Abstract
The aim of this study was identifying bacterial pathogens involved in meningitis, studying their antibiotic resistance profiles, investigating the antibiotic resistance genes as well as evaluating the use of various antibiotic combinations. Antibiotic susceptibility tests were evaluated according to CLSI guidelines. Antibiotic combinations were evaluated by calculating the Fractional Inhibitory Concentration (FIC) index. A total of 71 bacterial isolates were recovered from 68 culture positive CSF specimens. Sixty five of these isolates (91.5%) were recovered from single infection specimens, while 6 isolates (8.4%) were recovered from mixed infection specimens. Out of the 71 recovered isolates, 48 (67.6%) were Gram-positive, and 23 (32.4%) were Gram-negative. Thirty one of the Gram positive isolates were S. pneumoniae (64.6%, n = 48). Out of the recovered 71 isolates; 26 (36.6%) were multidrug-resistant (MDR) isolates of which, 18 (69.2%) were Gram-negative and 8 (30.8%) were Gram-positive. All MDR isolates (100%) showed resistance to penicillin and ampicillin, however, they showed lower resistance to meropenem (50%), levofloxacin (50%), amikacin (48%), pipercillin-tazobactam (45.8%). Most common antibiotic resistance genes were investigated including: tem (21.1%), shv (15.8%), ctx-m (15.8%) coding for TEM-, SHV, CTX-M extended-spectrum beta-lactamases (ESBLs), respectively; aac(6')-I b(26.3%) coding for aminoglycoside 6’-N-acetyltransferase type Ib ciprofloxacin resistant variant; and qnrA (5.3%) gene coding for quinolone resistance. The DNA sequences of the respective resistance genes of some selected isolates were PCR amplified, analyzed and submitted to the GenBank database under the accession numbers, KX214665, KX214664, KX214663, KX214662, respectively. The FIC values for ampicillin/sulbactam plus cefepime showed either additive or synergistic effect against ten tested Gram-negative MDR isolates, while doxycycline plus levofloxacin combination revealed synergism against two MDR Gram-positive isolates. The results indicate high prevalence of antibiotic resistance among MDR isolates. Therefore, new guidelines should be implemented in Egypt to rationalize the use and avoid the misuse and abuse of antimicrobial agents.
Introduction
Bacterial meningitis is a life threatening disease that is associated with significant mortality and morbidity [1]. Historically, three major pathogens account for most cases of bacterial meningitis, which includes: Neisseria (N.) meningitidis, Streptococcus (S.) pneumoniae, and Haemophilus (H.) influenzae, they accounted for about 75–80% of cases globally, while the majority of other cases accounted by Escherichia (E.) coli, Listeria (L.) monocytogenes, and Staphylococcus (S.) aureus [2], however bacterial pathogens causing meningitis are evolving constantly, as evidenced by the change in relative occurrence of pneumococcal meningitis in sub-Saharan [3]. The mortality from these bacterial varies from 3 to 21%, according to type of organism [4]. The incidence and case-fatality rates of bacterial meningitis vary according to country, region, pathogen, and age group. Without any treatment, the case-fatality rate can reach 70%, and one in five survivors of bacterial meningitis may be left with permanent disability including hearing loss, neurologic disability, or limb loss [5]. Routine vaccination against three most common causative bacterial pathogens had a considerable effect on the prevalence of bacterial meningitis. However, an estimated 1–2 million cases of bacterial meningitis occur worldwide every year, resulting in 180 000 deaths in children age from one to 59 months in 2010 [6,7]. The epidemiology of acute bacterial meningitis has changed markedly since the introduction of conjugate vaccines [4,8,9]. However, the disease continues to cause a heavy burden even in developed countries, causing substantial morbidity and mortality [1,8]. Early administration of antibiotics save lives, but the globally emerging multidrug resistant bacteria limits the effectiveness of many inexpensive and widely available antimicrobial drugs [10]. The molecular mechanisms of antibiotics resistance have been studied extensively and involved studying genetics and biochemical aspects of bacterial cell function [10–13]. However, most of these mechanisms can be disseminated by one or more distinctive gene transfer mechanisms [14]. Under selective pressure of certain antibiotics, bacterial variants evolve various mechanisms to survive in the presence of these antimicrobial agents. Drug resistant bacteria are usually multi-drug resistant against various structurally different drugs [15]. Although antimicrobial resistance is classically attributed to chromosomal mutations, resistance is mainly associated with extra-chromosomal elements acquired from other bacteria in the environment, such as plasmids. Multidrug-resistant bacteria will continue to persist and spread worldwide, causing clinical failure in treatment of infectious diseases and public health crises [11]. Therefore, in this study we aim to identify the most common bacterial pathogens together with their resistance profile against major antibiotics used in the empirical treatment of bacterial meningitis. In addition, we aimed to investigate the most common genes involved in bacterial resistance and to evaluate use of various antibiotic combinations for possible synergistic activities against the most clinically relevant pathogens causing bacterial meningitis particularly MDR.
Materials and methods
Sample collection
A total of 1337 cerebrospinal fluid (CSF) specimens from suspected cases of meningitis were collected over a2-year period from September 2013 to September 2015. From the culture positive CSF specimens, a total of 71 bacterial isolates were recovered from 3 different hospitals; (AbbassiaFeverHospital,54 isolates; Ain Shams University Hospital, 15 isolates and Ain Shams Specialized Hospital, 2 isolates). Microscopical examination of Gram-stained smears was performed. Characteristics of growth on mannitol salt agar, and results of coagulase and catalase tests, were used to identify the Gram-positive isolates as Staphylococcus aureus and Staphylococcus (S.) coagulase negative. Screening for methicillin resistance was done by agar disc diffusion method according to the CLSI using cefoxitin discs (FOX, 30μg). S. pneumoniae and Enterococcus were cultured on blood and chocolate agar. Optochin (OP) test and bile solubility test were performed to identify S. pneumoniae isolates, while Enterococci were identified according to growth characteristics on Bile Esculin Agar (BEA) and upon addition of Pyrrolidonyl Arylamidase (PYR) reagent. Gram negative isolates were cultured mainly on MacConkey’s agar. Several biochemical tests were performed including, triple sugar iron (TSI) test, oxidase test, citrate utilization, urease test, motility indole ornithine (MIO) test, lysine iron agar (LIA) and slide agglutination test, to identify different bacterial species. Identification of Enterobacteriaceae and Pseudomonas isolates were confirmed using API® 20E identification kit (BioMérieux, France).
Antimicrobial susceptibility testing and identification of multidrug-resistant (MDR) isolates
The Kirby-Bauer disk diffusion method was used to determine the susceptibility of the recovered clinical isolates to antimicrobial agents and it was carried out as recommended by the Clinical and Laboratory Standards Institute (CLSI) [16]. Isolates that were resistant to three or more classes of antimicrobials were considered as MDR isolates [17] and were selected for further study.
Determination of minimum inhibitory concentrations of some selected antimicrobial agents for multi-drug resistant isolates
The minimum inhibitory concentrations (MICs) of amikacin, ceftriaxone, cefepime, levofloxacin, gentamicin, ampicillin/sulbactam, ceftazidime, and imipenem were determined for MDR Gram-negative isolates, while for Gram-positive isolates gentamicin and ceftazidime were replaced by vancomycin and doxycycline. This was done by the micro-broth dilution technique described in the CLSI guidelines [16] and in triplicate where average MIC was calculated.
Antibiotic combinations
The value of the fractional inhibitory concentration (FIC) index as a predictor of synergy has been investigated according to the protocol described by Hsieh,et al. [18].
DNA extraction from MDR isolates
GeneJet plasmid miniprep kit (ThermoFisher Scientific, USA) was used to extract plasmid DNA from MDR isolates, however no bands were recovered. While chromosomal DNA was extracted from the tested clinical bacterial isolates using PrepMan Ultra Kit (ThermoFisher Scientific, USA) each according to its manufacturer specifications. The extracted DNA was analyzed using agarose gel electrophoresis [19].
Amplification of some resistance genes by PCR
Amplification of the selected antibiotic resistance genes were carried out via PCR using appropriate primers (Table 1) and either extracted plasmid or chromosomal DNA of tested MDR bacterial isolates as templates. Primers were synthesized by Invitrogen®, UK and supplied by Analysis Co., Egypt. Detection of the amplified products was done by agarose gel electrophoresis and the expected size of DNA fragment was determined as compared to DNA ladder (GeneRuler100bp, ThermoFisher Scientific, USA).
Table 1. Target genes and primers sequences, along with expected PCR product size.
| gene | Primer sequence (5'-3') | Expected product size (bp) | Ta (°C) | Reference |
|---|---|---|---|---|
| ctx-m | Pf−CGCTTTGCGATGTGCAG | 550 | 47 | [20] |
| Pr−ACCGCGATATCGTTGGT | ||||
| shv | Pf—GGTTATGCGTTATATTCGCC | 867 | 47 | [21] |
| Pr−TTAGCGTTGCCAGTGCTC | ||||
| tem | Pf—ATGAGTATTCAACATTTCCG | 867 | 47 | [21] |
| Pr−CTGACAGTTACCAATGCTTA | ||||
| aac(6')-Ib or aac(6')-Ib-cr | Pf—TTGCGATGCTCTATGAGTGG | 458 | 46 | [22] |
| Pr–CGTTTGGATCTTGGTGACCT | ||||
| qnr | Pf−GGAAGCCGTATGGATATTATTG | 660 | 51 | [23] |
| Pr−CTAATCCGGCAGCACTATTAC |
ctx-m, shv, tem, genes coding for extended-spectrum beta-lactamases (ESBLs); aac(6')-Ib gene coding for aminoglycoside 6’-N-acetyltransferase type Ib; aac(6')-Ib-cr gene coding for aminoglycoside 6’-N-acetyltransferase type Ib ciprofloxacin resistant variant; qnrA, gene coding for quinolone resistance, Ta, calculated annealing temperatures.
Sequencing of some selected PCR products
The PCR products were purified using GeneJETTM purification kit at Sigma Scientific Services Company, Egypt. PCR products were sent for sequencing at GATC, Germany using ABI3730 xl DNA sequencer. The obtained sequence files were assembled into the final consensus sequence using StadenPackage program version3 (http://staden.sourceforge.net/) [24].
Results
Data analysis
This study was conducted over a period of 2 years from September 2013 to September 2015 where a total of 1,337 CSF specimens were collected (according to Hospitals Official Daily Records). Depending on PMNLs/Lymphocytes ratios(obtained from cell count of conducted microscopically using Hemocytometer; (Assistent, Germany),WBC count (normal level 0–5 cell\mm3), protein level (measured by BCA protein assay with normal protein level: 15- 45mg\dl)and glucose levels (measured by Trinder method where the reference values for glucose level 45-75mg\dl), the specimens have been divided in to 4 categories: a) viral (lymph cells exceed PMNLs, protein>45mg\dl, glucose level: normal); b) zero (no cells found); c) turbid no growth (PMNLs exceeds lymphocytes and negative culture, glucose<40mg\dl, protein>50mg\dl); and d) bacterial (PMNLs exceed lymphocytes, glucose <40mg\dl, protein level >50mg\dl and positive culture growth). It has been found that the majority of meningitis cases over the two years period (n = 1337), were possibly viral meningitis (573 cases; 42.86%), followed by cases that have been probably misdiagnosed or have diseases that showed symptomatic similarities with meningitis in which cell count was reported “Zero” (467 cases; 34.93%), followed by cases that showed CSF findings suggesting bacterial meningitis yet showed no positive culture growth represented by letter “T” (turbid no growth; 229 cases; 17.13%), and finally, bacterial meningitis cases that showed positive culture growth represented by letter “B”(68; 5.08%) as shown in Fig 1.
Fig 1. Relative percentage of various CSF specimens collected during the study period (n = 1337).
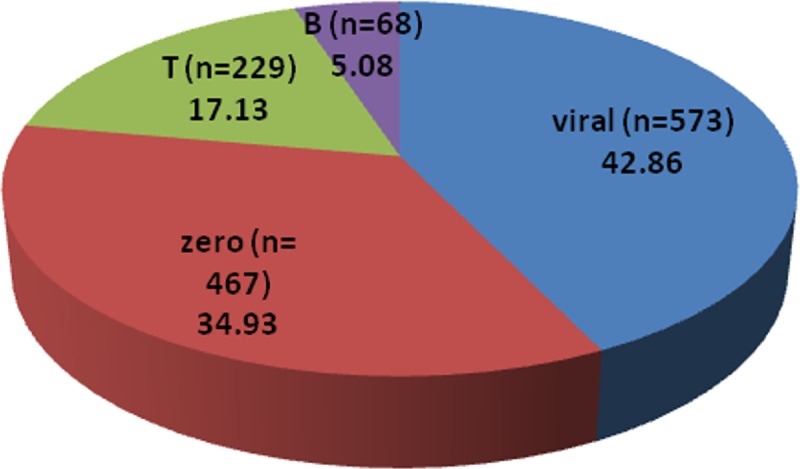
Out of the 68 culture positive specimens, 71 bacterial isolates were recovered from cerebrospinal fluid (CSF); 65 isolates (91.5%) were from specimens with single bacterial species, 6 isolates (8.4%) were from mixed culture. Specimens collected from males were 44(64.7%), while only 24 specimens were collected from females (35.3%). Regarding age, 5 specimens (7.35%) were from infants (age from 1–12 Months), 10 specimens (14.7%) were from children (>1–16 Years) and the rest of the specimens (77.9%) were from adults (>16Years). Using Gram-stain, 48 isolates (67.6%) were found to be Gram-positive and 23 isolates (32.4%) were found to be Gram-negative. The prevalence of different clinical bacterial isolates cultured from the 68 CSF specimens is delineated in Fig 2.
Fig 2. Prevalence of different clinical bacterial isolates cultured from the 68 CSF specimens.
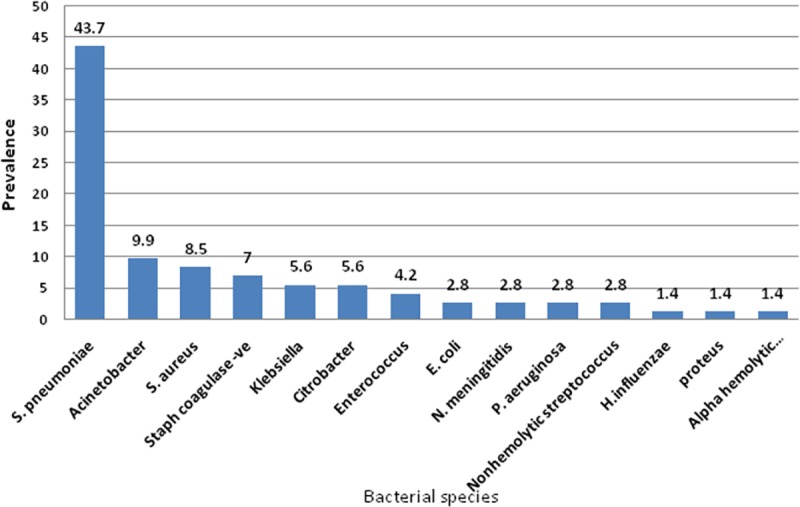
Prevalence was expressed as percentage from total count (n = 71).
Drug susceptibility test
The antimicrobial susceptibility patterns of the bacterial pathogens recovered from the collected CSF specimens of suspected cases of bacterial meningitis that showed positive cultural growth are shown in Table 2. Moreover, the antibiotic susceptibility patterns of the recovered Gram positive and Gram negative isolates are shown in Tables 3 and 4, respectively.
Table 2. Antimicrobial susceptibility patterns of meningitis pathogens.
| Antimicrobial Agent | Resistance profile | Total Tested isolates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitive | Intermediate | Resistant | |||||||||||
| Number of: | ≈%(b)) | Number of: | ≈%(b)) | Number of: | ≈%(b)) | ||||||||
| G+(a) | G-(a) | total | G+(a) | G-(a) | total | G+(a) | G-(a) | total | |||||
| Ampicillin/sulbactam | 31 | 4 | 35 | 64.8 | 1 | 2 | 3 | 5.6 | 3 | 13 | 16 | 29.6 | 54 |
| Piperacillin/tazobactam | 30 | 11 | 41 | 78.8 | 0 | 0 | 0 | 0.0 | 3 | 8 | 11 | 21.2 | 52 |
| Ciprofloxacin | 6 | 12 | 18 | 51.4 | 1 | 1 | 2 | 5.7 | 6 | 9 | 15 | 42.8 | 35 |
| Chloramphenicol | 38 | 7 | 45 | 68.2 | 1 | 0 | 1 | 1.5 | 5 | 15 | 20 | 30.3 | 66 |
| Penicillin-G | 26 | 0 | 26 | 38.8 | 0 | 0 | 0 | 0.0 | 21 | 20 | 41 | 61.2 | 67 |
| Cefotaxime | 30 | 6 | 36 | 65.5 | 0 | 0 | 0 | 0.0 | 5 | 14 | 19 | 34.5 | 55 |
| Vancomycin | 43 | nd | 43 | 89.5 | 0 | nd | 0 | 0.0 | 5 | nd | 5 | 10.4 | 48 |
| Ceftriaxone | 31 | 5 | 36 | 61.0 | 0 | 1 | 1 | 1.7 | 7 | 15 | 22 | 37.3 | 59 |
| Amoxicillin/clavulanate | 28 | 4 | 32 | 64.0 | 0 | 0 | 0 | 0.0 | 5 | 13 | 18 | 36.0 | 50 |
| Amikacin | 9 | 10 | 19 | 59.4 | 0 | 1 | 1 | 3.1 | 2 | 10 | 12 | 37.5 | 32 |
| Levofloxacin | 35 | 10 | 45 | 70.3 | 0 | 2 | 2 | 3.1 | 10 | 7 | 17 | 26.6 | 64 |
| Ampicillin | 28 | 2 | 30 | 48.4 | 0 | 0 | 0 | 0.0 | 13 | 19 | 32 | 51.6 | 62 |
| Gentamicin | 5 | 11 | 16 | 51.6 | 0 | 2 | 2 | 6.4 | 6 | 7 | 13 | 41.9 | 31 |
| Aztreonam | 0 | 2 | 2 | 33.3 | 0 | 0 | 0 | 0.0 | 0 | 4 | 4 | 66.7 | 6 |
| Meropenem | 32 | 13 | 45 | 78.9 | 0 | 0 | 0 | 0.0 | 3 | 9 | 12 | 21.1 | 57 |
nd, not determined; G+, Gram-positive isolates, G-, Gram-negative isolates, ≈%(b), percentage from total number.
Table 3. Antimicrobial susceptibility patterns of Gram-positive pathogens.
| S. aureus (N = 6) | |||
| Antimicrobial agent | Resistant (%) | Intermediate (%) | Sensitive (%) |
| Penicillin G | 100 | 0 | 0 |
| Ampicillin | 100 | 0 | 0 |
| Ceftriaxone | 50 | 0 | 50 |
| Cefoxitin | 0 | 0 | 100 |
| Levofloxacin | 16.7 | 0 | 83.3 |
| Ciprofloxacin (n = 5) | 40 | 0 | 60 |
| Chloramphenicol | 33.3 | 0 | 66.7 |
| Piperacillin / tazobactam | 0 | 0 | 100 |
| Ampicillin / sulbactam | 16.7 | 16.7 | 66.7 |
| Gentamicin | 50 | 0 | 50 |
| Amikacin | 0 | 0 | 100 |
| Meropenem | 0 | 0 | 100 |
| Vancomycin | 16.7 | 0 | 83.3 |
| Coagulase-negative staphylococci (N = 5) | |||
| Antimicrobial agent | Resistant (%) | Intermediate (%) | Sensitive (%) |
| Penicillin G | 100 | 0 | 0 |
| Ampicillin | 100 | 0 | 0 |
| Cefotaxime | 60 | 0 | 40 |
| Ceftriaxone | 60 | 0 | 40 |
| Levofloxacin | 100 | 0 | 0 |
| Ciprofloxacin | 60 | 0 | 40 |
| Chloramphenicol | 60 | 0 | 40 |
| Piperacillin / tazobactam | 60 | 0 | 40 |
| Ampicillin / sulbactam | 40 | 0 | 60 |
| Amoxicillin / clavulanate | 60 | 0 | 40 |
| Amikacin | 40 | 0 | 60 |
| Gentamicin | 60 | 0 | 40 |
| Meropenem | 60 | 0 | 40 |
| Vancomycin | 40 | 0 | 60 |
| Enterococcus sp. (N = 3) | |||
| Antimicrobial agent | Resistant (%) | Intermediate (%) | Sensitive (%) |
| Penicillin G | 66.7 | 0 | 33.3 |
| Ampicillin | 33.3 | 0 | 66.7 |
| Levofloxacin | 33.3 | 0 | 66.7 |
| Chloramphenicol | 0 | 33.3 | 66.7 |
| Ciprofloxacin | 33.3 | 33.3 | 33.3 |
| Vancomycin | 33.3 | 0 | 66.7 |
| Other Streptococci (N = 3) | |||
| Antimicrobial agent | Resistant (%) | Intermediate (%) | Sensitive (%) |
| Penicillin G (n = 2) | 50 | 0 | 50 |
| Ampicillin | 33.3 | 0 | 66.7 |
| Cefotaxime (n = 2) | 50 | 0 | 50 |
| Chloramphenicol | 0 | 0 | 100 |
| Ceftriaxone | 33.3 | 0 | 66.7 |
| Vancomycin | 33.3 | 0 | 66.7 |
N, total number of isolates, n, number of tested isolates, R, resistant, I, intermediate, S, sensitive, %, percentage
Table 4. Antimicrobial susceptibility patterns of Gram-negative isolates.
| Enterobacteriaceae (N = 11) | |||
| Antimicrobial agent | Resistant (%) | Intermediate (%) | Sensitive (%) |
| Penicillin G (n = 10) | 100 | 0 | 0 |
| Ampicillin | 90.9 | 0 | 9.1 |
| cefotaxime | 54.5 | 0 | 45.5 |
| Ceftriaxone | 63.6 | 0 | 36.4 |
| Levofloxacin | 27.3 | 18.2 | 54.5 |
| Ciprofloxacin (n = 10) | 20 | 10 | 70 |
| Chloramphenicol | 54.5 | 0 | 45.5 |
| Piperacillin / tazobactam | 27.3 | 0 | 72.7 |
| Ampicillin / sulbactam | 54.5 | 9.1 | 36.4 |
| Amoxicillin / clavulanate | 63.6 | 0 | 36.4 |
| Gentamicin | 18.2 | 18.2 | 63.6 |
| Amikacin | 36.4 | 0 | 63.6 |
| Meropenem | 27.3 | 0 | 72.7 |
| Acinetobacter spp (N = 7) | |||
| Antimicrobial agent | Resistant (%) | Intermediate (%) | Sensitive (%) |
| Penicillin G | 100 | 0 | 0 |
| Ampicillin | 100 | 0 | 0 |
| cefotaxime | 85.7 | 0 | 14.3 |
| Ceftriaxone | 85.7 | 14.3 | 0 |
| Levofloxacin (n = 6) | 66.7 | 0 | 33.3 |
| Ciprofloxacin | 85.7 | 0 | 14.3 |
| Chloramphenicol | 100 | 0 | 0 |
| Piperacillin / tazobactam | 71.4 | 0 | 28.6 |
| Ampicillin / sulbactam | 100 | 0 | 0 |
| Amoxicillin / clavulanate | 100 | 0 | 0 |
| Gentamicin | 71.4 | 0 | 28.6 |
| Amikacin | 71.4 | 14.3 | 14.3 |
| Meropenem (n = 6) | 66.7 | 0 | 33.3 |
| P. aeruginosa (N = 2) | |||
| Antimicrobial agent | Resistant (%) | Intermediate (%) | Sensitive (%) |
| Penicillin G (n = 1) | 100 | 0 | 0 |
| Piperacillin / tazobactam | 0 | 0 | 100 |
| Levofloxacin | 0 | 0 | 100 |
| Ciprofloxacin | 0 | 0 | 100 |
| Chloramphenicol | 100 | 0 | 0 |
| Gentamicin | 0 | 0 | 100 |
| Amikacin | 0 | 0 | 100 |
| Meropenem | 50 | 0 | 50 |
| Azetronam | 0 | 0 | 100 |
| N. meningitidis (N = 2) | |||
| Antimicrobial agent | Resistant (%) | Intermediate (%) | Sensitive (%) |
| Penicillin G | 100 | 0 | 0 |
| Ampicillin | 100 | 0 | 0 |
| Levofloxacin | 0 | 0 | 100 |
| Chloramphenicol | 0 | 0 | 100 |
| Cefotaxime | 100 | 0 | 0 |
| Ceftriaxone | 100 | 0 | 0 |
| Ciprofloxacin | 50 | 0 | 50 |
| Amikacin | 50 | 0 | 50 |
| Meropenem | 50 | 0 | 50 |
N, total number of isolates, n, number of tested isolates, R, resistant, I, intermediate, S, sensitive, %, percentage
Identification of the multidrug-resistant (MDR) isolates
Out of the 71 isolates, 26 isolates (36.6%) were MDR isolates. Of these, 18 isolates (69.2%) were Gram-negative and 8 isolates (30.8%) were Gram-positive. Based on yellowish growth on mannitol salt agar and results of coagulase test of Gram positive isolate, 4 isolates were identified as S. aureus (50%), 3 isolates were coagulase negative Staphylococci (37.5%) and one isolate was Enterococcus (12.5%). S. aureus isolates (n = 6) were screened for methicillin resistance using cefoxitin disc diffusion method. The results showed that the six S. aureus isolates were methicillin sensitive S. auerus. (identified as”MSSA”). According to the microscopical, cultural and biochemical characteristics of the 18 MDR Gram-negative isolates, 7 (38.9%), 4 (22.2%), 2 (11.1%), 1 (5.5%), 1 (5.5%), 1 (5.5%), and 1 (5.5%) isolates were identified as Acinetobacter spp, K. pneumoniae, E. coli, Citrobacter spp, P. aeruginosa, N. meningitidis and Proteus spp, respectively.
Antibiogram analysis of the MDR isolates
As shown in Fig 3, all selected MDR isolates (n = 26) showed resistance to penicillin and ampicillin, their relative prevalence of resistance to other tested agents is shown in Tables 5 and 6.
Fig 3. Prevalence of the antimicrobial resistance of the 26 tested MDR isolates to different antimicrobial agents.
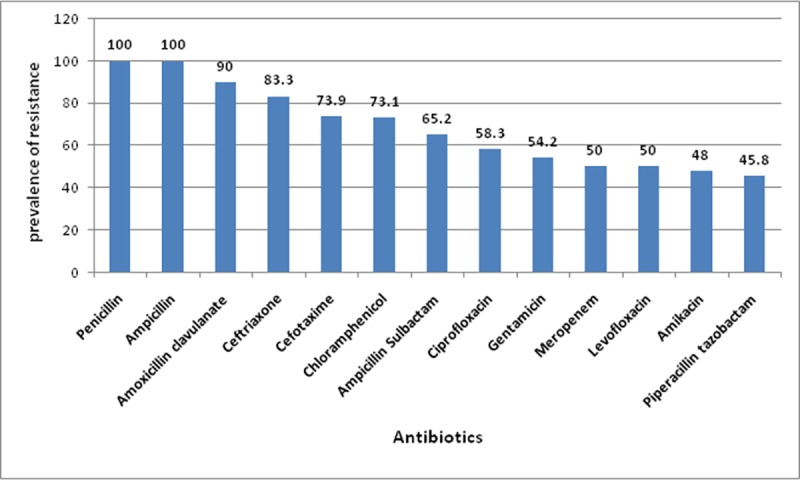
Prevalence was expressed as percent of resistant isolates relative to total tested isolates for each antimicrobial agent (n, 26).
Table 5. Antibiogram analysis of the MDR Gram negative isolates.
| Nr | isolate | Antibiotics | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AS | PT | Ci | CH | Pe | CT | Cx | AC | Ak | Le | No | Am | Ge | Az | Me | ||
| S 656 | K. pneumoniae | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| S 988 | K. pneumoniae | R | R | I | R | R | R | R | R | R | R | R | S | R | R | |
| S 414 | K. pneumoniae | S | S | S | R | R | R | R | R | S | S | S | R | S | S | |
| S 907 | K. pneumoniae | R | R | R | R | R | R | R | R | R | R | R | R | R | ||
| S 945 | Acinetobacter | R | R | R | R | R | R | R | R | R | R | R | R | R | R | |
| S 368 | Acinetobacter | R | R | R | R | R | R | R | R | R | R | R | R | R | R | |
| S 888 | Acinetobacter | R | R | R | R | R | R | R | R | R | R | R | R | R | R | |
| S 577 | Acinetobacter | R | R | R | R | R | R | R | R | R | R | R | R | R | R | |
| S 39 | Acinetobacter | R | S | S | R | R | S | I | S | S | S | R | S | S | ||
| S 306 | Acinetobacter | R | S | R | R | R | R | R | R | R | S | S | R | R | S | |
| S 130 | Acinetobacter | R | R | R | R | R | R | R | R | I | R | S | ||||
| S 414 | Citrobacter | I | S | S | S | R | S | R | R | S | S | S | R | S | R | S |
| S 85 | Citrobacter | S | S | S | S | R | S | S | S | S | S | S | R | S | R | S |
| S 85 | proteus | R | S | R | R | R | R | R | S | I | R | I | S | |||
| S 220 | N. meningitidis | R | S | R | R | R | R | R | ||||||||
| S 282 | E.coli | S | S | S | R | R | S | S | S | S | S | S | R | S | S | |
| S 215 | E.coli | R | S | S | S | R | R | R | R | R | I | S | R | I | S | |
| S 184 | P.aeruginosa | S | S | R | R | S | S | S | R | S | S | R | ||||
Abbreviations: AS, ampicillin-sulbactam; PT, Piperacillin-tazobactam; Ci, Ciprofloxacin; CH, Chloramphenicol; Pe, penicillin; CT, Cefotaxime; Cx, Ceftriaxone; AC, Amoxicillin -clavulanate; Ak, Amikacin; Le, Levofloxacin; No, Norfloxacin; Am, Ampicillin; Ge, Gentamicin, Az, Aztreonam; Me, Meropenem
Table 6. Antibiogram analysis of the MDR Gram positive isolates.
| Nr | Isolate | Antibiotics | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AS | PT | Ci | CH | Pe | CT | Va | Cx | AC | Ak | Le | Am | Ge | Me | ||
| S232 | S. aureus | R | S | R | S | R | S | R | R | S | R | R | R | S | |
| S353 | S. aureus | I | S | S | R | R | S | R | R | S | S | R | S | S | |
| S345 | S. aureus | S | S | S | S | R | S | S | S | R | S | S | R | R | S |
| S516 | S. aureus | S | S | R | R | R | S | R | S | S | R | R | S | ||
| S579 | S. coagulase-ve | R | R | R | R | R | R | S | R | R | S | R | R | R | R |
| S836 | S.coagulase-ve | S | R | R | R | R | R | R | R | R | R | R | R | R | R |
| S484 | S. coagulase-ve | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| S110 | Enterococcus | R | S | R | R | R | R | ||||||||
Abbreviations: AS, ampicillin-sulbactam; PT, Piperacillin-tazobactam; Ci, Ciprofloxacin; CH, Chloramphenicol; Pe, penicillin; CT, Cefotaxime; Va, vancomycin; Cx, Ceftriaxone; AC, Amoxicillin-clavulanate; Ak, Amikacin; Le, Levofloxacin; Am, Ampicillin; Ge, Gentamicin, Me, Meropenem
MIC results of some selected MDR isolates against some tested antimicrobial agents
The MIC results of some selected Gram negative and Gram positive MDR isolates are shown in Tables 7 & 8. The MDR K. pneumoniae and Acinetobacter spp isolates showed MIC values against ceftriaxone several fold shigher than 2μg/ml, therefore, they were considered as potential ESBLs producers according to the CLSI guidelines. For the MDR Gram negative isolates, the lowest resistance was observed to imipenem.
Table 7. MIC results for some selected MDR Gram-negative isolates (n = 15) against some selected antimicrobial agents.
| Isolate | Bacterial species | MIC value (μg/ml)against: | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AS | Cz | Cx | Am | Ge | Im | Cp | Le | ||
| S39 | Acinetobacter spp | 32\16 | 16 | 32 | 2 | 1 | 2 | 8 | 2 |
| R | I | I | S | S | S | S | S | ||
| S945 | Acinetobacter spp | 32\16 | 16 | 128 | 64 | 16 | 1 | 32 | 16 |
| R | I | R | R | R | S | R | R | ||
| S577 | Acinetobacter spp | 32\16 | 32 | 64 | 512 | 128 | 1 | 32 | 64 |
| R | R | R | R | R | S | R | R | ||
| S368 | Acinetobacter spp | 16\8 | 16 | 64 | 512 | 128 | 4 | 16 | 64 |
| I | I | R | R | R | S | I | R | ||
| S888 | Acinetobacter spp | 64\32 | 32 | 256 | 64 | 64 | 8 | 32 | 64 |
| R | R | R | R | R | I | R | R | ||
| S306 | Acinetobacter spp | 16\8 | 32 | 64 | 64 | - | 1 | 8 | 2 |
| I | R | R | R | - | S | S | S | ||
| S656 | K. pneumoniae | 256\128 | 32 | >64 | 64 | 16 | 16 | 32 | 16 |
| R | R | R | R | R | R | R | R | ||
| S988 | K. pneumoniae | 256\128 | 128 | 64 | 512 | 2 | 2 | 256 | 16 |
| R | R | R | R | S | I | R | R | ||
| S907 | K. pneumoniae | >256\128 | 128 | >64 | 64 | - | 8 | >256 | 8 |
| R | R | R | R | - | R | R | R | ||
| S282 | E. coli | 8\4 | 32 | 16 | 16 | 4 | 0.5 | 8 | 4 |
| S | R | R | S | S | S | S | I | ||
| S215 | E. coli | 16\8 | 256 | >64 | 512 | 32 | 0.5 | 256 | 8 |
| I | R | R | R | R | S | R | R | ||
| S85 | Citrobacter spp | 8\4 | 16 | 16 | 16 | 8 | 0.5 | 0.5 | 2 |
| S | R | R | S | I | S | S | S | ||
| S414 | Citrobacter spp | 16\8 | 64 | 64 | 16 | 4 | 0.5 | 16 | 4 |
| I | R | R | S | S | S | I | I | ||
| S85 | Proteus spp | 64\32 | 16 | 64 | 32 | 16 | 0.5 | 1 | 8 |
| R | R | R | I | R | S | S | R | ||
| S184 | P. aeruginosa | 4 | - | 4 | 0.5 | 8 | 4 | 1 | |
| S | - | S | S | I | S | S | |||
Abbreviations: AS, ampicillin-sulbactam; Cz, Ceftazidime; Cx, Ceftriaxone; Ak, Amikacin; Ge, Gentamicin, Im, Imipenem; Cp, Cefepime; Le, levofloxacin
Table 8. MIC results for some selected MDR Gram-positive isolates (n = 5) against some selected antimicrobial agents.
| Nr | Isolate | MIC value (μg/ml) against | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AS | Do | Cx | Ak | Va | Im | Cp | Le | ||
| S 516 | S. aureus | 8\4 | 64 | 64 | 16 | 8 | 2 | 8 | 2 |
| S | R | R | S | I | S | S | I | ||
| S 353 | S. aureus | 16\8 | 16 | 64 | 16 | 64 | 1 | 16 | 4 |
| I | R | R | S | R | S | I | R | ||
| S 345 | S. aureus | 2 | 16 | 2 | 4 | 2 | 0.25 | 8 | 1 |
| S | R | S | S | S | S | S | S | ||
| S 484 | S. coagulase-ve | 32\16 | 32 | 128 | 512 | 32 | - | 64 | >128 |
| R | R | R | R | R | - | R | R | ||
| S 836 | S. coagulase-ve | 64\32 | 16 | 512 | >512 | 256 | 1 | 16 | 32 |
| R | R | R | R | R | S | I | R | ||
Abbreviations: AS, ampicillin-sulbactam; Do, doxycycline;; Cx, Ceftriaxone; Ak, Amikacin; Va, vancomycin; Im, Imipenem; Cp, Cefepime; Le, Levofloxacin.
MIC results of certain antibiotic combinations for some selected MDR Gram-negative isolates (n = 10)
Ten MDR Gram-negative isolates were selected to be tested against four different antibiotic combinations, and fractional inhibitory concentration (FIC) values were calculated for each isolate. It was found that ampicillin/sulbactam+cefepime combination showed synergism against 8 tested isolates (80%) and additive effect against 2 isolates (20%). Results of the four tested antibiotic combinations including: ampicillin/sulbactam+cefepime; ampicillin/sulbactam+amikacin; ampicillin/sulbactam+ levofloxacin; amikacin + levofloxacin are shown in Tables 9, 10, 11 and 12, respectively.
Table 9. FIC values of Ampicillin/sulbactam plus Cefepime combination.
| Isolate code | Isolate | Ampicillin/sulbactam | Cefepime | Combination | FIC value | Interpretation | ||
|---|---|---|---|---|---|---|---|---|
| S85 | Proteus spp | 64/32 | R | 1 | S | 0.5/0.5 | 0.5 | Synergism |
| S215 | E. coli | 16/8 | I | 256 | R | 1/1 | 0.066 | Synergism |
| S414 | Citrobacter spp | 16/8 | I | 16 | I | 8/8 | 1 | Additive |
| S656 | K. pneumoniae | 256/128 | R | 32 | R | 16/16 | 0.562 | Additive |
| S907 | K. pneumoniae | >256/128 | R | >256 | R | 32/32 | 0.25 | Synergism |
| S988 | K.pneumoniae | 256/128 | R | 256 | R | 16/16 | 0.125 | Synergism |
| S888 | Acinetobacter spp | 64/32 | R | 32 | R | 8/8 | 0.375 | Synergism |
| S945 | Acinetobacter spp | 32/16 | R | 32 | R | 4/4 | 0.25 | Synergism |
| S368 | Acinetobacter spp | 16/8 | I | 16 | I | 4/4 | 0.5 | Synergism |
| S577 | Acinetobacter spp | 32/16 | R | 32 | R | 8/8 | 0.5 | Synergism |
Table 10. FIC values of Ampicillin/sulbactam plus amikacin combination.
| Isolate code | Isolate | Ampicillin/ sulbactam |
Amikacin | Combination |
FIC value | Interpretation | ||
|---|---|---|---|---|---|---|---|---|
| S85 | Proteus spp | 64/32 | R | 32 | I | 16/32 | 1.25 | Indifference |
| S215 | E. coli | 16/8 | I | 512 | R | 64/128 | 4.25 | Antagonism |
| S414 | Citrobacter spp | 16/8 | I | 16 | S | 2/4 | 0.375 | Synergism |
| S656 | K. pneumoniae | 256/128 | R | 64 | R | 128/256 | 4.5 | Antagonism |
| S907 | K. pneumoniae | >256/128 | R | 64 | R | 128/256 | 4.5 | Antagonism |
| S988 | K. pneumoniae | 256/128 | R | 512 | R | 64/128 | 0.5 | Synergism |
| S888 | Acinetobacter spp | 64/32 | R | 64 | R | 128/256 | 6 | Antagonism |
| S945 | Acinetobacter spp | 32/16 | R | 64 | R | 32/64 | 2 | Indifference |
| S368 | Acinetobacter spp | 16/8 | I | 512 | R | 128/256 | 8.5 | Antagonism |
| S577 | Acinetobacter spp | 32/16 | R | 512 | R | 128/256 | 4.5 | Antagonism |
Table 11. FIC values of Ampicillin/sulbactam plus Levofloxacin combination.
| code | Isolate | Ampicillin/ sulbactam | levofloxacin | Combination | FIC value | Interpretation | ||
|---|---|---|---|---|---|---|---|---|
| S85 | Proteus spp | 64/32 | R | 8 | R | 64/32 | 5 | Antagonism |
| S215 | E. coli | 16/8 | I | 8 | R | 64/32 | 8 | Antagonism |
| S414 | Citrobacter spp | 16/8 | I | 4 | I | 8/4 | 1.5 | Indifference |
| S656 | K.pneumoniae | 256/128 | R | 16 | R | 128/64 | 4.5 | Antagonism |
| S907 | K. pneumoniae | >256/128 | R | 8 | R | 32/16 | 2.125 | Indifference |
| S988 | K. pneumoniae | 256/128 | R | 16 | R | 32/16 | 1.125 | Indifference |
| S888 | Acinetobacter spp | 64/32 | R | 64 | R | 128/64 | 3 | Indifference |
| S945 | Acinetobacter spp | 32/16 | I | 16 | R | 64/32 | 4 | Indifference |
| S368 | Acinetobacter spp | 16/8 | I | 64 | R | 64/32 | 4.5 | Antagonism |
| S577 | Acinetobacter spp | 32/16 | R | 64 | R | 128/64 | 5 | Antagonism |
Table 12. FIC values of Amikacin plus Levofloxacin combination.
| Isolate code | Isolate | Amikacin | Levofloxacin | Combination | FIC value | Interpretation | ||
|---|---|---|---|---|---|---|---|---|
| S85 | Proteus spp | 32 | I | 8 | R | 128/32 | 8 | Antagonism |
| S215 | E. coli | 512 | R | 8 | R | 64/16 | 2.125 | Indifference |
| S414 | Citrobacter spp | 16 | S | 4 | I | 32/8 | 4 | Indifference |
| S656 | K. pneumoniae | 64 | R | 16 | R | 25/64 | 8 | Antagonism |
| S907 | K. pneumoniae | 64 | R | 8 | R | 64/16 | 3 | Indifference |
| S988 | K.pneumoniae | 512 | R | 16 | R | 64/16 | 1.125 | Indifference |
| S888 | Acinetobacter spp | 64 | R | 64 | R | 256/64 | 5 | Antagonism |
| S945 | Acinetobacter spp | 64 | R | 16 | R | 64/16 | 2 | Indifference |
| S368 | Acinetobacter spp | 512 | R | 64 | R | 256/64 | 1.5 | Indifference |
| S577 | Acinetobacter spp | 512 | R | 64 | R | 256/64 | 1.5 | Indifference |
R: resistance, I: intermediate, S: sensitive. FIC: fractional inhibitory conc. Synergism≤0.5, Additive≤1,Indifference >1and≤4.0, Antagonism>4.
MIC results of some selected antibiotic combination for two MDR Gram-positive isolates:
MDR Gram positive bacteria, 2 isolates were tested against five different antibiotic combinations and the FIC values were calculated for each combination. Synergism was observed with doxycycline+levofloxacin combination. Table 13 shows the results of the antibiotic combinations used.
Table 13. FIC values calculated for 5 different antibiotic combinations for MDR Gram-positive isolates.
| For Ampicillin/sulbactam plus Cefepime combination | ||||||
| Isolate code | Bacterial species | Ampicillin/ sulbactam | Cefepime | Combination | FIC value | Interpretation |
| S345 | S.aureus | 2/1 | 8 | 4/4 | 2.5 | Indifference |
| S836 | S. coagulase-ve | 64/32 | 16 | 8/8 | 0.62 | Additive |
| For Vancomycin plus Levofloxacin combination | ||||||
| Vancomycin | Levofloxacin | |||||
| S345 | S.aureus | 2/1 | 1 | 4/2 | 4 | Indifference |
| S836 | S. coagulase-ve | 256 | 32 | 128/64 | 2.5 | Indifference |
| For Doxycycline plus Levofloxacin combination | ||||||
| Doxycycline | Levofloxacin | |||||
| S345 | S.aureus | 16 | 1 | 0.25/0.25 | 0.26 | Synergism |
| S836 | S. coagulase-ve | 16 | 32 | 4/4 | 0.37 | Synergism |
| For Ampicillin/sulbactam plus Vancomycin combination | ||||||
| Ampicillin/ sulbactam | Vancomycin | |||||
| S345 | S.aureus | 2/1 | 2 | 4/4 | 4 | Indifference |
| S836 | S. coagulase-ve | 64/32 | 256 | 64/64 | 1.25 | Indifference |
| For Doxycycline plus Amikacin combination | ||||||
| Doxycycline | Amikacin | |||||
| S345 | S.aureus | 16 | 4 | 2/8 | 2.125 | Indifference |
| S836 | S. coagulase-ve | 16 | 512 | 4/16 | 0.28 | Synergism |
S. aureus, Staphylococcus aureus; S. coagulase-ve,coagulase negative Staphylococcus; R: resistant, I: intermediate, S:sensitive. FIC: fractional inhibitory conc. Synergism≤0.5, Additive≤1, Indifference> 1 and ≤4.0, Antagonism>4.
Detection of antibiotic resistance genes using PCR
Plasmid bands were not detected in any of tested 19 MDR isolates, however, when subjected to PCR gene detection, only 9 isolates gave positive results (47.36%).The PCR products of tem, shv, ctx-m obtained from chromosomal DNA of K. pneumoniae isolate S907 and the PCR product of aac(6') obtained from using chromosomal DNA of Acinetobacter spp, isolate S888 were analyzed, annotated and submitted into the GenBank database under accession numbers KX214665,KX214664,KX214663, KX214662, respectively. Prevalence of resistance genes investigated were: tem (21.1%), shv (15.8%), ctx-m (15.8%) coding TEM-, SHV, CTX-M extended spectrum beta-lactmases (ESBLs), respectively; aac(6')-Ib (26.32%) coding for aminoglycoside 6’-N-acetyltransferase type Ib ciprofloxacin resistant variant; and qnrA (5.3%) gene coding for quinolone resistance (Fig 4). The genotypic characteristics of the MDR resistant isolates were also analyzed (Table 14).
Fig 4. Prevalence of some selected antibiotic resistance genes among MDR bacterial pathogens.
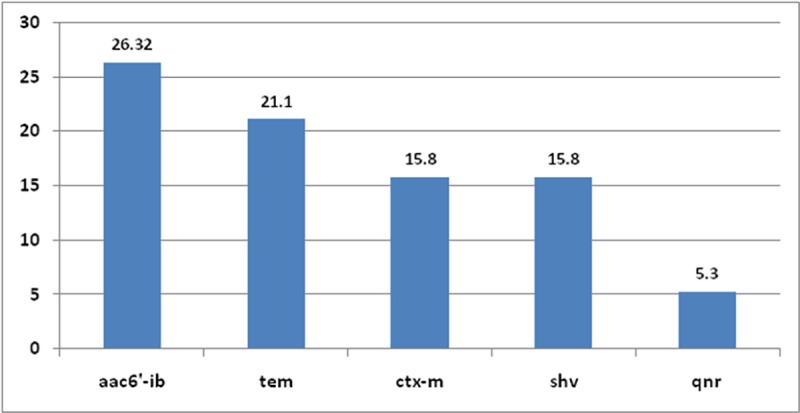
Table 14. Genotypic detection of resistant gene using chromosomal DNA.
| Specimen NO | Isolate | ctx-m | shv | tem | aac(6)-ib\ibcr | qnr |
|---|---|---|---|---|---|---|
| S 184 | P. aeruginosa | - | - | - | - | nd |
| S 656 | K. pneumoniae | - | + | - | - | - |
| S 907 | K. pneumoniae | + | + | + | - | - |
| S 988 | K. pneumoniae | - | - | - | - | - |
| S 414 | K. pneumoniae | - | - | - | nd | nd |
| S 888 | Acinetobacter spp | - | - | + | + | + |
| S 577 | Acinetobacter spp | - | + | - | + | - |
| S 368 | Acinetobacter spp | - | - | + | - | - |
| S 945 | Acinetobacter spp | + | - | + | + | - |
| S 306 | Acinetobacter spp | - | - | - | - | - |
| S130 | Acinetobacter spp | - | - | - | - | nd |
| S 414 | Citrobacter spp | - | - | - | nd | nd |
| S 282 | E. coli | - | - | - | nd | nd |
| S 215 | E. coli | + | - | - | - | - |
| S 85 | Proteus spp | - | - | - | - | - |
| S 220 | N. meningitidis | - | - | - | + | - |
| S836 | S. coagulase-ve | - | - | - | + | - |
| S 484 | S. coagulase-ve | - | - | - | - | - |
| S 579 | S. coagulase-ve | - | - | - | nd | - |
(-), absent, (+), present, nd, not detected.
Discussion
Bacterial meningitis continues to be a potentially life threatening disease with substantial morbidity and mortality throughout the world. It represents even more significant problem in many other regions of the world, especially in developing countries [1,25]. The uncontrolled use of antibiotics and their overuse lead to rapid and extensive spread of antimicrobial resistance [26]. Antimicrobial resistance (AMR) limits effective treatment and prevention of the increasing range of infections caused by bacteria, which represents critical threat to global public health, as new antimicrobial resistance mechanisms continue to emerge and spread globally [26]. It is obvious that bacterial pathogens will continue to develop resistance against currently available antibacterial agent either through developing newly genetic mutations or exchange of genetic information.
In the current study, a total of 71 clinical bacterial isolates were recovered from CSF specimens collected through two years period from patients with bacterial meningitis. These clinical specimens were CSF. Using Gram-stain, 48 isolates (67.6%) were found to be Gram-positive and 23 isolates (32.4%) were found to be Gram-negative, From 48 Gram positive isolates the majority were S. pneumoniae constituting 31 isolates (64.6%,n = 48) indicating that Gram positive bacteria mainly pneumococcal meningitis responsible for highest contribution to bacterial meningitis in Egypt. Comparing our results with a study conducted also in Egypt it was found that S. pneumoniae was recently described as the leading cause of bacterial meningitis [27–29], reflecting a change in disease epidemiology since N. meningitides was for a long time the main causative pathogen of bacterial meningitis [30–34].Results obtained in this study where Gram positive bacterial were of the highest prevalence, particularly Staphylococci and Streptococcus(S.) pneumoniae were in accordance to those obtained from previous studies conducted in USA hospital laboratories from 2000–2002 [35]as well as from a teaching hospital in Ghana[36].
In another prospective, multicenter, observational study of 156 consecutive adults hospitalized for pneumococcal meningitis, S.pneumoniae was found to be the most common bacterium isolated from adults with community- acquired meningitis, [37]. The antimicrobial susceptibility testing of the Gram-positive isolates collected in this study (n = 48) showed that the lowest resistance was observed to meropenem, ampicillin/sulbactam and piperacillin/tazobactam only 3 isolates (8.57%, 8.57%, 9.1%) were resistant to each. Contrary to that, the highest resistance was observed with penicillin where 21 isolates (44.7%) showed resistance. The antimicrobial susceptibility testing of the 23 Gram-negative isolates collected in this study showed that the lowest resistance was observed to levofloxacin and gentamicin. Thus both bacterial categories recorded low resistance pattern to meropenem, levofloxacin and piperacillin/tazobactam. The highest resistance was observed to penicillin and ampicillin where 41 isolates (61.2%) were resistant to penicillin and 32 isolates (51.6%) were resistant to ampicillin. Several studies reported that penicillin resistant S. pneumonia showing an increase in resistance pattern over time [28, 30, 38–40]. However, in the current study only 7 isolates out of 31 tested isolates of S. pneumoniae were resistant to penicillin.
In another study conducted in Lebanon, among 44 tested S. pneumoniae isolates using E-tests 35 isolates were resistant to penicillin [41]. Among the 71 collected isolates, 26 isolates (36.6%) were found to be resistant to three or more antimicrobial classes, 18 of these isolates were Gram-negative (69.2%), while 8 isolates were Gram-positive (30.8%). For MDR Gram negative isolates the highest resistance was observed to penicillin and ampicillin, the lowest resistance was observed to levofloxacin, gentamicin and piperacillin/tazobactam. For MDR Gram positive isolates the highest resistance was observed to penicillin and ampicillin and lowest resistance was observed to amikacin, piperacillin/tazobactam, and meropenem. It can be concluded that MDR organisms will evolve on a continuous basis, compromising antimicrobial efficacy and will represent a treatment challenge for microbial infections.
Multidrug-resistant Acinetobacter spp was the most prevalent Gram-negative MDR pathogen (n = 7/26; 27%). All tested isolates showed resistance to ampicillin/sulbactam, chloramphenicol, penicillin and ampicillin. However, resistance was absent to imipenem where none of the isolates (0%) were resistant. Comparing our results with a study conducted in India stating that since 1975, increasing antimicrobial resistance against Acinetobacter started to appear in almost all groups of antibiotics including the first and second generation cephalosporins. Initially Acinetobacter isolates retained partial susceptibility against the third and fourth generation cephalosporins, fluoroquinolones, aminoglycosides, and carbapenems, with almost 100% isolates retaining susceptibility towards imipenem [42–44]. By the late 1990s, carbapenems were the only valuable agents remaining that could treat many severe Acinetobacter infections. However, due to clonal spread of carbapenem resistance Acinetobacter baumannii strains, the therapeutic options are decreasing [45–47]. This resistance has been found to be attributed to various mechanisms [48].
Along with our study, a study conducted in USA revealed that the response of Acinetobacter spp isolates to the combination of cefepime and ampicillin/sulbactam showed that no antagonistic interactions were recorded. Nine isolates (26.5%) showed synergism (FIC, ≤0.5), 21 isolates (61.8%) showed partial synergism. While four isolates (11.8%) showed an additive effect. Cefepime MIC values were ≤8 mg/L to 85.3% of strains, while ampicillin/sulbactam MIC values were ≤16/8 mg/L (intermediate) for all tested isolates when these antibiotics were tested in combination [49]. In a study conducted in China to investigate the mechanism of drug resistance of carbapenems-resistant Acinetobacter baumannii (CRAB) in burn patients, the antimicrobial activity of drugs combination against these bacteria in vitro showed that, the synergic, additive, indifferent, and antagonistic effects were respectively observed in 40, 33, 6, and 15 strains applied with combination of amikacin and ampicillin/sulbactam [50]. While in the current study out of 4 tested Acinetobacter isolates none showed synergism, 3 isolates showed antagonism while 1 isolate showed indifference. In another study conducted in USA two clinical strains of carbapenemase (KPC)–producing K. Pneumoniae were investigated. Various combinations including amikacin, doripenem, levofloxacin, and rifampin were quantitatively assessed, time-kill studies were conducted using clinically relevant concentrations and it was found that, amikacin plus levofloxacin was found to have antagonistic effect, [51]. While in the current study out of 3 tested MDR K. pneumoniae, one isolate showed antagonism while the two other isolates showed indifference. This reveals the significance of avoiding empirical selection of antimicrobial combinations, particularly for infections involving MDR organisms in which high mortality may already be likely [52]
The detection of genetic determinants associated with MDR resistance isolates, plasmid bands were absent in 19 tested MDR isolates. While extracted DNA of 19 isolates (16 Gram negative and 3 Gram positive MDR isolates) using PrepMan Ultra kit in an attempt to detect resistance genes from bacterial chromosome. Out of MDR 19 isolates subjected to PCR gene detection only 9 isolates gave positive results (47.36%), other isolates showed negative results despite their positive resistance phenotype. In a study conducted also in china, MDR K. pneumoniae strains isolated from patients in intensive care units (ICUs) demonstrated β-lactamase genes, such as blaSHV (22/38), blaTEM (10/38) and blaCTX-M (7/38) [53]. While In the present study out of the four tested MDR K. pneumoniae, resistance genes were only detected in two isolates, both isolates harbored shv gene, while ctx-m and tem genes were detected in only one of them.
A population-based laboratory surveillance study of ESBL-producing E. coli infections conducted in Canada reported that 70% of the ESBL in E. coli isolated were of the CTX-M-type. [54]. A study by Jorgensen et al. conducted in 2007, indicated that CTX-M-type ESBLs had emerged in E. coli and other species of Enterobacteriaceae in USA [55]. Moreover, recent studies from Philadelphia, Chicago, and Pittsburgh revealed a high prevalence of CTX-M-producing E. coli ST131 in adult patients [56–58]. However, despite extensive studies of ESBL-producing microorganisms there is a lack of information regarding development and spread of ESBL-mediated resistance, specifically CTX-M, in Gram-negative infections [59]. In the present study from two MDR E. coli only one isolate expressed ctx-m gene. CTX gene was also found in one Acinetobacter and K.pneumoniae isolates. Quinolone resistance is mainly mediated through chromosomal mutations in bacterial topoisomerase genes or genes that regulate the expression of efflux pumps [60,61]. In a study conducted in the USA, 313 Enterobacteriaceae isolates including E. coli, K. pneumoniae and Enterobacter with ciprofloxacin MIC of ≥0.25 μg/ml and reduced susceptibility to ceftazidime, were tested to detect presence of aac(6′)-Ib and qnr genes. The results indicated the presence of aac(6′)-Ib in 50.5% of isolates, and of these isolates, 28% carried the cr variant associated with low-level ciprofloxacin resistance. Moreover, aac(6′)-Ib-cr gene proved to have geographic widespread, stability over time, most prevalent in E. coli and not associated with presence of qnr genes [62]. However in the current study aac(6′)-Ib was detected in 4 MDR Gram negative isolates and one coagulase negative Staphylococcus isolate, but it was absent in tested MDR K. pneumoniae and E. coli isolates. Plasmid-borne quinolone resistance qnr genes have been found in clinical isolates of Enterobacteriaceae [63]. In the current study qnr gene was detected in only one MDR Acinetobacter isolate, as qnr genes is most predominant in acquired plasmid rather than bacterial chromosome.
Conclusion
Piperacillin/tazobactam, levofloxacin, meropenem and ampicillin/sulbactam had a favorable sensitivity pattern among Gram-positive and Gram-negative pathogens; thus they are recommended for treatment of bacterial meningitis. MIC results showed that lowest resistance for MDR gram negative isolates was to imipenem, however its epiliptogenic side effect limits its use in meningitis especially in pediatrics. It was found that FIC values of ampicillin/sulbactam plus cefepime combination gave either synergism or additive effect against MDR Gram negative isolates, thus it is recommended to be used as a treatment option. While MDR Gram positive isolates showed synergism with doxycycline plus levofloxacin combination. Genotypic analysis detected that the antibiotic resistance of bacteria causing bacterial meningitis is mainly chromosomal mediated, several resistant genes were detected including ESBLs (tem, ctx-m, shv) accounting for resistance to β-lactam antibiotics, also aac(6')-Ib resistance gene accounting for resistance to aminoglycoside (amikacin). It is recommended to monitor drug-resistant isolates and consider rational use of antimicrobials agents in order to limit the spread and prevalence of the underlying resistance mechanisms.
Acknowledgments
Hereby we acknowledge and thank Microbiology and immunology department, faculty of Pharmacy, Ain Shams University for carrying out part of the practical experiments. We all thank all laboratory staff members of Abssia Fever Hospital, Ain Shams University Hospital, Ain Shams Specialized Hospital for providing us with CSF clinical specimens and all the required facilities.
Abbreviations
- FIC
fractional inhibitory concentration
- tem, shv, ctx-m
genes coded for TEM-, SHV, CTX-M extended spectrum beta-lactmases, respectively
- ESBLs
extended-spectrum beta-lactmases
- aac(6')-Ib
gene coded for aminoglycoside 6’-N-acetyltransferase type Ib ciprofloxacin resistant variant
- qnrA
gene coded for quinolone resistance
- CSF
cerebrospinal fluid
- MICs
minimum inhibitory concentrations
- Ta
calculated annealing temperatures
- AMR
Antimicrobial resistance
- PCR
polymerase chain reaction
Data Availability
All relevant data are within the manuscript.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Brouwer MC, Tunkel AR, van de Beek D.Epidemiology, diagnosis, and antimicrobial treatment of acute bacterial meningitis.J ClinMicrobiol.2010; 23(3):467–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rafi W, Chandramuki A, Mani R, Satishchandra P, Krishna S.Rapid diagnosis of acute bacterial meningitis: role of broad range 16S rRNA PCR.J Emerg Med.2010;38(2):225–30. 10.1016/j.jemermed.2008.02.053 [DOI] [PubMed] [Google Scholar]
- 3.Won H, Yang S, Gaydos C, Hardick J, Ramachandran P, Hsieh YH, et al. A broad range assay for rapid detection and etiologic characterization of bacterial meningitis: performance testing in samples from sub-Sahara. J Diagn Microbiol Infect Dis.2012;74(1): 22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van de Beek D, de Gans J, Spanjaard L, Weisfelt M, Reitsma JB, Vermeulen M. Clinical features and prognostic factors in adults with bacterial meningitis. N Engl J Med. 2004;351(18):1849–59. 10.1056/NEJMoa040845 [DOI] [PubMed] [Google Scholar]
- 5.Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. Meningococcal Disease. N Engl J Med.2001;344(18):1378–88. 10.1056/NEJM200105033441807 [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000.Lancet. 2012;379(9832):2151–61. 10.1016/S0140-6736(12)60560-1 [DOI] [PubMed] [Google Scholar]
- 7.Van de Beek D. Progress and challenges in bacterial meningitis. Lancet 2012; 380(9854):1623–4. 10.1016/S0140-6736(12)61808-X [DOI] [PubMed] [Google Scholar]
- 8.Thigpen MC, Whitney CG, Messonnier NE, Zell ER, Lynfield R, Hadler JL, et al. Bacterial meningitis in the United States, 1998–2007. N Engl J Med. 2011;364(21): 2016–25. 10.1056/NEJMoa1005384 [DOI] [PubMed] [Google Scholar]
- 9.Van de Beek D, deGans J, Tunkel AR, Wijdicks EF. Community-acquired bacterial meningitis in adults. N Engl J Me. 2006; 354(1):44–53. [DOI] [PubMed] [Google Scholar]
- 10.Van de Beek D, Brouwer MC,Thwaites GE, Tunkel AR. Advances in treatment of bacterial meningitis. Lancet. 2012;380(9854):1623–4. [DOI] [PubMed] [Google Scholar]
- 11.Alekshun MN, LevySB. Molecular mechanisms of antibacterial multidrug resistance.Cell 2007;128(6):1037–50. 10.1016/j.cell.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 12.Gale EF, Cundliffe E, Reynolds P E, Richmond MH, and Waring M. J. (ed.). The molecular basis of antibiotic action, 2nd ed. John Wiley, Chichester, United Kingdom, 1981. [Google Scholar]
- 13.Walsh C. Antibiotics: actions, origins, resistance ASM Press, Washington, DC, 2003. [Google Scholar]
- 14.Davies J, Davies D. Origins and Evolution of Antibiotic Resistance. Microbiol Mol Biol Rev. 2010;74(3):417–33. 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Varela MF. Molecular mechanisms of bacterial resistance to antimicrobial agents.Book; Microbial pathogens and strategies for combating them: science, technology and education, book edited by A. Méndez-Vilas, published: December2013, Formatex Research Center. Chapter 56 p 522–534. http://www.formatex.info/microbiology4/
- 16.CLSI document M100-S21. Performance Standards for Antimicrobials Susceptibility Testing; Twenty-First Informational Supplement. 2011.
- 17.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. : Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18(3): 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 18.Hsieh MH, Yu CM, Yu VL,Chow JW. Synergy assessed by checkerboard. A critical analysis. Diagn Microbiol Infect Dis. 1993;16(4):343–9. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Russell D. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press 2001; USA. ISBN: 0-879-69577-3.
- 20.Bonnet R, Dutour C, Sampaio JL, Chanal C, Sirot D, Labia R, et al. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240 à Gly.Antimicrob Agents Chemother.2001;45(8): 2269–2275. 10.1128/AAC.45.8.2269-2275.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasheed JK., Jay C, Metchock B, Berkowitz F, Weigel L, Crellin J, et al. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents Chemother. 1997;41(3): 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamed SM., Aboshanab KM, Elkhatib WF, Ashour MS. Aminoglycoside Resistance Patterns of Certain Gram Negative Uropathogens Recovered from Hospitalized Egyptian Patients.British Microbiol Res J 2013;3(4): 448–460. [Google Scholar]
- 23.Abdel-Aziz S, Aboshanab KM, Aboulwafa MM, Hassouna NA. Phenotypic and genotypic studies on antimicrobial resistance of lower respiratory tract bacterial pathogens.ActaMicrobiol.2015;6(1:1) [Google Scholar]
- 24.Staden R. The staden sequence analysis package. Molecular Biotechnology 1996;5(3): 233–41. [DOI] [PubMed] [Google Scholar]
- 25.Carbonnelle E. Laboratory diagnosis of bacterial meningitis: usefulness of various tests for the determination of the etiological agent. Med Mal Infect. 2009;39(7–8): 581–605. 10.1016/j.medmal.2009.02.017 [DOI] [PubMed] [Google Scholar]
- 26.Llor C, Bjerrum L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf.2014;5(6): 229–41. 10.1177/2042098614554919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ministry of Health and Population, Egypt. Enhanced Surveillance for Communicable Diseases, annual summary January- December 2000 report.[http://www.geis.fhp.osd.mil/GEIS/Training/EgyptSurv2000.htm]. US Department of Defense Global Emerging Infections Surveillance and Response System, 2000.
- 28.Youssef FG, El-Sakka H, Azab A, Eloun S, Chapman GD, Ismail T, et al. Etiology, antimicrobial susceptibility profiles, and mortality associated with bacterial meningitis among children in Egypt. Ann Epidemiol.2004;14(1):44–8. [DOI] [PubMed] [Google Scholar]
- 29.Afifi S, Wasfy MO, Azab MA, Youssef FG, Pimentel G, Graham TW, et al. Laboratory based surveillance of patients with bacterial meningitis in Egypt (1998–2004).Eur J Clin Microbiol Infect Dis.2007;26(5):331–40. 10.1007/s10096-007-0280-x [DOI] [PubMed] [Google Scholar]
- 30.Boctor WM.Statistical analysis of bacterial meningitis at embaba fever hospital.J Egypt Public Health Assoc.1969;44(4):253–9. [PubMed] [Google Scholar]
- 31.Girgis NI, Yassin MW, Sanborn WR, Burdick RE, El-Ela HA, Kent DC, et al. Ampicillin compared with penicillin and chloramphenicol combined in the treatment of bacterial meningitis. J Trop Med Hyg.1972;75(8):154–7. [PubMed] [Google Scholar]
- 32.Miner WF, Edman DC. Acute bacterial meningitis in Cairo, Arab Republic of Egypt, 1 January 1971 through 31 December 1975. Am J Trop Med Hyg 1978;27(5):986–94. [DOI] [PubMed] [Google Scholar]
- 33.Girgis NI, Sippel JE, Kilpatrick ME, Sanborn WR, Mikhail IA, Cross E, et al. Meningitis and encephalitis at the Abbassia fever hospital, Cairo, Egypt, from 1966 to 1989. Am J Trop Med Hyg.1993;48(1):97–107. [DOI] [PubMed] [Google Scholar]
- 34.Shaban L, and Siam R. Prevalence and antimicrobial resistance pattern of bacterial meningitis in Egypt. Ann Clin Microbiol Antimicrob. 2009;8:26 10.1186/1476-0711-8-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones ME, Draghi DC, Karlowsky JA, Sahm DF, Bradley JS. Prevalence of antimicrobial resistance in bacteria isolated from central nervous system specimens as reported by U.S. hospital laboratories from 2000 to 2002.Ann Clin Microbiol Antimicrob. 2004; 3:3 (http://www.ann-clinmicrob.com/content/3/1/3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owusu M, Nguah SB, Boaitey YA, Badu-Boateng E, Abubakr A, Lartey RA, et al. Aetiological agents of cerebrospinal meningitis: a retrospective study from a teaching hospital in Ghana. Ann ClinMicrobiolAntimicrob. 2012; 11:28 (http://www.ann-clinmicrob.com/content/11/1/28). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auburtin M, Wolff M, Charpentier J, Varon E, Le Tulzo Y,Girault C, et al. Detrimental role of delayed antibiotic administration and penicillin-non susceptible strains in adult intensive care unit patients with pneumococcal meningitis: The PNEUMOREA prospective multicenter study. J Crit Care Med.2006;34(11):2758–65. [DOI] [PubMed] [Google Scholar]
- 38.Ostroff SM, Harrison LH, Khallaf N, Assaad MT, Guirguis NI, Harrington S, et al. Resistance patterns of Streptococcus pneumoniae and Haemophilus influenzae isolates recovered in Egypt from children with pneumonia. The antimicrobial resistance surveillance study group. Clin Infect Dis.1996;23(5):1069–74. [DOI] [PubMed] [Google Scholar]
- 39.El Kholy A, Baseem H, Hall GS, Procop GW, Longworth DL.Antimicrobial resistance in Cairo, Egypt 1999–2000: A survey of five hospitals. Antimicrob Agents Chemother.2003;51(3):625–30. [DOI] [PubMed] [Google Scholar]
- 40.Borg MA, Tiemersma E, Scicluna E, Sande-Bruinsma N van de, de Kraker M, Monen J, et al. Prevalence of penicillin and erythromycin resistance among invasive Streptococcus pneumoniae isolates reported by laboratories in the southern and eastern mediterranean region. ClinMicrobiol Infect.2009;15(3):232–7. [DOI] [PubMed] [Google Scholar]
- 41.Taha N, Araj GF, Wakim RH, Kanj SS, Kanafani ZA, Sabra A, et al. Genotypes and serotype distribution of macrolide resistant invasive and non-invasive Streptococcus pneumoniae isolates from Lebanon. Ann Clin MicrobiolAntimicrob.2012; 11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergogne-Bérézin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev.1996;9(2):148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fournier PE, Richet H. The epidemiology and control of Acinetobacter baumanniiin health care facilities.Clin Infect Dis.2006;42(5):692–9. 10.1086/500202 [DOI] [PubMed] [Google Scholar]
- 44.Montefour K, Frieden J, Hurst S, Helmich C, Headley D, Martin M. Acinetobacter baumannii: an emerging multidrug-resistant pathogen in critical care. Crit Care Nurse.2008; 28(1):15–25. [PubMed] [Google Scholar]
- 45.Go ES, Urban C, Burns J, Kreiswirth B, Eisner W, Mariano N, et al. Clinical and molecular epidemiology of Acinetobacter infections sensitive only to polymyxin B and sulbactam.Lancet.1994;344(8933):1329–32. [DOI] [PubMed] [Google Scholar]
- 46.Bergogne-Bérézin E: The increasing significance of outbreaks of Acinetobacter spp: the need for control and new agents. J Hosp Infect.1995;30(1):441–52. [DOI] [PubMed] [Google Scholar]
- 47.Frnández-Cuenca F, Martínez-Martínez L, Conejo MC, Ayala JA, Perea EJ, Pascual A.Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother.2003;51(3):565–74. [DOI] [PubMed] [Google Scholar]
- 48.Manchanda V, Sanchaita S, Singh N. Multidrug Resistant Acinetobacter.J Glob Infect Dis.2010, 2(3):291–304. 10.4103/0974-777X.68538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sader HS, Jones RN. Comprehensive in vitro evaluation of cefepime combined with aztreonam or ampicillin/sulbactam against multi-drug resistant Pseudomonas aeruginosa and Acinetobacter spp. Int J Antimicrob Agents. 2005;25(5):380–4. 10.1016/j.ijantimicag.2005.01.011 [DOI] [PubMed] [Google Scholar]
- 50.Zhao C, Xie W, Zhang W, Ye Z, Wu H. Mechanism of drug resistance of carbapenems-resistant Acinetobacter baumannii and the application of a combination of drugs in vitro. J Zhonghua Shao Shang ZaZhi. 2014;30(2):166–70. [PubMed] [Google Scholar]
- 51.Hirsch EB,Guo B,Chang KT,Cao H,Ledesma KR,Singh M,Tam VH. Assessment of Antimicrobial Combinations for Klebsiella pneumoniae Carbapenemase–Producing K.pneumoniae. J Infect Dis.( 2013);207(5):786–793. [DOI] [PubMed] [Google Scholar]
- 52.Gasink LB, Edelstein PH, Lautenbach E, Synnestvedt M, Fishman NO. Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae.J Infect Control Hosp Epidemiol. 2009;30(12):1180.–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou XH, Song XY, Ma XB, Zhang SY, Zhang JQ. Molecular characterization of multidrug-resistant Klebsiella pneumoniae isolates. Braz J. Microbiol.2015;46(3):759–68. 10.1590/S1517-838246320140138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pitout JD, Hanson ND, Church DL, Laupland KB. Population-based laboratory surveillance for Escherichia coli-producing extended-spectrum beta-lactamases: importance of community isolates with blaCTX-M genes.Clin Infect Dis 2004;38(12):1736–41. 10.1086/421094 [DOI] [PubMed] [Google Scholar]
- 55.Lewis JS II, Herrera M, Wickes B, Patterson JE, Jorgensen JH. First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob Agents Chemother.2007;51(11):4015–21. 10.1128/AAC.00576-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGettigan SE, Hu B, Andreacchio K, Nachamkin I, Edelstein PH. Prevalence of CTX-M beta-lactamases in Philadelphia, Pennsylvania.J ClinMicrobiol.2009;47(9):2970–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sidjabat HE,Paterson DL, Adams-Haduch JM, Ewan L, Pasculle AW, Muto CA, et al. Molecular epidemiology of CTX-M-producing Escherichia coli isolates at a tertiary medical center in western Pennsylvania. Antimicrob. Agents Chemother. 2009; 53(11):4733–39. 10.1128/AAC.00533-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peirano G, Costello M, Pitout JD. Molecular characteristics of extended-spectrum beta-lactamase-producing Escherichia coli from the Chicago area: high prevalence of ST131 producing CTX-M-15 in community hospitals. Int J Antimicrob Agents. 2010;36(1):19–23. 10.1016/j.ijantimicag.2010.02.016 [DOI] [PubMed] [Google Scholar]
- 59.Chandramohan L, Revell PA. Prevalence and Molecular Characterization of Extended-Spectrum-β-Lactamase Producing Enterobacteriaceae in a Pediatric Patient Population. Antimicrob Agents Chemother.2012;56(9):4765–4770. 10.1128/AAC.00666-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hooper DC. Mechanisms of quinolone resistance.DrugResistUpdat.1999; 2(1):38–55. [DOI] [PubMed] [Google Scholar]
- 61.Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis.2000;31(2):S24–8. [DOI] [PubMed] [Google Scholar]
- 62.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6′)-Ib-cr Encoding a Ciprofloxacin-Modifying Enzyme.Antimicrob Agents Chemother. 2006;50(11):3953–3955. 10.1128/AAC.00915-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid.Lancet.1998;351(9105):797–9. 10.1016/S0140-6736(97)07322-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.


