Abstract
Aims: S-glutathionylation of cysteine residues, catalyzed by glutathione S-transferase Pi (GSTP), alters structure/function characteristics of certain targeted proteins. Our goal is to characterize how S-glutathionylation of proteins within the endoplasmic reticulum (ER) impact cell sensitivity to ER-stress inducing drugs.
Results: We identify GSTP to be an ER-resident protein where it demonstrates both chaperone and catalytic functions. Redox based proteomic analyses identified a cluster of proteins cooperatively involved in the regulation of ER stress (immunoglobulin heavy chain-binding protein [BiP], protein disulfide isomerase [PDI], calnexin, calreticulin, endoplasmin, sarco/endoplasmic reticulum Ca2+-ATPase [SERCA]) that individually co-immunoprecipitated with GSTP (implying protein complex formation) and were subject to reactive oxygen species (ROS) induced S-glutathionylation. S-glutathionylation of each of these six proteins was attenuated in cells (liver, embryo fibroblasts or bone marrow dendritic) from mice lacking GSTP (Gstp1/p2−/−) compared to wild type (Gstp1/p2+/+). Moreover, Gstp1/p2−/− cells were significantly more sensitive to the cytotoxic effects of the ER-stress inducing drugs, thapsigargin (7-fold) and tunicamycin (2-fold).
Innovation: Within the family of GST isozymes, GSTP has been ascribed the broadest range of catalytic and chaperone functions. Now, for the first time, we identify it as an ER resident protein that catalyzes S-glutathionylation of critical ER proteins within this organelle. Of note, this can provide a nexus for linkage of redox based signaling and pathways that regulate the unfolded protein response (UPR). This has novel importance in determining how some drugs kill cancer cells.
Conclusions: Contextually, these results provide mechanistic evidence that GSTP can exert redox regulation in the oxidative ER environment and indicate that, within the ER, GSTP influences the cellular consequences of the UPR through S-glutathionylation of a series of key interrelated proteins. Antioxid. Redox Signal. 26, 247–261.
Keywords: : endoplasmic reticulum, unfolded protein response, glutathione S-transferases, S-glutathionylation, dendritic cells, protein disulfide isomerase
Introduction
Glutathione S-transferase Pi (GSTP), initially isolated from the placenta, is ubiquitous in mammals (18) and expressed at particularly high levels in many cancers and drug-resistant tumor cells (46). Primarily, GSTP has been considered cytosolic, although nuclear (26) and mitochondrial (14) compartment localizations have been reported. Originally described as a phase II detoxification enzyme, GSTP has noncatalytic chaperone functions that regulate various kinases (46). It can also act as a glutathionylase in the post-translational S-glutathionylation of redox-sensitive cysteines in proteins (44, 52, 56). S-glutathionylation occurs on cysteine residues in low pKa environments in proteins, forming a disulfide bond with glutathione (GSH) (17, 58) in a dynamic and reversible cycle that can serve as a secondary level of regulation for a number of cell processes. Proteins with susceptible cysteines fall into functional clusters, two of which are protein folding and calcium transport pathways (47). The modification adds a tripeptide with a net negative charge, alters tertiary/quaternary structure, and influences protein–protein interactions (50, 58). When cells are exposed to reactive oxygen species (ROS), S-glutathionylation can protect proteins against irreversible oxidative damage and subsequent deglutathionylation can restore the protein to its native state (17, 47), providing a redox-mediated regulatory control cycle. Mice deficient in GSTP1/2 are more sensitive to certain chemical stresses that impact redox pathways (21) and also express phenotypes consistent with enhanced immunity and myeloproliferation (10, 39, 61).
Innovation.
Accurate folding of proteins in the endoplasmic reticulum (ER) is exquisitely dependent upon precise disulfide bond formation. As a consequence regulated redox conditions within the organelle are critical to controlling both the unfolded protein response and cell viability. Unexpectedly, we found that glutathione S-transferase P is an ER resident protein and catalyzes S-glutathionylation of a number of proteins that integrate protein-folding pathways. Interference with this process can alter sensitivity of cells to drugs that target ER functions.
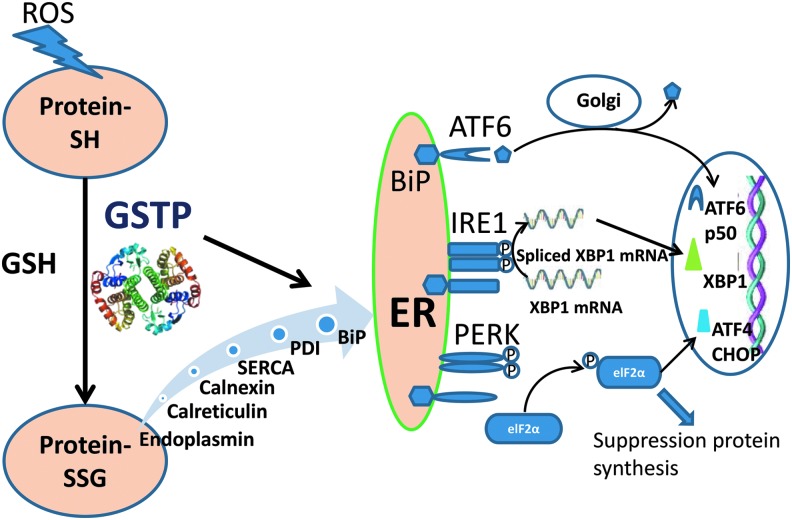
As the organelle responsible for protein folding newly synthesized, secreted, and membrane-bound proteins, the endoplasmic reticulum (ER) maintains a redox homeostasis that is shifted toward a more oxidized state when compared to the remainder of the cell (22). The ER also facilitates protein folding by sustaining higher calcium concentrations and distinct protein glycosylation enzymes (43, 55). Precisely how alterations in redox are transmitted to organelle components that govern disulfide bond formation, where GSH as the primary redox buffer, is not clear. Induction of the unfolded protein response (UPR) involves a shift toward a more reductive environment in the ER (35). Figure 1 illustrates how GSTP and GSH might adapt in expression and localization to regulate ER redox homeostasis with subsequent changes transmitted to the three main sensors of ER stress, namely, inositol-requiring protein-1 (IRE1), protein kinase-like ER kinase (PERK), and activating transcription factor 6 (ATF6). Activation of these proteins occurs via interaction(s) with immunoglobulin heavy chain-binding protein (BiP), which in the absence of ER stress complexes with and inhibits these sensors. Increased levels of unfolded proteins in the ER can trap free BiP, decrease the free steady-state levels of this chaperone, and release it from the sensors, activating them and initiating further signaling cascades, including activation of activating transcription factor 4 (ATF4), C/EBP homologous protein (CHOP), and phosphorylation of α subunit eukaryotic translation initiation factor 2 (eIF2α) (4, 45). A constant luminal calcium concentration is also necessary for correct protein folding, and alterations in ER calcium homeostasis can cause ER stress and activate the UPR. While calcium homeostasis is regulated through a complex series of reactions, the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) is the primary calcium pump that mediates Ca2+ uptake from the cytosol into the ER. Previous studies have shown that SERCA can be activated by S-glutathionylation (2) and it is logical that this protein will work in concert with other calcium binding proteins, such as calnexin, calreticulin, and endoplasmin, in regulating the UPR either to reestablish normal ER function or eliminate the cells through programmed cell death pathways.
FIG. 1.
Schematic of how GSTP-mediated S-glutathionylation of ER-resident proteins may influence the management of ER stress and UPR. ROS-induced oxidative stress causes protein oxidation. GSTP mediates S-glutathionylation of redox-sensitive cysteine containing proteins. This post-translational modification is known to change the structure and function of a variety of protein clusters [reviewed in (51)]. Currently, we found GSTP mediates S-glutathionylation of some ER-resident proteins (calnexin, calreticulin, endoplasmin, SERCA, protein disulfide isomerase (PDI), and BiP), which are involved in protein folding and ER calcium homeostasis. In turn, such changes need to be transmitted to the three main sensors of ER stress, IRE1, PERK, and ATF6. ATF, activating transcription factor; BiP, immunoglobulin heavy chain-binding protein; CHOP, C/EBP homologous protein; eIF2α, eukaryotic translation initiation factor 2α; ER, endoplasmic reticulum; GSTP, glutathione S-transferase Pi; IRE1, inositol-requiring protein-1; PERK, protein kinase-like ER kinase; PDI, protein disulfide isomerase; ROS, reactive oxygen species; SERCA, sarco/endoplasmic reticulum Ca2+-ATPase; UPR, unfolded protein response; XBP, x-box binding protein. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
A broad range of human pathologies have been associated with imbalanced protein folding and aberrant proteostasis. Rapid and sequential division of cancer cells requires high protein turnover and places the ER under high intrinsic stress. Partly as a consequence, it has proved to be a viable target for drugs that target the UPR (42), particularly in multiple myeloma. Although not front line clinical agents, thapsigargin (ThG) and tunicamycin (TuM) can induce ER stress and cause protein misfolding. ThG is a specific inhibitor of SERCA, altering ER Ca2+ homeostasis and interfering with the functional activities of calcium-dependent chaperones (36). TuM inhibits N-linked glycosylation leading to the accumulation of misfolded, nonglycosylated proteins in the ER. In either case, saturation of the ER folding capacity can, in turn, induce cell death through influencing the UPR. Our present study was designed to determine whether components of the S-glutathionylation cycle localize in the ER and whether these redox-dependent post-translational modifications influence component proteins involved in protein folding and cell survival.
Results
Identification of GSTP-mediated S-glutathionylated proteins
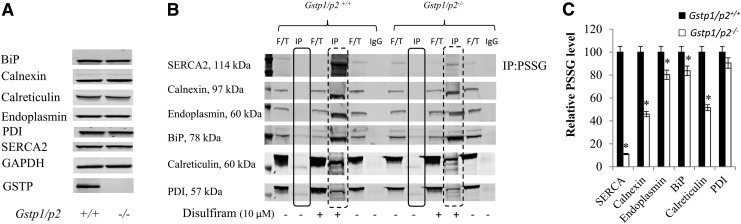
Comparative analysis of protein S-glutathionylation was enacted following treatment with disulfiram. Disulfiram possesses a reactive disulfide bond, which reacts readily with both protein and low-molecular-mass thiols, causing depletion of GSH and augmentation of mixed disulfides between GSH and protein thiols to form S-glutathionylated proteins (38). In the present study, liver lysates were freshly prepared, treated with disulfiram, and immediately subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under nonreducing conditions. Disulfiram induced S-glutathionylation in both Gstp1/p2+/+ and Gstp1/p2−/− mouse liver lysates, with higher levels found in Gstp1/p2+/+ samples (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/ars). GSTP-mediated S-glutathionylation was also shown in different subcellular compartments (Supplementary Fig. S1B). To identify those S-glutathionylated proteins that differed between the two, two gels were run simultaneously. One was transferred onto nitrocellulose membrane for anti-protein S-glutathionylation (PSSG) immunoblot (Supplementary Fig. S1A), and the other was stained with Brilliant Blue G-colloidal stain. Based on the immunoblot results, protein bands of interest, defined as those with S-glutathionylation levels that quantitatively differed between samples from Gstp1/p2+/+ and Gstp1/p2−/− mice (see arrows in Supplementary Fig. S1A), were excised from the Brilliant Blue G-colloidal stained gel and prepared for subsequent proteomic analyses. Both cytosolic and ER-resident proteins were identified. The latter included BiP, calnexin, calreticulin, endoplasmin, protein disulfide isomerase (PDI), and SERCA2 (Table 1). On the basis of these data and to confirm the S-glutathionylation of ER-resident proteins, immunoprecipitations (IPs) with anti-PSSG antibodies were performed. Basal expression levels of BiP, calnexin, calreticulin, endoplasmin, PDI, and SERCA2 were essentially similar for Gstp1/p2+/+ and Gstp1/p2−/− liver lysates (Fig. 2A), whereas disulfiram treatment induced significantly higher levels of S-glutathionylation in Gstp1/p2+/+ samples (Fig. 2B, C). Figure 2B shows the blots of these ER-resident proteins after IP with anti-PSSG antibodies. The basal S-glutathionylation levels of each ER-resident protein were low in both Gstp1/p2+/+ and Gstp1/p2−/− liver lysates (solid square in Fig. 2B), while the differences after disulfiram treatment were most marked for SERCA2, calreticulin, and calnexin in Gstp1/p2+/+ mice (dashed square in Fig. 2B). Densitometric quantification of the bands (dashed square in Fig. 2B) was compared between Gstp1/p2+/+ and Gstp1/p2−/− samples (Fig. 2C).
Table 1.
Proteomic Analysis of Endoplasmic Reticulum Proteins from Mouse Livers Following Disulfiram Treatment
| Accession number | Protein ID | Gene name | Unique peptides | MW (kDa) |
|---|---|---|---|---|
| Cytoplasmic proteins | ||||
| D3YX85 | Arf-GAP with SH3 domain, ANK repeat, and PH domain-containing protein 2 | Asap2 | 3 | 106 |
| Q60865 | Caprin-1 | Caprin1 | 2 | 73 |
| P07901 | Heat shock protein HSP 90-alpha | Hsp90aa1 | 4 | 90 |
| F8WJK8 | Hsc70-interacting protein | St13 | 6 | 40 |
| B1AXW5 | Peroxiredoxin 1 | Prx1 | 3 | 22 |
| Q3UR88 | Ras GTPase-activating protein-binding protein 1 | Gsbp1 | 4 | 44 |
| Q64374 | Regucalcin | Rgn | 6 | 33 |
| Endoplasmic reticulum proteins | ||||
| P20029 | 78 kDa glucose-regulated protein/immunoglobulin binding protein | Hspa5 | 4 | 78 |
| P35564 | Calnexin | Canx | 3 | 97 |
| P14211 | Calreticulin | Calr | 11 | 60 |
| P08113 | Endoplasmin | Hsp90b1 | 5 | 94 |
| O55143-2 | Isoform SERCA2A of sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | Atp2a2a | 8 | 114 |
| Q921X9 | Protein disulfide isomerase A5 | Pdia5 | 3 | 59 |
The proteomic analysis was designed to distinguish those proteins in Gstp1/p2+/+ and Gstp1/p2−/− samples with quantitatively different S-glutathionylation levels.
GSTP, glutathione S-transferase Pi; SERCA, sarco/endoplasmic reticulum Ca2+-ATPase.
FIG. 2.
GSTP-mediated ER protein S-glutathionylation in Gstp1/p2+/+ and Gstp1/p2−/− mouse liver lysates. Liver tissues were freshly prepared from Gstp1/p2+/+ and Gstp1/p2−/− mice. (A) Baseline levels of calnexin, calreticulin, endoplasmin, SERCA, PDI, and BiP in Gstp1/p2+/+ and Gstp1/p2−/− samples. (B) One milligram of liver lysates untreated or treated with disulfiram (10 μM, for 30 min at 37°C) was used for IP with protein A/G-agarose beads using mouse monoclonal anti-PSSG antibodies. Samples, including IP with anti-PSSG, IP with mouse IgG, and F/T after precipitation by A/G-agarose beads, were analyzed by subsequent nonreducing SDS-PAGE and blotted with anti-calnexin, calreticulin, endoplasmin, SERCA, PDI, or BiP antibodies. Solid squares show ER-resident protein levels after IP with anti-PSSG in untreated samples, and dashed squares show ER-resident protein levels after IP with anti-PSSG in disulfiram-treated samples. (C) Quantification of fluorescent intensities of the bands in dashed squares shown in (B). *Represents significant difference between Gstp1/p2+/+ and Gstp1/p2−/− mice. Data are representative blots from three independent experiments. F/T, flow-through; IgG, immunoglobulin G; IP, immunoprecipitation; PSSG, Protein S-glutathionylation; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
GSTP, ER-resident proteins, and S-glutathionylation
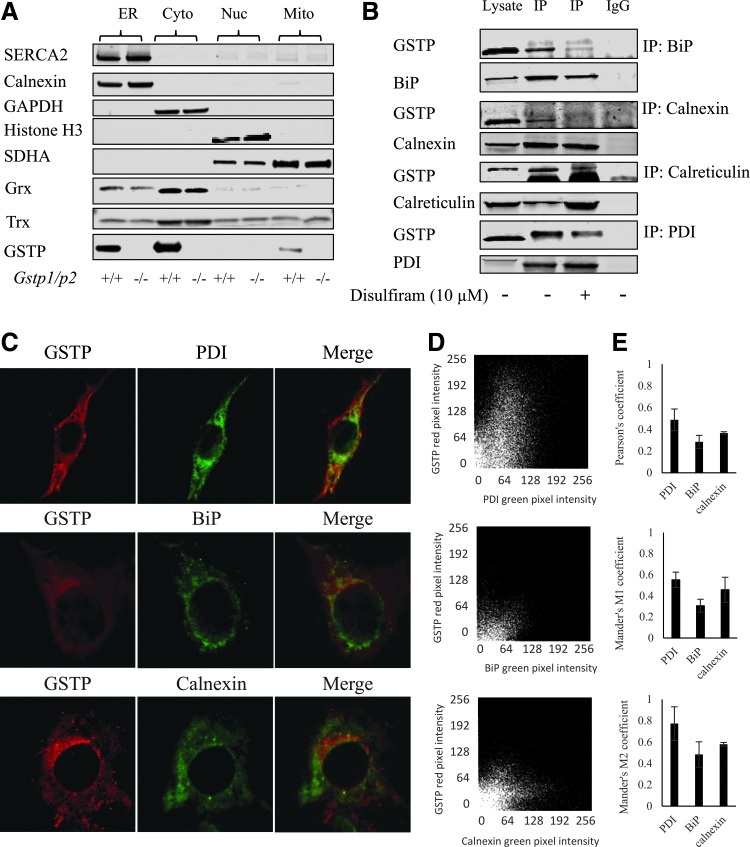
We used two independent approaches to demonstrate that GSTP is a resident protein in the ER. For the first, we used ER fractionation and organelle marker analyses of liver lysates from Gstp1/p2+/+ and Gstp1/p2−/− mice (Fig. 3A). SERCA and calnexin were used as ER markers, while GAPDH, succinate dehydrogenase A (SDHA), and histone H3 were markers for cytosol, mitochondria, and nucleus, respectively. We observed high expression levels of GSTP in the ER fraction. The relative expression levels of GSTP shown in Figure 3A for cytosol, ER, and mitochondria were 69%, 23%, and 8%, respectively. Other proteins that participate in the S-glutathionylation cycle, glutaredoxin and thioredoxin (Grx and Trx; Fig. 3A), also localized to the ER. Coimmunoprecipitation studies, where IP was enacted with ER-resident protein antibodies followed by blotting for GSTP, confirmed that GSTP interacts with BiP, calnexin, calreticulin, and PDI (Fig. 3B and Supplementary Fig. S2). To ensure accurate pull down of the ER proteins of interest, under each condition, we blotted for BiP, calnexin, calreticulin, or PDI to ensure quality control. With respect to the variable GSTP levels between the two IP samples, the affinity of GSTP for these ER-resident proteins was altered by S-glutathionylation. The first is liver lysate without disulfiram and the second with disulfiram treatment, each followed by IP with ER protein antibody of interest (BiP, calnexin, calreticulin, or PDI). In our second approach, we carried out immunofluorescence staining of GSTP and the ER-resident proteins in Gstp1/p2+/+ mouse embryo fibroblast (MEF) cells. Figure 3C shows proportional codistribution of GSTP with ER-resident proteins BiP, calnexin, and PDI. The results of fluorescence colocalization studies are also represented graphically in scatterplots where the intensity of one color is plotted against the intensity of the second for each pixel. The points of the scatterplot cluster for each of two probes (GSTP with PDI, BiP, or calnexin) around a straight line reflects the colocalization of GSTP with PDI, BiP, or calnexin. In addition, the degree of colocalization of the two probes for each pair was quantified by Pearson's correlation coefficient and Manders colocalization coefficient (8) (Fig. 3D). A higher degree for colocalization of GSTP with PDI was found with Pearson's coefficient 0.488 ± 0.100, compared to BiP and calnexin (0.285 ± 0.060 and 0.367 ± 0.012); similar values were shown using Manders coefficients (Fig. 3E). Such results are consistent with the concept that GSTP forms a protein complex with these resident proteins and could catalyze their GSTP-mediated S-glutathionylation in the ER compartment.
FIG. 3.
Colocalization and interaction of GSTP with ER-resident proteins. (A) Fresh liver tissue samples from Gstp1/p2+/+ and Gstp1/p2−/− mice were homogenized and subjected to subcellular fractionation by differential centrifugation and subsequent density gradient centrifugation. Each fraction was analyzed by SDS-PAGE and for the presence of specific organelle markers: (ER: SERCA2 and calnexin), (cytoplasmic: GAPDH), (nuclear: Histone H3), and (mitochondria: SDHA). (B) One milligram of liver lysates untreated or treated with disulfiram (10 μM, for 30 min at 37°C) was used for IP with BiP, calnexin, calreticulin, and PDI antibodies. Samples were analyzed by SDS-PAGE with the IgG controls and blotted for GSTP. Under each condition, we blotted for BiP, calnexin, calreticulin, or PDI to ensure quality control. (C) MEF cells from Gstp1/p2+/+ mice were fixed in 4% formaldehyde, incubated with GSTP and calnexin, PDI, or BiP antibodies at 4°C overnight, and then secondary antibodies conjugated with Fluor 488 or 594 were applied. No staining was detected in the absence of primary antibodies. Cells were imaged on an Olympus FV10i laser scanning confocal microscope. (D) Colocalization analyses of GSTP with PDI, BiP, or calnexin. Scatterplots represent the red and green pixel intensities of the corresponding two probes (GSTP with PDI, BiP, or calnexin) shown in (C). (E) Bar graphs represent the Pearson's coefficients and Manders M1 and M2 coefficients of the corresponding two probes from 30 single cells, analyzed by Fiji. MEF, mouse embryo fibroblast; SDHA, succinate dehydrogenase A. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Protective role of GSTP in ER stress caused by ThG or TuM
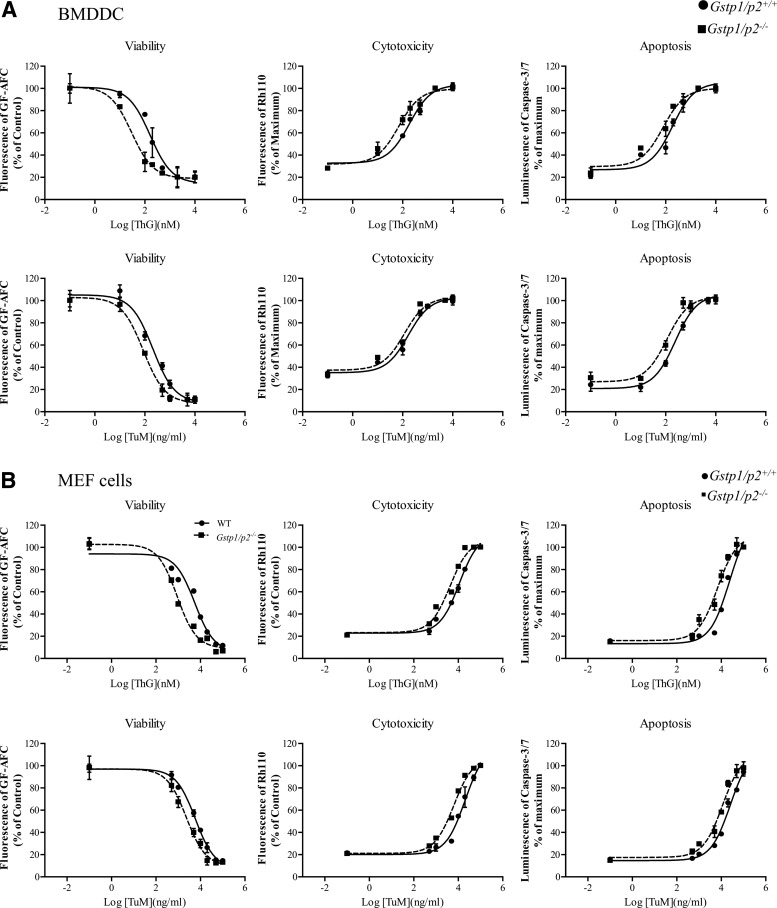
Since BiP, calnexin, calreticulin, endoplasmin, PDI, and SERCA are involved in the ER stress response, either through protein folding or through regulation of calcium homeostasis (Fig. 1), we assessed whether cell responses to either ThG or TuM were impacted by the presence or absence of GSTP. Using either MEF cells or bone marrow-derived dendritic cells (BMDDC) derived from Gstp1/p2+/+ and Gstp1/p2−/− animals, the viability, cytotoxicity, and levels of induced apoptosis were evaluated. As shown in Figure 4, both ThG (Fig. 4A) and TuM (Fig. 4B) elicited dose-dependent decreases in viability and increased cytotoxicity with caspase-3/7 activation. To evaluate the survival differences between Gstp1/p2+/+ and Gstp1/p2−/− cells, half-maximal effective concentrations were calculated (Table 2). Each of the drugs was more toxic to the cells lacking GSTP (Gstp1/p2−/−) (except for one case for TuM; see Table 2), and differences were generally more pronounced for ThG than for TuM. Generally, BMDDC were more sensitive to the drugs than MEF cells. Different levels of Gstp1/p2 expression were found in BMDDC compared to MEF cells (Supplementary Fig. S3). At maximal, an approximate sevenfold difference in sensitivity was shown for viability enacted by ThG.
FIG. 4.
Cytotoxicity of ThG or TuM in Gstp1/p2+/+ and Gstp1/p2−/− cells. BMDDC (A) and MEF (B) cells derived from Gstp1/p2+/+ or Gstp1/p2−/− mice were treated with drugs for 16 h at the concentrations indicated. Viability, cytotoxicity, and apoptosis were measured as described in “Materials and Methods.” Data are presented as % of control for viability or % of maximum effect for toxicity and apoptosis. Each drug concentration was measured in triplicate and three independent experiments were conducted. Means ± SDs were computed for each group. BMDDC, bone marrow-derived dendritic cells; ThG, thapsigargin; TuM, tunicamycin.
Table 2.
Comparative Toxicities of Thapsigargin or Tunicamycin in Bone Marrow-Derived Dendritic Cell and Mouse Embryo Fibroblast Cells From Either Gstp1/p2+/+ or Gstp1/p2−/− Mice
| Viability | Cytotoxicity | Apoptosis | ||||
|---|---|---|---|---|---|---|
| Cells | Thapsigargin (μM) | Tunicamycin (μg/ml) | Thapsigargin (μM) | Tunicamycin (μg/ml) | Thapsigargin (μM) | Tunicamycin (μg/ml) |
| Gstp1/p2+/+ BMDDC | 0.122 ± 0.028 | 0.209 ± 0.006 | 0.220 ± 0.037 | 0.150 ± 0.025 | 0. 215 ± 0.023 | 0.254 ± 0.009 |
| Gstp1/p2−/− BMDDC | 0.017 ± 0.001 | 0.092 ± 0.011 | 0.054 ± 0.02 | 0.119 ± 0.001a | 0.083 ± 0.01 | 0.144 ± 0.004 |
| Gstp1/p2+/+ MEF | 5.55 ± 0.26 | 5.87 ± 0.32 | 11.05 ± 0.23 | 20.55 ± 3.42 | 23.37 ± 0.56 | 25.06 ± 2.89 |
| Gstp1/p2−/− MEF | 0.904 ± 0.043 | 2.333 ± 0.112 | 4.27 ± 0.01 | 6.34 ± 0.13 | 6.80 ± 0.64 | 11.33 ± 0.46 |
The EC50 values for viability, toxicity, and apoptosis were simulated by software GraphPad Prism 6 (GraphPad Prism, Inc., La Jolla, CA).
Except for cytotoxicity effect by tunicamycin. In all other cases, the EC50 values between Gstp1/p2+/+ and Gstp1/p2−/− were significantly different, p < 0.01.
BMDDC, bone marrow-derived dendritic cells; EC50, half-maximal effective concentrations; MEF, mouse embryo fibroblast.
BMDDC and MEF cells from Gstp1/p2−/− mice are more susceptible to UPR following ThG or TuM
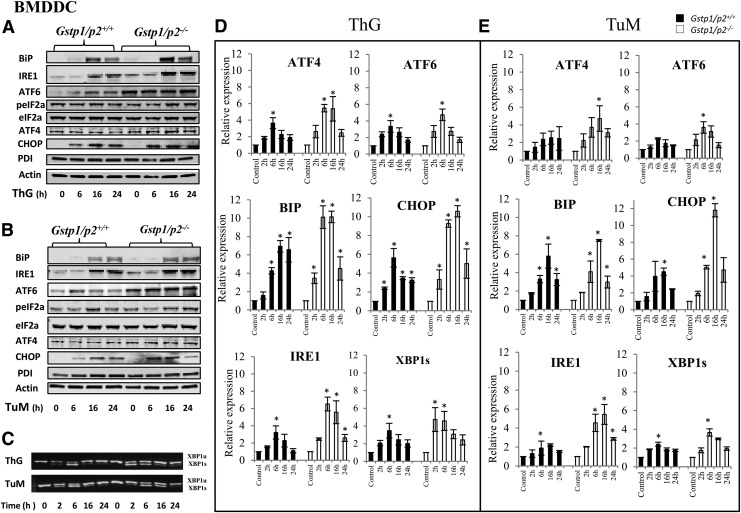
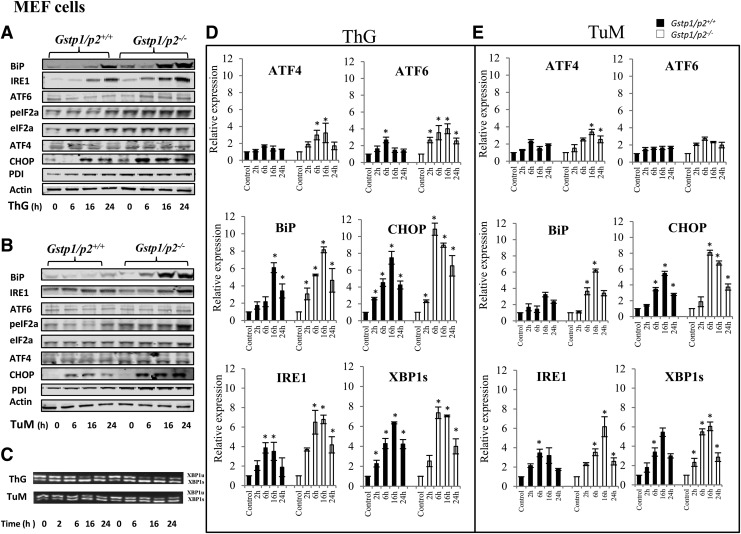
To understand these differential sensitivities between Gstp1/p2+/+ and Gstp1/p2−/− cells, we measured UPR gene expression (real-time polymerase chain reaction [PCR]) and protein levels (immunoblots) following treatments with either ThG or TuM in both BMDDC (Fig. 5 and Supplementary Fig. S4) and MEF cells (Fig. 6 and Supplementary Fig. S5). In these studies, two upstream proteins (BiP and PDI) and three downstream sensors of the UPR were evaluated (see Fig. 1, including IRE1, ATF6, and PERK pathways).
FIG. 5.
Drug-induced changes in UPR gene expression after ThG or TuM in BMDDC. Gstp1/p2+/+ and Gstp1/p2−/− BMDDC were treated with ThG (200 nM) or TuM (500 μg/ml) for certain times as indicated. Proteins were separated by SDS-PAGE and UPR proteins, including BiP, IRE1, ATF6, phospho-eIF2α, eIF2α, ATF4, PDI, and CHOP were evaluated by immunoblots. Even loading of proteins was confirmed by probing with anti-actin antibodies. Goat anti-mouse, goat anti-rabbit, and donkey anti-goat fluorescent secondary antibodies were used and immunoblots imaged on a two-channel IR fluorescent Odyssey CLx imaging system. Data are representative blots from three independent experiments (A, B). The activation of XBP1 was evaluated by normal PCR for unsliced and sliced XBP1(C). Relative gene expression levels were quantified by real-time-PCR. Bars represent the mean (±SD) from three independent experiments (D, E). *Represents significant difference between control and treatment by UPR inducer. IR, infrared; PCR, polymerase chain reaction; ThG, thapasigargin; TuM, tunicamycin.
FIG. 6.
Drug-induced changes in UPR gene expression after ThG or TuM in MEFs. Gstp1/p2+/+ and Gstp1/p2−/− MEF cells were treated with ThG (200 nM) or TuM (500 μg/ml) for certain times as indicated. Proteins were separated by SDS-PAGE and UPR proteins, including BiP, IRE1, ATF6, phosphor-eIF2α, eIF2α, ATF4, PDI, and CHOP evaluated by immunoblots. Even loading of proteins was confirmed by probing with anti-actin antibodies. Goat anti-mouse, goat anti-rabbit, and donkey anti-goat fluorescent secondary antibodies were used and immunoblots imaged on a two-channel IR fluorescent Odyssey CLx imaging system. Data are representative blots from three independent experiments (A, B). The activation of XBP1 was evaluated by normal PCR for unsliced and sliced XBP1(C). Relative gene expression levels were quantified by real-time PCR. Bars represent the mean (±SD) from three independent experiments (D, E). *Represents significant difference between control and treatment by UPR inducer.
In general, drug treatments produced a coordinated increase in most UPR proteins, except for PDI and phospho-eIF2a in BMDDC and PDI, ATF4, and ATF6 in MEF cells. Significant induction of BiP, IRE1, and CHOP was found in both BMDDC and MEF cells. For some UPR genes, response to drugs differed in distinct cell types, for example, ATF4, ATF6, and eIF2a (Figs. 5A, B, and 6A, B, and Supplementary Figs. S4 and S5). The basal levels of ATF4 and ATF6 were higher in Gstp1/p2−/− BMDDC, while the basal levels of PDI and phospho-eIF2a were higher in Gstp1/p2−/− MEF cells, implying that perhaps the stress levels were influenced by the cell type under study, but that the absence of GSTP is generally accompanied by constitutively enhanced ER stress. Consistent with the cell viability, cytotoxicity, and apoptosis assays, drug treatments induced stronger and faster UPRs for the majority of the UPR markers in Gstp1/p2−/− cells, for example, BiP, IRE1, ATF4, ATF6, and CHOP in BMDDC and BiP, IRE1, CHOP, and phospho-eIF2α in MEF cells. The effect of GSTP on UPR induced by ThG or TuM showed different profiles. ThG, as a SERCA inhibitor, was more effective in Gstp1/p2−/− cells. While in some instances, TuM showed no differences, for example, BiP and IRE1 in BMDDC and IRE1 in MEF cells.
The mRNA levels of UPR genes of interest were assessed by quantitative reverse-transcription-PCR in a time-dependent manner. Taking into account the protein turnover times, earlier time points for UPR genes at 2 h were also assessed. The spliced active form of XBP1 was also induced in both cell models (Figs. 5D and 6D). Results are similar to those found for proteins. Perhaps as a consequence of the different protein half-lives, the protein and RNA results did not always agree. Furthermore, inactive XBP1 and spliced XBP1 following ThG or TuM treatment were subject to normal PCR in both cell models (Figs. 5C and 6C). The data were essentially similar to those found by immunoblot in that responses were more pronounced in cells lacking GSTP.
Ca2+ localization and mobilization induced by ThG through GSTP-mediated S-glutathionylation
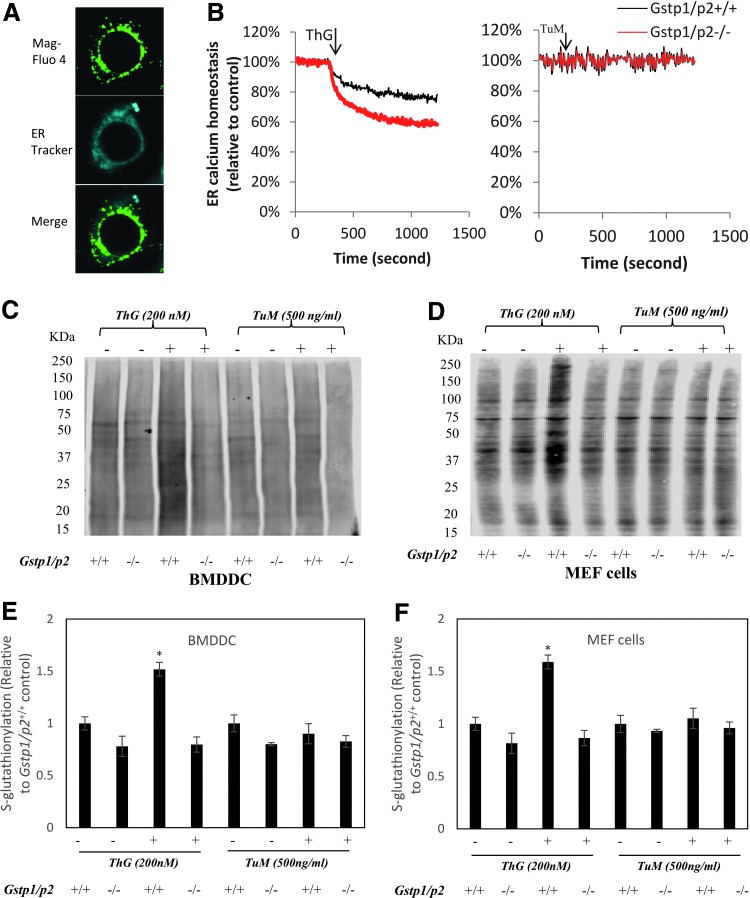
While both ThG and TuM induced ER stress, ThG produced more pronounced effects in Gstp1/p2−/− compared to Gstp1/p2+/+. Because ThG is a SERCA inhibitor, we hypothesized that the large difference in sensitivity to ThG between Gstp1/p2+/+ and Gstp1/p2−/− cells may be linked with altered ER calcium flux and/or homeostasis (Fig. 6). The ER-specific dye, Mag-Fluo 4 acetoxymethyl (AM) esters (49), was used as the calcium indicator in the ER. In practice, intact cells are first incubated with the Mag-fluo-4 AM ester that accumulates in intracellular organelles and the cytoplasm. The plasma membrane of the loaded cells is then permeabilized to release cytosolic indicator by saponin, leaving only the indicator trapped within the ER, confirmed by the ER-Tracker™ blue-white DPX dye (Fig. 7A). The arrow in Figure 7B indicates the addition point of ThG or TuM, after which ER calcium depletion was significantly decreased in Gstp1p2−/− BMDDC by ThG, but not TuM (Fig. 7B). The sensitivity of detection of protein S-glutathionylation by immunoblot is limited. A modified protocol using deglutathionylation catalyzed by Grx was used for the purpose of quantitative comparison. Following deglutathionylation by Grx, proteins were conjugated with either Maleimide-PEG11-Biotin (Fig. 7C, D) or thiol fluorescent probe IV (Fig. 7E, F). This allowed quantitative comparison between glutathionylation levels affected by either ThG or TuM. Each approach showed elevation of GSTP-mediated S-glutathionylation of cellular proteins in both BMDDC and MEF cells caused by ThG, but not TuM (Fig. 7C–F).
FIG. 7.
BMDDC were incubated with a low-affinity calcium indicator, 5 μM mag-fluo-4 at 37°C for 45 min or ER-Tracker™ blue-white DPX for 30 min at 1 μM. Then cells were washed with PBS twice to remove extracellular indicator, permeabilized by saponin (10 mg/ml), and incubated at 37°C for 5 min with shaking (permeabilization of cells was confirmed using trypan blue). Permeabilized cells were either fixed in 4% formaldehyde for fluorescence microscopy (A) or centrifuged and resuspended at a density of 1 million/ml. Two hundred thousand permeabilized cells were added to 96-well black-walled assay plate and ER calcium concentrations were subsequently monitored by fluorescence measurements at 490Ex/520Em by a SpectraMax M5 Multi-Mode Microplate Reader. ThG or TuM was added at the arrow (B). Global S-glutathionylation levels induced by ThG or TuM in BMDDC or MEF cells were quantified in two ways after deglutathionylation by glutaredoxin (see “Materials and Methods”). The regenerated free thiols were quantified either by immunoblotting using streptavidin secondary antibodies after labeling with maleimide-PEG11-Biotin (C, D) or directly by thiol fluorescent probe IV (E, F). *Represents significant difference between control and treatment by ThG. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Discussion
Modulation of redox homeostasis can directly influence the fate of a mammalian cell. Under oxidative stress, select cysteine residues in target proteins can undergo reversible modifications that alter structure/function and facilitate signaling pathways (11). The difference between too much and too little ROS will determine the fate of many pathways critical to survival (16). Many of these pathways are evolutionarily well conserved and cysteine is one of the least prevalent amino acids encoded by the human genome (25), where total cellular cysteine content correlates with the degree of biological complexity (32). The thiol group of cysteine permits a number of post-translational modifications (17), S-glutathionylation being favored within the oxidized environment of the ER (oxidation of cysteines to sulfenic acids (SOH) would facilitate protein S-glutathionylation). GSTP has been previously identified in three cellular compartments, the cytosol, nucleus, and mitochondria (14, 18, 26). Our present results show for the first time that it also localizes to the ER. GSTP has also been shown to be a substrate for glycosylation (29) and S-glutathionylation (52). Within the ER, we now show that GSTP interacts with and S-glutathionylates a cluster of proteins that between them contribute to the regulation of cell response to ER stress and impact downstream events in protein folding. Moreover, Grx and Trx, as deglutathionylating enzymes also localize to the ER, indicating that participants in the S-glutathionylation cycle (including GSH and oxidized GSH [GSSG]) are present in this organelle (9, 33). By comparing protein S-glutathionylation in cells from Gstp1/p2 wild-type and knockout mice, we were able to identify differences in a cluster of redox-active proteins found in the ER. In light of the different susceptibilities of these cells to various aspects of the UPR, their activities are likely controlled by S-glutathionylation. Among those, BiP, calnexin, calreticulin, endoplasmin, SERCA, and PDI have organelle-specific functions in controlling protein folding and the UPR. We have previously identified the modified cysteines in PDI and detailed the inhibitory effects of their S-glutathionylation on isomerase and chaperone activity (54, 57). This susceptible CxxC motif in PDI has structural homology with calnexin (30), which in turn is part of the calreticulin superfamily that has these two cysteines in similar beta sheet structures at molecular distances of ∼1.5 Å, a distance quite conducive to disulfide formation. The cysteine residues within calnexin are known to be critical in maintaining the structure and function of the protein (5). Earlier reports indicate that both calnexin and calreticulin are sensitive to disulfide-bonding during oxidative stress (6). S-glutathionylation of one of these residues impacts activity through inhibiting disulfide bond formation. In addition, S-glutathionylation of SERCA has been shown to regulate its function (2). The concentration of ER Ca is mainly regulated by SERCA. GSTP mediates SERCA S-glutathionylation, regulates SERCA activity, and thus affects ER Ca levels. Also, a number of chaperones such as calreticulin and calnexin bind Ca buffering in the ER (13). It is possible that GSTP-mediated S-glutathionylation of calreticulin and calnexin might regulate their Ca binding capacities, impacting Ca levels in the ER. It is known that ER Ca depletion induces the UPR as both the protein folding reactions and protein chaperone functions require high levels of Ca within this organelle. Therefore, GSTP might impact the UPR through regulating ER Ca levels through mediating S-glutathionylation of SERCA, calreticulin, and calnexin. It is also possible that GSTP-mediated S-glutathionylation of calreticulin and calnexin might alter their interactions with glycoproteins, directly regulating glycoprotein folding and thereby alter the UPR.
Although there is a degree of redundancy within the GST family, GSTP is the most highly expressed isozyme in MEF cells and BMDDC and influences the doubling time of these cells (39, 61) and remains the only isoform to exhibit thiolase activity (28, 51, 52). Our present results show a significant difference in levels of innate resistance of the Gstp1/p2+/+ and Gstp1/p2−/− cells to either of the UPR inducing drugs, ThG or TuM. While treatment of cells with either of these drugs produced ER stress and death via UPR-associated pathways, the mechanisms of action of these two agents are distinct. ThG is a sesquiterpene that inhibits the ER Ca2+ ATPase (SERCA), causing an initial depletion of intracellular Ca2+ stores and capacitive cellular uptake of extracellular Ca2+ (37). The Gstp1/p2+/+ BMDDC and MEF cells showed an approximate seven- to ninefold resistance to the drug compared with Gstp1/p2−/− cells, levels that correlate with greater sensitivity to induction of the UPR. The implication of such results is that drug response is facilitated through some component(s) of the SERCA Ca2+ transport pathways and that these are linked to ER homeostasis. Earlier studies in prostate cancer cell lines suggested that ThG caused cell death through altered Ca2+ flux that was associated with downstream changes in the transcriptional regulation of apoptosis inducing genes (48). Our results indicate a link with S-glutathionylation of proteins involved in Ca2+ homeostasis. In particular, SERCA, calnexin, calreticulin, and endoplasmin were each differentially post-translationally modified in the GSTP Gstp1/p2+/+ cells. The higher levels of S-glutathionylation of these proteins in Gstp1/p2+/+ cells may initially serve to protect their functionalities, while maintaining ER Ca2+ homeostasis, preventing UPR and providing a survival advantage, which would explain the quite marked levels of intrinsic resistance of the Gstp1/p2+/+ cells to ThG. The protective effects of GSTP expression (S-glutathionylation) observed following ThG treatment are significant because the propagation of signal transduction pathways via oscillations in cytosolic free Ca2+ concentrations is driven by the release of Ca2+ from the ER. It is also important to recognize that this degree of natural resistance is high for cells that have never previously been exposed to drug selection.
TuM is also cytotoxic through initiation of a UPR, but primarily through interference with the process of N-linked glycosylation of proteins. The Gstp1/p2+/+ cells express approximately two- to sixfold innate resistance to this drug. While quantitatively less than ThG, the fact that resistance is still expressed by the Gstp1/p2+/+ cells would imply that the effect may be UPR related, but not necessarily directly linked with SERCA and Ca2+ flux changes. The earlier studies did not identify either the functional importance or the type of the glycosylation of GSTP, but it is possible that TuM might have a direct effect upon the transfer of GSTP from the cytosol to the ER, perhaps impacting S-glutathionylation efficiency and response to drug. Deletion of the chaperone domain of calnexin increased sensitivity to TuM (7), suggesting that calnexin may be a sentinel protein in regulating sensitivity to TuM. Moreover, ER calcium depletion could cause ripple effects to manifest other stresses such as decreased protein glycosylation (31). Thus, the observed differences in sensitivity to ThG and TuM likely involve both distinct and/or overlapping functionalities.
From the point of view of functional relevance, dendritic cells (DC) play a role in the initiation, maintenance, resolution of an immune response acting as a bridge from the innate to adaptive immune systems. Several UPR genes have been found to play a role in the development and survival of DC. These include ER stress-induced transcription factor CHOP, crucial for BMDDC IL-23 expression (12), and XBP1, essential for survival of plasmacytoid and conventional DC (23). Activation of the UPR is not limited to an overload in protein folding capacity and, in fact, alternative ways to trigger ER stress sensors exist but are not well understood (24, 62). In vivo studies show that constitutive activation of the IRE-1 pathways is present in CD8α+ DC as well as developing B and T cells (34). In the present study, we show that markers of the UPR are activated in nonstressed BMDDC derived from Gstp1/p2-deficient mice. These findings support our previous studies showing that ablation of GSTP causes altered redox homeostasis and higher levels of both myeloproliferation and cell migration (60, 61). The co-opting of UPR in the absence of ER stress remains speculative but it appears that GSTP expression and UPR collectively play a role in the development and differentiation of DC.
Emerging evidence suggests that disruption in redox homeostasis can link with aberrant UPR and causally contribute to human disease pathologies. The ER has physical and functional connections with other organelles that can expedite the bidirectional transfer of lipids, calcium, and other molecules (41). In consequence, disruptions in redox signaling through alterations in levels of ROS or reactive nitrogen species can resonate back to the ER (illustrated in Fig. 1). Our present data show that GSTP and its mediation of specific S-glutathionylation of ER-resident proteins can act in a regulatory manner, impacting UPR pathways and sensitivity to drug and stress response.
Materials and Methods
Mice
C57BL/6 wild-type mice were purchased from Jackson Laboratory (Bar Harbor, ME). Gstp1/p2−/− mice were generated as described earlier (20). The mice were bred and kept in the Association for Assessment and Accreditation of Laboratory Animal Care—certified animal facility of the Medical University of South Carolina (MUSC). All of the mice were used at ∼8–12 weeks of age. The Institutional Animal Care and Use Committee of MUSC approved all of the experimental procedures used in this study.
Primary cells and culture conditions
BMDDC were generated according to previous reported procedures (61). Bone marrow cells (4–5 × 105/ml, 10 ml/plate) were plated in RPMI-1640 (HyClone, Logan, UT), supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin (all from Mediatech, Manassas, VA), and 20 ng/ml recombinant mouse granulocyte macrophage colony-stimulating factor (GM-CSF) (BioAbChem, Ladson, SC) (DC medium) into 100-mm culture dishes (Sarstedt, Newton, NC). Fresh DC medium was added on day 4 and was gently replaced by fresh DC medium containing 10 ng/ml recombinant mouse GM-CSF on day 7. Immature BMDDC (nonadherent and loosely adherent cells) were used in experiments on day 8.
Extraction of the MEF cells from Gstp1/p2+/+ or Gstp1/p2−/− mice and the establishment of immortalized cultures were described previously (39). Cells were maintained in Eagle's Minimum Essential Medium (EMEM) (ATCC, Manassas, VA) containing 10% FBS, 1% nonessential amino acid, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Antibodies
The following antibodies were used for immunoblots: rabbit polyclonal anti-ATF4 (from Aviva Systems Biology, San Diego, CA), mouse monoclonal anti-ATF6 (from Stratagene, La Jolla, CA), goat polyclonal anti-BiP (from R&D System, Minneapolis, MN), rabbit polyclonal anti-calnexin and rabbit polyclonal anti-Trx 1 (Novus Biologicals, Littleton, CO), rabbit polyclonal anti-GSTP (MBL, Woburn, MA), mouse monoclonal anti-PSSG (Virogen, Watertown, MA), mouse monoclonal anti-SERCA2 (from Santa Cruz Biotechnology, Dallas, TX), rabbit polyclonal anti-BiP, chicken polyclonal anti-calreticulin, rabbit polyclonal anti-Histone H3, mouse monoclonal anti-PDI (all from Thermo Scientific, Rockford, IL), rabbit polyclonal anti-beta actin, goat polyclonal anti-endoplasmin, mouse monoclonal anti-GAPDH, rabbit polyclonal to Grx 1, rabbit polyclonal anti-PDI (all from Abcam, Cambridge, MA), rabbit polyclonal anti-BiP, mouse monoclonal anti-CHOP, mouse monoclonal anti-eIF2α, rabbit polyclonal anti-phospho-eIF2α (Ser51), rabbit monoclonal anti-IRE1, rabbit monoclonal anti-PDI, rabbit monoclonal anti-SDHA, rabbit monoclonal anti-SERCA2 (all from Cell Signaling Technology), IRDye 800CW Goat anti-Mouse immunoglobulin G (IgG), IRDye 800CW Goat anti-rabbit IgG, IRDye 800CW Donkey anti-goat IgG, IRDye 800CW Donkey anti-chicken IgG, IRDye 800CW Streptavidin, IRDye 680RD Goat anti-mouse IgG, and IRDye 680RD Goat anti-Rabbit IgG, IRDye 680RD Donkey anti-goat IgG (all from LI-COR, Lincoln, NE).
Immunoblotting
Total soluble protein was quantitated by bicinchoninic acid protein assay (Pierce, Rockford, IL). Cell lysates were resolved in an SDS-loading buffer (80 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.02% bromophenol blue, [±]5 mM tris (2-carboxyethyl) phosphine) and heated to 95°C for 5 min. Equal amounts of protein were electrophoretically separated by SDS-PAGE (BioRad, Hercules, CA) and transferred onto low fluorescent polyvinylidene fluoride membranes (Millipore, Billerica, MA) or nitrocellulose membranes (BioRad, Hercules, CA) by the Trans-Blot Turbo Transfer System (BioRad, Hercules, CA). PVDF or nitrocellulose membranes were incubated in the Odyssey blocking buffer (LI-COR) for 1 h to reduce nonspecific binding and then probed with appropriate primary antibodies (diluted in Odyssey blocking buffer) at 4°C overnight. Immunoblots were then developed with infrared (IR) fluorescence IRDye secondary antibodies (LI-COR) at a dilution of 1:15,000, imaged with a two-channel (red and green) IR fluorescent Odyssey CLx imaging system (LI-COR) and quantified with Image Studio 4.0 software (LI-COR).
Identification of GSTP-mediated S-glutathionylated proteins
Fresh liver tissue samples from Gstp1/p2+/+ and Gstp1/p2−/− mice were washed twice with ice-cold phosphate-buffered saline (PBS), cut into small pieces, and homogenized in ice-cold 1% NP-40 lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 1 mM ethylenediaminetetraaceticacid [EDTA]) plus protease and phosphatase inhibitors (Roche Diagnostics, Indianapolis, IN). Liver lysates/supernatants were collected after spinning at 21,100 g for 10 min at 4°C. Thirty micrograms of control or disulfiram-treated (10 μM, 30 min at 37°C, used as a positive control for S-glutathionylation, from Sigma-Aldrich, St. Louis, MO) liver lysates was then separated by SDS-PAGE under nonreducing conditions. One gel was transferred onto nitrocellulose membrane for anti-PSSG Western blot and the other gel was stained with Brilliant Blue G-colloidal stain (Sigma-Aldrich) following the standard protocol. Protein bands of interest (those that showed differences in S-glutathionylation levels between Gstp1p2+/+ and Gstp1p2−/−) were excised and subject to digestion. The peptides were analyzed and identified by mass spectrometry at the Proteomics Core Facility of the MUSC.
Detection of S-glutathionylated proteins
One milligram of control or disulfiram-treated (10 μM, 30 min at 37°C) liver lysate from Gstp1/p2+/+ and Gstp1/p2−/− mice was incubated with 5 μg mouse anti-PSSG antibody overnight at 4°C. The antibody–antigen complexes were then IP by incubating with 40 μl protein A/G-agarose beads (Santa Cruz Biotechnology) overnight at 4°C. The flow-throughs were collected by brief centrifugation (500 g × 5 min) and 20 μg of protein was resolved in the SDS-loading buffer. Immunoprecipitates were washed three times with ice-cold 1% NP-40 lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 1 mM ethylenediaminetetraacetic acid (EDTA)] to remove nonspecific bound proteins and the bound immunoprecipitates were solubilized in the SDS-loading buffer. The flow-throughs and the immunoprecipitates were then subjected to SDS-PAGE and probing of the immunoblots with goat anti-BiP, rabbit anti-calnexin, chicken anti-calreticulin, goat anti-endoplasmin, rabbit anti-PDI, and rabbit-SERCA2 antibodies. As a reagent control, liver lysates were incubated with 5 μg mouse IgG2a isotype control (Thermo Scientific) and subjected to the same procedures.
The global S-glutathionylation levels induced by ThG or TuM in BMDDC or MEF cells were quantified in two ways after deglutathionylation by Grx as reported (1) with some modifications. Briefly, Gstp1/p2+/+ and Gstp1/p2−/− BMDDC (1 × 106 cells/ml) were suspended in the RPMI-1640 medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. MEF cells (1 × 106 cells) were seeded on 100-mm culture dishes in 10 ml EMEM containing 10% FBS, 1% nonessential amino acid, 100 U/ml penicillin, and 100 μg/ml streptomycin per dish, and cultured for 2 days. Cells were then exposed to 200 nM ThG or 500 ng/ml TuM (both from Sigma-Aldrich) at 37°C for 30 min. Cell pellets were washed once with ice-cold PBS and then solubilized by ice-cold 1% Triton lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton, 1 mM ethylenediaminetetraacetic acid EDTA, 1 mM ethylene glycol tetraacetic acid [EGTA]) plus protease and phosphatase inhibitors. Cell lysates/supernatants were collected after spinning at 16,000 g for 10 min at 4°C. Free thiols were blocked by adding 40 mM N-ethylmaleimide for 30 min, and the glutathionylated thiols were then deglutathionylated by adding 1 mM GSH, 1 mM NADPH, 35 μg/ml GSSG reductase, and 13.5 μg/ml Grx and incubated at 37°C for 30 min. Excess reagents were removed by Bio-Spin 6 columns (BioRad, Hercules, CA) following the manufacturer's protocol. The regenerated free thiols were quantified either directly by thiol fluorescent probe IV (EMD Millipore, Taunton, MA) or by western blotting using the streptavidin secondary antibody (LI-COR) after labeling with Maleimide-PEG11-Biotin (Thermo Scientific).
Coimmunoprecipitation analysis of protein–protein interaction
The IP procedure was similar as mentioned earlier. Briefly, 500 μg of control or disulfiram-treated (10 μM, 30 min at 37°C) liver lysate from Gstp1/p2+/+ and Gstp1/p2−/− mice was immunoprecipitated using Bip, calnexin, calreticulin, or PDI antibody, and the precipitated proteins were subjected to SDS-PAGE and subsequent immunoblotting for GSTP. The IP efficiencies were confirmed by immunoblotting for BiP, calnexin, calreticulin, or PDI antibody as well. As a reagent control, liver lysates were incubated with the corresponding IgG isotype controls and subjected to the same procedures.
Immunofluorescence
MEF cells were fixed in 4% formaldehyde and immunofluorescence staining applied according to previous reported protocols (59). After fixation, the cells were incubated with GSTP, calnexin, PDI, or BiP antibody at 4°C overnight, and then, a secondary antibody conjugated with Fluor 488 or 594 was applied. No staining was detected when the primary antibodies were omitted. Slides were imaged on an Olympus FV10i laser scanning confocal microscope (Olympus, Tokyo, Japan).
Subcellular fractionation
Fresh liver tissue samples from Gstp1/p2+/+ and Gstp1/p2−/− mice were homogenized and subjected to subcellular fractionation by differential centrifugation and subsequent density gradient centrifugation. In brief, liver tissue samples were washed twice with ice-cold PBS, cut into small pieces, resuspended in ice-cold homogenizing buffer (10 mM HEPES [pH 7.5], 250 mM sucrose, 1 mM EDTA, 25 mM KCl) plus a protease inhibitor cocktail, and homogenized with Wheaton glass tissue grinder. The homogenate was passed through a 40-μm cell strainer and centrifuged at 1000 g for 10 min at 4°C. The nuclear pellet was washed once with ice-cold homogenizing buffer, resuspended in 60% (w/v) sucrose in homogenizing buffer, and purified by sucrose density gradient centrifugation (100,000 g for 60 min 4°C). The postnuclear supernatant was centrifuged at 12,000 g for 15 min at 4°C. The mitochondrial pellet was washed once, resuspended in homogenizing buffer, layered over two solutions of 20% and 52% Percoll in homogenizing buffer, and purified by Percoll density gradient centrifugation (37,000 g for 30 min at 4°C, mitochondrial band at the lower interface). The postmitochondrial supernatant was centrifuged at 100,000 g for 60 min at 4°C. The supernatant was used as the cytoplasmic fraction and the pellet as the ER fraction. The ER, mitochondrial and nuclear pellets were solubilized in ice-cold 1% Triton lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton, 1 mM EDTA, 1 mM EGTA) plus a protease inhibitor cocktail and sonicated three times for 20 s. The lysates/supernatants were collected after spinning at 16,000 g for 10 min at 4°C and used for immunoblotting.
ApoTox-Glo triplex assay
BMDDC were seeded on 96-well plates at a density of 20,000 cells in 100 μL DC medium per well. MEF cells (5000 cells) were seeded on 96-well plates in 100 μl EMEM per well and cultured for 2 days. Increasing drug concentrations (ThG: 1–2000 nM; TuM: 1–5000 ng/ml) were added to a final volume of 200 μl and the plates incubated at 37°C for 16 h. The ApoTox-Glo Triplex Assay (Promega, Madison, WI) was used to measure cell viability, cytotoxicity, and apoptosis. Briefly, viability and cytotoxicity were measured by fluorescent signals produced when either live or dead cell proteases cleave added substrates glycyl-phenylalanyl-aminofluorocoumarin (GF-AFC) (viability) and bis-alanylalanyl-phenylalanyl-rhodamine 110 (bis-AAF-R110) (cytotoxicity). Proportional fluorescence of the cleaved products distinguishes the two. GF-AFC can enter cells and is only cleavable by live-cell proteases, which becomes inactive when cell membrane activity is lost; bis-AAF-R110 cannot enter the cell and is cleaved only by dead-cell proteases leaked from cells lacking membrane integrity. Each cleaved substrate has a distinct excitation and emission spectrum. Apoptosis is measured by the addition of a luminogenic caspase-3/7 substrate, which is cleaved in apoptotic cells to produce a luminescent signal. Fluorescence (400Ex/505Em for viability, 485Ex/520Em for cytotoxicity) and luminescence (apoptosis) were measured with SpectraMax M5 Multi-Mode Microplate Readers (Molecular Devices, Sunnyvale, CA). The IC50 values for viability, toxicity, and apoptosis were simulated by software GraphPad Prism 6 (GraphPad Prism, Inc., La Jolla, CA).
RNA isolation and quantitative real-time reverse transcription-PCR
Total RNA was prepared using the Isolate II RNA Mini Kit (Bioline USA, Inc., Taunton, MA) and cDNA was then generated with the iScriptTM cDNA Synthesis Kit (Bio-Rad, Hercules, CA) according to the manufacturers' protocols. Subsequently, quantification of gene expression was performed in duplicates using iQ™ SYBR® Green supermix (Bio-Rad, Hercules, CA) with detection on a MyiQ™ Real-Time PCR System (Bio-Rad, Hercules, CA). The reaction cycles used were 95°C for 5 min, and then, 40 cycles at 95°C for 15 s and 58°C for 1 min followed by melt curve analysis. The following primers were used: ATF4, forward 5′-ACTCTGCTGCTTACATTACTC-3′, reverse 5′-TAGGACTCTGGGCTCATAC-3′; ATF6, forward 5′-GCTACCACCCACAACAAG-3′, reverse 5′-TTCATAGTCCTGCCCATT-3′; BiP, forward 5′-TGCAGCAGGACATCAAGTTC-3′, reverse 5′-ATGTCTTTGTTTGCCCACCT-3′; CHOP, forward 5′-ACCTTCACTACTCTTGACCCTG-3′, reverse 5′-GACCACTCTGTTTCCGTTT-3′; GAPDH, forward, 5′-CCCAGCAAGGACACTGAGCAA-3′, reverse 5′-AGGCCCCTCCTGTTATTATGG-3′; IRE1, forward 5′-GACGGTCCCACAACAGAT-3′, reverse 5′-CTCAGCAGACACTTTCCCT-3′; XBP1u, forward 5′-TCCGCAGCACTCAGACTATGT-3′, reverse 5′-ATGCCCAAAAGGATATCAGACTC-3′; XBP1s, forward 5′-GAGTCCGCAGCAGGTG-3′, reverse 5′-GTGTCAGAGTCCATGGGA-3′; and XBP1u/s, forward 5′-ACACGCTTGGGAATGGACAC-3′, reverse 5′-CCATGGGAAGATGTTCTGGG-3′. Relative gene expression quantification was based on the comparative threshold cycle (CT) method (2−ΔΔCt) with normalization of the raw data to the included housekeeping gene (GAPDH).
ER calcium assay
BMDDC were incubated with a low-affinity calcium indicator, Mag-Fluo-4 AM (5 μM; from Life Technology, Eugene, OR), at 37°C for 45 min. For the ER-Tracker™ blue-white DPX dye, incubation times were 30 min at 1 μM following the manufacturer's protocol (Life Technology). Cells were washed with PBS twice to remove extracellular indicator, permeabilized by adding saponin (10 mg/ml), and incubated at 37°C for 5 min with shaking. Then, permeabilized cells were either fixed in 4% formaldehyde for Olympus FV10i laser scanning confocal microscopy or centrifuged and resuspended at a density of 1 million/ml. Two hundred thousand permeabilized cells (200 μl per well) were added to 96-well black-walled assay plates for further analysis. Measuring the fluorescence at 490Ex/520Em by SpectraMax M5 Multi-Mode Microplate Readers monitored ER calcium concentrations.
Statistical analysis
Colocalization of proteins was analyzed by Fiji (http://fiji.sc/Documentation). Student's t tests were used to analyze significant differences. p values <0.01 were regarded as statistically significant. Data are expressed as means ± standard deviations with n equal to the number of animals/group examined under each condition.
Supplementary Material
Abbreviations Used
- AM
acetoxymethyl
- ATF4
activating transcription factor 4
- ATF6
activating transcription factor 6
- BiP
immunoglobulin heavy chain-binding protein
- bis-AAF-R110
bis-alanylalanyl-phenylalanyl-rhodamine 110
- BMDDC
bone marrow-derived dendritic cells
- CHOP
C/EBP homologous protein
- CT
threshold cycle
- DC
dendritic cells
- DTT
dithiothreitol
- EC50
half-maximal effective concentrations
- EDTA
ethylenediaminetetraacetic acid
- EGTA
ethylene glycol tetraacetic acid
- eIF2α
eukaryotic translation initiation factor 2α
- EMEM
Eagle's Minimum Essential Medium
- ER
endoplasmic reticulum
- F/T
flow-through
- FBS
fetal bovine serum
- GF-AFC
glycyl-phenylalanyl-aminofluorocoumarin
- GM-CSF
granulocyte macrophage colony-stimulating factor
- Grx
glutaredoxin
- GSH
glutathione
- GSSG
oxidized glutathione
- GSTP
glutathione S-transferase Pi
- IgG
immunoglobulin G
- IP
immunoprecipitation
- IR
infrared
- IRE1
inositol-requiring protein-1
- MEF
mouse embryo fibroblasts
- MUSC
Medical University of South Carolina
- NEM
N-ethylmaleimide
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PDI
protein disulfide isomerase
- PERK
protein kinase-like ER kinase
- PSSG
protein S-glutathionylation
- ROS
reactive oxygen species
- SDHA
succinate dehydrogenase A
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SERCA
sarco/endoplasmic reticulum Ca2+-ATPase
- ThG
thapsigargin
- Trx
thioredoxin
- TuM
tunicamycin
- UPR
unfolded protein response
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA08660, CA117259, NCRR P20RR024485—COBRE in Oxidants, Redox Balance and Stress Signaling) and support from the South Carolina Centers of Excellence program and was conducted in a facility constructed with the support from the National Institutes of Health, Grant Number C06 RR015455 from the Extramural Research Facilities Program of the National Center for Research Resources. Supported, in part, by the Drug Metabolism and Clinical Pharmacology shared Resource, Hollings Cancer Center, Medical University of South Carolina. J.Z. was supported by the Swedish Research Council (No. 524-2011-6998). The authors thank Jennifer Bestman from Cell and Molecular Imaging Core of the COBRE for the technical support and data analysis of colocalization.
Author Contributions
Conception and experimental design: Z.-W.Y., J.Z., D.M.T., K.D.T; Development of methodology: Z.-W.Y., J.Z., Y.M.; Analysis and interpretation of data: D.M.T., K.D.T.; Writing article: Z.-W.Y., J.Z., D.M.T., K.D.T; Technical and material support: Y.M., T.A.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Abend JR, Uldrick T, and Ziegelbauer JM. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J Virol 84: 12139–12151, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, and Cohen RA. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat Med 10: 1200–1207, 2004 [DOI] [PubMed] [Google Scholar]
- 3.This reference has been deleted.
- 4.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, and Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2: 326–332, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Coe H, Schneider JD, Dabrowska M, Groenendyk J, Jung J, and Michalak M. Role of cysteine amino acid residues in calnexin. Mol Cell Biochem 359: 271–281, 2012 [DOI] [PubMed] [Google Scholar]
- 6.Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, and Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem 279: 21749–21758, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Delom F, Emadali A, Cocolakis E, Lebrun JJ, Nantel A, and Chevet E. Calnexin-dependent regulation of tunicamycin-induced apoptosis in breast carcinoma MCF-7 cells. Cell Death Differ 14: 586–596, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Dunn KW, Kamocka MM, and McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol 300: C723–C742, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Findlay VJ, Townsend DM, Morris TE, Fraser JP, He L, and Tew KD. A novel role for human sulfiredoxin in the reversal of glutathionylation. Cancer Res 66: 6800–6806, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gate L, Majumdar RS, Lunk A, and Tew KD. Increased myeloproliferation in glutathione S-transferase pi-deficient mice is associated with a deregulation of JNK and Janus kinase/STAT pathways. J Biol Chem 279: 8608–8616, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Go YM. and Jones DP. Thiol/disulfide redox states in signaling and sensing. Crit Rev Biochem Mol Biol 48: 173–181, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodall JC, Wu C, Zhang Y, McNeill L, Ellis L, Saudek V, and Gaston JS. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proc Natl Acad Sci U S A 107: 17698–17703, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorlach A, Klappa P, and Kietzmann T. The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxid Redox Signal 8: 1391–1418, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Goto S, Kawakatsu M, Izumi S, Urata Y, Kageyama K, Ihara Y, Koji T, and Kondo T. Glutathione S-transferase pi localizes in mitochondria and protects against oxidative stress. Free Radic Biol Med 46: 1392–1403, 2009 [DOI] [PubMed] [Google Scholar]
- 15.This reference has been deleted.
- 16.Grek C. and Townsend DM. Protein disulfide isomerase superfamily in disease and the regulation of apoptosis. Endoplasm Reticulum Stress Dis 1: 4–17, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grek CL, Zhang J, Manevich Y, Townsend DM, and Tew KD. Causes and consequences of cysteine S-glutathionylation. J Biol Chem 288: 26497–26504, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes JD, Flanagan JU, and Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol 45: 51–88, 2005 [DOI] [PubMed] [Google Scholar]
- 19.This reference has been deleted.
- 20.Henderson CJ, Smith AG, Ure J, Brown K, Bacon EJ, and Wolf CR. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc Natl Acad Sci U S A 95: 5275–5280, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson CJ. and Wolf CR. Knockout and transgenic mice in glutathione transferase research. Drug Metab Rev 43: 152–164, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Hwang C, Sinskey AJ, and Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257: 1496–1502, 1992 [DOI] [PubMed] [Google Scholar]
- 23.Iwakoshi NN, Pypaert M, and Glimcher LH. The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J Exp Med 204: 2267–2275, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janssens S, Pulendran B, and Lambrecht BN. Emerging functions of the unfolded protein response in immunity. Nat Immunol 15: 910–919, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol 295: C849–C868, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamada K, Goto S, Okunaga T, Ihara Y, Tsuji K, Kawai Y, Uchida K, Osawa T, Matsuo T, Nagata I, and Kondo T. Nuclear glutathione S-transferase pi prevents apoptosis by reducing the oxidative stress-induced formation of exocyclic DNA products. Free Radic Biol Med 37: 1875–1884, 2004 [DOI] [PubMed] [Google Scholar]
- 27.This reference has been deleted.
- 28.Klaus A, Zorman S, Berthier A, Polge C, Ramirez S, Michelland S, Seve M, Vertommen D, Rider M, Lentze N, Auerbach D, and Schlattner U. Glutathione S-transferases interact with AMP-activated protein kinase: evidence for S-glutathionylation and activation in vitro. PloS One 8: e62497, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuzmich S, Vanderveer LA, and Tew KD. Evidence for a glycoconjugate form of glutathione S-transferase pI. Int J Pept Protein Res 37: 565–571, 1991 [DOI] [PubMed] [Google Scholar]
- 30.Maattanen P, Kozlov G, Gehring K, and Thomas DY. ERp57 and PDI: multifunctional protein disulfide isomerases with similar domain architectures but differing substrate-partner associations. Biochem Cell Biol 84: 881–889, 2006 [DOI] [PubMed] [Google Scholar]
- 31.Merksamer PI, Trusina A, and Papa FR. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding- functions. Cell 135: 933–947, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miseta A. and Csutora P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol Biol Evol 17: 1232–1239, 2000 [DOI] [PubMed] [Google Scholar]
- 33.Mishra M, Jiang H, Wu L, Chawsheen HA, and Wei Q. The sulfiredoxin-peroxiredoxin (Srx-Prx) axis in cell signal transduction and cancer development. Cancer Lett 366: 150–159, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osorio F, Tavernier SJ, Hoffmann E, Saeys Y, Martens L, Vetters J, Delrue I, De Rycke R, Parthoens E, Pouliot P, Iwawaki T, Janssens S, and Lambrecht BN. The unfolded-protein-response sensor IRE-1alpha regulates the function of CD8alpha+ dendritic cells. Nat Immunol 15: 248–257, 2014 [DOI] [PubMed] [Google Scholar]
- 35.Pani G, Giannoni E, Galeotti T, and Chiarugi P. Redox-based escape mechanism from death: the cancer lesson. Antioxid Redox Signal 11: 2791–2806, 2009 [DOI] [PubMed] [Google Scholar]
- 36.Peters LR. and Raghavan M. Endoplasmic reticulum calcium depletion impacts chaperone secretion, innate immunity, and phagocytic uptake of cells. J Immunol 187: 919–931, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Putney JW, Jr, Broad LM, Braun FJ, Lievremont JP, and Bird GS. Mechanisms of capacitative calcium entry. J Cell Sci 114: 2223–2229, 2001 [DOI] [PubMed] [Google Scholar]
- 38.Rossi R, Giustarini D, Dalle-Donne I, and Milzani A. Protein S-glutathionylation and platelet anti-aggregating activity of disulfiram. Biochem Pharmacol 72: 608–615, 2006 [DOI] [PubMed] [Google Scholar]
- 39.Ruscoe JE, Rosario LA, Wang T, Gate L, Arifoglu P, Wolf CR, Henderson CJ, Ronai Z, and Tew KD. Pharmacologic or genetic manipulation of glutathione S-transferase P1-1 (GSTpi) influences cell proliferation pathways. J Pharmacol Exp Therap 298: 339–345, 2001 [PubMed] [Google Scholar]
- 40.This reference has been deleted.
- 41.Rutkowski DT. and Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol 189: 783–794, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito S, Furuno A, Sakurai J, Sakamoto A, Park HR, Shin-Ya K, Tsuruo T, and Tomida A. Chemical genomics identifies the unfolded protein response as a target for selective cancer cell killing during glucose deprivation. Cancer Res 69: 4225–4234, 2009 [DOI] [PubMed] [Google Scholar]
- 43.Sevier CS. and Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev 3: 836–847, 2002 [DOI] [PubMed] [Google Scholar]
- 44.Shen H, Schultz MP, and Tew KD. Glutathione conjugate interactions with DNA-dependent protein kinase. J Pharmacol Exp Ther 290: 1101–1106, 1999 [PubMed] [Google Scholar]
- 45.Shen J, Chen X, Hendershot L, and Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 3: 99–111, 2002 [DOI] [PubMed] [Google Scholar]
- 46.Tew KD. Redox in redux: emergent roles for glutathione S-transferase P (GSTP) in regulation of cell signaling and S-glutathionylation. Biochem Pharmacol 73: 1257–1269, 2007 [DOI] [PubMed] [Google Scholar]
- 47.Tew KD. and Townsend DM. Glutathione-s-transferases as determinants of cell survival and death. Antioxid Redox Signal 17: 1728–1737, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tombal B, Weeraratna AT, Denmeade SR, and Isaacs JT. Thapsigargin induces a calmodulin/calcineurin-dependent apoptotic cascade responsible for the death of prostatic cancer cells. Prostate 43: 303–317, 2000 [DOI] [PubMed] [Google Scholar]
- 49.Tovey SC, Sun Y, and Taylor CW. Rapid functional assays of intracellular Ca2+ channels. Nat Protoc 1: 259–263, 2006 [DOI] [PubMed] [Google Scholar]
- 50.Townsend DM. S-glutathionylation: indicator of cell stress and regulator of the unfolded protein response. Mol Interv 7: 313–324, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Townsend DM, Lushchak VI, and Cooper AJ. A comparison of reversible versus irreversible protein glutathionylation. Adv Cancer Res 122: 177–198, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Townsend DM, Manevich Y, He L, Hutchens S, Pazoles CJ, and Tew KD. Novel role for glutathione S-transferase pi. Regulator of protein S-glutathionylation following oxidative and nitrosative stress. J Biol Chem 284: 436–445, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.This reference has been deleted.
- 54.Townsend DM, Manevich Y, He L, Xiong Y, Bowers RR, Jr, Hutchens S, and Tew KD. Nitrosative stress-induced s-glutathionylation of protein disulfide isomerase leads to activation of the unfolded protein response. Cancer Res 69: 7626–7634, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Anken E. and Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol 40: 191–228, 2005 [DOI] [PubMed] [Google Scholar]
- 56.Ward NE, Stewart JR, Ioannides CG, and O'Brian CA. Oxidant-induced S-glutathiolation inactivates protein kinase C-alpha (PKC-alpha): a potential mechanism of PKC isozyme regulation. Biochemistry 39: 10319–10329, 2000 [DOI] [PubMed] [Google Scholar]
- 57.Xiong Y, Manevich Y, Tew KD, and Townsend DM. S-glutathionylation of protein disulfide isomerase regulates estrogen receptor alpha stability and function. Int J Cell Biol 2012: 273549, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong Y, Uys JD, Tew KD, and Townsend DM. S-glutathionylation: from molecular mechanisms to health outcomes. Antioxid Redox Signal 15: 233–270, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye Z-W, Ghalali A, Högberg J, and Stenius U. Silencing p110β prevents rapid depletion of nuclear pAkt. Biochem Biophys Res Commun 415: 613–618, 2011 [DOI] [PubMed] [Google Scholar]
- 60.Ye ZW, Zhang J, Townsend DM, and Tew KD. Oxidative stress, redox regulation and diseases of cellular differentiation. Biochim Biophys Acta 1850: 1607–1621, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Ye ZW, Gao P, Reyes L, Jones EE, Branham-O'Connor M, Blumer JB, Drake RR, Manevich Y, Townsend DM, and Tew KD. Glutathione S-transferase P influences redox and migration pathways in bone marrow. PLoS One 9: e107478, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, and Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124: 587–599, 2006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.