Abstract
Single-cell microscopy provides a powerful tool to visualize cellular and subcellular processes in wild-type and mutant cells by observing fluorescently tagged proteins. Here, we describe three simple methods to visualize fission yeast cells: gelatin slides, coverslip-bottom dishes, and tetrad fluorescence microscopy. These imaging methods and data analysis using free software make it possible to quantify protein localization, dynamics, and concentration with high spatial and temporal resolution. In fission yeast, the actomyosin contractile ring is essential for cytokinesis. We use the visualization and quantification of contractile ring proteins as an example to demonstrate how to use these methods.
Keywords: Contractile ring, Cytokinesis, Fimbrin, Fission yeast, Gelatin, GFP, ImageJ, Microscopy, Quantification, Septin, Tetrad fluorescence microscopy
1 Introduction
The discovery and modification of the GFP (Green Fluorescent Protein) have revolutionized biomedical research and triggered the development of highly automated live cell fluorescence microscopy [1 – 6]. Analysis of fluorescence images and time-lapse movies has redefined the understanding of many biological processes. The unicellular fission yeast Schizosaccharomyces pombe is a particularly useful organism to study cytokinesis in live cells. The actomyosin contractile ring is essential for cytokinesis in fission yeast. Characteristics such as less genetic redundancy, regular cell size and shape, and efficient homologous recombination make S. pombe ideal for observing and studying contractile ring proteins during cytokinesis [7 – 9].
Visualization of a protein in S. pombe involves gene targeting to tag the protein of interest with a fluorescent protein at one of its termini, ideally expressed at its endogenous level [10]. A variety of options are available for choosing fluorescent proteins and techniques for imaging. Here, we demonstrate the need to choose the best monomeric fluorescent proteins with two examples. Depending on the duration and temperature of imaging, we describe how to visualize fission yeast cells using gelatin slides [11 – 14] and coverslip-bottom dishes [15, 16]. Visualization using gelatin slides is ideal for counting protein molecules and making short time-lapse movies (1–2 h) at lower temperatures. Using coverslip-bottom dishes allows imaging for longer durations (>2 h) and at higher temperatures.
We also describe a novel microscopy technique called “tetrad fluorescence microscopy” that we have developed to unequivocally determine the phenotypes of deletions of essential genes and synthetic lethal interactions [16, 17]. The technique is a promising new approach to determine the cause of cell death in synthetic lethal double mutants or in deletions of essential genes by following the progenies of a meiotic event. Using both a tetrad dissection microscope and a sensitive fluorescence microscope, this fusion microscopy technique can distinguish the possible causes of cell death in cells expressing certain fluorescent markers with a much higher certainty and resolution than the traditional random spore assays. Because spore germination and growth are often highly variable, imaging cells with known genotypes shortly after spore germination simplifies the process necessary to obtain images of cell polarization and the first few mitotic divisions [16].
Then we discuss some basic consideration on microscopy settings to obtain the best images. Finally, we briefly explain how to quantify the timing of contractile ring proteins using established methods and free software ImageJ [9, 11, 18]. We hope that these detailed methods are useful for studies of cytokinesis and other cellular processes.
1.1 How to Choose the Best Fluorescent Proteins for Imaging
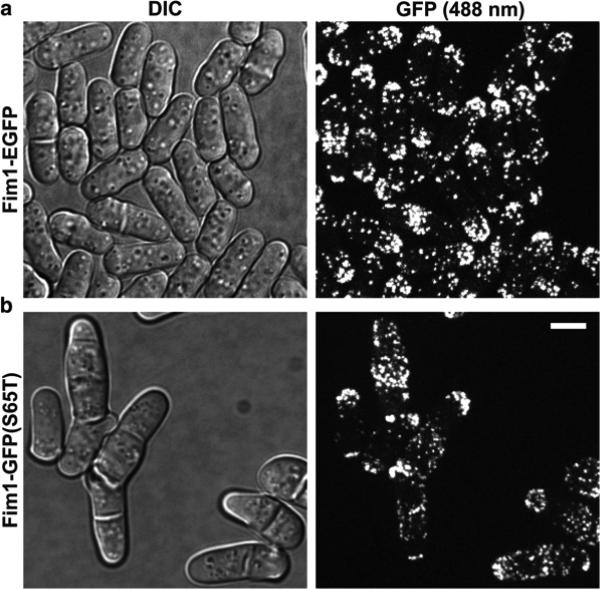
The published guides for choosing suitable fluorescent proteins should be followed [19 – 21]. Here, we emphasize two important considerations. Besides maturing rapidly and having a bright signal, ideal fluorescent proteins should be thermal stable and functional when fused with the protein of interest. A good example to illustrate this point is by comparing the tags GFP (S65T) and EGFP(F64L, S65T). The plasmid pFA6a-GFP(S65T)-kanMX6 for gene targeting was described in [10, 22, 23] and widely distributed/used in yeast community. At 36 °C, cells expressing fimbrin Fim1 tagged with enhanced GFP (EGFP) were normal (Fig. 1a). However, cells expressing fimbrin Fim1-GFP(S65T) had severe cytokinesis defects such as accumulating multiple and/or misplaced/aberrant septa (Fig. 1b). Thus, GFP(S65T) should not be used except for some special experiments.
Fig. 1.
Confocal microscopy of (a) fimbrin Fim1-EGFP and (b) Fim1- GFP (S65T). DIC (Differential Interference Contrast) images are shown to the left and the corresponding fluorescence images to the right. The fluorescence images are a maximum intensity projection of 21 slices with 0.3 μm spacing. Cells were grown in YE5S liquid medium for ~36 h at 25 °C, then shifted to 36 °C for 10 h before imaging at 36 °C. Cells were imaged using coverslip-bottom dish at 36 °C. Bar, 5 μm
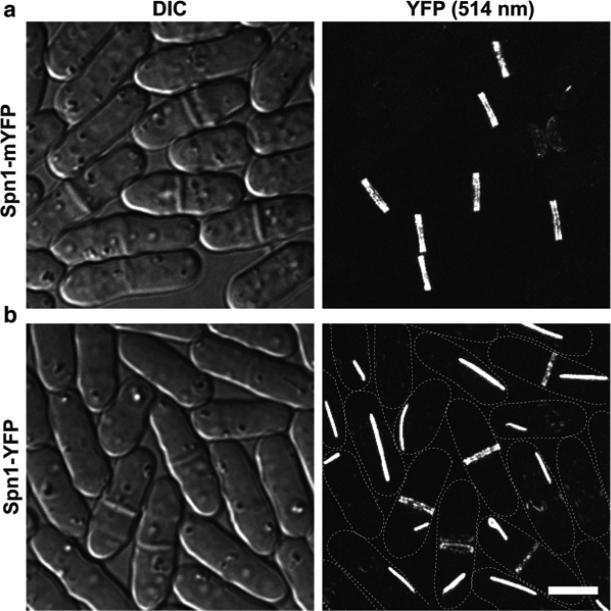
Another important consideration is the difference between regular (often forming a weak dimer) and monomeric fluorescent proteins. For example, YFP (Yellow fluorescent protein) is a weak dimer and mYFP (monomeric YFP) has a dimer interface breaking mutation A206K [20, 24]. Septin Spn1 tagged with mYFP showed normal septin localization to the double ring at the division site (Fig. 2a). In contrast, Spn1-YFP formed atypical structures such as bars or spirals in the cytoplasm in addition to its normal division site localization (Fig. 2b). Thus, monomeric fluorescent proteins should be used whenever possible to avoid artificial dimerization of the tagged proteins.
Fig. 2.
Confocal microscopy of (a) septin Spn1-mYFP and (b) Spn1-YFP. DIC images are shown to the left and the corresponding fluorescence images to the right. The fluorescence images are a maximum intensity projection of 31 slices with 0.2 μm spacing. The dash lines mark the cell boundary in (b). Cells were cultured in YE5S liquid medium for ~48 h at 25 °C and imaged on slides with YE5S medium at 24 °C. Bar, 5 μm
2 Materials
The recipes for the standard fission yeast media can be found at PombeNet of the Forsburg lab at http://www-bcf.usc.edu/~forsburg/media.html. The S. pombe strains used in this study are listed in Table 1.
Table 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| JW811 | h-spn1-YFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | This study |
| JW1092 | h-spn1-mYFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | This study |
| JW1143 | h-fim1-GFP(S65T)-kanMX6 ade6-M210 leu1-32 ura4-D18 | This study |
| JW1211 | h+fim1-EGFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | This study |
| JW6716 | kanMX6-Pmyo2-mEGFP-myo2 sad1-mCherry-natMX6 ade6-M210 leu1-32 ura4-D18 | This study |
2.1 Materials for Microscopy Using Gelatin Slides
Edinburgh minimal medium + Supplements (EMM5S) liquid medium (minimal medium with low autofluorescence).
Yeast extract medium + Supplements (YE5S) agar plates and YE5S liquid medium (rich medium with slightly higher autofluorescence).
10× n-propyl-gallate (n-PG) stock solution: 50 μM n-propylgallate in EMM5S or YE5S. Keep at −20 °C in the dark.
Gelatin from porcine skin.
Valap: 1:1:1 of Parafin, Vaseline, and Lanoline. Melt equal weight of each component completely at ~80 °C, mix thoroughly, and aliquot into 1.5 ml microcentrifuge tubes before solidification. Keep at room temperature.
Regular glass slides and 22 mm No. 1.5 square coverslip.
Fluorescence microscope.
2.2 Materials for Microscopy Using Coverslip-Bottom Dishes
YE5S agar plates and YE5S liquid media.
Delta TPG 35-mm culture dish with No. 1.5 coverslip bottom (0.17 mm thick, round).
18 mm × 18 mm No. 1.5 square coverslip.
Fluorescence microscope.
2.3 Materials for Tetrad Fluorescence Microscopy
Sporulation medium + Supplements (SPA5S) plates for crosses and sporulation.
YE5S agar plates with flat surface for tetrad dissection: 25 ml melted YE5S agar in each 100 × 15 mm petri dish. Lay out singly on a flat surface before agar solidification. Dry for 2 days at room temperature and then keep at 4 °C for storage.
Tetrad dissection microscope.
No.1.5 coverslips: 22 mm × 50 mm and 24 mm × 60 mm
Rectangular plastic frame to prevent agar medium from drying: the dimensions of the homemade frame are 50 mm × 23 mm × 3 mm (outer edge) and 40 mm × 17 mm × 3 mm (inner edge).
Fluorescence microscope with an automated stage.
2.4 Software for Quantification and Data Analysis
Image acquisition: Volocity Beta 6.3.1 or UltraVIEWERS or others.
Quantification and data analyses: ImageJ (NIH) at http://imagej.nih.gov/ij/.
3 Methods
3.1 Preparing Cells for Microscopy
Wake up cells from −80 °C stocks and grow on a YE5S plate for 2 days at 25 °C (or other plates and temperatures dependent on the strains). Re-streak cells on the plate twice.
Inoculate a strain in YE5S liquid medium (or other medium) in a baffled 50 ml flask and grow cells in 25 °C water bath with shaking. Prior to microscopy, cells are usually grown in YE5S liquid for ~48 h (see Note 1). The optical density (OD) of cells is measured at 595 nm. The OD595 should always be maintained at less than 0.5 (1 × 10 7 cells/ml) to assure that cells are growing in the log phase. Thus, the culture needs to be diluted using fresh YE5S medium every 9–15 h.
Spin down 1 ml of the cells at 3000 rpm (approximately 850 × g) for 30 to 60 s. Alternatively, spin cells for 30 s, rotate the microcentrifuge tube 180°, and then spin for another 30 s. Remove the supernatant.
Wash the cells in 1 ml fresh EMM5S to reduce background fluorescence (see Note 2), and spin down the cells as described in step 3. Remove the supernatant.
Add 900 μl of fresh EMM5S and 100 μl of 10× n-PG stock solution made in EMM5S (thawed and warmed up to room temperature) to resuspend the cells and keep the cells in the dark for several minutes (see Note 3). Spin down the cells as described in step 3 just before the slide is ready.
Remove extra supernatant and resuspend the cells in the remaining 20–100 μl (the volume dependent on cell concentration) of medium.
3.2 Microscopy Using a Gelatin Slide
Add 0.9 ml of EMM5S (or YE5S if the autofluorescence from the medium is not a concern) liquid medium to a microcentrifuge tube.
Add 0.25 g of gelatin to the 0.9 ml of EMM5S. Mix immediately by shaking vigorously so that gelatin will not clog at the top of the tube.
Place the tube at 65 °C hot plate (dry bath) for ~5 min to melt gelatin.
Add 100 μl of 10× n-PG stock solution in EMM5S (or in YE5S if YE5S is used in step 1 ) to the tube (see Note 4). Invert the tube a few times to mix thoroughly. Keep the tube at 65 °C and cover with foil. Wait for approximately 20 min until all air bubbles disappear.
Wipe slides clean. Then slowly take 100 to 150 μl of the melted EMM5S + gelatin + n-PG using a 1 ml pipette and add to the center of the slide. Avoid introducing air bubbles on the slide (see Note 5). Place another clean slide on top of the gelatin medium and clip one small paper binder on each end of the slides (see Note 6). The two slides should stagger a little bit to aid the eventual slide separation. Keep the clipped slides in the dark for 15–20 min to solidify the gelatin medium (see Note 7). Make two or three slides for each strain to ensure at least one is good.
Keep the microcentrifuge tube with extra gelatin medium at room temperature in the dark (see Note 8).
When the cell sample is ready to be loaded onto the slide (see Note 9), slowly separate two slides from one side by removing one binder. Separate the slides with a razor blade if necessary. Then remove the other binder. If no cracks appear in the middle of the solidified gelatin then the slide is usable since the entire gelatin pad will stick to one of the slides.
Pipet 5 μl of the concentrated cells from step 6 of ‘Preparing cells for microscopy’ to the center of the gelatin pad. Cover the cells with a 22 mm No. 1.5 square coverslip. Gently tap the coverslip with the blunt end of a 1 ml pipet tip to spread the cells evenly and avoid cell overlaps.
Seal the edges of the coverslip properly using Valap melted at 65 °C to prevent the slide from drying out.
Observe the slide on a microscope.
3.3 Microscopy Using a Coverslip-Bottom Dish
If cells are grown and being observed at temperatures other than the room temperature, all used materials (dish, agar plate, liquid YE5S, n-PG, tips, microcentrifuge tubes, etc.) should be pre-warmed to the desired temperature (see Note 10).
Cut YE5S agar medium in a circle shape with a diameter of ~20 mm using a sterile razor blade from a YE5S plate (see Note 11). To be consistent from experiments to experiments, the YE5S agar plate for tetrad dissection is recommended. It is better to add n-PG to a final concentration of 5 μM when making the plate.
Place 2–10 μl of concentrated cells washed with 900 μl YE5S plus 100 μl 5 μM n-PG in YE5S onto the center of the cover-slip of the dish.
Place pre-cut agar on top of the cells.
Place an 18 mm × 18 mm No. 1.5 coverslip on top of the agar to slow down the evaporation.
Gently tap on the top of the 18 mm × 18 mm coverslip to spread cells evenly with the blunt end of a 1 ml pipet tip.
Cover the dish with its lid and observe the dish on a microscope.
3.4 Tetrad Fluorescence Microscopy
Tetrad dissection is carried out from a 2- or 3-day-old cross as usual with one modification. Place the spores 2.5 mm apart from each other in both X and Y axes using the micromanipulator and glass-fiber needle (see Note 12).
Grow the dissected tetrads at 25 °C or other desired temperatures for 12–24 h or longer if necessary (see Note 13).
Preparing the agar plates with dissected tetrads for fluorescence microscopy: Since locating single or a few cells derived from germinating spores under a high magnification microscope can be a challenge, we mark the tetrads for imaging by punching holes in the agar near the cells using the dissection needle under the tetrad dissection microscope (see Note 14).
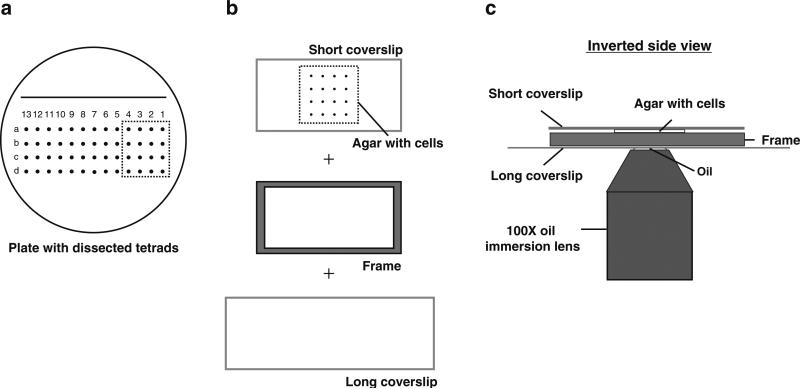
Preparing slides for the tetrad fluorescence microscopy: Typically, imaging 3 or 4 tetrads on a fluorescence microscope with an automated stage would yield images with enough temporal and spatial resolution (Fig. 3a). We cut an agar piece with 3–4 tetrads using the 22 mm × 50 mm coverslip. Then insert the coverslip underneath the agar and scoop up the agar piece that has been cut (tetrads facing up) (Fig. 3b, top). Center the agar on the coverslip and place the plastic frame around it (Fig. 3b). Then cover the tetrads with a slightly bigger 24 mm × 60 mm coverslip. Make sure the bigger coverslip is resting evenly on the agar without any air bubbles (see Note 15).
Invert the entire setup (tetrads facing down), clamp and rest it on slide holder over an objective lens (immersion oil added) for 5–10 min before imaging (Fig. 3c).
Use the marks in the agar to focus and to assist in finding colonies. Alternatively, the automated stage can be used to find the colonies based on the regular spacing between them.
Save each colony coordinates using the imaging software so they can be revisited for imaging at each time point.
Fig. 3.
Illustration of tetrad fluorescence microscopy. (a) Tetrads were dissected onto a YE5S plate made for tetrad dissection. (b) Four tetrads were cut out from the plate and placed (cells facing up) on a 22 mm × 50 mm glass coverslip. A plastic frame is then placed around the agar piece followed by a longer glass coverslip (24 mm × 60 mm) over the agar. (c) The entire setup from (b) was inverted and imaged using a 100× oil immersion lens
3.5 Image Acquisition
Settings of image acquisition depend on the imaging system, camera, acquisition software, and signals of the samples. Using appropriate microscopy settings is important to obtain a good fluorescence image. Laser power or other excitation lights must be adjusted so that they do not cause excessive photobleaching and phototoxicity. Whenever possible, use lower excitation power and longer exposure times without sacrificing the temporal resolution to obtain decent images. One method to boost the signal artificially is to adjust gain/sensitivity setting that amplifies the signals at the camera/detector. Another way to improve image brightness while slightly compromising spatial resolution is to adjust the binning. For regular CCD camera (like Hamamastu ORCA AG), we use 2 × 2 binning due to its small pixel size. However, binning is not recommended for EMCCD camera due to its bigger pixel size.
3.6 Quantification of Microscopy Data
A tremendous amount of informative and quantitative data can be extracted from microscopy images, like absolute global and local protein concentrations and timing of cytokinesis events. We and others have discussed how to count protein molecules extensively elsewhere [14, 17, 19, 25 – 28]. Here, we use quantification of the timing of contractile-ring proteins during cytokinesis as an example to explain how to analyze microscopy data using free software ImageJ (see Note 16).
Download and install ImageJ from http://imagej.nih.gov/ij/. Choose the version for your operating system. You can also download many useful Plugins from the website.
Open the microscopy data file in ImageJ as a TIFF file by importing it as “Image sequence” or “Bio-Formats” (see Note 17). The images are usually grouped in separate channels, like DIC and fluorescence channels.
Combine different Z-slices at the same time point using “Z projection” function in the Stacks option under Image tab (see Note 18). Typically this action gives a better signal for analysis. However, depending on the purpose, sometimes single slice is needed for analysis as well.
Open the “Brightness/Contrast” function in the Adjust option under Image tab. This allows the user to adjust the image. “Auto” option will usually give a good contrast of the image. If needed, “Set” option allows the user to manually enter the minimum and maximum displayed values (see Note 19).
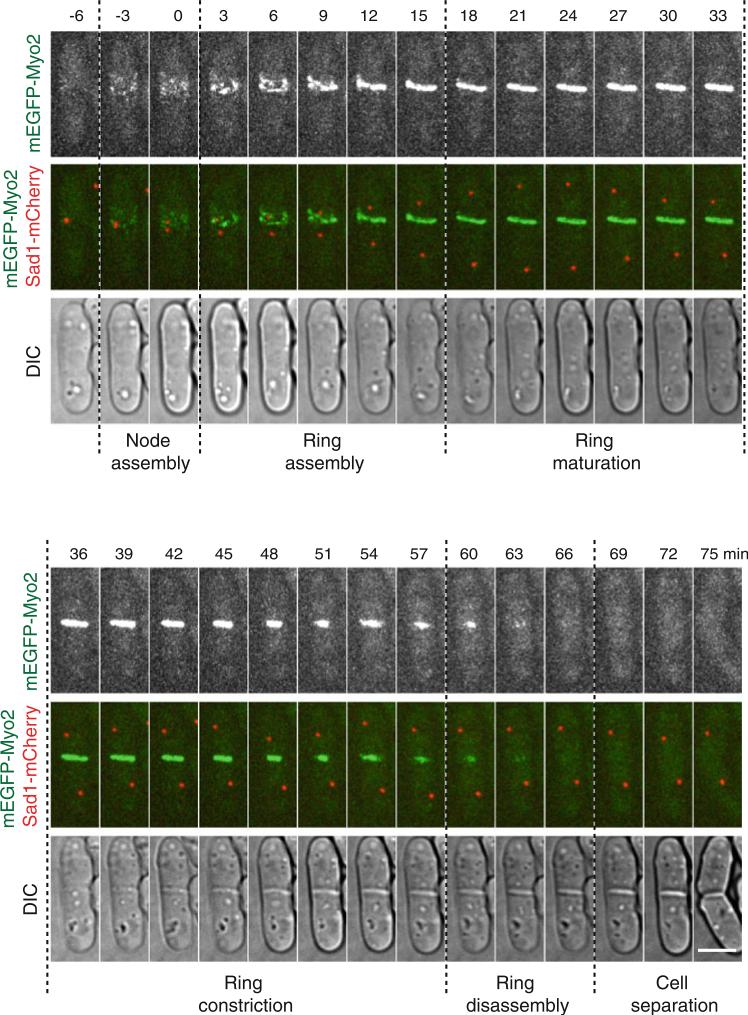
- Quantify the timing of contractile ring proteins by comparing it to different markers. The contractile ring protein, myosin II heavy chain Myo2, is a commonly used marker because the distinct stages of cytokinesis are easily distinguishable (Fig. 4). The timings of other proteins can be compared to the stages defined by Myo2. We usually use the spindle pole body protein Sad1 as an accurate marker for the cell-cycle stages (Fig. 4). Spindle pole body separation marks the entry of mitosis and is generally considered as time zero (see Note 20).
-
(a)Cytokinesis node assembly: This stage starts from the appearance of cytokinesis nodes at the division site to the time point just before nodes begin to condense around time zero (see Note 21).
-
(b)Contractile ring assembly: This stage starts from the beginning of node condensation to the formation of a compact ring without lagging nodes.
-
(c)Contractile ring maturation: This stage begins when a compact ring is formed until the ring starts to constrict.
-
(d)Contractile ring constriction: This stage begins when the contractile ring starts to constrict and ends when the ring has constricted to a dot (with the highest Myo2 pixel intensity). The Myo2 fluorescence intensity at the division site drops rapidly right after the end of ring constriction. The septum forms during ring constriction.
-
(e)Contractile ring disassembly: This stage is from the completion of ring constriction to the disappearance of Myo2 signal at the division site. This is a narrow definition of ring disassembly since ring components also disassemble during ring constriction.
-
(a)
Fig. 4.
Stages of cytokinesis in fi ssion yeast. Wild-type cells expressing mEGFP-Myo2 and Sad1-mCherry at their endogenous levels were used. The time course of a representative cell from a movie with 90 s interval is shown. The fluorescence images are a maximum intensity projection of 13 slices with 0.5 μm spacing. Time zero marks SPB separation. The dotted lines separate the various stages of cytokinesis. Bar, 5 μm
Acknowledgement
We would like to thank Valerie Coffman, Damien Laporte, I-Ju Lee, Ning Wang, Yanfang Ye, and Yihua Zhu for developing and improving the various microscopy techniques currently used in the lab. We thank Tom Pollard for yeast strains. The work in the Wu lab was supported by the National Institute of General Medical Sciences of the NIH grant GM086546 and American Cancer Society grant RSG-13-005-01-CCG to J.-Q.W.
Footnotes
The long culture time is necessary to ensure data consistency. For mutants that pick up a suppressor quickly, cells are usually grown in liquid medium for a shorter time.
This wash step is most useful for epifluorescence microscopy or for proteins with low abundance. It is not absolutely required for microscopy with spinning disk confocal microscope. When imaging in a dish with YE5S agar, the cells need to be washed only once with 900 μl of YE5S + 100 μl of 10× n-PG made in YE5S. Then directly proceed to step 6.
n-PG (Sigma P-3130) protects cells from free radicals during microscopy. It is light sensitive. An aliquot can be kept at room temperature in the dark for 1 day. Stocks should be kept at −20 °C for long-term storage.
Medium with gelatin is viscous. Do not use a pipette to mix.
A trick to pipet the liquid onto the slide without introducing bubbles is to avoid dispensing the last tiny drop of liquid in the pipette tip. If the bubbles are still introduced, they can be sucked up using the pipette tip. Because the gelatin medium is viscous, do not use a 200 μl pipette to dispense it.
When putting on the paper clips on both ends of the slides, it is better to hold one end while clipping the other. This ensures that the gelatin pad is even and not torn apart.
If the slide is kept longer than 30 min before use, the slides can be put in an airtight container with wet paper towels to prevent the gelatin pad from drying out.
The leftover EMM5S + gelatin + n-PG should be removed from the 65 °C hot plate after making the slides. Prolonged incubation at 65 °C may damage the vitamins and other components in the medium. In addition, it takes only a few minutes to re-melt the medium.
Begin preparing cells for imaging ~10 min after preparing the slides and keeping them in the dark.
When imaging cells at a temperature other than the room temperature, all the steps should be done on a surface warmed to that temperature. We use a small portable incubator that has been preheated for this purpose. The microcentrifuge tube with cells is kept in a beaker with water at the desired temperature during the collection and wash.
Before cutting, you can draw a circle on the bottom of the petri dish using a coin (e.g. a five-cent coin struck by the United States Mint with a diameter ~21 mm) as the guide.
Typically spores from tetrads are placed 5 mm apart. However, it takes too long for the automated stage to travel that distance over ten times at each time point during time-lapse imaging so 2.5 mm spacing is used.
The growth time depends on when the phenotype shows up in the mutant. For example, if the mutant dies during the first cell division or cell cycle, shorter incubation times are appropriate.
These holes in the medium are easily visible to the eye and under the microscope. Different patterns may be used to mark the location of the cells/colonies/spores.
Place the coverslip on the agar from one end and then release it slowly to prevent the formation of air bubbles. At this point the punched holes on the agar medium become less visible to eyes. Thus, it is helpful to make a mark on the coverslip using a permanent marker. Do not shift the coverslip once it has been placed on the agar. The frame is designed to be slightly thinner than the agar. Alternatively, one can use an 18 × 18 mm coverslip to scoop up the agar with tetrads and then invert it onto a coverslip-bottom dish for imaging.
Fiji is an image processing package that is akin to an advanced version of ImageJ and can also process super-resolution microscopy data. It is a free software and can be downloaded from www.Fiji.sc/Fiji.
“Image sequence” import is used for TIFF files. “Bio- Formats” import can be used for library files from Volocity multi-file library.
Typically, the Z-slices are combined using “Max Intensity” projection. If the signal intensity needs to be quantified or compared, “Sum Slices” projection should be used.
The “Brightness/Contrast” window shows a peak for each image. Typically, the minimum intensity value is displayed at the left bottom of the peak.
The time of arrival of various contractile ring proteins to the division site can be measured relative to the distance of SPB separation. The maximum SPB separation marks the end of anaphase B.
For some node proteins, the signal at the division site may start with one dot or the dot signal may appear and then disappear. It is hard to define the signal appearance with just one dot, so typically the appearance of signal is defined as the appearance of ~1/3 of the total number of nodes that exist at the beginning of node condensation for this certain protein. Due to higher autofluorescence at the GFP channel than YFP channel, GFP-tagged proteins may “appear” later than YFP-tagged same proteins (see Fig. 4 and [11, 14]).
References
- 1.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 2.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 3.Shimomura O. Structure of the chromophore of Aequorea green fluorescent protein. FEBS Lett. 1979;104:220–222. doi: 10.1016/0014-5793(79)80818-2. [Google Scholar]
- 4.Deerinck TJ, Martone ME, Lev-Ram V, Green DP, Tsien RY, Spector DL, Huang S, Ellisman MH. Fluorescence photooxidation with eosin: a method for high resolution immunolo calization and in situ hybridization detection for light and electron microscopy. J Cell Biol. 1994;126:901–910. doi: 10.1083/jcb.126.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean KM, Palmer AE. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat Chem Biol. 2014;10:512–523. doi: 10.1038/nchembio.1556. doi: 10.1038/nchembio.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao L, Shao L, Higgins CD, Poulton JS, Peifer M, Davidson MW, Wu X, Goldstein B, Betzig E. Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluores-cent specimens. Cell. 2012;151:1370–1385. doi: 10.1016/j.cell.2012.10.008. doi: 10.1016/j.cell.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts-Galbraith RH, Gould KL. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 2008;22:3082–3088. doi: 10.1101/gad.1748908. doi: 10.1101/gad.1748908. [DOI] [PubMed] [Google Scholar]
- 8.Laporte D, Zhao R, Wu J-Q. Mechanisms of contractile-ring assembly in fission yeast and beyond. Semin Cell Dev Biol. 2010;21:892–898. doi: 10.1016/j.semcdb.2010.08.004. doi: 10.1016/j.semcdb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee I-J, Coffman VC, Wu J-Q. Contractile-ring assembly in fission yeast cytokinesis: recent advances and new perspectives. Cytoskeleton. 2012;69:751–763. doi: 10.1002/cm.21052. doi: 10.1002/cm.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe . Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AIDYEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 11.Wu J-Q, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 12.Wu J-Q, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. doi: 10.1083/ jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffman VC, Nile AH, Lee I-J, Liu H, Wu J-Q. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laporte D, Coffman VC, Lee I-J, Wu J-Q. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y-H, Ye Y, Wu Z, Wu J-Q. Cooperation between Rho-GEF Gef2 and its binding partner Nod1 in the regulation of fission yeast cytokinesis. Mol Biol Cell. 2013;24:3187–3204. doi: 10.1091/mbc.E13-06-0301. doi: 10.1091/mbc.E13-06-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee I-J, Wang N, Hu W, Schott K, Bähler J, Giddings TH, Jr, Pringle JR, Du L-L, Wu J-Q. Regulation of spindle pole body assembly and cytokinesis by the centrin-binding protein Sfi1 in fission yeast. Mol Biol Cell. 2014;25:2735–2749. doi: 10.1091/mbc.E13-11-0699. doi: 10.1091/mbc.E13-11-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffman VC, Sees JA, Kovar DR, Wu J-Q. The formins Cdc12 and For3 cooperate during contractile ring assembly in cytokinesis. J Cell Biol. 2013;203:101–114. doi: 10.1083/jcb.201305022. doi: 10.1083/jcb.201305022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang N, Lo Presti L, Zhu Y-H, Kang M, Wu Z, Martin SG, Wu J-Q. The novel proteins Rng8 and Rng9 regulate the myosin-V Myo51 during fission yeast cytokinesis. J Cell Biol. 2014;205:357–375. doi: 10.1083/jcb.201308146. doi: 10.1083/jcb.201308146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coffman VC, Wu J-Q. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem Sci. 2012;37:499–506. doi: 10.1016/j.tibs.2012.08.002. doi: 10.1016/j.tibs.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 21.Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr Opin Struct Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae . Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. doi: 10.1002/(SICI)1097-1061(1997 09 15)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 23.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AIDYEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 24.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdo-mains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 25.Wu J-Q, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 26.Coffman VC, Wu J-Q. Every laboratory with a fluorescence microscope should consider counting molecules. Mol Biol Cell. 2014;25:1545–1548. doi: 10.1091/mbc.E13-05-0249. doi: 10.1091/mbc.E13-05-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joglekar AP, Salmon ED, Bloom KS. Counting kinetochore protein numbers in budding yeast using genetically encoded fluorescent proteins. Methods Cell Biol. 2008;85:127–151. doi: 10.1016/S0091-679X(08)85007-8. doi: 10.1016/S0091-679X(08)85007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coffman VC, Wu P, Parthun MR, Wu J-Q. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]