Abstract
Background
Natural transformation enables acquisition of adaptive traits and drives genome evolution in prokaryotes. Yet, the selective forces responsible for the evolution and maintenance of natural transformation remain elusive since taken-up DNA has also been hypothesized to provide benefits such as nutrients or templates for DNA repair to individual cells.
Results
We investigated the immediate effects of DNA uptake and recombination on the naturally competent bacterium Acinetobacter baylyi in both benign and genotoxic conditions. In head-to-head competition experiments between DNA uptake-proficient and -deficient strains, we observed a fitness benefit of DNA uptake independent of UV stress. This benefit was found with both homologous and heterologous DNA and was independent of recombination. Recombination with taken-up DNA reduced survival of transformed cells with increasing levels of UV-stress through interference with nucleotide excision repair, suggesting that DNA strand breaks occur during recombination attempts with taken-up DNA. Consistent with this, we show that absence of RecBCD and RecFOR recombinational DNA repair pathways strongly decrease natural transformation.
Conclusions
Our data show a physiological benefit of DNA uptake unrelated to recombination. In contrast, recombination during transformation is a strand break inducing process that represents a previously unrecognized cost of natural transformation.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-017-0953-2) contains supplementary material, which is available to authorized users.
Keywords: Competence, Natural transformation, Horizontal gene transfer, Bacterial evolution, DNA repair, DprA
Background
Naturally transformable bacteria take up DNA from the environment upon entering a physiological state called competence [1, 2]. If sufficient homology is present in the taken-up DNA it may be integrated in the recipient genome and in turn combine beneficial alleles in the same genetic background, provide novel traits, and remove deleterious mutations [2–6]. Conversely, natural transformation may also cause harmful changes such as acquisition of deleterious mutations present in external DNA [7]. It is clear that natural transformation shapes bacterial genomes, and the mechanistic insights to DNA uptake and integration is well described for a number of bacterial species, reviewed in [2]. Yet, the selective forces responsible for the evolution and maintenance of natural transformation are still debated [6, 8, 9].
Natural transformation is considered a driving force of bacterial evolution [2, 10, 11], and mathematical models support that natural competence has evolved and is maintained as a mechanism for genome plasticity that lead to increased environmental adaption rates [12, 13]. In contrast, natural transformation has also been hypothesised to support genome stability [14–16] and to remove deleterious mobile genetic elements from chromosomes [17]. Experimental evolution approaches using various bacterial species competent for natural transformation including Helicobacter pylori, Acinetobacter baylyi and Streptococcus pneumoniae have revealed that the impact of natural transformation on rates of adaptive evolution can be beneficial [18, 19], neutral [20], and/or context dependent [21, 22]. Taken together, these reports suggest that the widely accepted idea that natural transformation accelerates bacterial adaptation lacks generality.
Although natural transformation contributes to genome evolution [2], the immediate effects of DNA uptake and recombination in individual cells remain unclear and several non-mutually exclusive hypotheses have been proposed. One hypothesis suggests that taken-up DNA may be used as a source of phosphate, nitrogen, carbon, or nucleotides [8, 23, 24]. Observations that expression of competence genes is controlled by carbon catabolite repression in Escherichia coli [25], Haemophilus influenzae [25, 26], Vibrio cholerae [27] and Streptococcus gordonii [28] support this hypothesis. More specific links between intracellular availability of nucleotides, their precursors and natural competence have been found in H. influenzae where purine depletion activates the competence activator sxy [23], and in Vibrio cholerae competence is repressed by exogenous levels of cytidine [29].
An alternative hypothesis suggests that taken-up DNA can be used as template for repair of genomic DNA damages [9, 30]. Four reports by Michod and co-workers provided experimental evidence in favour of this explanation in Bacillus subtilis [9, 31–33]. Increased transformation rates were observed when homologous DNA, but not heterologous DNA, was added after exposure of cells to UV-light [33]. However, other reports in B. subtilis and H. influenzae [34, 35], as well as in S. pneumoniae [36, 37] rather suggest that competence induction may be a general stress response. This is further supported by observations that competence-specific transcriptional regulators in S. peumoniae and B. subtilis control genes whose functions are linked to stress response (reviewed in [2]). The induction of competence by DNA damaging agents in S. pneumoniae [38], Legionella pneumophila [39] and H. pylori [40] also favour the “DNA-repair hypothesis” for the evolution and maintenance of natural transformation.
In the genus Acinetobacter competence is not regulated by DNA damages [41, 42] and in A. baylyi maximum competence is reached during exponential growth-phase [43]. Under optimal conditions up to 25% of all cells in a competent A. baylyi culture are transformed [44]. During transformation competent cells bind extracellular double-stranded DNA [4, 45] and a single-stranded DNA fragment is transported into the cytoplasm [1, 4]. While some bacterial species e.g., H. influenzae and species in the family of Neisseriaceae selectively take up isogenic DNA [46, 47], other species such as B. subtilis and A. baylyi transfer DNA of any source into the cytoplasm [44, 48]. Homologous recombination is then initiated by DprA-mediated loading of RecA onto the single-stranded DNA [49, 50] and by subsequent RecA-mediated strand invasion and integration of the donor DNA into the recipient genome. Previous reports on A. baylyi indicate that genomic integration frequently cause single-strand breaks in the transforming cell that can turn into double-strand breaks when unrepaired [51, 52].
In this study, we used A. baylyi to determine the immediate benefits and costs of DNA-uptake and -integration under both benign and stressful conditions. Our data show that uptake of both homologous and heterologous DNA increased fitness of competent A. baylyi cells. The fitness increment was independent of DNA damage and did not involve genomic integration of the acquired DNA. We further show that genomic integration of homologous DNA both reduced transformant fitness when exposed to UV-induced stress, and under benign conditions strongly depended on the RecBCD and RecFOR DNA strand break repair pathways.
Methods
Strains, media and DNA
The strains used in this study (Additional file 1: Table S1) are derivatives of Acinetobacter baylyi strain JV28 [53] and were constructed using standard molecular biology techniques [54]. Plasmids and primers used in this study are listed in supplementary tables (Additional file 1: Table S2 and Table S3). JV28 is derived from A. baylyi ADP1 (GenBank NC_005966) [55]. Strain LCQ2 was constructed by crossing the trpE + wildtype allele into JV28 as described [56]. KOM130 was constructed by replacing the comB-F operon of JV28 with the ΔcomB-F::dhfr1 allele [20, 22] by natural transformation with DNA isolated from strain ADP1200Com- [22]. Cotransformation of unwanted markers was excluded phenotypically (TrpE−) or by PCR (lifO-lipB::aphA3′). The dprA gene (ACIAD0209) of A. baylyi was inactivated by replacing an internal 349-bp fragment of dprA with an aacC1 (gentamicin resistance) gene using Splicing by Overlap Extension PCR [57]; see Additional file 2 for details]. The ∆uvrA (ACIAD3455), ∆recF (ACIAD0003) and ∆recR (ACIAD2312) mutations were constructed using the plasmid pGT41 as described [58, 59], see Additional file 2. The ΔuvrA strain KOM141 was obtained by replacing uvrA + of JV28 by the ΔuvrA::(nptII sacB) allele, and subsequent replacement of ΔuvrA::(nptII sacB) by the ΔuvrA allele. The recO (ACIAD2578) deletion allele was constructed similarly using a modified pGT41 plasmid (pGT33), and the plasmids containing the ΔrecO::(nptII sacB) and ΔrecO alleles were provided by J. de Vries (University of Oldenburg, Germany). Similar to ΔuvrA, the ∆recF, ∆recR and ∆recO alleles were each crossed into the JV28 strain, giving strains MKD1, MKD2 and KOM82, respectively. The ∆recR and ∆recF alleles were subsequently inserted into the KOM82 mutant, resulting in strain MKD3 (∆recFOR). Finally, the ∆recBCD allele [52] was crossed into each of the MKD3, MKD1, KOM82 and MKD2 mutants, giving the strains MKD6, MKD4, KOM86 and MKD5, respectively. All strains were verified phenotypically (sensitivity to UV irradiation; transformation by 2 kbp donor DNA molecules) and by PCR. The donor DNA plasmid pSBP1 was constructed by inserting a PCR product of the lifO-lipB::aphA3 insertion allele [60] (amplified with ACIAD3308-f/ACIAD3309-down) of ADP1200Com + KanR [22] into the HincII site of the pUC19 plasmid vector conferring kanamycin resistance.
Bacterial cultures were incubated at 30 °C in Luria Bertani broth (LB) (BD Difco™, USA). Transformants were scored on LB agar plates supplemented with kanamycin (Sigma-Aldrich, Germany) at 50 mg l−1 or 10 mg l−1 (plating efficiencies were indistinguishable), and trimethoprim at 250 mg l−1.
Genomic and plasmid DNA was isolated using QIAGEN Genomic-tip (QIAGEN, Germany) and the QIAGEN genomic DNA or plasmid DNA purification kit, respectively, according to the manufacturer’s instructions.
Pairwise competitions
Overnight cultures of a transformation-proficient test strain (LCQ2 or NH29) and the DNA uptake-deficient competitor KOM130 were grown in ten ml for 16 h at 30 °C. The cells were pelleted, re-suspended and diluted 100-fold (approximately 107 cells ml−1) in PBS [54] pre-warmed to 30 °C. One ml aliquots of each suspension were mixed, transferred to a sterile Petri dish and irradiated with ultraviolet (UV) light using a germicidal lamp. One ml of the irradiated cells was transferred to tubes containing one ml of pre-warmed double-strength LB (2 × LB) with genomic DNA [isogenic (ADP1200Com + KanR) or heterologous (salmon sperm); 2 μg ml−1] or with DNase 1 (100 μg ml−1) and aerated in the dark for 24 h at 30 °C. The cells were plated in appropriate dilution on LB medium with and without trimethoprim (250 mg l−1; in some control experiments: 10 mg l−1 kanamycin) after two and 24 h. The plates were incubated at 30 °C for 24 h, colonies were counted, and the titers at both time points for the test strain (Tt-2 and T t-24) and the competitor strain (C t-2 and C t-24) were calculated. The relative fitness w of the test strain was calculated as w = ln(T t-24/T t-2)/ln(C t-24/C t-2).
UV survival measurements
Overnight cultures of A. baylyi were treated as described for the pairwise competitions but without mixing with a competitor and irradiated using a germicidal lamp. Control assays were treated accordingly without irradiation (UV0). One ml of the cells was then mixed with one ml pre-warmed 2 × LB, amended with pSBP1 plasmid DNA (0.1 μg ml−1; 1800 molecules per cell) linearized with XmnI (New England Biolabs) or with homologous genomic DNA from strain ADP1200Com + KanR (2 μg ml−1; 50 genome equivalents per cell), and aerated without light for two hours before DNase 1 (100 μg ml−1) was added. pSBP1 is pUC19 with a two kbp insert from the A. baylyi chromosome with a KanR marker (aphA3) cloned in the centre of the insert (Additional file 1: Table S1). In some control experiments without DNA, DNase 1 was added at the start. When the ΔuvrA strain was used, ten assays (one ml per assay) were irradiated separately and pooled after irradiation of the cells. Cells were plated in appropriate dilution on LB with (survival transformant titer) and without kanamycin (10 mg l−1; survival recipient titer) and incubated for 24 h at 30 °C. Colonies were counted, the titers were determined, and the recipient and transformant survival frequencies were calculated as frequencies relative to those at UV0.
Natural transformation experiments
The preparation of competent cells and the transformation assays were performed as described previously [58]. Briefly, the A. baylyi strains were grown in a shaker in LB broth at 30 °C to 1 × 109 cells ml−1 (determined with a Neubauer hemocytometer) and stored at −80 °C as a concentrated stock (1 × 1010 cells ml−1 in LB with 10% glycerol) until use. For transformation assays, a freshly thawed bacterial suspension was diluted to 2.5 × 108 cells ml−1 in LB containing 100 ng ml−1 donor DNA (isolated from A. baylyi KH12), aerated for two hours at 30 °C and plated in appropriate dilution on LB medium (recipient titer) and LB with 10 mg ml−1 kanamycin (transformants). The colonies were counted after 16 to 40 h at 30 °C. Transformation frequencies were calculated as transformants per recipient. No kanamycin-resistant colonies were obtained in control experiments without donor DNA.
Statistical analysis
The relative fitness data were analysed using Two-ways ANOVA, with relative fitness as response variable and competitor strain (two levels), UV-exposure to cells (two levels) and DNA-availability (three levels) as explanatory variables. We also subjected the data to post hoc tests (with Tukey HSD correction) using the same model specifications. Confidence limits (95%) for the group means were obtained using formula based estimation.
The survival data were analysed by regression using a linear Mixed-model controlling for dependencies among the replicates from the same overnight culture used in a series of increasing UV-exposure. In our model, we used relative survival as response and UV-fluence (continuous) and total population or transformants (two levels) as explanatory variables, whereas the overnight culture was included as a random term. The observed trends were linearized by log transforming relative survival and the linearity of the trend was diagnosed, and confirmed, by generalized additive modelling [61]. The transformation frequencies of the set of recombinational repair deficient (ΔrecBCDFOR) mutants were analysed by one-way ANOVA followed by post hoc tests with TukeyHSD correction. All analyses were performed with the statistical software R [62] and the additional R-packages lme and mgcv.
Results
Uptake of exogenous DNA is beneficial regardless of sequence homology, DNA damage and recombination
We investigated the general fitness effects of DNA uptake in mixed culture competition experiments with the transformation-proficient A. baylyi wildtype strain (LCQ2) and its non-transformable ΔcomB-F derivative (Additional file 1: Table S1). The comB-F operon encodes genes required for type IV pilus formation, which is essential for transport of free DNA into the periplasm [20, 22, 56] and for spreading growth on semi-solid surfaces (Additional file 1: Fig. S1). To test for differences between benign and DNA damaging conditions, the mixed cultures were treated with or without initial pulses of UV irradiation, and were amended either with homologous DNA, heterologous DNA, or DNase. The UV-dose applied was 216 J m−2, which generates one photoproduct every 420 bp on average [63] and killed approximately 98% of the cells. In the absence of DNA, the wildtype strain displayed a relative fitness (w) reduction compared with the ΔcomB-F mutant (Fig. 1a) [w = 0.88 (0.83–0.92, 95% CI)]. A mixed culture serial transfer experiment confirmed that the fitness reduction was due to the absence of type IV pilus components in the ΔcomB-F competitor (Additional file 1: Fig. S2).
Fig. 1.
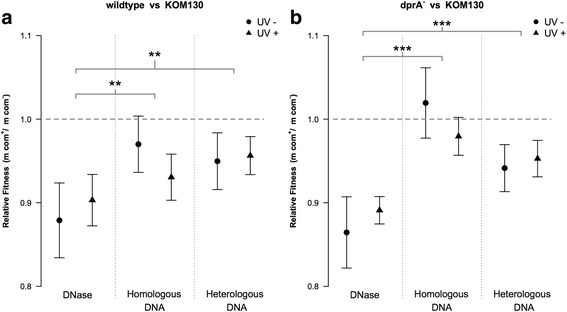
Relative fitness of wildtype strain (a) and its ∆dprA derivative (b) grown in head-to-head competitions against the DNA uptake-deficient ∆comB-F mutant. The competitions were treated with a combination of an initial UV pulse of 216 J m−2 or no UV irradiation with a subsequent amendment of no DNA (DNase), homologous DNA or heterologous DNA. In each group n = 13. In the control competitions with no UV and no DNA both the wildtype and its ∆dprA derivative displayed a reduced fitness relative to KOM130 of w = 0.88 (0.83–0.92, 95% CI) and w = 0.86 (0.82–0.91, 95% CI), respectively, due to the biological cost of type IV pilus expression (Additional file 1: Fig. S1). Error bars indicate 95% confidence intervals. Significant differences between the treatment groups (pooled results of both UV- and UV+ competitions) are indicated by asterisks (** P < 0.001, *** P < 0.0001). A two-way ANOVA of all mean values is provided in (Additional file 1: Table S4)
The addition of homologous or heterologous DNA increased the fitness of the wildtype compared to the DNA-free experiments (both P < 0.002) (Fig. 1a). The fitness effects of added DNA to the competitions were independent of UV-exposure (P = 0.83). Together these two results provide a strong argument in favour of a general benefit of DNA uncoupled from recombination.
To further investigate whether the observed fitness increase was independent of recombination, we used a DNA uptake-proficient but transformation-deficient mutant lacking the dprA gene [Additional file 1: Table S1] in head-to-head competitions with the ΔcomB-F mutant. Absence of DprA abolished homologous recombination during natural transformation in A. baylyi as reported for S. pneumoniae [49] (detection limit 1 × 10−9 transformants/recipient). The ΔdprA mutant displayed the same cost of type IV pilus formation as the wildtype [w = 0.86 (0.82–0.91, 95% CI); Fig. S3]. As seen with the wildtype, addition of both heterologous and homologous DNA increased the fitness of the ΔdprA strain relative to the DNA uptake-deficient competitor ΔcomB-F (both P < 0.001) regardless of UV-exposure (Fig. 1b). These data show that under the experimental conditions the DNA uptake machinery is costly to the cell. This cost can be partially compensated by addition of DNA to the competing cultures. However, the benefit of added DNA is unlinked to DNA recombination and thus DNA damage repair. This observation is further supported by the fact that A. baylyi takes up DNA from any source [44, 56], suggesting the benefit of competence is independent of DNA sequence homology.
Homologous DNA reduces relative survival of transformants after UV-exposure
The competition experiments described above demonstrated a clear fitness increase for DNA-uptake-proficient cells regardless of the cells’ ability to use that DNA for recombination under benign or genotoxic conditions. However, the fitness benefit observed might obscure an additional benefit for the transformant fraction of cells in the culture, especially under DNA damaging conditions.
To test that, we investigated the effect of increasing intracellular UV damages in DNA on survival in the transformant fraction compared with the total population in monoculture experiments. We added a short donor DNA substrate (linearized pSBP1; [Additional file 1: Table S1]) containing a chromosomal A. baylyi segment (2 kbp) with a kanamycin resistance marker inserted in the center (lifO-lipB::aphA3) [22]. Single-locus homologous recombination is largely independent of the chromosomal position [64]. pSBP1 DNA can only be used for recombination at the lifO-lipB locus, and recombination results in kanamycin resistance. The donor DNA substrate was added in excess (0.1 μg ml−1), and the survival of transformants following UV irradiation from 0 to 216 J m−2 was quantified. The results showed an initial benefit of added pSBP1 DNA at low UV doses on survival of the transformant fraction (Fig. 2), similar to what was observed in H. influenzae [34]. However, at higher UV-doses (≥144 J m−2) that increased the likelihood of UV damages generated in the vicinity of the marker insertion site, this benefit was lost (Fig. 2). The result suggests that recombination with taken-up DNA interfered with repair of UV damages at the recombination locus, which is further supported by a steeper decline in survival of the transformant fraction with increasing UV-doses (Fig. 2 and [Additional file 1: Fig. S4 A]).
Fig. 2.
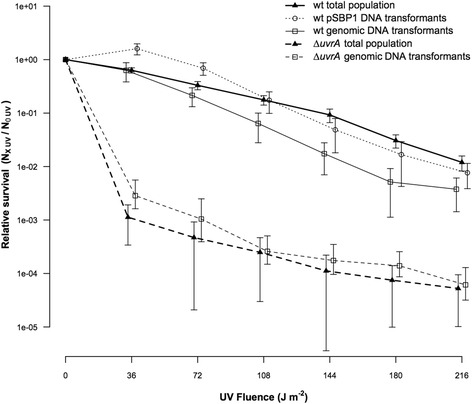
Relative survival of the pooled total population of the wildtype (closed triangles and bold solid line, n = 16) and the transformant fractions obtained with either pSBP1 DNA (0.1 μg ml−1; open circles with dotted line, n = 10) or with homologous genomic DNA (2 μg ml−1; open squares with solid line, n = 6) with increasing levels of UV irradiation. We also determined the relative survival of the total population (closed triangles and bold dashed line, n = 10) and the transformant fraction in the ∆uvrA mutant obtained with homologous DNA (2 μg ml−1; open squares and dashed line, n = 10). Error bars denote the 95% confidence intervals. The transformation frequencies without UV irradiation were (7.7 ± 1.4) × 10−4 (pSBP1 DNA) and (2.5 ± 2.8) × 10−2 (genomic DNA) for the wildtype; and (1.9 ± 1.0) × 10−2 (genomic DNA) for the ∆uvrA mutant. Mean initial titers (CFU/ml) for the wildtype and the ∆uvrA mutant were 1.8 × 107 and 4.2 × 107, respectively
We hypothesized that fully homologous DNA as donor DNA would be even more disadvantageous because it can recombine and interfere with UV-damages genome-wide. We tested this by adding genomic A. baylyi donor DNA containing the lifO-lipB::aphA3 marker (2 μg ml−1). The donor DNA substrate can undergo phenotypically silent recombinations at any chromosomal locus, and we tracked the integration of the lifO-lipB::aphA3 marker as a proxy for the genome wide transformation frequency. Transformants survived less than the total population from low to high UV-doses (72–216 J m−2) (P < 0.001, Fig. 2 and [Additional file 1: Fig. S4 B]). The UV-survival curves of the total populations with pSBP1 or genomic DNA were indistinguishable and these data were pooled (Fig. 2). Control experiments without DNA revealed no difference in kill rates of the total population (Additional file 1: Fig. S4 D).
UV damages in bacteria can be repaired by nucleotide excision repair (NER) [65]. During NER, the UvrA-directed UvrBC excinuclease [59] cleaves one strand on either side of a UV- lesion in double-stranded DNA. Excision of the incised fragment with the lesion by helicase II is finally followed by fill-in synthesis by DNA polymerase I and sealing of the nick by DNA ligase. With increasing UV-doses, more NER repair sites would remain temporarily unfinished, and gaps and nicks would accumulate. In addition, recombination attempts with taken up DNA will add to the number of single strand breaks, as reported in [52]. We hypothesised that the decrease of transformant survival with increasing UV-doses was caused by interference of NER with transformational recombination (Fig. 3).
Fig. 3.
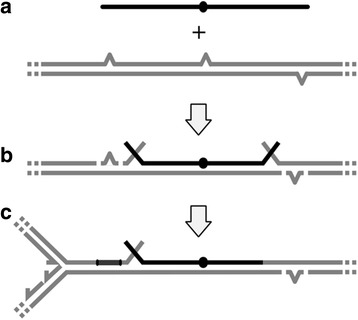
Formation of DNA double strand-breaks conferred by nucleotide excision repair during natural transformation. a: Grey lines: genomic DNA; grey spikes: UV-induced lesions. Black line: taken-up DNA single-strand with genetic marker (oval). b: Generation of DNA single-strand-breaks by UvrAB-directed, UvrBC-initiated cleavage of the damage-containing strand upstream and downstream of the lesions, and by RecA-mediated strand invasion of the taken-up DNA and cleavage of the displaced DNA strand. c: Partial repair of single-strand breaks: 1. by removal of the lesion-containing single-strand fragment (left lesion), fill-in synthesis, and covalent ligation (dark grey; catalysed by UvrD, DNA polymerase I, and DNA ligase, respectively); and 2. by covalent joining of the invaded taken-up strand with the genomic DNA at one side (downstream of the marker). Single-strand breaks persist when the UV lesion-containing strands are not removed (right lesion) and when an invaded DNA strand remains unligated at one end (upstream of the marker). One-sided ligation of the invaded DNA strand is common (see text). Following DNA replication (indicated by a replication fork approaching from the left end), single-strand breaks are converted into potentially lethal double-strand breaks
To test this hypothesis, we knocked out the key NER gene uvrA and investigated relative survival of transformants of the ΔuvrA strain with increasing UV-doses as outlined above. As expected, UvrA-deficiency resulted in greatly reduced survival when exposed to UV irradiation (Fig. 2). However, the ΔuvrA transformant fraction did not display any detrimental effect with increasing UV doses as observed in the wildtype (Fig. 2), and the kill rates of transformants in the absence of UvrA were the same as that of the total population (P = 0.79, [Additional file 1: Fig. S4 C]). This result suggests that in absence of NER, genome-wide recombination with homologous chromosomal DNA provided no additional burden, but also no benefit, to uptake-proficient cells. Cumulatively, NER and recombination with taken-up DNA become a burden to competent cells with increasing levels of DNA damages, due to generation of nicks and gaps that turn into DNA double-strand breaks when unrepaired. Consequently, successful integration of recombination attempts during natural transformation in the wildtype would depend on DNA strand-break repair functions.
Genomic integration of taken-up DNA strongly depends on DNA repair
Two studies on RecA-mediated recombination in A. baylyi [51, 52] suggested that both ends of a donor DNA molecule are integrated through two independent ligation events. While one end is covalently joined to the genomic DNA, the opposite end frequently remains temporally unligated, resulting in a single-strand break that can turn into a double-strand break following DNA replication. These reports [51, 52] are in agreement with the observed interaction between NER of UV-damages and recombination on transformant survival which suggest that incomplete recombination attempts during transformation may represent an additional source of DNA damages that rely on DNA strand break repair (Fig. 3). Recombinational DNA strand break repair is initiated in bacteria through two major pathways, the RecBCD and the RecFOR pathways of recombination [66, 67]. DNA single-strand breaks such as nicks or gaps can be repaired through the latter pathway, which requires among others the RecF, RecO and RecR gene products [68]. The RecFOR proteins in concert process gaps and generate RecA-loaded DNA 3′-ends for strand invasion and thus can initiate homologous reciprocal strand transfer and can also restore stalled and broken replication forks [68]. Alternatively, nicks and gaps can turn into double-strand breaks when unprocessed, and these damages are mainly repaired by the RecBCD pathway with the RecBCD heterotrimeric complex as the key repair function [69]. RecBCD processes DNA double-strand ends and generates a recombinogenic RecA-loaded 3′-end that initiates reciprocal strand exchange with a template for repair of the double-strand break [69]. To test the hypothesis that natural transformation depends on DNA strand break repair for successful integration of acquired DNA, we deleted components of the RecFOR pathway and the RecBCD pathway in A. baylyi and determined the effects of these deficiencies on natural transformation by homologous chromosomal DNA. The results are shown in Fig. 4. Deletions of recF, recO and recR in combination or alone overall had little or no effect on natural transformation frequencies compared with wildtype, as observed previously in B. subtilis and S. pneumoniae [70, 71], although in the ΔrecR mutant it was decreased 3.3-fold (P = 0.007). Deletion of the recBCD operon, while highly detrimental to viability, did not affect transformation frequency (P = 0.94). A recent finding that RecR protein has a role in both RecBCD and RecFOR recombination pathways in mycobacteria [72] supports this observation. In contrast, in a ΔrecBCD ΔrecFOR mutant transformation was decreased approximately 10-fold (P < 0.001), and this decrease was strongly synergistic (about 5-fold higher than expected from the decreases in frequency of the single mutants, Fig. 4), confirming earlier reports on H. pylori [73]. This observation was confirmed using ΔrecF, ΔrecO or ΔrecR mutants of the ΔrecBCD strain (all p-values < 0.001; Fig. 4), with the ΔrecBCD ΔrecF double mutant yielding particularly low transformation frequencies (approximately 80-fold decreased compared with wildtype). It is conceivable that in this mutant, the functional RecOR proteins [74] generate recombination intermediates that cannot be resolved in the absence of RecF and RecBCD. An ANOVA table for multiple comparisons of means of the transformation frequencies is provided in (Additional file 1: Table S5). In summary, these results suggest that the majority of natural transformation events depends on DNA strand-break repair and support our hypothesis that taken-up DNA causes damages such as strand-breaks following recombination attempts. It can be hypothesized that amendment of DNA is detrimental to growing cells impaired in recombinational DNA repair. However, comparisons of serial transfer experiments of the ΔrecBCD ΔrecFOR mutant with and without DNA suggest that the hypothetical detrimental effect of added DNA on cell survival is masked by the initially observed beneficial effect of DNA (Fig. 1) on cell growth (Additional file 1: Fig. S5).
Fig. 4.
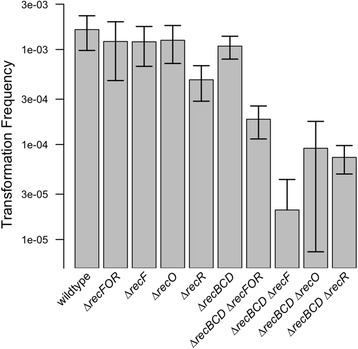
Natural transformation frequencies of A. baylyi wildtype and DNA recombination-impaired mutants. Transformation frequencies were obtained in liquid transformation experiments with genomic homologous DNA (0.1 μg ml−1) containing a kanamycin resistance marker gene and are given as means with 95% confidence intervals. The initial recipient titers are listed in (Additional file 1: Table S4)
Discussion
In this report we show that the costs of the DNA uptake machinery (comB-F gene products) in A. baylyi is offset by the amendment of DNA in competition experiments. The beneficial effect of added DNA was independent of UV-exposure to cells, DNA sequence homology, and unlinked to recombination. These results are inconsistent with a role of acquired DNA in strand-break repair as originally proposed by Bernstein and co-workers [30]. The high biological cost of type IV pilus formation (up to 14% reduction in relative fitness) suggests that the ability to take up DNA could rapidly be lost in naturally transformable populations. To this end, reduced transformability have been previously shown in A. baylyi during experimental evolution but this is not necessarily dependent on loss of DNA uptake functions since the cost of pilus formation appears to be medium-specific [20, 22]. It is also important to stress that the type IV pilus is involved in motility (Additional file 1: Fig. S1) suggesting that other selective forces may be responsible for the maintenance of central components of the DNA uptake machinery [75].
Early reports favoured the DNA repair hypothesis in B. subtilis [9, 31–33], and it has been demonstrated that B. subtilis displayed increased transformation frequencies during UV-exposure [33]. However, DNA damage does not induce the competence machinery in B. subtilis [35], and more recent reports have made a clear distinction between natural transformation and the physiological state of competence. For example, competence for natural transformation is induced by genotoxic stress in S. pneumoniae [38] and it was recently demonstrated that competence development following DNA-damaging stress was beneficial in the presence but not the absence of DNA in S. pneumoniae [36]. However, direct evidence demonstrating recombination between incoming donor DNA and damaged areas of the genome was not presented and it is not clear if this benefit was due to other processes, as also pointed out by the authors [36]. Interestingly, competence induction was also beneficial in the presence of non-DNA damaging stress, suggesting a general benefit that extends beyond recombination [36, 37]. Competence induction in S. pneumoniae involves quorum sensing of an extracellular competence-stimulating-peptide (CSP) for a short time during log phase and includes transcriptional regulation by ComX of high number of genes not only limited to DNA uptake and recombination [37]. This is consistent with our finding that taken up DNA increases relative fitness in A. baylyi. These results also provide support for a previous report on H. influenzae, where an increased survival of the whole population was demonstrated in the presence of both fully and partially homologous DNA [34]. Moreover, we show that the benefit of DNA is also present in an A. baylyi strain lacking a functional recombination mediator DprA. Since DprA loads taken-up single-stranded DNA with RecA, and the nucleoprotein filament subsequently initiates recombination [49], the result reported here strongly suggests that the fitness benefit of DNA uptake is not linked to recombination, but rather the uptake of free DNA.
The beneficial effect of DNA uptake does not necessarily exclude a role of the taken-up DNA for repair purposes in the recombining fraction of the competent population. We investigated the effect of exogenous DNA on transformant survival with increasing UV doses. The results revealed that recombination with taken-up DNA is costly. These data provide additional support for the proposed mechanistic model for, and consequences of, integration of DNA into the chromosome following uptake in A. baylyi [51, 52]. Kickstein and colleagues provided evidence in favour of separate integration of 5′- and 3′- donor DNA ends during recombination in A. baylyi, suggesting frequent occurrence of DNA single-strand breaks. If replication forks reach joints between donor and recipient DNA prior to ligation, a double-strand-break would occur [52]. This suggests that natural transformation in A. baylyi strongly depends on DNA strand break repair functions to ensure viability. In our DNA damage experiments we used UV-irradiation to induce intracellular DNA lesions that can lead to single- and double-strand breaks during DNA repair [76]. Consequently, the DNA repair machinery will be saturated due to multiple strand-breaks formed as a response to DNA damage. It is conceivable that the saturation affects initiation of recombinational repair (conferred by RecBCD or by RecFOR), but it is also possible that the repair processes exhaust the pool of cellular RecA protein which is required for initial strand invasion and for repair of strand-breaks due to incomplete insertions of the donor DNA. It can be hypothesized that the repair functions titrate RecA protein away from DprA and thus reduce transformation due to RecA shortage. However, it has been shown that DNA uptake in A. baylyi is initiated immediately after exposure to DNA [44], and in our UV experiments the cells were still in lag phase (before initiation of replication) when DNA was added. The integration of acquired DNA into the genome then is likely to cause additional stress in the recipient cells (Fig. 3).
Three lines of evidence presented in this report support this hypothesis. First, we demonstrate in UV-time-kill experiments that the transformant fraction is more susceptible to UV-killing than the total population. Second, UV-exposure reduced transformant survival due to interference between NER and natural transformation (Fig. 2). Third, under benign experimental conditions we demonstrate that the majority of natural transformation events depend on DNA repair functions as demonstrated in recBCDFOR mutants (Fig. 4). Taken together, the data presented here and previously [51, 52, 73] strongly suggest that recombination during natural transformation is costly and represents an additional hurdle that the selective forces responsible for the maintenance of natural transformation must overcome. Consequently, our findings are inconsistent with the DNA repair hypothesis for the evolution and maintenance of natural transformation in A. baylyi [9, 30].
If DNA uptake is beneficial and unlinked to recombination (which is detrimental), what causes this benefit? Taken-up DNA is rapidly degraded by single-strand-specific DNA exonucleases (RecJ and ExoX) in A. baylyi [56]. It is thus possible that the recipient cells can directly access this pool of nucleotides and incorporate them into the DNA biosynthesis pathways as suggested in previous reports [8, 77]. Our results would add to the recent finding that purine depletion induced the H. influenzae competence activator sxy [23], suggesting that DNA can be utilized as a nutrient or as building blocks in DNA metabolism.
Conclusions
Our data suggest that the selective pressures responsible for the maintenance of natural transformation are not linked to recombination following DNA uptake. Our findings rather suggest that recombination is a DNA damaging process and that the positive effect of DNA uptake on A. baylyi fitness is due to free cytoplasmic DNA where it may be utilized as nutrition or as building blocks for DNA metabolism. In contrast, our data suggest that recombination during transformation is a strand break inducing process that represents a previously unrecognized cost of natural transformation. Our results add new insights to the costs and benefits that play a selective role in the maintenance of natural transformation.
Acknowledgements
We are grateful to J. de Vries for providing the pBlue-recO11 and pBlue-recO10 plasmids We thank two anonymous reviewers for constructive criticism and suggestions that improved the manuscript.
Funding
This work was supported by the Tromsø Research Foundation (grant number A5870 to P.J.J.), UIT The Arctic University of Norway and the Research Council of Norway (grant number 204263/F20 to P.J.J.). N.H. was partially supported by the European Research Council (grant number 281357). K.H. was supported by the Danish National Research Foundation.
The funding bodies had no role in the writing process, study design and collection, analysis and interpretation of data.
Availability of data and materials
Datasets and R-code for the statistical models is provided upon request.
Authors’ contributions
PJJ and KH conceived the study. NH, VS, KB-P, MML and ALGU conducted the experiments. NH, KH and PJJ supervised the project. NH, VS, KH and PJJ designed the experiments and analysed the data. VS and RP conducted the statistical analyses. VS and NH created the Figures. NH, VS, KH, ALGU and PJJ wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- bp
Base pairs
- CI
Confidence interval
- HSD
Honest significant difference
- LB
Luria-Bertani
- NER
Nucleotide excision repair
- PBS
Phosphate-buffered saline
- UV
Ultraviolet
Additional files
Supplementary Figures and Tables. (DOCX 1.33 mb)
Supplementary Methods. (DOCX 13.6 kb)
Contributor Information
Nils Hülter, Email: nhuelter@ifam.uni-kiel.de.
Vidar Sørum, Email: vidar.a.sorum@uit.no.
Kristina Borch-Pedersen, Email: kristina.borch-pedersen@nmbu.no.
Mikkel M. Liljegren, Email: m.m.liljegren@ibv.uio.no
Ane L. G. Utnes, Email: ane.utnes@uit.no
Raul Primicerio, Email: raul.primicerio@uit.no.
Klaus Harms, Email: klaus.harms@spdir.net.
Pål J. Johnsen, Email: paal.johnsen@uit.no
References
- 1.Chen I, Christie PJ, Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310(5753):1456–60. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol. 2014;12(3):181–96. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 3.Johnsborg O, Eldholm V, Havarstein LS. Natural genetic transformation: prevalence, mechanisms and function. Res Microbiol. 2007;158(10):767–78. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58(3):563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3(9):711–21. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 6.Vos M. Why do bacteria engage in homologous recombination? Trends Microbiol. 2009;17(6):226–32. doi: 10.1016/j.tim.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Redfield RJ. Evolution of bacterial transformation - is sex with dead cells ever better than no sex at all. Genetics. 1988;119(1):213–21. doi: 10.1093/genetics/119.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redfield RJ. Do bacteria have sex? Nat Rev Genet. 2001;2(8):634–9. doi: 10.1038/35084593. [DOI] [PubMed] [Google Scholar]
- 9.Michod RE, Wojciechowski MF, Hoelzer MA. DNA-repair and the evolution of transformation in the Bacterium Bacillus-Subtilis. Genetics. 1988;118(1):31–9. doi: 10.1093/genetics/118.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science. 2015;347(6217):63–7. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 11.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, Bentley SD, Hanage WP, Lipsitch M. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet. 2013;45(6):656–63. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levin BR, Cornejo OE. The population and evolutionary dynamics of homologous gene recombination in bacterial populations. PLoS Genet. 2009;5(8):e1000601. doi: 10.1371/journal.pgen.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moradigaravand D, Engelstadter J. The evolution of natural competence: disentangling costs and benefits of sex in bacteria. Am Nat. 2013;182(4):E112–26. doi: 10.1086/671909. [DOI] [PubMed] [Google Scholar]
- 14.Ambur OH, Engelstadter J, Johnsen PJ, Miller EL, Rozen DE. Steady at the wheel: conservative sex and the benefits of bacterial transformation. Phil Trans R Soc B. 2016;371:20150528. [DOI] [PMC free article] [PubMed]
- 15.Mell JC, Redfield RJ. Natural competence and the evolution of DNA uptake specificity. J Bacteriol. 2014;196(8):1471–83. doi: 10.1128/JB.01293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treangen TJ, Ambur OH, Tonjum T, Rocha EP. The impact of the neisserial DNA uptake sequences on genome evolution and stability. Genome Biol. 2008;9(3):R60. doi: 10.1186/gb-2008-9-3-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croucher NJ, Mostowy R, Wymant C, Turner P, Bentley SD, Fraser C. Horizontal DNA transfer mechanisms of bacteria as weapons of intragenomic conflict. Plos Biol. 2016;14(3):e1002394. doi: 10.1371/journal.pbio.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baltrus DA, Guillemin K, Phillips PC. Natural transformation increases the rate of adaptation in the human pathogen helicobacter pylori. Evolution. 2008;62(1):39–49. doi: 10.1111/j.1558-5646.2007.00271.x. [DOI] [PubMed] [Google Scholar]
- 19.Perron GG, Lee AE, Wang Y, Huang WE, Barraclough TG. Bacterial recombination promotes the evolution of multi-drug-resistance in functionally diverse populations. Proc Biol Sci. 2012;279(1733):1477–84. doi: 10.1098/rspb.2011.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bacher JM, Metzgar D, de Crecy-Lagard V. Rapid evolution of diminished transformability in Acinetobacter baylyi. J Bacteriol. 2006;188(24):8534–42. doi: 10.1128/JB.00846-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelmoer DJ, Donaldson I, Rozen DE. Conservative sex and the benefits of transformation in Streptococcus pneumoniae. PLoS Pathog. 2013;9(11):e1003758. doi: 10.1371/journal.ppat.1003758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Utnes AL, Sorum V, Hulter N, Primicerio R, Hegstad J, Kloos J, Nielsen KM, Johnsen PJ. Growth phase-specific evolutionary benefits of natural transformation in Acinetobacter baylyi. ISME J. 2015;9(10):2221–31. doi: 10.1038/ismej.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinha S, Mell J, Redfield R. The availability of purine nucleotides regulates natural competence by controlling translation of the competence activator Sxy. Mol Microbiol. 2013;88(6):1106–19. doi: 10.1111/mmi.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart GJ, Carlson CA. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–35. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- 25.Cameron AD, Redfield RJ. Non-canonical CRP sites control competence regulons in Escherichia coli and many other gamma-proteobacteria. Nucleic Acids Res. 2006;34(20):6001–14. doi: 10.1093/nar/gkl734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wise EM, Alexander SP, Powers M. Adenosine 3′-5′-cyclic monophosphate as a regulator of bacterial transformation. Proc Natl Acad Sci U S A. 1973;70(2):471–4. doi: 10.1073/pnas.70.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blokesch M. Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ Microbiol. 2012;14(8):1898–912. doi: 10.1111/j.1462-2920.2011.02689.x. [DOI] [PubMed] [Google Scholar]
- 28.Zheng L, Chen Z, Itzek A, Herzberg MC, Kreth J. CcpA regulates biofilm formation and competence in Streptococcus gordonii. Mol Oral Microbiol. 2012;27(2):83–94. doi: 10.1111/j.2041-1014.2011.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antonova ES, Bernardy EE. Hammer Natural competence in Vibrio cholerae is controlled by a nucleoside scavenging response that requires CytR-dependent anti-activation. Mol Microbiol. 2012;86(5):1215–31. doi: 10.1111/mmi.12054. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein H, Byerly HC, Hopf FA, Michod RE. Origin of sex. J Theor Biol. 1984;110(3):323–51. doi: 10.1016/S0022-5193(84)80178-2. [DOI] [PubMed] [Google Scholar]
- 31.Hoelzer MA, Michod RE. DNA-repair and the evolution of transformation in bacillus-subtilis.3. Sex with damaged DNA. Genetics. 1991;128(2):215–23. doi: 10.1093/genetics/128.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michod RE, Wojciechowski MF. DNA-repair and the evolution of transformation.4. DNA-damage increases transformation. J Evol Biol. 1994;7(2):147–75. doi: 10.1046/j.1420-9101.1994.7020147.x. [DOI] [Google Scholar]
- 33.Wojciechowski MF, Hoelzer MA, Michod RE. DNA-repair and the evolution of transformation in bacillus-subtilis.2. Role of inducible repair. Genetics. 1989;121(3):411–22. doi: 10.1093/genetics/121.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mongold JA. DNA repair and the evolution of transformation in Haemophilus influenzae. Genetics. 1992;132(4):893–8. doi: 10.1093/genetics/132.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redfield RJ. Evolution of natural transformation: testing the DNA repair hypothesis in Bacillus subtilis and Haemophilus influenzae. Genetics. 1993;133(4):755–61. doi: 10.1093/genetics/133.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelmoer DJP, Rozen DE. Competence increases survival during stress in streptococcus pneumoniae. Evolution. 2011;65(12):3475–85. doi: 10.1111/j.1558-5646.2011.01402.x. [DOI] [PubMed] [Google Scholar]
- 37.Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol. 2006;60:451–75. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 38.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006;313(5783):89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- 39.Charpentier X, Kay E, Schneider D, Shuman HA. Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J Bacteriol. 2011;193(5):1114–21. doi: 10.1128/JB.01146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dorer MS, Fero J, Salama NR. DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog. 2010;6(7):e1001026. doi: 10.1371/journal.ppat.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hare JM, Ferrell JC, Witkowski TA, Grice AN. Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of acinetobacter baumannii and acinetobacter baylyi. Plos One. 2014;9(4):e93861. doi: 10.1371/journal.pone.0093861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rauch PJG, Palmen R, Burds AA, Gregg-Jolly LA, van der Zee JR, Hellingwerf KJ. The expression of the Acinetobacter calcoaceticus recA gene increases in response to DNA damage independently of RecA and of development of competence for natural transformation. Microbiol-Uk. 1996;142:1025–32. [DOI] [PubMed]
- 43.Palmen R, Vosman B, Kok R, van der Zee JR, Hellingwerf KJ. Characterization of transformation-deficient mutants of acinetobacter-calcoaceticus. Mol Microbiol. 1992;6(13):1747–54. [DOI] [PubMed]
- 44.Palmen R, Vosman B, Buijsman P, Breek CK, Hellingwerf KJ. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J Gen Microbiol. 1993;139(2):295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 45.Smith HO, Danner DB, Deich RA. Genetic-transformation. Annu Rev Biochem. 1981;50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- 46.Frye SA, Nilsen M, Tonjum T, Ambur OH. Dialects of the DNA uptake sequence in Neisseriaceae. PLoS Genet. 2013;9(4):e1003458. doi: 10.1371/journal.pgen.1003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mell JC, Hall IM, Redfield RJ. Defining the DNA uptake specificity of naturally competent Haemophilus influenzae cells. Nucleic Acids Res. 2012;40(17):8536–49. doi: 10.1093/nar/gks640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Provvedi R, Dubnau D. ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol Microbiol. 1999;31(1):271–80. doi: 10.1046/j.1365-2958.1999.01170.x. [DOI] [PubMed] [Google Scholar]
- 49.Mortier-Barriere I, Velten M, Dupaigne P, Mirouze N, Pietrement O, McGovern S, Fichant G, Martin B, Noirot P, Le Cam E, et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130(5):824–36. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 50.Quevillon-Cheruel S, Campo N, Mirouze N, Mortier-Barriere I, Brooks MA, Boudes M, Durand D, Soulet AL, Lisboa J, Noirot P, et al. Structure-function analysis of pneumococcal DprA protein reveals that dimerization is crucial for loading RecA recombinase onto DNA during transformation. Proc Natl Acad Sci U S A. 2012;109(37):E2466–75. doi: 10.1073/pnas.1205638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harms K, Wackernagel W. The RecBCD and SbcCD DNases suppress homology-facilitated illegitimate recombination during natural transformation of Acinetobacter baylyi. Microbiol-Sgm. 2008;154:2437–45. doi: 10.1099/mic.0.2008/018382-0. [DOI] [PubMed] [Google Scholar]
- 52.Kickstein E, Harms K, Wackernagel W. Deletions of recBCD or recD influence genetic transformation differently and are lethal together with a recJ deletion in Acinetobacter baylyi. Microbiology. 2007;153:2259–70. doi: 10.1099/mic.0.2007/005256-0. [DOI] [PubMed] [Google Scholar]
- 53.de Vries J, Heine M, Harms K, Wackernagel W. Spread of recombinant DNA by roots and pollen of transgenic potato plants, identified by highly specific biomonitoring using natural transformation of an Acinetobacter sp. Appl Environ Microbiol. 2003;69(8):4455–62. doi: 10.1128/AEM.69.8.4455-4462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E, Maniatis T. Molecular cloning : a laboratory manual. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, Labarre L, Cruveiller S, Robert C, Duprat S, Wincker P, et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32(19):5766–79. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Overballe-Petersen S, Harms K, Orlando LA, Mayar JV, Rasmussen S, Dahl TW, Rosing MT, Poole AM, Sicheritz-Ponten T, Brunak S, et al. Bacterial natural transformation by highly fragmented and damaged DNA. Proc Natl Acad Sci U S A. 2013;110(49):19860–5. doi: 10.1073/pnas.1315278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8(5):528–35. [PubMed] [Google Scholar]
- 58.Harms K, Schon V, Kickstein E, Wackernagel W. The RecJ DNase strongly suppresses genomic integration of short but not long foreign DNA fragments by homology-facilitated illegitimate recombination during transformation of Acinetobacter baylyi. Mol Microbiol. 2007;64(3):691–702. doi: 10.1111/j.1365-2958.2007.05692.x. [DOI] [PubMed] [Google Scholar]
- 59.Orren DK, Sancar A. The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex. Proc Natl Acad Sci U S A. 1989;86(14):5237–41. doi: 10.1073/pnas.86.14.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kok RG, Young DM, Ornston LN. Phenotypic expression of PCR-Generated random mutations in a Pseudomonas putida gene after its introduction into an Acinetobacter chromosome by natural transformation. Appl Environ Microb. 1999;65(4):1675–80. doi: 10.1128/aem.65.4.1675-1680.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crawley MJ. Statistical computing. An introduction to data analysis using S-plus. 1. Chichester: Wiley; 2002. [Google Scholar]
- 62.R core team . R: A language and environment for statistical computing. 321. Vienna: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 63.Courcelle CT, Chow KH, Casey A, Courcelle J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci U S A. 2006;103(24):9154–9. doi: 10.1073/pnas.0600785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ray JL, Harms K, Wikmark OG, Starikova I, Johnsen PJ, Nielsen KM. Sexual isolation in Acinetobacter baylyi is locus-specific and varies 10,000-fold over the genome. Genetics. 2009;182(4):1165–81. doi: 10.1534/genetics.109.103127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990;54(1):18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58(3):401–65. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. MMBR. 1999;63(4):751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell. 2003;11(5):1337–47. doi: 10.1016/S1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 69.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. MMBR. 2008;72(4):642–71. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carrasco B, Fernandez S, Asai K, Ogasawara N, Alonso JC. Effect of the recU suppressors sms and subA on DNA repair and homologous recombination in Bacillus subtilis. Mol Genet Genomics. 2002;266(5):899–906. doi: 10.1007/s00438-001-0616-7. [DOI] [PubMed] [Google Scholar]
- 71.Johnston C, Mortier-Barriere I, Granadel C, Polard P, Martin B, Claverys JP. RecFOR is not required for pneumococcal transformation but together with XerS for resolution of chromosome dimers frequently formed in the process. PLoS Genet. 2015;11(1):e1004934. doi: 10.1371/journal.pgen.1004934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gupta R, Shuman S, Glickman MS. RecF and recr play critical roles in the homologous recombination and single-strand annealing pathways of mycobacteria. J Bacteriol. 2015;197(19):3121–32. doi: 10.1128/JB.00290-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang G, Lo LF, Maier RJ. The RecRO pathway of DNA recombinational repair in Helicobacter pylori and its role in bacterial survival in the host. DNA Repair (Amst) 2011;10(4):373–9. doi: 10.1016/j.dnarep.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakai A, Cox MM. RecFOR and RecOR as distinct reca loading pathways. J Biol Chem. 2009;284(5):3264–72. doi: 10.1074/jbc.M807220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bakkali M. Could DNA, uptake be a side effect of bacterial adhesion and twitching motility? Arch Microbiol. 2013;195(4):279–89. doi: 10.1007/s00203-013-0870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bonura T, Smith KC. Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K-12. J Bacteriol. 1975;121(2):511–7. doi: 10.1128/jb.121.2.511-517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Redfield RJ. Genes for breakfast: the have-your-cake-and-eat-it-too of bacterial transformation. J Hered. 1993;84(5):400–4. doi: 10.1093/oxfordjournals.jhered.a111361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets and R-code for the statistical models is provided upon request.


