Abstract
In the past few decades, live cell microscopy techniques in combination with fluorescent tagging have provided a true explosion in our knowledge of the inner functioning of the cell. Dynamic phenomena can be observed inside living cells and the behavior of individual molecules participating in those events can be documented. However, our preference for simple or easy model systems such as cell culture, has come at a cost of chasing artifacts and missing out on understanding real biology as it happens in complex multicellular organisms. We are now entering a new era where developing meaningful, but also tractable model systems to study biological phenomenon dynamically in vivo in a mammal is not only possible; it will become the gold standard for scientific quality and translational potential.1,2 A study by Oddoux et al. describing the dynamics of the microtubule (MT) cytoskeleton in skeletal muscle is one example that demonstrates the power of developing in vivo/ex vivo models.3 MTs have long attracted attention as targets for cancer therapeutics 4 and more recently as mediators of Duchene muscular dystrophy.5 The muscle fiber MT cytoskeleton forms an intricate rectilinear lattice beneath the sarcolemma and is essential for the structural integrity of the muscle. Cultured cells do not develop such a specialized organization of the MT cytoskeleton and our understanding of it has come from static snapshots of muscle sections.6 In this context, the methodology and the findings reported by Oddoux et al. are a significant step forward.
Keywords: intravital, microscopy, microtubules, cytoskeleton, muscle, fiber, muscular, dystrophy
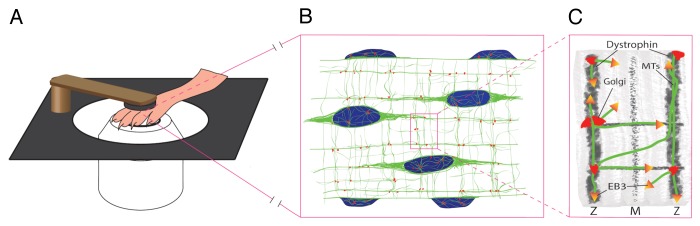
The researchers combined high-resolution intravital microscopy, live imaging of ex vivo muscle fibers and super-resolution microscopy showing that MTs are nucleated largely at the Golgi elements and describing how MT dynamics lead to the organization of a stable grid-like cytoskeletal network. To visualize the dynamics of the MT network they electroporated cDNA encoding GFP- and mCherry-tagged MT and Golgi markers into the mouse flexor digitorum brevis (FDB) muscle in situ. Then the researchers imaged the FDB muscle in vivo by exposing it and stabilizing the foot on the microscope (Fig. 1A). Alternatively, the muscle fibers were isolated and imaged in a short-term ex vivo culture, before the effects of denervation were evident. Time-lapse microscopy with GFP-tubulin showed a rather stable network of MTs with some events of elongation and shrinking of single MTs suggesting dynamic instability. However, imaging of a EB3-GFP tag, which marks growing tips of MTs revealed a much more dynamic picture of the MT cytoskeleton. Numerous EB3-GFP puncta were primarily moving longitudinally and transversely across the muscle fiber at about 8.6 μm/min. This reported growth rate was on a par with that of mammalian cells in culture7 and D. melongaster neurons in vivo.8 The MT growth rates measured in the FDB in vivo were somewhat slower than in cultured muscle fibers, which may be due to lower temperatures in situ. The growing tips of muscle MTs move along the tracks in either the same or opposite direction in a rectilinear fashion reminiscent of an aerial view of cars on two-way streets. This pointed to the existence of underlying tracks that guide MT polymerization in either direction. FRAP experiments performed on muscle fibers expressing GFP-tubulin supported this idea. In several cases bleached MTs regrew indicating new MT polymerization along pre-existing MTs. This was the first experimental evidence that bundles of multiple MTs form the rectilinear lattice in muscle fibers. To show this unequivocally, the authors turned to gated STED super resolution microscopy. With the resolution of 65 nm they were able to distinguish individual MTs forming bundles of 2–4 often twisting around each other which provided us with a more complete model of the muscle MT cytoskeleton (Fig. 1B). Nonetheless, how this stable network of MTs is generated still remained an open question.
Figure 1. Intravital microscopy setup and model of MT cytoskeleton in skeletal muscle. (A) diagram showing the intravital setup for imaging mouse FDB muscle. The foot of an anesthetized mouse is stabilized on the coverslip by a clamp. (B) Sketch of a muscle fiber showing surface MTs (green) Golgi elements (red) and nuclei (blue). (C) Model of MT lattice organization showing MTs that are nucleated from the Golgi and guided by costameric dystrophin bands. One sarcomere width is shown as noted by the Z-lines and the M-band (labeled as Z and M respectively).
It was shown previously that nuclei acquire MT nucleation factors during myogenesis.9-11 However, through live imaging of muscle fibers expressing EB3-GFP Oddoux et al. observed that the MT growing tips originate from many secondary locations along the lattice, not just from the nuclei. They hypothesized that the Golgi may harbor the nucleators of MTs as Golgi elements are commonly localized at the intersections of MT bundles. Indeed, nocodazole washout experiments revealed that newly polymerized asters of MTs form at the Golgi elements confirming an additional MT nucleation site. They also found that γ-tubulin which nucleates MTs is localized at the Golgi and proposed that it may be anchored to the Golgi membranes by pericentrin. The live imaging experiments showed that Golgi are completely static in muscle fibers suggesting that Golgi elements position MT nucleation (Fig. 1B). This is in contrast to the dogma based on mammalian cell culture studies, that MTs position the Golgi complex. The new data from live imaging of muscle fibers is supported by previous work which suggested that in muscle fibers the Golgi complex is positioned along the Z lines and is tethered to the ER/sarcoplasmic reticulum via ER exit sites (ERES).6,12 However, Golgi positioning alone could not explain the rectilinear nature of the sub-sarcolemmal MT cytoskeleton. Thus, Oddoux et al. invoked a role for dystrophin in their model for the organization of the MT cytoskeleton in skeletal muscle. Dystrophin is a cytolinker-like protein that connects intermediate filaments, actin filaments, MTs and the sarcolemma via dystroglycan binding.13 Dystrophin is thus an essential element of the costamere, which is a functional component of the striated muscle cells that couples the sarcomere with the cytoskeleton, plasma membrane and the extracellular matrix. This coupling to the sarcomere occurs at the Z-lines, hence all the costameric proteins including dystrophin localize to transverse bands adjacent to Z-lines. Costamere- and Z-line-associated actin filaments and actin regulators such as tropomyosins are also critical in preserving muscle fiber integrity.14,15 MTs underlie the pathology of Duchene muscular dystrophy5 and their transverse localization in muscle fibers of the mdx mouse is compromised.13,16 In their study Oddoux et al. show that MTs elongating at an oblique angle abruptly change their direction, presumably after encountering a costameric band of dystrophin. Based on their data and previous work the authors propose a model for the sub-sarcolemmal MT cytoskeleton, which is both dynamic and persistent, is nucleated largely from the Golgi elements and its rectilinear arrangement is guided by dystrophin (Fig. 1C).
The methodologies developed in this study are as important as the findings. Oddoux et al. were able to visualize and assess the kinetics of the MT cytoskeleton in a living mouse. Such techniques are primed to launch new studies and their potential for translational applications is extremely exciting. For example, the dynamics of the MT cytoskeleton in conjunction with other structural components can now be investigated in mouse models of muscular dystrophy. The MT cytoskeleton density and rigidity has been implicated in Duchene muscular dystrophy,5 thus targeting MTs or their interacting partners maybe a viable therapeutic strategy. In vivo assays pioneered in the Ralston laboratory may serve as an excellent means to measure on-target drug effects in animal models of disease. The use of intravital microscopy to observe the impact of therapeutics at a molecular level will open the door to defining dose-response relationships in terms of blood levels of drugs and corresponding pathological outcomes. Finally, on-target and off-target effects of anti-microtubule agents used in cancer chemotherapy could be evaluated using similar models based on high-resolution intravital microscopy.
References
- 1.Masedunskas A, Appaduray M, Hardeman EC, Gunning PW. . What makes a model system great? A case for intravital microscopy for studying the actin cytoskeleton. IntraVital 2013; 2:e26287; http://dx.doi.org/ 10.4161/intv.26287 [DOI] [Google Scholar]
- 2.Weigert R, Porat-Shliom N, Amornphimoltham P. . Imaging cell biology in live animals: ready for prime time. J Cell Biol 2013; 201:969 - 79; http://dx.doi.org/ 10.1083/jcb.201212130; PMID: 23798727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oddoux S, Zaal KJ, Tate V, Kenea A, Nandkeolyar SA, Reid E, Liu W, Ralston E. . Microtubules that form the stationary lattice of muscle fibers are dynamic and nucleated at Golgi elements. J Cell Biol 2013; 203:205 - 13; http://dx.doi.org/ 10.1083/jcb.201304063; PMID: 24145165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kavallaris M. . Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer 2010; 10:194 - 204; http://dx.doi.org/ 10.1038/nrc2803; PMID: 20147901 [DOI] [PubMed] [Google Scholar]
- 5.Khairallah RJ, Shi G, Sbrana F, Prosser BL, Borroto C, Mazaitis MJ, Hoffman EP, Mahurkar A, Sachs F, Sun Y, et al. . Microtubules underlie dysfunction in duchenne muscular dystrophy. Sci Signal 2012; 5:ra56; http://dx.doi.org/ 10.1126/scisignal.2002829; PMID: 22871609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ralston E, Lu Z, Ploug T. . The organization of the Golgi complex and microtubules in skeletal muscle is fiber type-dependent. J Neurosci 1999; 19:10694 - 705; PMID: 10594053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rusan NM, Fagerstrom CJ, Yvon AM, Wadsworth P. . Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol Biol Cell 2001; 12:971 - 80; http://dx.doi.org/ 10.1091/mbc.12.4.971; PMID: 11294900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ori-McKenney KM, Jan LY, Jan YN. . Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron 2012; 76:921 - 30; http://dx.doi.org/ 10.1016/j.neuron.2012.10.008; PMID: 23217741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bugnard E, Zaal KJ, Ralston E. . Reorganization of microtubule nucleation during muscle differentiation. Cell Motil Cytoskeleton 2005; 60:1 - 13; http://dx.doi.org/ 10.1002/cm.20042; PMID: 15532031 [DOI] [PubMed] [Google Scholar]
- 10.Tassin AM, Maro B, Bornens M. . Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol 1985; 100:35 - 46; http://dx.doi.org/ 10.1083/jcb.100.1.35; PMID: 3880758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaal KJ, Reid E, Mousavi K, Zhang T, Mehta A, Bugnard E, Sartorelli V, Ralston E. . Who needs microtubules? Myogenic reorganization of MTOC, Golgi complex and ER exit sites persists despite lack of normal microtubule tracks. PLoS One 2011; 6:e29057; http://dx.doi.org/ 10.1371/journal.pone.0029057; PMID: 22216166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaisto T, Metsikkö K. . Distribution of the endoplasmic reticulum and its relationship with the sarcoplasmic reticulum in skeletal myofibers. Exp Cell Res 2003; 289:47 - 57; http://dx.doi.org/ 10.1016/S0014-4827(03)00231-3; PMID: 12941603 [DOI] [PubMed] [Google Scholar]
- 13.Prins KW, Humston JL, Mehta A, Tate V, Ralston E, Ervasti JM. . Dystrophin is a microtubule-associated protein. J Cell Biol 2009; 186:363 - 9; http://dx.doi.org/ 10.1083/jcb.200905048; PMID: 19651889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kee AJ, Schevzov G, Nair-Shalliker V, Robinson CS, Vrhovski B, Ghoddusi M, Qiu MR, Lin JJ, Weinberger R, Gunning PW, et al. . Sorting of a nonmuscle tropomyosin to a novel cytoskeletal compartment in skeletal muscle results in muscular dystrophy. J Cell Biol 2004; 166:685 - 96; http://dx.doi.org/ 10.1083/jcb.200406181; PMID: 15337777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlahovich N, Schevzov G, Nair-Shaliker V, Ilkovski B, Artap ST, Joya JE, Kee AJ, North KN, Gunning PW, Hardeman EC. . Tropomyosin 4 defines novel filaments in skeletal muscle associated with muscle remodelling/regeneration in normal and diseased muscle. Cell Motil Cytoskeleton 2008; 65:73 - 85; http://dx.doi.org/ 10.1002/cm.20245; PMID: 17968984 [DOI] [PubMed] [Google Scholar]
- 16.Percival JM, Gregorevic P, Odom GL, Banks GB, Chamberlain JS, Froehner SC. . rAAV6-microdystrophin rescues aberrant Golgi complex organization in mdx skeletal muscles. Traffic 2007; 8:1424 - 39; http://dx.doi.org/ 10.1111/j.1600-0854.2007.00622.x; PMID: 17714427 [DOI] [PubMed] [Google Scholar]