Abstract
Activation of primary CD4+ T cells induces the CD155, but not the CD112 ligands for the natural killer (NK) cell activation receptor (aNKR) CD226 [DNAX accessory molecule-1 (DNAM-1)]. We hypothesize that HIV productively infects activated CD4+ T cells and makes itself vulnerable to NK cell-mediated lysis when CD155 on infected T cells engages DNAM-1. The primary objective of this study is to determine whether CD155 alone or together with NKG2D ligands triggers autologous NK cell lysis of HIV-infected T cells and whether HIV modulates CD155. To determine whether HIV modulates this activation ligand, we infected “activated” CD4+ T cells with HIV in the absence or presence of Nef and/or Vpu and determined by flow cytometry whether they modulated CD155. To determine if CD155 alone, or together with NKG2D ligands, triggered NK cell lysis of autologous HIV-infected T cells, we treated purified NK cells with DNAM-1 and/or NKG2D blocking antibodies before the addition of purified autologous HIV-infected cells in cytolytic assays. Finally, we determined whether DNAM-1 works together with NKG2D as an NK cell coactivation receptor (caNKR) or whether they work independently as aNKRs to induce an NK cell lytic response. We demonstrate that HIV and specifically Nef and/or Vpu do not modulate CD155 on infected primary T cells; and both CD155 and NKG2D ligands synergize as aNKRs to trigger NK cell lysis of the infected cell.
Keywords: : natural killer cells, HIV, Nef, Vpu, cytotoxic
Introduction
Natural killer (NK) cells function as innate “sentinels” of the immune system. These lymphocytes circulate through the blood and lymph or reside within tissues in search of abnormal or virus-infected cells to rapidly eliminate them before the development of systemic disease. NK cells respond within minutes to hours following infection or transformation of cells and do not require prior recognition of components from abnormal or pathogen-infected cells.1–5
NK cells destroy infected and tumorigenic cells following activation receptor (aNKR)-mediated recognition of molecules that are expressed on the infected or abnormal cell surface.6–9 Activation ligands are induced to be expressed as a result of activation (e.g., CD155/CD112),10 as a result of “cellular stress” (e.g., ULBPs),8 or expressed as cell surface pathogen products (e.g., influenza hemagglutinin).7 Engagement of a single aNKR is insufficient to trigger a cytotoxic response by the NK cell. A coactivation receptor (caNKR) must also be triggered simultaneously with an aNKR in order for an NK cell to mediate its effector functions.11–13
In addition to expression of ligands to aNKRs and caNKRs major histocompatibility complex class I (MHC-I) molecules are another element that must be overcome for an NK cell to effectively respond to a target cell. MHC-I molecules on normal target cells trigger inhibitory receptors on NK cells (iNKRs), thus preventing them from functioning even when aNKRs are triggered simultaneously.14–17
We and other investigators demonstrated that ligands to the NK cell aNKR NKG2D are induced by HIV-1 Vpr. Cell surface expression of these ligands on HIV-infected T cells trigger NK cells to lyse the infected cells.18,19
In our previous studies, we demonstrated that even though Nef downmodulates HLA-A and -B,20 and Vpr induces NKG2D activation ligands,18 HIV-1 avoids lysis by NK cells by downmodulating NTB-A through Vpu.21 We showed that a mutated Vpu, which could not downmodulate NTB-A, greatly enhanced NK cell lysis of the infected T cells.21 The lysis of T cells infected with a virus containing the mutated Vpu was reversed in the presence of blocking antibodies to NTB-A.21 These findings indicate that HIV-1 Vpu primarily affects NTB-A and not other NK cell activation ligands.
However, it remains to be determined if other NK cell aNKR ligands trigger NK cells to lyse the HIV-infected cells. We did not observe the presence of ligands to NCRs NKp30, NKp44, and NKp46 on primary HIV-infected T cells.22 Recently, CD155 was shown to be present on HIV-infected T cells and could be a potential ligand that triggers NK cell lysis of the infected cell.23 This study also reported that the HIV-1 viral proteins Nef and Vpu antagonize cell surface expression of CD155 and allow HIV to evade NK cell lysis.23 A subsequent study reported that Vpu downmodulates CD155 through its transmembrane residues' (A10xxxA14xxxA18xxxW22) interaction with CD155 and sequesters it within the cell, thereby preventing its presentation on the cell surface.24
The discrepancy between our previous studies21 and those by other investigators23 may lie in the differences of HIV-1-infected target cells used. We used purified HIV-infected T cells as target cells in our cytotoxic assays,25 while the studies by Matusali et al.23 utilized HIV-infected Jurkat T-cell lines as target cells in their lytic assay. HIV-infected Jurkat cells may express different types and levels of NK cell activation ligands not seen with primary CD4+ T cells. Therefore, we wanted to determine whether Vpu and/or Nef modulated CD155 on the natural host cell of HIV, primary CD4+ T-cell blasts, to avoid lysis by autologous NK cells.
In this study, we demonstrate that (1) activated primary CD4+ T cells express CD155; (2) HIV and specifically Nef and/or Vpu do not modulate CD155 on infected primary T cells; and (3) CD155 and NKG2D ligands trigger NK cell lysis of the infected cell, but this response appears optimal when the caNKR NTB-A is triggered simultaneously. Therefore, despite the presence of NK cell activation ligands, CD155 and ULBP-1/-2, on HIV-infected cells, downmodulation of NTB-A by Vpu has consequences in the ability of NK cells to lyse HIV-infected T cells.
Materials and Methods
Primary cells and cell lines
All primary cells (i.e., NK cells and CD4+ T cells) used in this study were isolated from peripheral blood of healthy HIV-1-uninfected donors after informed written consent was acquired in accordance with the Declaration of Helsinki and the policies of the Institutional Review Board at Rush University Medical Center, Chicago, IL. NK cells and CD4+ T cells were isolated from the peripheral blood in separate blood draws and stimulated in vitro as described in Ref.25 The P815 mouse lymphoblast-like mastocytoma cell line (ATCC) was maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated (56°C, 30 min) fetal bovine serum (FBS) and penicillin/streptomycin (Mediatech).
HIV infection of primary CD4+ T cells
Freshly isolated primary CD4+ T cells were activated using anti-CD3/anti-CD28 mAb coupled to magnetic beads (Miltenyi Biotech) for 72 h before infection with an HIV-1NL4-3 strain in which HIV-1 envelope is deleted (DHIV3). We also infected CD4+ T cells with the same strain of virus, which lacked Vpu, Nef, or Nef and Vpu. These envelope-defective viruses were VSV-G pseudotyped. A replication-incompetent virus was used since Vpu and Nef could impact the replication capacity of HIV-1 within CD4+ T cells. Infection was performed by spin inoculation with a MOI50 = 1 as described in Ref.26 Following the infection, the cells were cultured in the RPMI complete medium with 200 U/ml recombinant IL-2 (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, deposited by Dr. Maurice Gately; Hoffmann-La Roche, Inc.).
Flow cytometry reagents and antibodies
Uninfected and infected T cells were first incubated with the viability dye AquaDead LIVE/DEAD (Life Technologies) before being surface stained with fluorochrome-conjugated anti-CD155 [Biolegend (clone SKII.4)], anti-CD112 [Biolegend (clone TX31)], anti-NTB-A [Biolegend (clone 292811)], anti-HLA-DR [Biolegend (clone L423)], anti-HLA-A, -B, and -C [Biolgend (clone W6/32)], or anti-CD4 [BDIS (clone RPA-T4)]. Cells were subsequently permeabilized and fixed using Perm/Fix (BD Biosciences) and intracellularly stained for HIV-1 p24 with fluorochrome-conjugated anti-HIV-1 Gag p24 antibodies [Beckman-Coulter (clone KC57)]. HIV-1 p24 negative (p24−) and HIV-1 p24 positive (p24+) were collected (2 × 105) on FACSLSRII (BD Biosciences) and analyzed using FlowJo software (TreeStar). FACSLSRII was a generous gift from the James B. Pendelton Charitable Trust.
Relative NTB-A and CD155 surface expression were calculated as follows: [median fluorescent intensity (MFI) of NTB-A or CD155 on p24+ cells−MFI of isotype of p24+ cells]/(MFI NTB-A or CD155 on p24− cells−MFI of isotype of p24− cell) × 100. NTB-A and CD155 on uninfected cells were set at 100%. In some studies we evaluated DNAM-1 [Biolegend (clone: 11AE)] expression of NK cell marker (CD56+ CD3− CD14− CD19−) (vendor and clones of antibodies to CD56, CD3, CD14, and CD19 were similar to those used in our previous study21).
CD107a degranulation and chromium release assays
NK cells were obtained from PBMCs of same donor as the CD4+ T cells. NK cells were obtained from separate blood drawn and was done 6–9 days after isolating CD4+ T cells because it takes 7–10 days to stimulate and infect CD4+ T cells with HIV-1. One day before the degranulation and cytotoxic assays, fresh NK cells were isolated from PBMC using immunomagnetic beads (Miltenyi). After isolation, NK cells were cultured overnight in either a plain medium or in a medium containing 200 U/ml IL-2. The resulting purified and cultured NK cells were placed in RPMI-1640 and 10% FBS and exposed to target cells.
HIV-infected cells were isolated from uninfected cells in bulk culture before addition to NK cells as described.25 Briefly, HIV-infected cells were treated with anti-CD4 Ab coupled to magnetic beads (Invitrogen) at a ratio of 10 beads per cell. The cells were incubated at 4°C for 1 h. The cells bound to the beads were removed with a magnet and the remaining cells suspension treated with another round of anti-CD4 Ab coupled to magnetic beads for 1 h at 4°C at a bead to cell ratio of 10:1. The resulting cells, after immunomagnetic bead separations of CD4+ T cells, were shown to be infected by HIV as demonstrated by the presence of HIV-1 p24 antigen within the unbound cells, which was determined as described.25
NK cell cytotoxic effector functions (both CD107a Degranulation and Chromium Release Assays) following exposure to target cells were evaluated as previously described in extensive detail.25 Before the coculture of target cells with NK cells, the NK cells were treated at 4°C for 30 min with 10 μg/ml of the following antibodies: anti-human DNAM-1 antibody [R&D Systems (clone:102511) or BDIS (clone: DX11)] and/or anti-human NKG2D antibody [R&D Systems (clone:149810) or BDIS (clone: 1D11)].
In addition, we determined the extent to which purified NK cells degranulated (surface CD107a expression) in response to antibodies directed to CD16 [BDIS (clone: 3G8)], DNAM-1 [BDIS (clone: DX11)], NTB-A [R&D Systems (clone:292811)], NKG2D [BDIS (clone:1D11)] or a combination of the antibodies. After treatment of P815 cells with antibodies for 30 min at 4°C, the NK cells were added to the culture. NK cells were cocultured with antibody-labeled P815 cells for 4 h. As a control, we cocultured NK cells with P815 cells without antibody. NK cells with target cells were labeled with fluorochrome-conjugated anti-CD107a Ab [BDIS (clone H4A3) along with CD56, CD3, CD14, and CD19 (all BDIS antibodies)]. NK cells (CD56+ CD3− CD14− CD19− cells) were collected (2 × 105) on FACSLSRII (BD Biosciences) and analyzed using FlowJo software (TreeStar).
Statistical analyses
Nonparametric statistical analysis of the data in the study was done using Mann–Whitney U-test to determine statistical significance between two groups. p values ≤.05 were considered statistically significant.
Results and Discussion
Activated primary CD4+ T cells express the DNAM-1 ligand CD155, but not CD112, and surface levels of CD155 is not modulated by HIV-1 Nef and/or Vpu
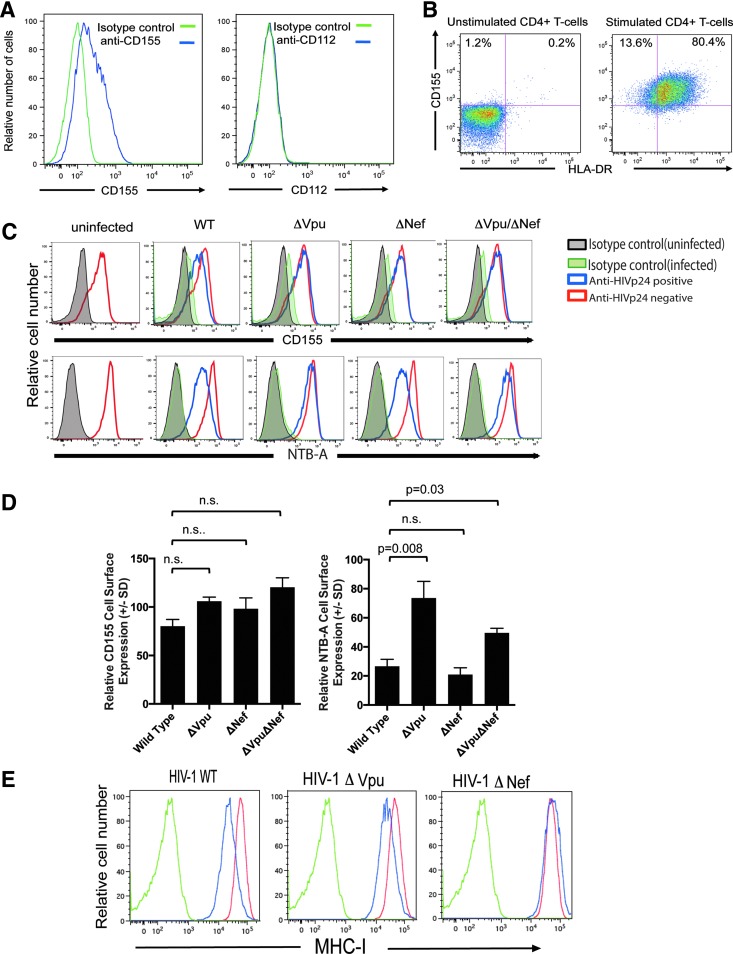
When evaluating the expression of the DNAM-1 ligands CD155 (i.e., polio virus receptor) and CD112 (i.e., Nectin-2), we found that only CD155 was expressed on anti-CD3 and CD28 mAb-stimulated primary CD4+ T cells (Fig. 1A). CD155 was present mostly (∼80%) on “activated” CD4+ T cells as indicated by coexpression of HLA-DR (Fig. 1B).
FIG. 1.
DNAM-1 ligand CD155, but not CD112, is expressed on activated CD4+ T cell and is not modulated by Nef or Vpu during HIV infection. (A) Expression of CD155 (blue line), but not CD112 (blue line), on anti-CD3 and -CD28 antibody-treated primary CD4+ T cells. Isotype controls (green line) for the CD155 and CD112 Ab are provided. This is an example of two experiments. (B) Untreated and anti-CD3 and anti-CD28 antibody-treated primary CD4+ T cells were stained with anti-CD155 and anti-HLA-DR. Numbers in quadrant one and two represent the frequency of CD4+ T cells that stained with anti-CD155 or anti-CD155 and anti-HLA-DR. This is an example of two experiments. (C) Anti-CD3 and -CD28 antibody-treated primary CD4+ T cells were infected with VSV-G-pseudotyped DHIV3 (WT), DHIV3 lacking Vpu (ΔVpu), DHIV3 lacking Nef (ΔNef), or DHIV3 lacking both Vpu and Nef (ΔVpu ΔNef) for 3 days and then surface stained for CD155 or NTB-A. The cells were then permeabilized and stained intracellularly for anti-HIV-1 p24 (red line). As a control, uninfected Anti-CD3 and -CD28 antibody-treated primary CD4+ T cells were evaluated (blue line). Isotype controls for both infected (orange line) and uninfected cells (green line) are provided. This is an example of five different subjects. (D) The mean relative surface expression of CD155 or NTB-A ±SD of five different subjects. Formula to determine relative levels of CD155 and NTB-A is provided in Material and Methods section. The statistical significance of the relative CD155 of NTB-A surface expression was determined using the Mann–Whitney U-test. p > .05 was not statistically significant. (E) HLA-A-A, -B, and -C expression on anti-CD3 and -CD28 antibody-treated primary-uninfected CD4+ T cells (blue line) or anti-CD3 and -CD28 antibody-treated primary CD4+ T cells infected (red line) with VSV-G-pseudotyped DHIV3 WT, with VSV-G pseudotyped DHIV3 lacking Vpu (ΔVpu), and with VSV-G-pseudotyped DHIV3 lacking Nef (ΔNef). Isotype control antibody-stained cells are also provided (green line). DNAM-1, DNAX accessory molecule-1; SD, standard deviation; WT, wild type.
Infection of primary anti-CD3 and -CD28 Ab-stimulated CD4+ T cells with HIV did not downmodulate CD155 even though it significantly downmodulated NTB-A from the cell surface (Fig. 1C). Moreover, there was no statistically significant increase in CD155 surface levels when HIV-1 Vpu and/or Nef were removed from the virus (Fig. 1C, D). In contrast, loss of Vpu greatly affected NTB-A surface expression [(p < .05); Fig. 1C, D] and loss of Nef from HIV led to the increased expression of MHC-I (Fig. 1E). Thus, it appears that the activation of CD4+ T cells induces CD155 on the cell surface and HIV-1 infection of CD4+ T cells does not significantly impact CD155 surface expression, but does result in the decrease of both MHC-I and NTB-A expression on the infected cell surface.
Since HIV-1-infected CD4+ primary T cells express CD155, we next wanted to determine if the presence of CD155 contributed to NK cell lysis of infected CD4+ T cells. To test this, we utilized a blocking antibody against DNAM-1 on NK cells to impair its interaction with CD155 on autologous HIV-infected cells and then compared the contribution of DNAM-1 to NK cell lysis of autologous HIV-infected CD4+ T cells with that of NKG2D. We18 and other investigators19 have previously demonstrated that HIV-1 Vpr induces NKG2D ligands (i.e., ULBP-1 and -2) on primary HIV-infected T cells. As a control, we also evaluated the ability of NK cells to lyse uninfected CD4+ T cells.
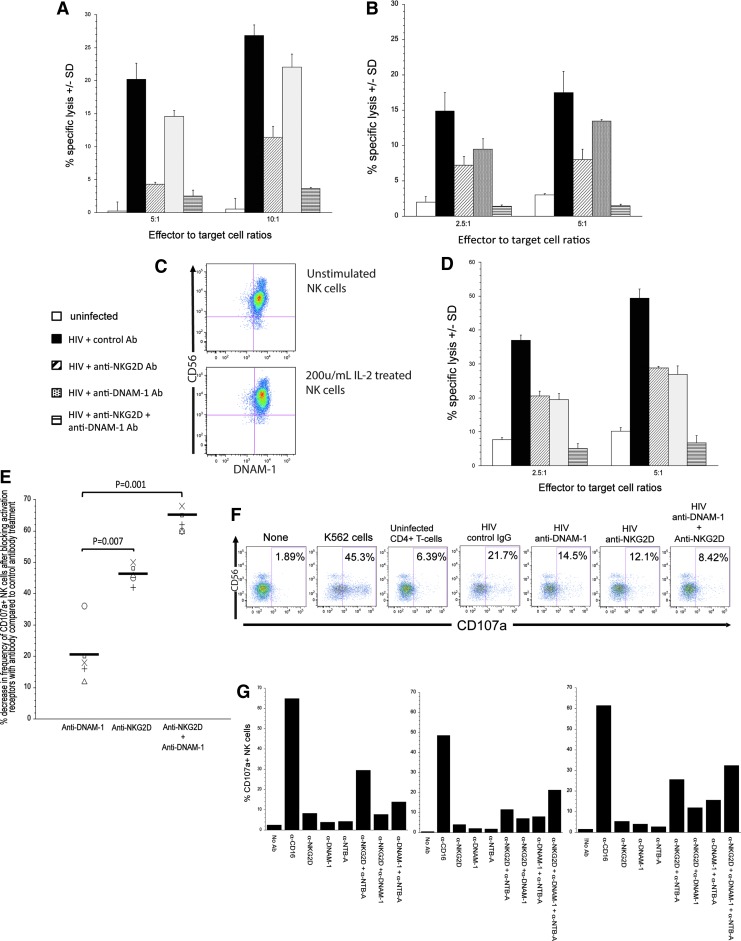
As shown in Figure 2A and B, NK cells from two different donors were able to lyse HIV-infected cells 15–25-fold higher in the presence of a control antibody in comparison to NK cell lysis of uninfected target cells. As expected, blocking NKG2D on NK cells led to a decrease in their ability to lyse autologous HIV-infected cells (Fig. 2A, B). In contrast, blocking DNAM-1 led to a modest decrease in the lysis of HIV-infected cells (Fig. 2A, B). However, when both DNAM-1 and NKG2D are prevented from interacting with their ligands on the infected cell surface, the ability to lyse HIV-infected T cells dropped close to or at levels observed with NK cell lysis of uninfected cells. Thus, it appears that CD155 and ULBP-1/-2 work in concert with one another to trigger optimal primary NK cell lysis of autologous HIV-infected cells.
FIG. 2.
NK cells lyse autologous HIV-infected T cells when DNAM-1 and NKG2D are triggered by HIV-infected T cells. (A, B) anti-CD3 and -CD28 antibody-treated primary CD4+ T cells were infected with VSV-G-pseudotyped DHIV3 for 3 days. The infected cells were then separated from the uninfected cells using anti-CD4 beads. The unattached cells that remained behind were used as target cells. As a control target cell, we utilized anti-CD3 and -CD28 antibody-treated primary CD4+ T cells (white bar). Target cells were labeled with 51Cr for 2 h and then mixed with NK cells at effector cell to target cell (E:T) ratios of 2.5:1, 5:1, or 10:1. In some groups, NK cells were incubated with blocking antibodies to DNAM-1 (hashed line bar), NKG2D (gray bar), DNAM-1, and NKG2D (horizontal line bar) or control antibody of similar isotype as blocking antibody (black bar). After 4 h, fluids were removed and the remaining 51Cr in the fluid was measured. Each group was done in triplicate. The mean % specific lysis ±SD was determined as described in Material and Methods section. Each Figure (A, B) represents data from experiments involving two separate donors. (C) Purified NK cells cultured in a medium or 200 U/ml of IL-2 were stained with CD56 and DNAM-1. (D) Similar to A and B except NK cells were treated overnight with 200 U/ml of IL-2 before their use in the 51Cr release assay and involved a different donor. (E) Ability of NK cells to degranulate when exposed to autologous HIV-infected T cells (NK cell:HIV-infected cell ratio of 2:1) when exposed to anti-DNAM-1, anti-NKG2D, or a combination of both blocking antibody when compared with NK cells exposed to Ig of irrelevant specificity. Each symbol represents a different donor. Bars represent mean percent decrease in frequency of CD107a+ NK cells in the presence of blocking antibody to DNAM-1 and/or NKG2D compared to frequency of CD107a+ NK cells treated with antibody of irrelevant specificity. The statistical difference between treatment groups was determined using the Mann–Whitney U-test. (F) Degranulation of NK cells from a donor involved in the study presented in (E) following exposure to target cells in the presence of control of blocking antibodies. NK cell degranulation following exposure to control target cells (i.e., uninfected CD4+ T cells and K562 cells) is displayed. (G) Degranulation of NK cells when triggered with antibodies to DNAM-1, NTB-A, NKG2D, or a combination of the various antibodies. Antibody-labeled P815 cells were treated with NK cells for 4 h and the extent to which the NK cells degranulated was measured by determining the frequency of NK cells that expressed CD107a on their surface. Each figure represents data from experiments involving three separate donors. NK, natural killer.
Next, we wanted to determine what affect stimulation of NK cells with IL-2 would have on the ability of NK cells to lyse HIV-infected T cells in the presence or absence of DNAM-1 signaling. This is important given that, on IL-2-treated NK cells, cis-binding of DNAM-1 to LFA-1 leads to intracellular signaling.27 We noted no change to the frequency of NK cells expressing DNAM-1 before and after stimulation with IL-2 (Fig. 2C). We noted that the level of NK cell-mediated lysis of HIV-infected cells was similar whether it used DNAM-1 or NKG2D to trigger lysis (Fig. 2D). However, the combination of triggering both receptors gave the best lytic response as indicated by our study, where we found that blocking the interaction of DNAM-1 and NKG2D on NK cells with its ligands on the infected cells led to a level of lysis comparable to NK cell lysis to uninfected T cells.
Using another assay (degranulation assay) to evaluate the role of DNAM-1 on NK cell cytotoxic response to HIV-infected cells, we demonstrate that blocking DNAM-1 had only a marginal impact on the ability of NK cells to degranulate in response to autologous HIV-infected cells in comparison to blocking NKG2D (p = .07) (Fig. 2E, F). However, when blocking DNAM-1 is paired with blocking NKG2D, NK cell degranulation occurred to a lesser degree compared with blocking either receptor alone (p = .001 anti-DNAM-1 vs. anti-NKG2D and anti-DNAM-1 and p = .003 anti-NKG2D vs. anti-NKG2D and anti-DNAM-1).
Although both DNAM-1 and NKG2D are important for lysis of HIV-infected cells, it remains to be determined if they work independently or act as coreceptors. To determine if DNAM-1 and NKG2D require each other for cell lysis or are just separate aNKRs that work independent of one another, we utilized a redirected lysis assay where we triggered NKG2D and DNAM-1 on NK cells using anti-NKG2D or anti-DNAM-1 antibodies as ligand mimics, either alone or combined, in the presence of P815 cells (to crosslink the Ig molecules bound to the receptors). By doing so, we were able to determine if NKG2D and DNAM-1 act independent of one another, using another caNKR (e.g., NTB-A) or are caNKRs for one another. As a positive control for this study for coactivation of NK cells leading to degranulation, we triggered NKG2D with NTB-A with specific antibodies.21
Although, it is ideal to use primary cells as targets for this study, there are too many possible combinations of ligands (both ligands to aNKRs and caNKRs) on HIV-infected primary cells to decipher the specific role of NKG2D and DNAM-1 in eliciting the cytolytic response. Moreover, additional ligands on HIV-infected cells involved in eliciting NK cell cytolysis remain to be determined.
As predicted,21 triggering NKG2D and NTB-A leads to NK cell degranulation (Fig. 2G). In addition to NK cell degranulation occurring when NKG2D and NTB-A are engaged at the same time, triggering DNAM-1 and NTB-A simultaneously leads to NK cell degranulation above the additive levels of degranulation when either were triggered alone (Fig. 2G). Based on the data presented in our study, it appears that DNAM-1 and NKG2D act independent of one another as aNKRs for NK cell killing of HIV-infected cells, instead requiring another caNKR such as NTB-A in order for NK cells to degranulate (Fig. 2G). Thus, downmodulation of NTB-A by Vpu (Fig. 1C, D) would most likely dampen the maximal capacity of NK cells to kill HIV-infected cells even when both NKG2D and DNAM-1 were triggered on NK cells.21
One explanation for the discrepant results between the previous studies23 and this investigation may be the fact that we used only purified infected primary CD4+ T cells, whereas the previous studies were done using infected Jurkat cell lines as target cells in their lytic assay. In addition to this, we utilized MFI of anti-CD155 staining for primary infected cells and their mutants since the histograms do not follow a normal, but rather skewed, binomial distribution (Fig. 1C). While we did see a statistically significant difference in the ability of Vpu to modulate NTB-A, we found no statistical difference in the ability of HIV to modulate CD155 through Vpu and/or Nef.
It is of interest that activated uninfected CD4+ T cells express CD155 and yet are not lysed to the same extent as HIV-infected cells. This is likely due to expression of MHC-I molecules, which trigger inhibitory receptors on NK cells to prevent lysis. Although HIV downmodulates HLA-A and -B on primary CD4+ T cells, HLA-C remains on the infected cell surface. HLA-E is also present on the infected cell surface and triggers inhibitory receptors expressed on NK cells. However, we noted that HLA-E on HIV-infected cells is unable to inhibit NK cell lysis.28 Thus, while NK cells have the capacity to kill HIV-infected cells when triggered by CD155, HIV-1s downmodulation of NTB-A dampens the synergistic signal critical for DNAM-1-induced NK cell degranulation.
Although our study focuses on the role of CD155 on triggering the activation of NK cells through DNAM-1, there is a need to determine the role that CD155 will have on triggering the inhibitory receptor, T cell, immunoglobulin, and intracellular tyrosine inhibitory motifs (TIGIT) on NK cells.29,30 TIGIT is an immune cell-specific immunoglobulin superfamily receptor of the CD28 family, which is present on the cell surface of 20%–90% of human NK cells with a median of 64%.31 Similar to DNAM-1, TIGIT binds to CD112 and CD155; however, TIGIT affinity for CD112 and CD155 is higher compared with DNAM-1.32 Moreover, TIGIT is often coexpressed with DNAM-1 on NK cells,31 making it likely that inhibition of NK cell function will dominate over activation when the NK cells encounter a target cell expressing CD155.
Although TIGIT prevents NK cells from lysing normal primary fibroblasts,29 it has not been determined if TIGIT modulates NK cell responses against autologous HIV-infected cells. The primary focus in studies evaluating the role of inhibitory receptor on NK cell response to HIV-infected cells is on iNKR responses to MHC class I molecules.20,28,33–36 Since some KIRs may not be engaged due to modulation of HLA-A and -B by Nef33,37 or the inability of HLA-E to trigger the iNKR NKG2A/CD9428 and as we show in this study, HIV does not modulate CD155 on CD4+ T cells, it is likely that TIGIT may contribute to modulating NK cell response to HIV-infected cells.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Takasugi M, Mickey MR, Terasaki PI: Reactivity of lymphocytes from normal persons on cultured tumor cells. Cancer Res 1973;33:2898–2902 [PubMed] [Google Scholar]
- 2.Oldham RK, Siwarski D, McCoy JL, Plata EJ, Herberman RB: Evaluation of a cell-mediated cytotoxicity assay utilizing 125 iododeoxyuridine-labeled tissue-culture target cells. Natl Cancer Inst Monogr 1973;37:49–58 [PubMed] [Google Scholar]
- 3.Kiessling R, Klein E, Wigzell H: “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol 1975;5:112–117 [DOI] [PubMed] [Google Scholar]
- 4.Ching C, Lopez C: Natural killing of herpes simplex virus type 1-infected target cells: Normal human responses and influence of antiviral antibody. Infect Immun 1979;26:49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez C, Kirkpatrick D, Fitzgerald PA, et al. : Studies of the cell lineage of the effector cells that spontaneously lyse HSV-1 infected fibroblasts (NK(HSV-1)). J Immunol 1982;129:824–828 [PubMed] [Google Scholar]
- 6.Bauer S, Groh V, Wu J, et al. : Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 1999;285:727–729 [DOI] [PubMed] [Google Scholar]
- 7.Mandelboim O, Lieberman N, Lev M, et al. : Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 2001;409:1055–1060 [DOI] [PubMed] [Google Scholar]
- 8.Gasser S, Orsulic S, Brown EJ, Raulet DH: The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 2005;436:1186–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandt CS, Baratin M, Yi EC, et al. : The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med 2009;206:1495–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottino C, Castriconi R, Pende D, et al. : Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med 2003;198:557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryceson YT, March ME, Barber DF, Ljunggren HG, Long EO: Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J Exp Med 2005;202:1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryceson YT, March ME, Ljunggren HG, Long EO: Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood 2006;107:159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HS, Das A, Gross CC, Bryceson YT, Long EO: Synergistic signals for natural cytotoxicity are required to overcome inhibition by c-Cbl ubiquitin ligase. Immunity 2010;32:175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ljunggren HG, Karre K: In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today 1990;11:237–244 [DOI] [PubMed] [Google Scholar]
- 15.Bix M, Liao NS, Zijlstra M, Loring J, Jaenisch R, Raulet D: Rejection of class I MHC-deficient haemopoietic cells by irradiated MHC-matched mice. Nature 1991;349:329–331 [DOI] [PubMed] [Google Scholar]
- 16.Ciccone E, Pende D, Viale O, et al. : Involvement of HLA class I alleles in natural killer (NK) cell-specific functions: Expression of HLA-Cw3 confers selective protection from lysis by alloreactive NK clones displaying a defined specificity (specificity 2). J Exp Med 1992;176:963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purdy AK, Campbell KS: Natural killer cells and cancer: Regulation by the killer cell Ig-like receptors (KIR). Cancer Biol Ther 2009;8:2211–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward J, Davis Z, DeHart J, et al. : HIV-1 Vpr triggers natural killer cell-mediated lysis of infected cells through activation of the ATR-mediated DNA damage response. PLoS Pathog 2009;5:e1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richard J, Sindhu S, Pham TN, Belzile JP, Cohen EA: HIV-1 Vpr up-regulates expression of ligands for the activating NKG2D receptor and promotes NK cell-mediated killing. Blood 2010;115:1354–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonaparte MI, Barker E: Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood 2004;104:2087–2094 [DOI] [PubMed] [Google Scholar]
- 21.Shah AH, Sowrirajan B, Davis ZB, et al. : Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe 2010;8:397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward J, Bonaparte M, Sacks J, et al. : HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood 2007;110:1207–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matusali G, Potesta M, Santoni A, Cerboni C, Doria M: The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J Virol 2012;86:4496–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolduan S, Reif T, Schindler M, Schubert U: HIV-1 Vpu mediated downregulation of CD155 requires alanine residues 10, 14 and 18 of the transmembrane domain. Virology 2014;464–465:375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis ZB, Ward JP, Barker E: Preparation and Use of HIV-1 infected primary CD4+ T-cells as target cells in natural killer cell cytotoxic assays. J Vis Exp 2011;pii, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Doherty U, Swiggard WJ, Malim MH: Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol 2000;74:10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tahara-Hanaoka S, Shibuya K, Onoda Y, et al. : Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int Immunol 2004;16:533–538 [DOI] [PubMed] [Google Scholar]
- 28.Davis ZB, Cogswell A, Scott H, et al. : A conserved HIV-1-derived peptide presented by HLA-E renders infected T-cells highly susceptible to attack by NKG2A/CD94-bearing natural killer cells. PLoS Pathog 2016;12:e1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanietsky N, Simic H, Arapovic J, et al. : The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci U S A 2009;106:17858–17863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Zhang H, Li M, et al. : Recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT suppresses granule polarization and cytotoxicity of NK cells. Cell Death Differ 2013;20:456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Hou H, Wu S, et al. : TIGIT expression levels on human NK cells correlate with functional heterogeneity among healthy individuals. Eur J Immunol 2015;45:2886–2897 [DOI] [PubMed] [Google Scholar]
- 32.Stanietsky N, Rovis TL, Glasner A, et al. : Mouse TIGIT inhibits NK-cell cytotoxicity upon interaction with PVR. Eur J Immunol 2013;43:2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen GB, Gandhi RT, Davis DM, et al. : The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 1999;10:661–671 [DOI] [PubMed] [Google Scholar]
- 34.Alter G, Martin MP, Teigen N, et al. : Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med 2007;204:3027–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alter G, Heckerman D, Schneidewind A, et al. : HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 2011;476:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song R, Lisovsky I, Lebouche B, Routy JP, Bruneau J, Bernard NF: HIV protective KIR3DL1/S1-HLA-B genotypes influence NK cell-mediated inhibition of HIV replication in autologous CD4 targets. PLoS Pathog 2014;10:e1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Gall S, Erdtmann L, Benichou S, et al. : Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 1998;8:483–495 [DOI] [PubMed] [Google Scholar]