Abstract
Background
Riboflavin is the precursor of important redox cofactors such as flavin mononucleotide (FMN) and flavin adenine dinucleotide, required for several biological processes. Vibrio cholerae, a pathogenic bacterium responsible for the cholera disease, possesses the ability to biosynthesize de novo as well as to uptake riboflavin through the riboflavin biosynthetic pathway (RBP) and the RibN importer, respectively. The intra-organism relationship between riboflavin biosynthesis and uptake functions has not been studied.
Results
This work determined the transcriptional organization of RBP genes and ribN in V. cholerae through reverse transcription polymerase chain reaction and analyzed their expression when growing with or without extracellular riboflavin using real time PCR. The RBP is organized in three transcriptional units, the major one containing ribD, ribE, ribA and ribH together with genes involved in functions not directly related to riboflavin biosynthesis such as nrdR and nusB. In addition, two independent monocistronic units contain ribA2 and ribB, the later conserving a putative FMN riboswitch. The ribN gene is encoded in operon with a gene coding for a predicted outer membrane protein and a gene encoding a protein with a glutaredoxin domain. Regulation analysis showed that among these transcriptional units, only ribB is negatively regulated by riboflavin and that its repression depends on the RibN riboflavin importer. Moreover, external riboflavin highly induced ribB transcription in a ΔribN strain. Also, a genomic database search found a negative correlation between the presence of nrdR and nusB and the FMN riboswitch in bacterial RBP operons.
Conclusions
Growing in the presence of riboflavin downregulates only a single element among the transcriptional units of riboflavin supply pathways. Thus, endogenous riboflavin biosynthesis seems to be negatively regulated by extracellular riboflavin through its specific effect on transcription of ribB in V. cholerae.
Electronic supplementary material
The online version of this article (doi:10.1186/s13099-017-0159-z) contains supplementary material, which is available to authorized users.
Keywords: Vibrio cholerae, Riboflavin, Transporter, Biosynthesis, Gene regulation
Background
Flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), the main derivatives of riboflavin, are cofactors for enzymes mediating many redox reactions in the cell [1]. Globally, up to 17% of enzymes dependent on cofactors use flavins [2]. Flavoenzymes are involved in a mixture of biological processes such as vitamin, fat and carbohydrate metabolism, photosensitization and oxidative stress response. In bacteria, flavins per se are also involved in extracellular processes such as assimilatory iron reduction, extracellular respiratory chain and symbiotic interactions [1, 3–6].
There are two ways bacteria can obtain riboflavin, one of them being the riboflavin biosynthetic pathway (RBP) and the other the riboflavin importer systems. The RBP uses guanosine-5-triphosphate (GTP) and ribulose 5-phosphate as precursors for the biosynthesis of riboflavin. Generally, the RBP consists of five enzymes, GTP cyclohydrolase II (RibA), a bifunctional pyrimidine deaminase/reductase (RibD), 3,4-dihydroxy-2-butanone-4-phosphate (3,4-DHBP) synthase (RibB), 6,7-dimethyl-8-ribityllumazine (lumazine) synthase (RibH) and riboflavin synthase (RibE) [1, 7, 8]. To date, nine riboflavin transport systems have been identified in bacteria. This include the energy-coupling factor system-RibU, RfuABCD and RibXYZ, which are members of the ATP binding cassette family of transporters, and the RfnT, RibM, RibN, RibZ and RibV transporters [9–17]. The transcriptional organization of the RBP genes largely differ among bacteria species. While some species arrange the full pathway genes in a single operon, others have the genes scattered in different transcriptional units along the chromosome. In addition, operons with RBP genes may also contain genes not directly related to riboflavin biosynthesis. Particularly, the global regulators nrdR and nusB, encoding a transcriptional repressor of nucleotide reductases and other genes and a factor of the bacterial antiterminator complex, respectively, have been found genetically associated to RBP in various species [14, 16, 18–22]. In some bacteria, RBP and riboflavin importer genes conserve the FMN riboswitch [9, 12, 15, 16, 23–25]. This is a genetic element encoded in RNA leader regions, which downregulates transcription and/or translation by adopting alternative expression-permissive or expression-repressive secondary structures in response to FMN binding [25, 26]. Thus, many riboflavin supply genes seem to be regulated in response to intracellular flavin levels.
Despite the fact that some bacteria lack the RBP and fulfill their riboflavin demands through riboflavin uptake, in many organisms the RBP and riboflavin importer genes coexist [10, 15, 27, 28]. Recently, a search on a set of fully sequenced bacterial genomes showed that most bacteria with a riboflavin importer also encode the RBP. Moreover, some species conserve two different families of riboflavin importers besides the RBP [12]. Additionally, bacteria may encode duplicated or multiplicated orthologs of some RBP enzymes. The complexity of bacterial riboflavin supply pathways seem to respond to species-specific riboflavin needs. For example, a duplicated RibH ortholog in Brucella abortus is specifically associated to survival inside the host and the ribBA gene of Sinorhizobium meliloti is specialized in the production of riboflavin targeted for secretion [29, 30]. This has led to the hypothesis that riboflavin biosynthesis has a modular structure in bacteria [30]. Although it is possible that riboflavin importers substitute for the RBP in riboflavin prototrophs when environmental riboflavin is available, the way intraspecies riboflavin provision pathways coordinate to accomplish flavin requirements in bacteria has been scarcely studied.
Vibrio cholerae is an aquatic gammaproteobacteria that causes cholera, a human pandemic disease characterized by acute watery diarrhea, which can lead to death in a short term if untreated. Normally thriving in sea and estuarine waters, the environmental cycle of V. cholerae includes biofilm formation in biotic and abiotic surfaces and the entrance into the metabolically quiescent viable but non-culturable state under unfavorable conditions [31]. After human consumption, V. cholera expresses a series of virulence factors in human intestinal tract, most notably the cholera toxin and the toxin-coregulated pilus. These factors contribute to host colonization and diarrhea development [32].
Vibrio cholerae is a riboflavin prototroph and it also has the ability to scavenge riboflavin through the RibN riboflavin importer [14, 33]. Given the wide range of conditions comprising its life cycle, it is likely that V. cholerae faces variable riboflavin concentrations. This feature makes it an interesting species to study the interconnections between riboflavin biosynthesis and uptake. The present work determined the transcriptional organization of the RBP and ribN genes in V. cholerae. In addition, to gain insights into the cues governing the interrelation between riboflavin biosynthesis and transport, we investigated the effect of extracellular riboflavin on the expression of the transcriptional units encoding the riboflavin supply pathways.
Results
Transcriptional organization of riboflavin supply pathways in V. cholerae
We searched the genome of V. cholerae N16961 in the Kyoto encyclopedia of genes and genomes (KEGG) [34] for RBP genes and ribN. Results show that this strain conserves a cluster of contiguous ribD, ribE, ribA-COG3236 and ribH genes in chromosome 1 (Fig. 1a). This cluster is localized between nrdR and nusB. The ribA-COG3236 gene fusion encodes a protein with GTP cyclohydrolase II (RibA) activity fused to a protein domain belonging to the cluster of orthologous groups (COG) 3236. COG3236 modifies early riboflavin biosynthesis intermediates, cleaving N-glycosidic bonds presumably to diminish their reactivity when overproduced. The COG3236 is often found fused to RBP enzymes in plants and bacteria [35]. An alignment performed by us determined that the V. cholerae fusion shares 89% identity with the experimentally characterized ortholog in Vibrio vulnificus. Importantly, the intergenic region between ribA-COG3236 and ribH is relatively large (264 bp) and conserves an apparently incomplete FMN riboswitch, as detected by the RIBEX software [36]. Thus, it is not possible to predict hiterto if these genes form an operon. Genes ribB and a second ortholog of ribA, hereafter referred to as ribA2, were identified in different regions of chromosome 2 and 1, respectively. The ribB gene conserves an FMN riboswitch. Overall, the distribution and assortment of RBP genes in V. cholerae resembles the predicted distribution of such genes in V. parahaemolyticus [22]. In addition, the ribN gene is localized in chromosome 2 between the VCA1008 and VCA1011 open reading frames, encoding a putative outer membrane protein and a protein containing a glutaredoxin domain, respectively.
Fig. 1.
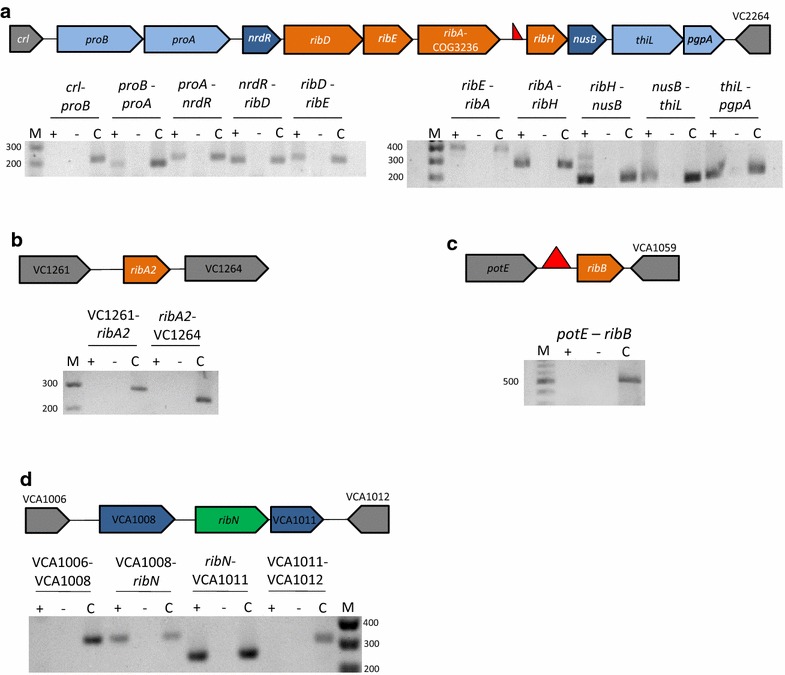
Assessment of the transcriptional organization of rib genes in V. cholerae. PCR reactions were carried on cDNA from V. cholerae with primers designed to amplify the indicated gene junctions of rib genes and their flanking genes in the loci encoding ribD, ribE, ribA-COG3236 and ribH (a), ribA2 (b), ribB (c) and ribN (d). Each reaction was independently performed three times with the same results. + Template cDNA derived from RT-PCR with reverse transcriptase, − template cDNA derived from RT-PCR without reverse transcriptase (negative control), C PCR with chromosomal DNA as template (positive control), M molecular weight marker (basepairs). Isosceles triangle over the regulatory region indicates FMN riboswitch. Half Isosceles triangle indicates truncated FMN riboswitch
To experimentally determine the transcriptional organization of the identified riboflavin supply genes, a reverse transcriptase polymerase chain reaction (RT-PCR) analysis was performed. This carried PCR reactions on complementary DNA (cDNA) obtained from V. cholerae RNA with primers designed to amplify putative gene junctions. In this approach, positive PCR amplifications indicate the joint of coding sequences in the same messenger RNA. Results showed that the main RBP operon extends from proB to pgpA, also including the genes proA, nrdR, ribD, ribE, ribA-COG3236, ribH, nusB and thiL. All of the RT-PCR reactions produced amplicons of the expected size for these gene junctions, while negative controls, consisting on templates obtained without reverse transcriptase, did not yield amplicons. Positive controls, using chromosomal DNA as template, yielded amplicons in all cases, including junctions of genes not forming operon (Fig. 1a). A similar analysis showed that ribA2 and ribB genes comprise monocistronic untis (Fig. 1b, c). Likewise, results showed that ribN forms an operon with VCA1008 and VCA1011 (Fig. 1d).
Effect of riboflavin on the expression of riboflavin supply genes
We assessed the effect of extracellular riboflavin on the expression of the four transcriptional units identified. Bacteria were grown in T minimal medium with or without riboflavin and the relative expression among these two conditions of ribD, ribA2, ribB and ribN was assessed by real time PCR (Fig. 2). This assay also included the housekeeping gyrB gene unrelated to riboflavin provision as a control. Results showed that ribB transcription was reduced by two-thirds when growing in the presence of riboflavin. Although in this experiment the expression of ribD and ribA2 was somewhat reduced in the presence of riboflavin, this effect was not statistically significant. Meanwhile, ribN expression was not affected. To determine if the negative regulatory effect of riboflavin on ribB depends on the activity of the RibN riboflavin importer, we constructed a ribN null mutant and assessed its ribB expression. Unexpectedly, the elimination of ribN caused a sevenfold increase in the expression of ribB when growing in the presence of riboflavin when compared to the WT without riboflavin (Fig. 3). This overexpression is indeed dependent on extracellular riboflavin, as the expression of ribB in the ∆ribN mutant is similar to that of the WT when no riboflavin is added to the medium. Moreover, the expression of ribB on the ∆ribN is increased by 19 fold by the presence of riboflavin. This overexpression was prevented by the complementation of the ∆ribN strain with a plasmid encoding ribN (pBribNVch). In all cases, ribD expression remained around the same irrespective of the presence of riboflavin. These results show not only that RibN is necessary to repress ribB in response to riboflavin, but that extracellular riboflavin highly induces the expression of this gene when the RibN riboflavin importer is absent.
Fig. 2.
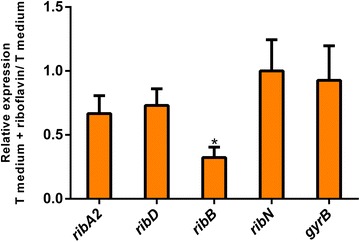
Relative expression of riboflavin supply pathways genes in V. cholerae growing in media with and without riboflavin. V. cholerae was grown in T minimal media and relative expression of the ribD, ribA2, ribB, ribN and gyrB genes in cultures with and without added riboflavin was determined by real time PCR. Media and standard deviation from three independent experiments are shown. * Statistical difference compared to the gyrB control (p < 0.05) using analysis of variance and Tukey HSD post hoc test
Fig. 3.
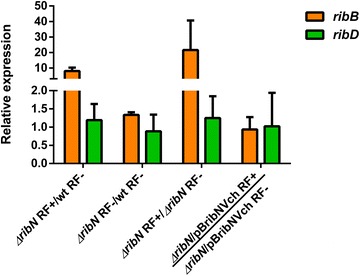
Effect of ribN deletion on expression of ribB and ribD. Relative ribB and ribD expression in ∆ribN with riboflavin and WT without riboflavin, both strains without riboflavin and ∆ribN and its complemented derivative growing with or without riboflavin as indicated, determined by real time PCR. Media and standard deviation from three independent experiments are shown. RF riboflavin
The presence of the FMN riboswitch negatively correlates with nrdR and nusB in bacterial RBP operons
The expression profiles defined here suggest that only ribB is significantly downregulated in response to extracellular riboflavin. The presence of genes involved in functions different to riboflavin biosynthesis in the main operon, most notably the nrdR and nusB global regulators, may preclude its systematic regulation by riboflavin levels. As association of FMN riboswitch and nrdR and nusB to RBP genes is widespread in bacteria [12, 16, 37, 38], we seek to estimate the co-occurrence of these genetics elements in RBP genes. For this purpose, we searched the bacterial genomes in the Prokaryotic Operon Data Base (ProOpDB) for the presence of the FMN riboswitch. ProOpDB contains the predicted transcriptional organization of a set of fully sequenced prokaryotic genomes [39]. 651 predicted bacterial transcriptional units displayed by the ProOpDB contained the FMN riboswitch (element RF 00050) (Additional file 1). While almost all of them clustered RBP and/or riboflavin importer genes, none of them included nrdR and only five of them included nusB. On the other side, a search for the transcriptional organization of nrdR homologues in the database shows its association with RBP genes in 86 transcriptional units, none of them conserving the FMN element (Additional file 2). Overall, ProOpDB search results support the hypothesis that riboflavin biosynthesis is not repressed by flavins through the FMN riboswitch when the operon encodes additional genes not directly related to riboflavin biosynthesis such as the global regulators NrdR and NusB.
Discussion
Results showed that riboflavin provision pathways in V. cholerae are encoded in four transcriptional elements. The main RBP operon includes the genes ribD, ribE, ribA-COG3236 and ribH, together with genes involved in proline biosynthesis (proBA), thiamin biosynthesis (thiL) and a phosphatidylglycerophosphatase (pgpA) in addition to nusB and nrdR. In spite of its large upstream intergenic region, ribH and downstream genes are part of the operon. Noteworthy, this region conserves a truncated riboswitch. This may hint to a relatively recent integration of this segment to the operon. Two other RBP components, ribB and ribA2 were found to be encoded monocistronically. The conservation of a FMN riboswitch only in ribB suggested a differential regulation for the RBP components a priori. Later results showed a negative correlation between nrdR and nusB and the FMN riboswitch in RBP genes. This is an interesting finding as it starts to define patterns in the apparently highly variable ways bacteria transcriptionally arrange their RBP genes [28]. Finally, RibN was found encoded in operon with a putative outer membrane protein (VCA1008) and a protein with a domain of unknown function fused to a glutaredoxin-like domain (VCA1011). Glutaredoxins are enzymes involved in redox homeostasis in bacteria [40]. Thus, it is possible that imported riboflavin is related to functions of VCA1008 and VCA1011.
Riboflavin needs in bacteria are highly diverse and seem to be quite dependent on species specific traits. While some bacteria may fulfill all of its riboflavin needs through the RBP and others rely solely on riboflavin uptake, many have conserved both functions. The selection constraints driving this phenomenon are not known. It has been pointed out that riboflavin biosynthesis requires more metabolic energy than its transport inside the cell [41]. Hence, activation of riboflavin uptake and biosynthesis halt when growing in nutrient rich environments would help save energy in a prototroph species. This could be the case for V. cholerae. Growing on riboflavin rich media downregulated ribB. Noteworthy, RibB activity is the only one lacking on the otherwise full RBP encoded in the main riboflavin biosynthetic operon. Thus, it seems that the extracellular availability of riboflavin diminishes endogenous riboflavin biosynthesis at the level of 3,4-DHBP synthesis. Nonetheless, ribB transcription was not completely abolished. This may reflect that some ribB expression is still afforded in the presence of extracellular riboflavin or that the ribB FMN riboswitch also acts at the translational level to fully shut down expression, as recently shown for the FMN riboswitch in ribB from Escherichia coli [25]. It is intriguing why constitutive expression of the rest of the RBP enzymes is maintained in the presence of extracellular riboflavin. One explanation for this may be the participation of these enzymes in other metabolic pathways. Indeed, RibA has been involved in folate biosynthesis in Chlamydia trachomatis [42] and it has been suggested a participation of RibD and RibA in toxoflavin biosynthesis in Pseudomonas protegens [43].
RibN was necessary for external riboflavin to repress ribB. Moreover, extracellular riboflavin highly induced the expression of ribB after deletion of ribN. Importantly, this result may imply that the presence of extracellular riboflavin triggers riboflavin-dependent traits inside the cell. In WT strain, these emergent riboflavin demands may be fulfilled through the RibN uptake activity. However, the ∆ribN strain may suffer a high reduction of intracellular flavin levels in such conditions, which could account for ribB overexpression.
Overall, results indicate that V. cholerae riboflavin provision pathways components are encoded in four transcriptional units. These units are differentially regulated by riboflavin, which may putatively reduce riboflavin biosynthesis through repression of RibB transcription. Although the transcription of the ribN riboflavin importer gene was not affected, the presence of riboflavin may trigger responses that increase the intracellular flavin requirements. Further experimental work is needed to effectively determine if endogenous riboflavin biosynthesis is diminished when growing in the presence of riboflavin, and to assess the bacterial physiological traits activated in response to it.
Methods
Growth conditions, RNA extraction, retrotranscription and PCR analysis
Vibrio cholerae N16961 strain and its ∆ribN derivative were grown overnight in LB plates at 37 °C. 5 ml of LB broth were inoculated with these overnight cultures and incubated at 37 °C in an orbital shaker at 150 rpm until they reached an OD600 nm of 1.0. Next, cultures were centrifuged, supernatant discarded, pellet washed two times with T minimal medium [44] and resuspended in 1 ml of fresh T medium. 10 µl of these resuspensions were used to inoculate 10 ml of T medium and T medium plus 2 µM riboflavin. Cultures were incubated at 37 °C with 180 rpm shaking until an OD600 nm of 0.8. Next, 3 ml of these cultures were pelleted and subjected to RNA extraction using the Thermo Scientific Genejet RNA purification kit according to manufacturer´s instructions. RNA extracts were digested with Turbo DNA-free DNAase at 37 °C for 1 h. cDNA synthesis was performed with AffinityScript QPCR cDNA Synthesis kit (Agilent Technologies) according to manufacturer’s instructions. A control reaction with no reverse transcriptase was included for each sample. PCR reactions using primers designed to amplify gene junctions were performed on these cDNA. Primers’ sequences are detailed in Additional file 3.
Real time PCR
Real time PCR assays were performed using cDNA obtained as previously described as templates and using the Brilliant II SYBR Green QPCR Master Mix kit in a One-Step Applen Biosystems (Life Technologies) thermocycler. Relative expression was calculated using the ∆∆Ct method as developed by Livak et al. [45]. 16s RNA subunit gene was used for normalization. Primers used for each gene for the real time PCR assay are detailed in Additional file 3.
Construction of ribN mutant and complementation plasmid
Deletion of ribN in Vibrio cholerae was performed with homologous recombination using PCR products as reported before [46]. Briefly, WT strain was transformed with plasmid pKD46 encoding λ phage Red recombinase system. Next, a PCR product containing a kanamycin resistant cassette flanked by sites homologous to ribN 5′ and 3′ ends, using primers ribNH1P1and ribNH2P2 (Additional file 3) and plasmid pKD4 as template was obtained. The PCR product was electroporated into WT strain carrying pKD46, recombination system expression induced with arabinose and recombinants selected in LB+kanamycin plates. Positive clones were tested for the replacement of ribN by the kanamycin cassette by PCR using primers VCA1008FwHindIII and grxRv, flanking the insertion site. One of the positive clones was electroporated with plasmid pCP20, encoding FRT recombinase. After growth in nonselective LB media, clones were analyzed for the loss of the kanamycin cassette by testing antibiotic sensibility and PCR using primers VCA1008FwHindIII and grxRv. To construct a complementation plasmid with ribN, this gene was amplified from V. cholerae genomic DNA with PCR using primers VCA1008FwHindIII and ribNRvFLAGPstI. The resulting product was cloned in the pGEM T Easy commercial cloning vector (Promega). Next, ribN was excised and subcloned into the PstI/XbaI sites of pBBR1 MCS 1 vector. This rendered the pBribNVch plasmid. The loss of expression of ribN transcript in the ∆ribN strain as well as the rescue of expression by the pBribNVch plasmid were corroborated by end point RT-PCR with primers ribNFw and ribNRv (Additional file 4).
Authors’ contributions
ISC performed reverse transcriptase and real time PCR experiments, discussed results and helped writing the paper. AT constructed the ribN mutant and discussed results. AFF helped in RNA extractions, discussed results and helped preparing the manuscript. VAG conceived the study, analyzed results, searched the ProOpDB and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional files.
Funding
This study was funded by CONICYT FONDECYT Regular (Chile), Grant Number 1150818.
Additional files
Additional file 1. ProOpDB output for the search of the FMN riboswitch (RF00050), visualized with GeConT [47]. Top of the figure shows the putative transcriptional organization of elements containing the FMN riboswitch in bacterial genomes. Bottom lists the clusters of orthologous groups (COG) contained in genes depicted in the top. Note that the COG1327, clustering nrdR homologs, is absent from the list.
Additional file 2. ProOpDB output for the search of the COG1327 (nrdR), visualized with GeConT. Top of the figure shows the putative organization of transcriptional units containing the COG1327. Bottom lists the COGs contained in genes depicted in the top.
Additional file 3. List of primers used in this study.
Additional file 4. Lack of expression of ribN in the V. cholerae ∆ribN strain and complementation by the pBribNVch plasmid. The presence of the ribN messenger RNA was assessed by end-point real time PCR using primers ribNFw and ribNRv and cDNA from the indicated strains. +, template cDNA derived from RT-PCR with reverse transcriptase; −, template cDNA derived from RT-PCR without reverse transcriptase (negative control); C, PCR with genomic DNA as template (positive control).
Contributor Information
Ignacio Sepúlveda Cisternas, Email: sepulveda.ignacio@gmail.com.
Alexia Torres, Email: ale.ntm6@gmail.com.
Andrés Fuentes Flores, Email: andres.a.e.fuentes@gmail.com.
Víctor Antonio García Angulo, Phone: +56 2 29789628, Email: v.garcia.angulo@gmail.com.
References
- 1.Abbas CA, Sibirny AA. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol Mol Biol Rev MMBR. 2011;75:321–360. doi: 10.1128/MMBR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monteverde DR, Gómez-Consarnau L, Suffridge C, Sañudo-Wilhelmy SA. Life’s utilization of B vitamins on early Earth. Geobiology. 2016;15:3–18. doi: 10.1111/gbi.12202. [DOI] [PubMed] [Google Scholar]
- 3.Nakabachi A, Ishikawa H. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol. 1999;45:1–6. doi: 10.1016/S0022-1910(98)00104-8. [DOI] [PubMed] [Google Scholar]
- 4.Crossley RA, Gaskin DJH, Holmes K, Mulholland F, Wells JM, Kelly DJ, et al. Riboflavin biosynthesis is associated with assimilatory ferric reduction and iron acquisition by Campylobacter jejuni. Appl Environ Microbiol. 2007;73:7819–7825. doi: 10.1128/AEM.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashoori M, Saedisomeolia A. Riboflavin (vitamin B2) and oxidative stress: a review. Br J Nutr. 2014;111:1985–1991. doi: 10.1017/S0007114514000178. [DOI] [PubMed] [Google Scholar]
- 6.Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci USA. 2008;105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer M, Bacher A. Biosynthesis of flavocoenzymes. Nat Prod Rep. 2005;22:324–350. doi: 10.1039/b210142b. [DOI] [PubMed] [Google Scholar]
- 8.Haase I, Gräwert T, Illarionov B, Bacher A, Fischer M. Recent advances in riboflavin biosynthesis. Methods Mol Biol Clifton NJ. 2014;1146:15–40. doi: 10.1007/978-1-4939-0452-5_2. [DOI] [PubMed] [Google Scholar]
- 9.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogl C, Grill S, Schilling O, Stülke J, Mack M, Stolz J. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J Bacteriol. 2007;189:7367–7375. doi: 10.1128/JB.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deka RK, Brautigam CA, Biddy BA, Liu WZ, Norgard MV. Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes. mBio. 2013;4:e00615. doi: 10.1128/mBio.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutiérrez-Preciado A, Torres AG, Merino E, Bonomi HR, Goldbaum FA, García-Angulo VA. Extensive identification of bacterial riboflavin transporters and their distribution across bacterial species. PLoS ONE. 2015;10:e0126124. doi: 10.1371/journal.pone.0126124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodionova IA, Li X, Plymale AE, Motamedchaboki K, Konopka AE, Romine MF, et al. Genomic distribution of B-vitamin auxotrophy and uptake transporters in environmental bacteria from the Chloroflexi phylum. Environ Microbiol Rep. 2014;7:204–210. doi: 10.1111/1758-2229.12227. [DOI] [PubMed] [Google Scholar]
- 14.García Angulo VA, Bonomi HR, Posadas DM, Serer MI, Torres AG, Zorreguieta A, et al. Identification and characterization of RibN, a novel family of riboflavin transporters from Rhizobium Leguminosarum and other proteobacteria. J Bacteriol. 2013;195:4611–4619. doi: 10.1128/JB.00644-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grill S, Yamaguchi H, Wagner H, Zwahlen L, Kusch U, Mack M. Identification and characterization of two Streptomyces davawensis riboflavin biosynthesis gene clusters. Arch Microbiol. 2007;188:377–387. doi: 10.1007/s00203-007-0258-1. [DOI] [PubMed] [Google Scholar]
- 16.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess CM, Slotboom DJ, Geertsma ER, Duurkens RH, Poolman B, van Sinderen D. The riboflavin transporter RibU in Lactococcus lactis: molecular characterization of gene expression and the transport mechanism. J Bacteriol. 2006;188:2752–2760. doi: 10.1128/JB.188.8.2752-2760.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKethan BL, Spiro S. Cooperative and allosterically controlled nucleotide binding regulates the DNA binding activity of NrdR. Mol Microbiol. 2013;90:278–289. doi: 10.1111/mmi.12364. [DOI] [PubMed] [Google Scholar]
- 19.Naveen V, Hsiao C-D. NrdR transcription regulation: global proteome analysis and its role in Escherichia coli viability and virulence. PLoS ONE. 2016;11:e0157165. doi: 10.1371/journal.pone.0157165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bubunenko M, Court DL, Al Refaii A, Saxena S, Korepanov A, Friedman DI, et al. Nus transcription elongation factors and RNase III modulate small ribosome subunit biogenesis in Escherichia coli. Mol Microbiol. 2013;87:382–393. doi: 10.1111/mmi.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stagno JR, Altieri AS, Bubunenko M, Tarasov SG, Li J, Court DL, et al. Structural basis for RNA recognition by NusB and NusE in the initiation of transcription antitermination. Nucleic Acids Res. 2011;39:7803–7815. doi: 10.1093/nar/gkr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brutinel ED, Dean AM, Gralnick JA. Description of a riboflavin biosynthetic gene variant prevalent in the phylum Proteobacteria. J Bacteriol. 2013;195:5479–5486. doi: 10.1128/JB.00651-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun EI, Leyn SA, Kazanov MD, Saier MH, Jr, Novichkov PS, Rodionov DA. Comparative genomics of metabolic capacities of regulons controlled by cis-regulatory RNA motifs in bacteria. BMC Genom. 2013;14:597. doi: 10.1186/1471-2164-14-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ott E, Stolz J, Lehmann M, Mack M. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 2009;6:276–280. doi: 10.4161/rna.6.3.8342. [DOI] [PubMed] [Google Scholar]
- 25.Pedrolli D, Langer S, Hobl B, Schwarz J, Hashimoto M, Mack M. The ribB FMN riboswitch from Escherichia coli operates at the transcriptional and translational level and regulates riboflavin biosynthesis. FEBS J. 2015;282:3230–3242. doi: 10.1111/febs.13226. [DOI] [PubMed] [Google Scholar]
- 26.Serganov A. The long and the short of riboswitches. Curr Opin Struct Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.García Angulo VA, Bonomi HR, Posadas DM, Serer MI, Torres AG, Zorreguieta A, et al. Identification and characterization of RibN, a novel family of riboflavin transporters from Rhizobium leguminosarum and other proteobacteria. J Bacteriol. 2013;195:4611–4619. doi: 10.1128/JB.00644-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Angulo VA. Overlapping riboflavin supply pathways in bacteria. Crit Rev Microbiol. 2017;43:196–209. doi: 10.1080/1040841X.2016.1192578. [DOI] [PubMed] [Google Scholar]
- 29.Bonomi HR, Marchesini MI, Klinke S, Ugalde JE, Zylberman V, Ugalde RA, et al. An atypical riboflavin pathway is essential for Brucella abortus virulence. PLoS ONE. 2010;5:e9435. doi: 10.1371/journal.pone.0009435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yurgel SN, Rice J, Domreis E, Lynch J, Sa N, Qamar Z, et al. Sinorhizobium meliloti flavin secretion and bacteria–host interaction: role of the bifunctional RibBA protein. Mol Plant Microbe Interact MPMI. 2014;27:437–445. doi: 10.1094/MPMI-11-13-0338-R. [DOI] [PubMed] [Google Scholar]
- 31.Conner JG, Teschler JK, Jones CJ, Yildiz FH. Staying alive: vibrio cholerae’s cycle of environmental survival, transmission, and dissemination. Microbiol Spectr. 2016;4. doi:10.1128/microbiolspec.VMBF-0015-2015. [DOI] [PMC free article] [PubMed]
- 32.Muanprasat C, Chatsudthipong V. Cholera: pathophysiology and emerging therapeutic targets. Future Med Chem. 2013;5:781–798. doi: 10.4155/fmc.13.42. [DOI] [PubMed] [Google Scholar]
- 33.Chao MC, Pritchard JR, Zhang YJ, Rubin EJ, Livny J, Davis BM, et al. High-resolution definition of the vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res. 2013;41:9033–9048. doi: 10.1093/nar/gkt654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frelin O, Huang L, Hasnain G, Jeffryes JG, Ziemak MJ, Rocca JR, et al. A directed-overflow and damage-control N-glycosidase in riboflavin biosynthesis. Biochem J. 2015;466:137–145. doi: 10.1042/BJ20141237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abreu-Goodger C, Merino E. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 2005;33:W690–W692. doi: 10.1093/nar/gki445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doerks T, Andrade MA, Lathe W, von Mering C, Bork P. Global analysis of bacterial transcription factors to predict cellular target processes. Trends Genet TIG. 2004;20:126–131. doi: 10.1016/j.tig.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.GR-1619R. [DOI] [PubMed] [Google Scholar]
- 39.Taboada B, Ciria R, Martinez-Guerrero CE, Merino E. ProOpDB: prokaryotic operon database. Nucleic Acids Res. 2012;40:D627–D631. doi: 10.1093/nar/gkr1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loi VV, Rossius M, Antelmann H. Redox regulation by reversible protein S-thiolation in bacteria. Front Microbiol. 2015;6:187. doi: 10.3389/fmicb.2015.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaehme M, Slotboom DJ. Diversity of membrane transport proteins for vitamins in bacteria and archaea. Biochim Biophys Acta BBA Gen Subj [Internet]. http://www.sciencedirect.com/science/article/pii/S0304416514001639. Accessed 28 June 2014. [DOI] [PubMed]
- 42.Adams NE, Thiaville JJ, Proestos J, Juárez-Vázquez AL, McCoy AJ, Barona-Gómez F, et al. Promiscuous and adaptable enzymes fill “holes” in the tetrahydrofolate pathway in Chlamydia species. mBio. 2014;5:e01378–01314. doi: 10.1128/mBio.01378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philmus B, Shaffer BT, Kidarsa TA, Yan Q, Raaijmakers JM, Begley TP, et al. Investigations into the biosynthesis, regulation, and self-resistance of Toxoflavin in Pseudomonas protegens Pf-5. Chembiochem Eur J Chem Biol. 2015;16:1782–1790. doi: 10.1002/cbic.201500247. [DOI] [PubMed] [Google Scholar]
- 44.Wyckoff EE, Payne SM. The Vibrio cholerae VctPDGC system transports catechol siderophores and a siderophore-free iron ligand. Mol Microbiol. 2011;81:1446–1458. doi: 10.1111/j.1365-2958.2011.07775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Guerrero CE, Ciria R, Abreu-Goodger C, Moreno-Hagelsieb G, Merino E. GeConT 2: gene context analysis for orthologous proteins, conserved domains and metabolic pathways. Nucleic Acids Res. 2008;36:W176–W180. doi: 10.1093/nar/gkn330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Additional files.


