ribonucleoprotein complex | down | 0.0001 |
| Response to stress | down | 0.0001 |
| Response to endogenous stimulus | up | 0.0001 |
| Cell death | up | 0.0001 |
| Death | up | 0.0001 |
| Meiosis | down | 0.0001 |
| Dioxygenase activity | up | 0.0001 |
| Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen | up | 0.0001 |
| Isoprenoid biosynthetic process | down | 0.0001 |
| Carbon–carbon lyase activity | down | 0.0001 |
| Lipid digestion | down | 0.0001 |
| Nitric oxide metabolic process | down | 0.0001 |
| Nitric oxide biosynthetic process | down | 0.0001 |
| Cellular homeostasis | down | 0.0001 |
| Positive regulation of locomotion | down | 0.0001 |
| Positive regulation of cell motility | down | 0.0001 |
| Sulfur compound biosynthetic process | up | 0.0001 |
| Proton-transporting ATP synthase complex, catalytic core F(1) | down | 0.0001 |
| Mitochondrial proton-transporting ATP synthase complex | down | 0.0002 |
| Muscle development | up | 0.0002 |
| Integrator complex | up | 0.0002 |
| Caspase inhibitor activity | down | 0.0002 |
| Fatty acid binding | down | 0.0002 |
| Isoprenoid metabolic process | down | 0.0002 |
| Blood vessel morphogenesis | up | 0.0002 |
| Vasculogenesis | up | 0.0002 |
| Kinetochore | down | 0.0002 |
| Low-density lipoprotein binding | down | 0.0002 |
| Cell structure disassembly during apoptosis | up | 0.0002 |
| Endoplasmic reticulum lumen | down | 0.0002 |
| Negative regulation of multicellular organismal process | down | 0.0002 |
| Intestinal absorption | down | 0.0002 |
| Organelle outer membrane | down | 0.0002 |
| Proton-transporting two-sector ATPase complex, catalytic domain | down | 0.0002 |
| Regulation of transforming growth factor beta receptor signaling pathway | down | 0.0002 |
| Clathrin adaptor complex | down | 0.0002 |
| Fatty acid metabolic process | down | 0.0003 |
| Replication fork | down | 0.0003 |
| Lipoprotein binding | down | 0.0003 |
| Insemination | up | 0.0003 |
| Behavior | down | 0.0003 |
| Leukocyte differentiation | up | 0.0003 |
| Single-stranded RNA binding | down | 0.0003 |
| Histone acetyltransferase complex | up | 0.0003 |
| Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen | up | 0.0003 |
| Hemopoiesis | up | 0.0003 |
| Outer kinetochore of condensed | down | 0.0003 |
| chromosome | ||
| Response to virus | up | 0.0003 |
| Vasculature development | up | 0.0003 |
| DNA-directed RNA polymerase complex | up | 0.0003 |
| Perinuclear region of cytoplasm | down | 0.0003 |
| Sterol binding | down | 0.0003 |
| Generation of precursor metabolites and energy | down | 0.0003 |
| Copulation | up | 0.0004 |
| Spindle localization | down | 0.0004 |
| Establishment of mitotic spindle localization | down | 0.0004 |
| Establishment of spindle localization | down | 0.0004 |
| Neural crest cell development | down | 0.0004 |
| GTPase inhibitor activity | down | 0.0004 |
Notes: Functional groupings of the modulated genes were determined using Gene Ontology classifications in NextBio. The top 200 biogroups most significantly regulated by AZA (at 3 µM) are shown.
Table S5.
Top 200 biogroups modulated by decitabine (DAC) in H1299 cells
| H1299 cells treated with 3 μM DAC (48 hours)
| |||
|---|---|---|---|
| Biogroup name | Direction | P value | |
| Cofactor binding | down | 4.70E-12 | |
| Lipid metabolic process | down | 5.10E-12 | |
| Cell differentiation | up | 2.50E-08 | |
| Coenzyme binding | down | 3.70E-08 | |
| Transcription | up | 1.00E-07 | |
| Inner ear development | up | 2.20E-07 | |
| Cell fate determination | up | 2.50E-07 | |
| Fatty acid metabolic process | down | 2.90E-07 | |
| Collagen binding | up | 4.90E-07 | |
| Oxidoreductase activity, acting on the CH–OH group of donors, NAD or NADP as acceptor | down | 1.20E-06 | |
| Enzyme inhibitor activity | up | 1.30E-06 | |
| Nucleosome | up | 1.30E-06 | |
| Aldehyde metabolic process | down | 1.40E-06 | |
| Sensory organ development | up | 2.30E-06 | |
| Oxidoreductase activity, acting on CH–OH group of donors | down | 2.50E-06 | |
| Response to external stimulus | up | 2.80E-06 | |
| Hormone biosynthetic process | up | 3.00E-06 | |
| CoA–ligase activity | down | 4.00E-06 | |
| Insulin-like growth factor binding | up | 4.20E-06 | |
| Response to stress | up | 4.40E-06 | |
| Mitochondrion | down | 5.20E-06 | |
| Acid–thiol ligase activity | down | 6.30E-06 | |
| Proteinaceous extracellular matrix | up | 7.40E-06 | |
| Transcription repressor activity | up | 7.90E-06 | |
| Extracellular matrix | up | 8.60E-06 | |
| Muscle cell differentiation | up | 9.40E-06 | |
| Response to wounding | up | 9.50E-06 | |
| Alcohol metabolic process | down | 9.90E-06 | |
| Enzyme regulator activity | up | 9.90E-06 | |
| Neurotransmitter metabolic process | up | 1.00E-05 | |
| Muscle fiber development | up | 1.10E-05 | |
| Skeletal muscle fiber development | up | 1.10E-05 | |
| Peroxisome | down | 1.10E-05 | |
| Microbody | down | 1.10E-05 | |
| Ligase activity, forming carbon-sulfur bonds | down | 1.40E-05 | |
| Regulation of epidermis development | up | 2.00E-05 | |
| Cell fate commitment | up | 2.20E-05 | |
| Hormone metabolic process | up | 2.20E-05 | |
| Sterol metabolic process | down | 2.30E-05 | |
| Inflammatory response | up | 2.30E-05 | |
| Oxidoreductase activity, acting on the CH–CH group of donors, NAD or NADP as acceptor | down | 2.60E-05 | |
| Death | up | 3.20E-05 | |
| Cell death | up | 3.20E-05 | |
| Steroid metabolic process | down | 3.50E-05 | |
| Lipid biosynthetic process | down | 3.80E-05 | |
| Lyase activity | down | 4.10E-05 | |
| Glycosaminoglycan binding | up | 4.50E-05 | |
| Polysaccharide binding | up | 5.30E-05 | |
| Pyridoxal phosphate binding | down | 5.50E-05 | |
| Muscle development | up | 6.70E-05 | |
| Nitrogen compound biosynthetic process | down | 6.90E-05 | |
| Protease inhibitor activity | up | 7.60E-05 | |
| Endopeptidase inhibitor activity | up | 7.60E-05 | |
| Phosphatase activator activity | up | 8.00E-05 | |
| Phenol metabolic process | up | 8.40E-05 | |
| Epidermis development | up | 8.50E-05 | |
| Regulation of neurotransmitter levels | up | 9.20E-05 | |
| Pattern binding | up | 9.60E-05 | |
| Cofactor catabolic process | down | 9.80E-05 | |
| Positive regulation of developmental process | up | 1.00E-04 | |
| Vitamin binding | down | 0.0001 | |
| Oxidoreductase activity, acting on the CH–CH group of donors | down | 0.0001 | |
| Amino acid derivative metabolic process | up | 0.0001 | |
| Chromatin assembly | up | 0.0001 | |
| Protein–DNA complex assembly | up | 0.0001 | |
| Amino acid derivative biosynthetic process | up | 0.0001 | |
| Positive regulation of cell differentiation | up | 0.0001 | |
| Acute inflammatory response | up | 0.0001 | |
| Dopamine metabolic process | up | 0.0001 | |
| Growth factor binding | up | 0.0002 | |
| Endothelial cell development | down | 0.0002 | |
| Transcription corepressor activity | up | 0.0002 | |
| Keratinocyte differentiation | up | 0.0002 | |
| Ectoderm development | up | 0.0002 | |
| Cellular respiration | down | 0.0002 | |
| RNA polymerase II transcription elongation factor activity | up | 0.0002 | |
| Angiogenesis | up | 0.0003 | |
| Calcium-dependent phospholipid binding | down | 0.0003 | |
| Suckling behavior | down | 0.0003 | |
| Oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | down | 0.0003 | |
| Germ-line sex determination | down | 0.0003 | |
| RNA polymerase II transcription factor activity | up | 0.0003 | |
| Protein kinase inhibitor activity | up | 0.0004 | |
| Translation activator activity | up | 0.0004 | |
| Regulation of Notch signaling pathway | up | 0.0004 | |
| Fatty acid biosynthetic process | down | 0.0004 | |
| Kinase inhibitor activity | up | 0.0004 | |
| Regulation of cell differentiation | up | 0.0004 | |
| ER-Golgi intermediate compartment | up | 0.0005 | |
| UDP-glycosyltransferase activity | down | 0.0005 | |
| Cell maturation | up | 0.0005 | |
| Cell surface | down | 0.0005 | |
| Inner ear receptor cell fate commitment | up | 0.0005 | |
| Organic acid biosynthetic process | down | 0.0005 | |
| Negative regulation of signal transduction | up | 0.0006 | |
| Hydro–Lyase activity | down | 0.0006 | |
| Epidermal cell differentiation | up | 0.0007 | |
| Inner ear morphogenesis | up | 0.0007 | |
| Regulation of cell growth | up | 0.0007 | |
| Response to bacterium | up | 0.0007 | |
| Blood vessel morphogenesis | up | 0.0007 | |
| Carbohydrate metabolic process | down | 0.0007 | |
| Lipoprotein binding | down | 0.0007 | |
| Multicellular organismal movement | down | 0.0008 | |
| Aromatic compound metabolic process | up | 0.0008 | |
| Amine biosynthetic process | down | 0.0008 | |
| tRNA binding | down | 0.0008 | |
| Cell migration | up | 0.0008 | |
| Transcription elongation factor complex | up | 0.0009 | |
| Glutathione peroxidase activity | up | 0.0009 | |
| Oxidoreductase activity, acting on the CH–NH group of donors, NAD or NADP as acceptor | down | 0.0009 | |
| Primary sex determination | down | 0.0009 | |
| Negative regulation of cell differentiation | up | 0.001 | |
| Defense response to bacterium | up | 0.001 | |
| Nuclear envelope | down | 0.001 | |
| Ear morphogenesis | up | 0.0011 | |
| Ligase activity, forming carbon–oxygen bonds | down | 0.0011 | |
| Ligase activity, forming aminoacyl–tRNA and related compounds | down | 0.0011 | |
| Generation of precursor metabolites and energy | down | 0.0011 | |
| Developmental maturation | up | 0.0011 | |
| Peripheral nervous system development | down | 0.0011 | |
| Extracellular matrix structural constituent | up | 0.0012 | |
| Oxidoreductase activity, acting on the aldehyde or oxo group of donors | down | 0.0012 | |
| Biogenic amine metabolic process | up | 0.0012 | |
| Epidermis morphogenesis | up | 0.0013 | |
| Endothelial cell differentiation | down | 0.0013 | |
| Endoplasmic reticulum | down | 0.0013 | |
| Carbon–oxygen lyase activity | down | 0.0013 | |
| Germ cell development | up | 0.0014 | |
| Cell proliferation | down | 0.0015 | |
| Peroxidase activity | up | 0.0015 | |
| Oxidoreductase activity, acting on peroxide as acceptor | up | 0.0015 | |
| Specific RNA polymerase II transcription factor activity | up | 0.0015 | |
| Positive regulation of programmed cell death | up | 0.0016 | |
| Response to nutrient levels | down | 0.0016 | |
| Myeloid cell differentiation | up | 0.0016 | |
| Lung development | up | 0.0017 | |
| Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | down | 0.0017 | |
| Vasculature development | up | 0.0017 | |
| Respiratory tube development | up | 0.0018 | |
| Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen | down | 0.0018 | |
| Dioxygenase activity | down | 0.0018 | |
| Negative regulation of enzyme activity | up | 0.0019 | |
| Odontogenesis | up | 0.0019 | |
| Positive regulation of locomotion | down | 0.0019 | |
| Positive regulation of cell motility | down | 0.0019 | |
| Cell cycle | up | 0.0019 | |
| Anatomical structure formation | up | 0.002 | |
| Cell growth | up | 0.0021 | |
| Regulation of cell size | up | 0.0021 | |
| NAD binding | down | 0.0021 | |
| Steroid biosynthetic process | down | 0.0022 | |
| Response to extracellular stimulus | down | 0.0022 | |
| Endocytosis | down | 0.0023 | |
| Membrane invagination | down | 0.0023 | |
| Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen | down | 0.0025 | |
| Musculoskeletal movement | down | 0.0025 | |
| Brain development | up | 0.0026 | |
| Immune system development | up | 0.0027 | |
| Negative regulation of cell growth | down | 0.0028 | |
| Negative regulation of cell size | down | 0.0028 | |
| Envelope | down | 0.0029 | |
| Negative regulation of developmental process | up | 0.0029 | |
| Chromatin | up | 0.003 | |
| mRNA binding | up | 0.0032 | |
| Antioxidant activity | up | 0.0033 | |
| Selenium binding | up | 0.0035 | |
| Receptor signaling protein serine/threonine kinase activity | down | 0.0036 | |
| Outer membrane-bounded periplasmic space | down | 0.0036 | |
| Cell envelope | down | 0.0036 | |
| Axon | up | 0.0036 | |
| Transferase activity, transferring hexosyl groups | down | 0.0038 | |
| Energy derivation by oxidation of organic compounds | down | 0.0038 | |
| Response to reactive oxygen species | up | 0.0038 | |
| Meiosis | up | 0.0038 | |
| Meiotic cell cycle | up | 0.0038 | |
| Enzyme activator activity | up | 0.0039 | |
| Hemopoiesis | up | 0.0043 | |
| Positive regulation of biosynthetic process | up | 0.0043 | |
| Regulation of transforming growth factor beta receptor signaling pathway | down | 0.0043 | |
| FAD binding | down | 0.0044 | |
| Negative regulation of growth | down | 0.0045 | |
| Defense response to Gram-positive bacterium | up | 0.0048 | |
| Sensory perception of light stimulus | down | 0.0048 | |
| Catabolic process | down | 0.0049 | |
| Hemopoietic or lymphoid organ development | up | 0.005 | |
| Positive regulation of protein metabolic process | up | 0.0051 | |
| Intermediate filament | up | 0.0051 | |
| Ras GTPase activator activity | down | 0.0052 | |
| Response to nutrient | down | 0.0053 | |
| Protein kinase regulator activity | up | 0.0053 | |
| Carbohydrate binding | up | 0.0054 | |
| Calcium ion binding | down | 0.0055 | |
| Oxidoreductase activity, acting on sulfur group of donors | down | 0.0056 | |
| Response to hypoxia | down | 0.0057 | |
| Transferase activity, transferring aldehyde or ketonic groups | up | 0.0058 | |
| Positive regulation of multicellular organismal process | up | 0.0059 | |
| Xenobiotic transporter activity | up | 0.0063 | |
| Xenobiotic-transporting ATPase activity | up | 0.0063 | |
| Phospholipase inhibitor activity | up | 0.0064 | |
Notes: Functional groupings of the modulated genes were determined using Gene Ontology classifications in NextBio. The top 200 biogroups most significantly regulated by DAC (at 3 µM) are shown.