Abstract
Mutations in PTEN-induced putative kinase 1 (PINK1) and parkin cause autosomal-recessive Parkinson’s disease through a common pathway involving mitochondrial quality control. Parkin inactivation leads to accumulation of the parkin interacting substrate (PARIS, ZNF746) that plays an important role in dopamine cell loss through repression of proliferator-activated receptor gamma coactivator-1α (PGC-1α) promoter activity. Here we show that PARIS, links PINK1 and parkin in a common pathway that regulates dopaminergic neuron survival. PINK1 interacts with and phosphorylates serines 322 and 613 of PARIS to control its ubiquitination and clearance by parkin. PINK1 phosphorylation of PARIS alleviates PARIS toxicity as well as repression of PGC-1α promoter activity. Conditional knockdown of PINK1 in adult mouse brains leads to a progressive loss of dopaminergic neurons in the substantia nigra that is dependent on PARIS. Together, these results uncover a function of PINK1 to direct parkin-PARIS regulated PGC-1α expression and dopaminergic neuronal survival.
Keywords: Parkin, PINK1, PARIS, ZNF746, Ubiquitin, PGC-1α, Parkinson’s disease
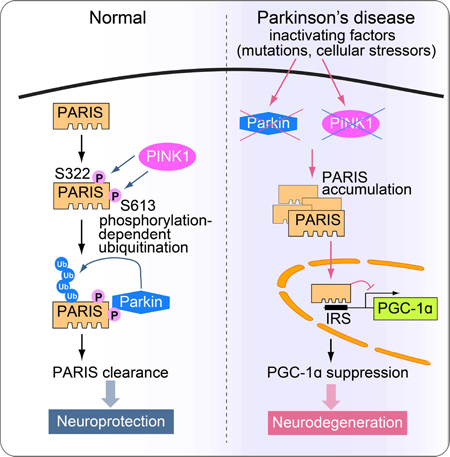
Graphical abstract
In Brief (eTOC blurb)
Lee et al. demonstrate that PARIS is a common substrate of PINK1 and parkin. PINK1 phosphorylation of PARIS primes it for ubiquitination and clearance by parkin. Thus, dysfunction of either PINK1 or parkin converges onto PARIS accumulation, which leads to PGC-1α repression and dopamine neuron loss.
INTRODUCTION
Mutations in the E3 ligase PARKIN (Kitada et al., 1998) or the serine-threonine kinase PINK1 (phosphatase and tensin (PTEN) homolog-induced putative kinase 1) (Valente et al., 2004) cause autosomal recessive Parkinson’s disease (PD) (Corti et al., 2011; Martin et al., 2011). PINK1 and parkin interact in a poorly understood genetic pathway important for dopamine (DA) neuronal survival (Clark et al., 2006; Park et al., 2006; Yang et al., 2006). Recently, a number of co-substrates for PINK1 and parkin have been identified tying these proteins to multiple aspects of mitochondrial quality control (Pickrell and Youle, 2015; Scarffe et al., 2014; Winklhofer, 2014). PARIS (ZNF746) is a pathologic parkin substrate, which is increased in sporadic and familial PD brains and is responsible for DA neuronal loss in mouse models of parkin inactivation (Shin et al., 2011; Siddiqui et al., 2015; Siddiqui et al., 2016; Stevens et al., 2015). PARIS accumulation represses the transcriptional coactivator, peroxisome proliferator-activated receptor gamma coactivator-1-alpha (PGC-1α), which is critical for DA neuron survival (Ciron et al., 2015; Jiang et al., 2016; Mudo et al., 2012; Zheng et al., 2010).
Since parkin and PINK1 are thought to regulate DA neuronal survival in a common pathway (Corti and Brice, 2013; Rochet et al., 2012; Scarffe et al., 2014), and regulation of PARIS by parkin is critical for DA cell survival (Shin et al., 2011; Siddiqui et al., 2015; Siddiqui et al., 2016; Stevens et al., 2015; Winklhofer, 2014), we investigated whether PINK1 plays any role in the regulation of PARIS. Here we show that PINK1 is a priming kinase for parkin-mediated PARIS ubiquitination and clearance. PINK1 depletion in adult mouse brains leads to PARIS accumulation, PGC-1α repression, and progressive DA neuron loss that is PARIS dependent. Identification of PARIS as a PINK1 substrate provides a molecular mechanism linking PINK1 and parkin to DA neuronal loss in PD.
RESULTS
PARIS interacts with PINK1 and parkin
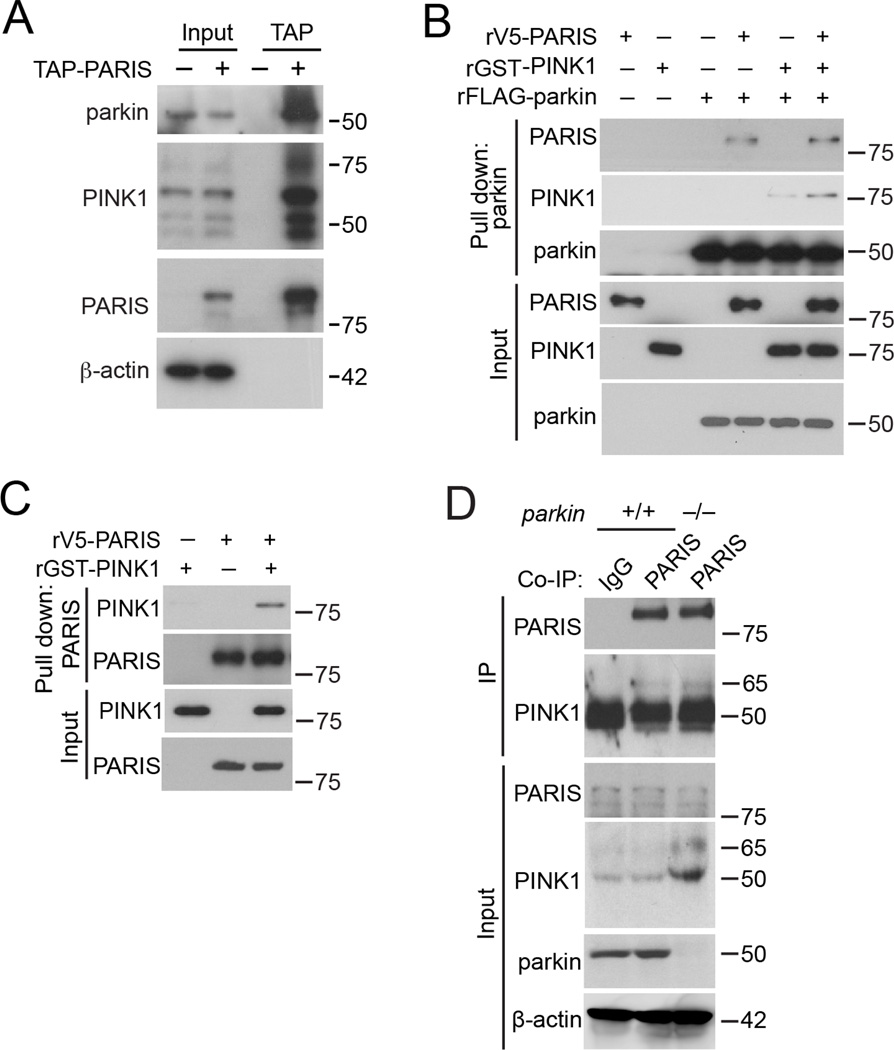
Interaction of PINK1 and PARIS was first suggested by tandem affinity purification of PARIS from SH-SY5Y cells which pulls down both endogenous parkin and PINK1 (Figure 1A). An in vitro pull down assay using an anti-parkin antibody was conducted which showed co-immunoprecipitation of both PARIS and PINK1 (Shiba et al., 2009; Shin et al., 2011). Notably, addition of recombinant PINK1 enhances the association of these three proteins (Figure 1B). In the absence of parkin, an N-terminal V5-tagged recombinant PARIS (rV5-PARIS) pulls down GST-tagged recombinant PINK1 (rGST-PINK1) (Figure 1C) suggesting that PINK1 directly interacts with PARIS.
Figure 1. PARIS interacts with both PINK1 and Parkin.
(A) Tandem affinity purification of PARIS with endogenous parkin and PINK1 in SH-SY5Y cells. (B) Recombinant V5-tagged PARIS (rV5-PARIS), recombinant GST-PINK1 (rGST-PINK1), and recombinant FLAG-tagged parkin (rFLAG-parkin) pull-downs using an anti-parkin antibody (n = 2). (C) Pull-down of rGST-PINK1 and rV5-PARIS with a PARIS antibody (n = 2). (D) Co-immunoprecipitation of endogenous PARIS with PINK1 from parkin+/+ and parkin−/− mouse brain tissue (n = 2). See also Figure S1.
To further characterize this interaction SH-SY5Y cells were transfected with N-terminal FLAG-tagged PARIS (FLAG-PARIS) or deletion mutants and N-terminal GFP-tagged PINK1 (GFP-PINK1). GFP-PINK1 co-immunoprecipitates PARIS as well as the Krüppel-associated box (KRAB) domain containing N-terminal fragment (Figure S1A). PARIS co-immunoprecipitates both the ~65 kDa and ~55 kDa forms of PINK1 (Figure S1B), while PD-linked mutant L347P-PINK1 and kinase inactive K219M-PINK1 exhibit reduced interaction with PARIS (Figure S1C). Co-immunoprecipitation of PARIS with deletion mutants of N-terminal GFP-tagged PINK1 (N, amino acids (aa) 1-270; C, aa 268-581) reveals that the N-terminal half of PINK1 is sufficient for the PARIS interaction (Figure S1D). Attempts to generate smaller fragments of GFP-PINK1 were hampered by protein instability.
The interaction of PARIS with PINK1 was also evaluated in mouse brain. Immunoprecipitation of PARIS pulls down endogenous PINK1 (Figure 1D). Parkin, is not required for the interaction of PARIS and PINK1, since immunoprecipitation of PARIS from both wild type (WT) and parkin−/− brains pulls down PINK1 (Figure 1D). These experiments suggest a direct interaction of PINK1 with PARIS.
PINK1 phosphorylates PARIS
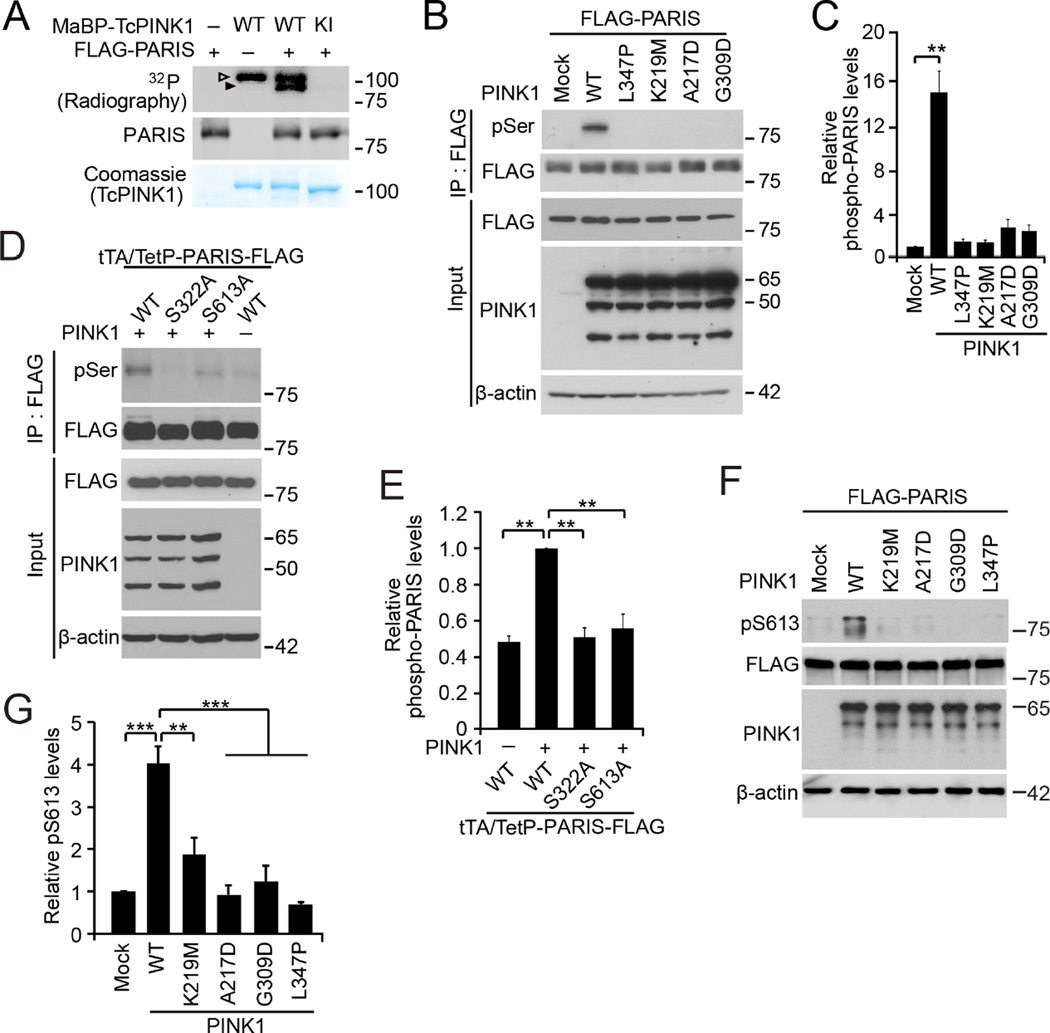
Maltose-binding protein fused Tribolium castaneum PINK1 (MaBP-TcPINK1), possesses both strong kinase activity and high homology with human PINK1 (Woodroof et al., 2011). Wild type TcPINK1 markedly increases immunopurified FLAG-PARIS phosphorylation as detected by 32P, whereas kinase inactive TcPINK1 (D359A) fails to phosphorylate PARIS (Figure 2A). Moreover, despite lower kinase activity, recombinant human PINK1 purified from E. coli phosphorylates immunopurified FLAG-PARIS as detected by a phosphoserine antibody, while a kinase inactive PINK1 (K219M) does not (data not shown). Consistent with these in vitro assays, PINK1 overexpression in SH-SY5Y cells phosphorylates FLAG-PARIS as detected by a phosphoserine immunoblot of FLAG immunoprecipitates (Figures 2B and 2C). The L347P, A217D, G309D familial PD mutants of PINK1 and the K219M kinase inactive PINK1 fail to phosphorylate FLAG-PARIS (Figures 2B and 2C).
Figure 2. PINK1 phosphorylates PARIS.
(A) In vitro kinase assay using [32P]-ATP of PARIS phosphorylation by maltose-binding protein tagged Tribolium castaneum PINK1 (MaBP-TcPINK1). Phosphorylated TcPINK1 and PARIS are indicated by empty and filled arrowheads, respectively (n = 2). KI, kinase inactive mutant TcPINK1 (D359A). (B) PARIS phosphorylation of PARIS IPs via a phosphoserine antibody by WT, L347P, A217D, G309D or K219M PINK1 in SH-SY5Y cells. (C) Quantification of relative phosphorylation levels of PARIS (n = 3). (D) Serine phosphorylation of WT and phosphomutant (S322A and S613A) PARIS by PINK1 in SH-SY5Y cells. (E) Quantification of relative phosphorylation levels of PARIS WT and phosphomutants (n = 3). (F) PINK1 WT and mutant phosphorylation at S613 of PARIS determined by Immunoblots using pS613-PARIS antibodies for anti-FLAG immunoprecipitates from SH-SY5Y cells. (G) Quantification of relative S613 phosphorylation normalized to total immunoprecipitated PARIS levels (n = 3 per group). Quantified data (Figures 2C, 2E, and 2G) are expressed as mean ± s.e.m., **P < 0.01, ***P < 0.001, analysis of variance (ANOVA) test followed by Tukey HSD post-hoc analysis. See also Figure S2.
Phosphorylation of PARIS by PINK1 was identified by mass spectrometry at S106, S322, S359, and S613 and T603. Of these S322 and S613 were conserved among mammals (human, monkey, chimpanzee, rat and mouse) (Figures S2A–S2C). Site directed mutagenesis of S322 or S613 to alanine (S322A or S613A) reduces the phosphorylation of PARIS by PINK1 (Figures 2D and 2E). To characterize the role of S613 phosphorylation of PARIS, we raised and purified rabbit antibodies against a PARIS peptide containing phosphoserine 613 (pS613). The purified rabbit antibody to pS613-PARIS is highly specific for S613 phosphorylation (Figures S2D and S2E). Interestingly, it appears that S322A substitution (S322A-PARIS) affects PARIS S613 phosphorylation, suggesting a synergistic interaction in the phosphorylation of these two serine residues (Figures S2D and S2E). Phosphorylation of PARIS S613 is markedly enhanced by PINK1 coexpression, whereas kinase deficient PINK1 mutants fail to phosphorylate PARIS (Figures 2F and 2G).
PINK1 is required for ubiquitination and degradation of PARIS by parkin
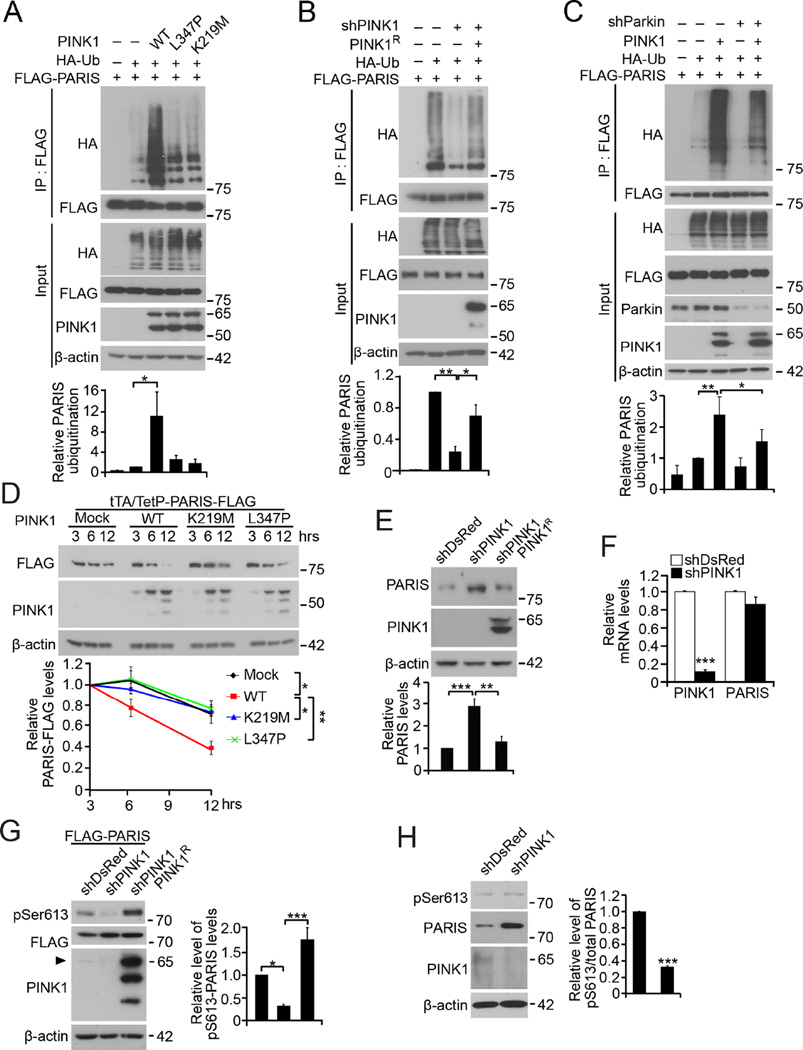
To determine if PINK1 regulates the ubiquitination of PARIS by parkin, SH-SY5Y cells were transfected with PINK1, HA-ubiquitin and FLAG-PARIS. PINK1 enhances the endogenous ubiquitination of PARIS, while L347P and K219M PINK1 mutants have no effect on PARIS ubiquitination (Figure 3A) indicating the importance of PINK1 kinase activity. HA-ubiquitin signal is not measurable in the absence of FLAG-PARIS expression indicating that the ubiquitination is specific for FLAG-PARIS (Figure S3A).
Figure 3. PINK1 is required for Parkin ubiquitination and degradation of PARIS.
(A) PARIS ubiquitination with PINK1 or mutant (L347P and K219M) overexpression by HA western blot of FLAG immunoprecipitates from transfected SH-SY5Y cells. Graph shows quantification of HA-ubiquitin normalized to total PARIS (n = 4). (B) PARIS ubiquitination after knockdown of PINK1 in SH-SY5Y cells. Graph shows relative ubiquitination normalized to total PARIS. PINK1R is shRNA resistant PINK1 (n = 3). (C) PARIS ubiquitination with PINK1 overexpression and co-transfection of shRNA-parkin. Graph shows PARIS ubiquitination normalized to total PARIS (n = 4; 3 for no HA-Ub). (D) Pulse chase analysis of PARIS-FLAG with PINK1 or kinase deficient mutants, K219M and L347P overexpression. PARIS-FLAG was induced by the Tet-off system for 24 hours, and the degradation of PARIS-FLAG was monitored after doxycycline treatment. Graph shows quantification of PARIS-FLAG normalized to β-actin (n = 4, statistical comparison done at 12 h). (E) Western blot of endogenous PARIS in SH-SY5Y cells 2 days after transfection with PINK1 shRNA with and without shRNA-resistant PINK1 expression. Graph shows quantification of endogenous PARIS normalized to β-actin (n = 5). (F) Quantification of PINK1 and PARIS mRNA normalized to GAPDH in SH-SY5Y cells transfected with shPINK1 or shDsRed control (n = 3) (see Figure 5A for PINK1 mRNA). (G) PARIS S613 phosphorylation in SH-SY5Y cells transfected with FLAG-PARIS and shRNA to PINK1 or shDsRed. Endogenous human PINK1 is indicated with an arrow head. Graph shows relative quantification of pS613 PARIS normalized to FLAG-PARIS (n = 4; 3 for shPINK1/PINK1R). (H) Representative western blots showing accumulation of endogenous PARIS and concomitant reduction of relative S613 PARIS phosphorylation by PINK1 depletion. Graph shows relative quantification of pS613 PARIS normalized to total PARIS (n = 3). Quantified data expressed as mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, a two-tailed student t test for Figures 3F and 3H and analysis of variance (ANOVA) test followed by Tukey HSD post-hoc analysis for the others. See also Figure S3.
To determine if endogenous PINK1 is required for PARIS ubiquitination, PINK1 was knocked down by shRNA (Figures 3B, S3B and S3C) resulting in reduced ubiquitination of PARIS (Figure 3B). Ubiquitination is restored by expression of shRNA resistant PINK1 (PINK1R) (Figure 3B, S3B, and S3C), thus controlling for potential off-target effects. To evaluate whether endogenous parkin contributes to the enhanced ubiquitination of PARIS following PINK1 phosphorylation, parkin was knocked down by shRNA attenuating this enhancement (Figure 3C).
To investigate the impact of PARIS phosphorylation on its proteasomal degradation the tetracycline sensitive PARIS expression system (TetP-PARIS-FLAG) was used to assess steady state levels of PARIS-FLAG in PINK1 overexpression. Twenty-four hours after induction of PARIS-FLAG with tTA, doxycycline was added to block further PARIS-FLAG expression. PINK1 overexpression accelerates the degradation of PARIS, while kinase deficient L347P and K219M PINK1 do not (Figure 3D).
To evaluate the effect of endogenous PINK1 on endogenous PARIS levels we knocked down PINK1 in SH-SY5Y cells which lead to a three-fold increase in PARIS. ShRNA resistant PINK1 (PINK1R) restores PARIS to baseline (Figure 3E). PARIS mRNA does not change in response knockdown of PINK1 (Figure 3F). These results suggest that endogenous PINK1 even under basal conditions possesses sufficient catalytic activity to regulate its potential substrates. Supporting this notion, basal phosphorylation of both PARIS and PINK1 were detected at appreciable levels by phosphorylated protein enrichment (Figure S3E and S3F). The phosphorylation of PARIS and PINK1, measured by column binding, was enhanced by treatment with the mitochondrial depolarizing agent, carbonyl cyanide m-chlorophenyl hydrazone (CCCP) and abolished by phosphatase treatment of cell lysates indicating that binding of proteins to the column requires phosphorylation (Figure S3E and S3F).
Ubiquitin is the most extensively characterized PINK1 substrate (Fiesel et al., 2015; Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014; Ordureau et al., 2014; Wauer et al., 2015a). We used the phosphorylation of ubiquitin as a measure of PINK1 activity. Phosphorylated ubiquitin is detected in the postnuclear, mitochondrial and cytoplasmic compartments, and the phosphorylation is enhanced by CCCP treatment (Figure S3G). Conversely, transfection of SH-SY5Y cells with siRNA to PINK1 leads to a reduction in ubiquitin phosphorylation (Figure S3H). This would suggest that PINK1 is catalytically active under basal conditions.
PINK1 knockdown also reduces PARIS phosphorylation in the cytoplasm (Figure S3H). FLAG-PARIS is phosphorylated at S613 by endogenous PINK1 since depletion of PINK1 reduces S613 phosphorylation (Figure 3G). Introduction of PINK1R, an shRNA resistant PINK1, restores the phosphorylation of FLAG-PARIS, thus controlling for potential off-target effects (Figure 3G). Endogenous PARIS is phosphorylated by PINK1 since PINK1 knockdown reduces the relative phosphorylation of endogenous PARIS leading to PARIS accumulation (Figure 3H). These results taken together demonstrate that PINK1 has basal catalytic activity in the absence of wide scale mitochondrial depolarization and that PARIS is a physiologic phosphosubstrate of PINK1.
PINK1 regulates parkin-mediated ubiquitination and degradation of PARIS via phosphorylation
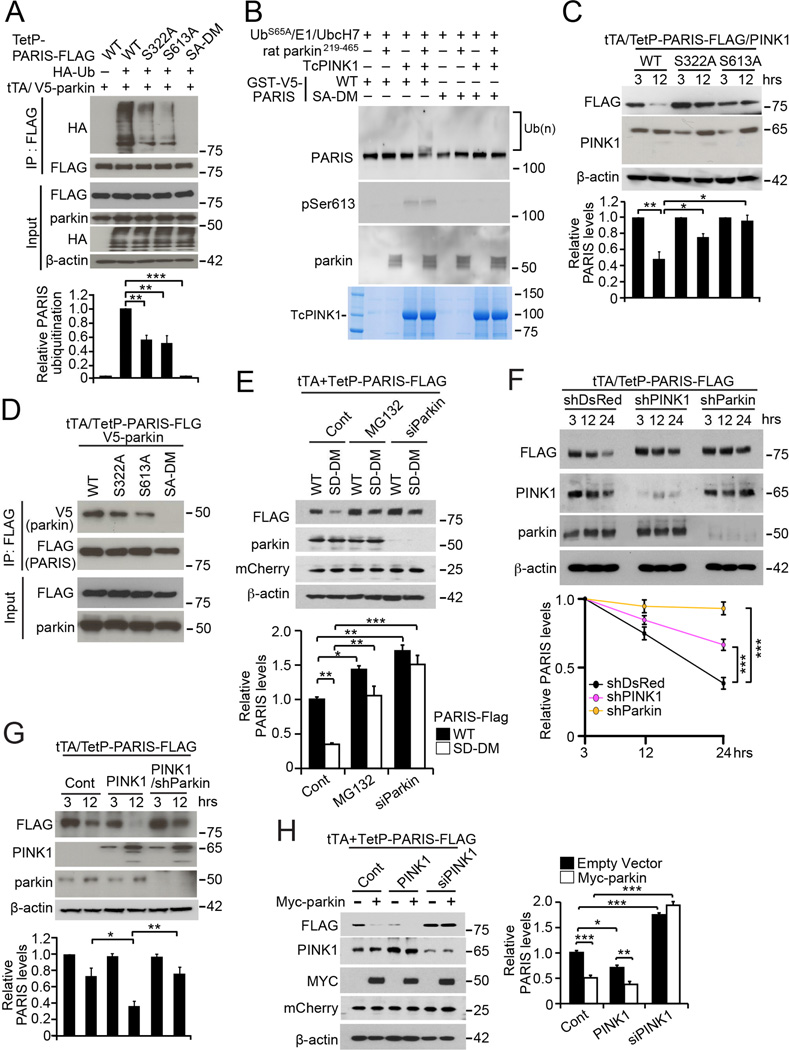
PARIS ubiquitination and degradation are regulated by parkin (Shin et al., 2011). A ubiquitination assay in SH-SY5Y cells reveals that PARIS ubiquitination is markedly enhanced by parkin overexpression (Figure S4A). The role of PARIS phosphorylation by PINK1 in PARIS turnover was assessed using pulse chase experiments. SH-SY5Y cells were transfected with TetP-PARIS-FLAG or TetP-PARIS-FLAG mutants (S322A or S613A or S322/613A), HA-ubiquitin, parkin and tTA. Parkin robustly ubiquitinates PARIS, but in S322A and S613A PARIS mutants this effect is reduced (Figure 4A), while S322/613A PARIS (SA-DM) is not ubiquitinated (Figure 4A), linking PARIS S322/S613 phosphorylation by PINK1 to subsequent ubiquitination by parkin.
Figure 4. PINK1 phosphorylation of PARIS is required for Parkin ubiquitination of PARIS.
(A) Immunoblots of parkin ubiquitination of PARIS WT and S322A, S613A, and S322,613A (SA-DM)) phosphorylation mutants. Bar graph shows quantification of the levels of ubiquitination of immunoprecipitated PARIS normalized to the levels of PARIS (n = 3). (B) Immunoblot analysis of an in vitro parkin ubiquitination assay with UbS65A and rat Parkin219–465 of WT or SA-DM PARIS in the presence of TcPINK1. Recombinant PARIS phosphorylation and ubiquitination was monitored by Immunoblot using pSer613 and PARIS antibodies respectively. Similar results were obtained in two independent experiments. (C) Immunoblot of pulse chase analysis of WT and phosphorylation mutant PARIS-FLAG in the presence of PINK1 in SH-SY5Y cells. Bar graph of quantification of relative WT and phosphorylation mutant PARIS-FLAG levels normalized to β-actin in the indicated time points (n = 3). (D) Immunoblots of anti-FLAG immunoprecipitates from SH-SY5Y cells expressing V5-parkin and WT or phosphodeficient S322A, S613A, and SA-DM PARIS. (E) Immunoblots of WT and the phosphomimetic form of FLAG-PARIS (SD-DM) in response to MG132 in SH-SY5Y cells. mCherry was used for transfection control. Bar graph of quantification of relative PARIS-FLAG levels normalized to mCherry (n = 3 per group). (F) Immunoblots of pulse chase analysis of PARIS-FLAG levels in the setting of either PINK1 or parkin knockdown in SH-SY5Y cells. Line graph of quantification of relative PARIS-FLAG levels normalized to β-actin (n = 3 per group). (G) Immunoblots of pulse chase analysis of PARIS-FLAG levels in the presence of PINK1 overexpression with or without knockdown of parkin in SH-SY5Y cells. Bar graph of quantification of relative PARIS-FLAG levels normalized to β-actin (n = 4). (H) Immunoblot of PARIS-FLAG degradation by Myc-parkin in the setting of PINK1 overexpression or knockdown in SH-SY5Y cells. mCherry was used as a transfection control. Bar graph of quantification of relative PARIS-FLAG levels normalized to mCherry (n = 3 per group). Quantified data (Figures 4A, 4C, 4E, 4F, 4G, and 4H) are expressed as mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, analysis of variance (ANOVA) test followed by Tukey HSD post-hoc analysis. See also Figure S4.
Additionally, an in vitro ubiquitination reaction using recombinant ubiquitin, E1, UbcH7 as an E2, PARIS WT or SA-DM together with TcPINK1 and rat full-length parkin showed increased activation of parkin by addition of TcPINK1 (Figure S4B), consistent with previous reports (Fiesel et al., 2015; Kazlauskaite et al., 2015; Kumar et al., 2015; Sauve et al., 2015; Wauer et al., 2015a; Yamano et al., 2015). In contrast, phosphorylation mutant PARIS SA-DM fails to be phosphorylated and ubiquitinated by TcPINK1/parkin even in the presence of parkin activation (Figure S4B).
Recent in vitro studies have shown that TcPINK1 can phosphorylate ubiquitin and parkin, enhancing parkin activity (Fiesel et al., 2015; Kane et al., 2014; Kazlauskaite et al., 2014; Kazlauskaite et al., 2015; Koyano et al., 2014; Kumar et al., 2015; Ordureau et al., 2014; Sauve et al., 2015; Wauer et al., 2015a; Wauer et al., 2015b; Yamano et al., 2015). We observe strong phosphorylation of both PARIS and parkin by TcPINK1 in vitro (Figure S4C–S4E). Phosphorylation-deficient ubiquitin (S65A-Ub) and constitutively active parkin (rat parkin fragment 219–465) were used to examine the effect of PARIS phosphorylation on ubiquitination independent of parkin or ubiquitin phosphorylation. Rat parkin enhances PARIS ubiquitination but fails to ubiquitinate the PARIS phosphorylation mutant (Figure 4B), demonstrating the necessity of PARIS S322/S613 phosphorylation for parkin-mediated ubiquitination.
The impact of PARIS phosphorylation on its steady state levels was assessed using TetP-PARIS-FLAG or TetP-PARIS-FLAG mutants (S322A or S613A). PINK1 overexpression reduces steady state levels of PARIS-FLAG by greater than 50%, whereas S322A PARIS-FLAG is only reduced by about 30% and S613A PARIS-FLAG is not reduced (Figure 4C). We examined whether this difference might be due to a change in the binding of mutant PARIS for N-terminal V5-tagged parkin (V5-parkin). S322A or S613A mutations diminished the parkin interaction, while double mutation (S322/613A) abolished PARIS’s interaction with parkin (Figure 4D), suggesting that phosphorylation of PARIS by PINK1 promotes parkin interaction.
To mimic phosphorylation of PARIS, plasmids with serine to aspartate substitution at S322 or/and S613 (S322D, S613D, S322/613D (SD-DM)) were generated and the stability of the proteins assessed. Cellular levels of S322D, S613D and SD-DM are significantly lower than WT PARIS (Figure S4F), and SD-DM PARIS was stabilized by the proteasome inhibitor, MG132, or parkin knock down (Figure 4E). These changes in protein expression are not due to transcriptional alterations because mCherry, expressed from the same transcript via internal ribosomal entry site (IRES), did not show noticeable differences among the groups (Figure S4G and Figure 4E).
The physiologic role of endogenous parkin and PINK1 in PARIS clearance were assessed using a tetracycline responsive PARIS expression system (TetP-PARIS-FLAG) to monitor steady state levels of PARIS-FLAG after PINK1 or parkin knockdown. With control shRNA, PARIS-FLAG decrease by ~60% by 24 h after doxycycline treatment (Figure 4F). PINK1 knockdown delays PARIS degradation by ~50% while parkin knockdown almost completely prevents PARIS clearance (Figure 4F) demonstrating that PARIS clearance is regulated by both endogenous PINK1 and parkin. Further, parkin knockdown prevents the reduction in PARIS seen with PINK1 overexpression (Figure 4G), while PINK1 knockdown abolished parkin-mediated degradation of PARIS (Figure 4H). Thus, it seems ubiquitination and degradation of PARIS by parkin is regulated by PINK1 phosphorylation at S322 and S613.
PINK1-parkin interaction has been extensively studied in CCCP-induced mitophagy, where catalytically active PINK1 is stabilized on damaged mitochondria (Narendra et al., 2010; Vives-Bauza et al., 2010). We report that CCCP treatment of SH-SY5Y cells leads to accumulation of PINK1 and a gradual decrease in PARIS (Figure S4H and S4I). This reduction of endogenous PARIS is parkin and PINK1 dependent since knockdown of either prevents PARIS degradation (Figure S4J and S4K). The cellular distribution of PARIS after CCCP treatment was evaluated using confocal immunofluorescence. FLAG-PARIS is localized to the nucleus and to a lesser extent the cytosol (Figure S4L). CCCP treatment for 2 h redistributes PARIS to the cytoplasm where it partially colocalizes with mitochondria (Figure S4L). Following CCCP treatment, PINK1 and parkin colocalize with PARIS at the mitochondria (Figure S4M and S4N), indicating that, under depolarizing conditions, PARIS can localize to mitochondria where it is targeted for degradation by PINK1 and parkin.
PINK1 regulates PARIS repression of PGC-1α and cell toxicity
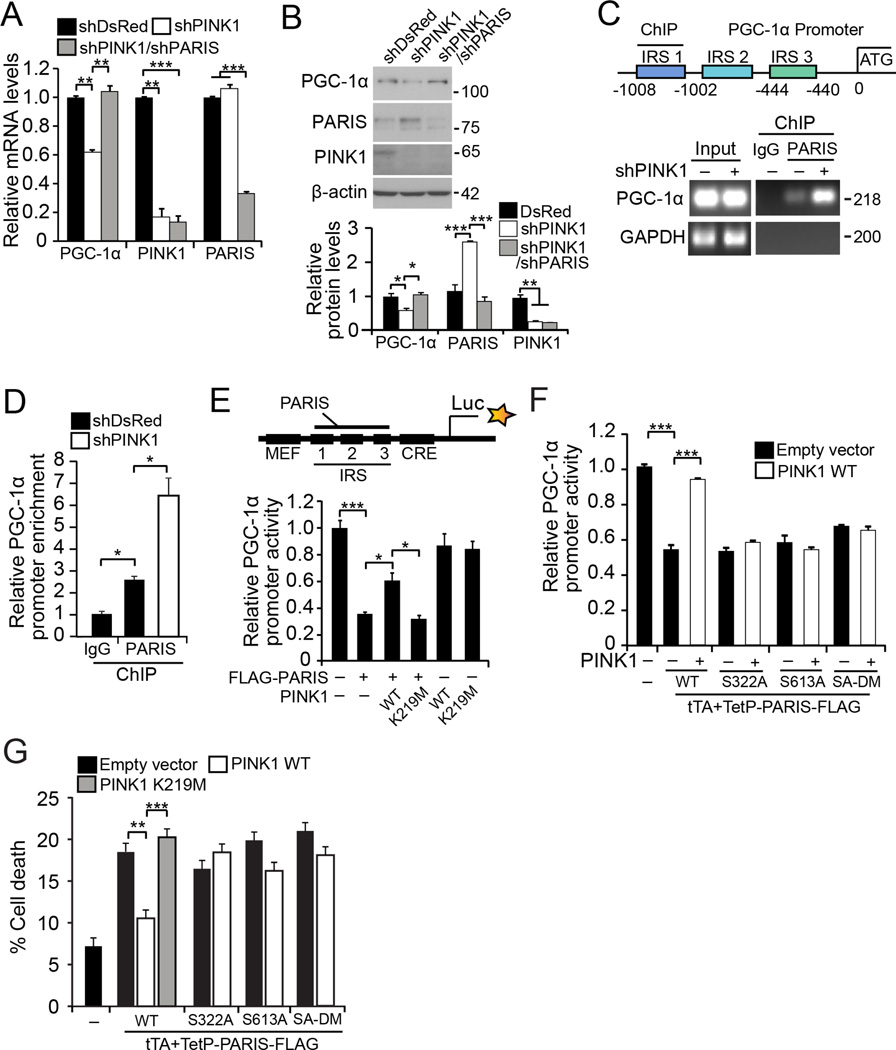
PARIS accumulation downstream of parkin inactivation leads to PGC-1α repression (Shin et al., 2011; Siddiqui et al., 2015; Siddiqui et al., 2016; Stevens et al., 2015). We sought to determine whether PINK1 alters the expression of the PGC-1α. PGC-1α mRNA was monitored using RT-qPCR following shRNA knockdown of PINK1 and/or PARIS in SH-SY5Y cells. PINK1 knockdown reduces PGC-1α mRNA by 40% and simultaneous knockdown of PARIS restores PGC-1α mRNA (Figure 5A). Similarly, knockdown of PINK1 reduces PGC-1α protein by 50% whereas knockdown of PARIS restores basal PGC-1α (Figure 5B). Consistent with PGC-1α repression, PINK1 knockdown in SH-SY5Y cells results in PARIS enrichment at the insulin response sequence (IRS) region of the PGC-1α promoter in a chromatin immunoprecipitation assay (Figure 5C and 5D). As previously reported, PARIS substantially reduces PGC-1α promoter activity in a luciferase assay using a 1 kb human PGC-1α luciferase promoter construct (Shin et al., 2011) (Figure 5E). PINK1 overexpression partially prevents this reduction, and PINK1 kinase activity is required for this reduction as the kinase inactive K219M PINK1 mutant has no effect (Figure 5E). Neither PINK1 or K219M PINK1 overexpression directly affect PGC-1α promoter activity (Figure 5E). Notably, repression of PGC-1α by PARIS phosphorylation mutants (S322A or S613A or SA-DM) could not be rescued by PINK1 overexpression. (Figure 5F).
Figure 5. PINK1 regulates PGC-1α via PARIS.
(A) Relative quantification of PGC-1α, PINK1, and PARIS mRNA after PINK1 and PARIS knockdown in SH-SY5Y cells by RT-qPCR normalized to GAPDH (n = 4). (B) PGC-1α, PARIS and PINK1 protein in SH-SY5Y cells transfected with indicated shRNA (n = 4). Bottom shows quantification normalized to β-actin (C) Chromatin immunoprecipitation (ChIP) with PARIS antibodies or IgG in SH-SY5Y cells transfected with shDsRed or shPINK1. Occupancy of the PGC-1α promoter by PARIS was determined by RT-qPCR of PARIS immunoprecipitates using primers for the PGC-1α IRS1 site. Representative PGC-1α and GAPDH PCR are shown as a negative control for anti-PARIS ChIP DNA. IRS, insulin response sequence (D) Quantification of PGC-1α DNA after shDsRed or shPINK1 transfection normalized to input (n = 4). (E) Quantification of PGC-1α promoter activity in a stable luciferase SH-SY5Y cell line transfected with PARIS and PINK1 or kinase inactive (K219M) PINK1 (n = 3). Top shows schematic of the PGC-1α luciferase construct used to generate stable cell lines (IRS, insulin response sequence; CRE, cAMP response element; MEF, muscle enhancer factor; Luc, luciferase expression). (F) Relative quantification of PGC-1α promoter activity in the stable luciferase SH-SY5Y cell line transfected with PARIS or phosphorylation mutants and PINK1 (n = 3). (G) Percent cell death in SH-SY5Y cells transfected with PARIS or phosphodeficient mutants and PINK1 determined by trypan blue exclusion assay (n = 12 ; 3 for S322A and S613A; 5 for SA-DM). Quantified data (Figures 5A, 5B, 5D–5G) expressed as mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, analysis of variance (ANOVA) test followed by Tukey HSD post-hoc analysis. See also Figure S5 and Table S1.
PARIS accumulation and PGC-1α repression are key molecular events in DA cell death (Shin et al., 2011; Stevens et al., 2015). To determine if PARIS toxicity is influenced by PINK1 phosphorylation, cell death was measured following transient overexpression of PARIS in SH-SY5Y cells. With about 60% transfection efficiency (Figure S5A, and S5B), overexpression of PARIS results in approximately 19% cell death compared with 7% cell death in controls. PINK1 overexpression attenuates PARIS toxicity, while K219M PINK1 has no effect (Figure 5G). PARIS S322A, S613A and SA-DM results in more than 16% cell death in SH-SY5Y cells, but PINK1 overexpression was unable to rescue the toxicity of the PARIS phosphorylation defective mutants (Figure 5G).
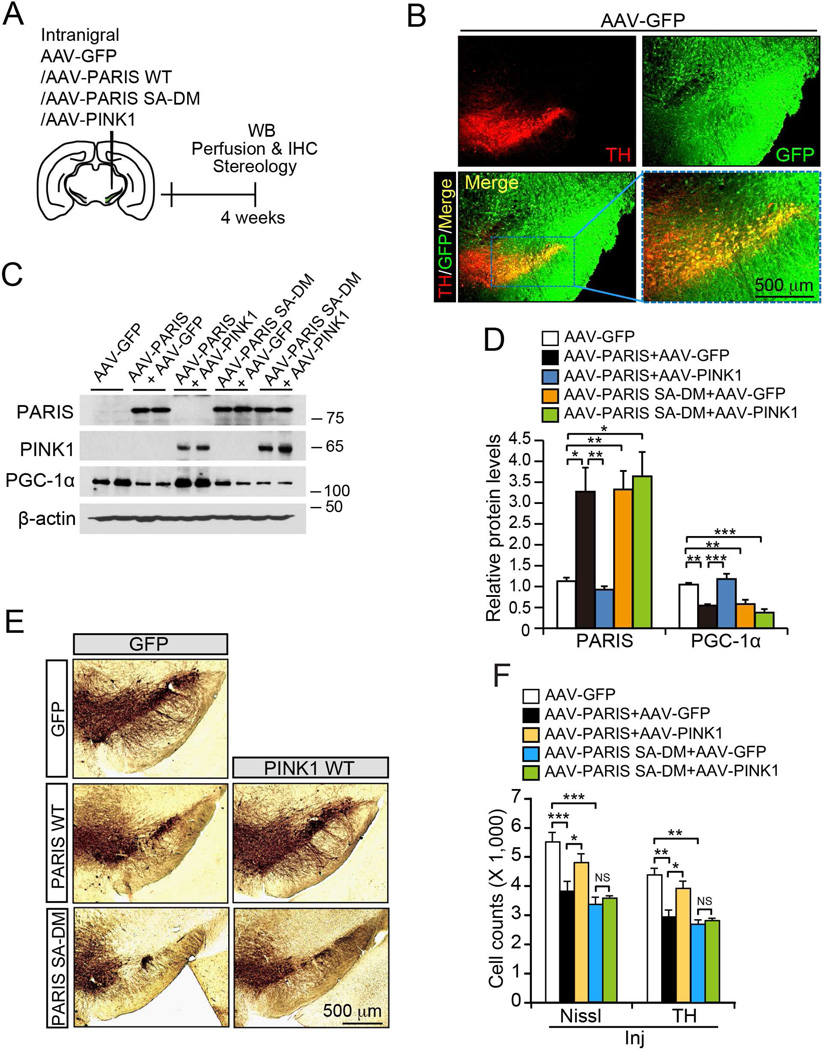
To further evaluate the ability of PINK1 to rescue PARIS toxicity in vivo, we used recombinant adeno-associated virus (rAAV) to overexpress PARIS and PINK1 in mouse VM (Figure 6A and 6B). Consistent with our previous report (Shin et al., 2011), rAAV-PARIS leads to a three-fold increase in PARIS, similar to levels seen in sporadic PD and adult conditional parkin KO mice, repression of PGC-1α (Figure 6C and 6D) (Shin et al., 2011) and loss of DA neurons (Figure 6E and 6F). Co-expression of PINK1 reduced PARIS and reversed PGC-1α repression (Figure 6C and 6D) preventing the loss of DA neurons due to PARIS overexpression (Figure 6E and 6F). The protective effect of PINK1 is dependent on PARIS S322 and S613 phosphorylation because PINK1 failed to prevent PGC-1α repression and DA neuron toxicity induced by phosphorylation deficient mutant PARIS (Figure 6C-6F).
Figure 6. PINK1 rescues PARIS-induced dopaminergic cell loss in vivo.
(A) Schematic of SN virus injection schedule in 2 month old C57/B6 mice. (B) Representative immunofluorescence images of injection quality. Scale bar = 500 mm. (C) Western blots from mouse VM stereotaxically injected with AAV-GFP, or AAV-PARIS, or AAV-PARIS SA-DM with AAV-hPINK1 or AAV-GFP. (D) Quantification of relative PARIS and PGC-1α band densities normalized to β-actin. (n = 3). (E) Representative immunohistochemistry of tyrosine hydroxylase (TH) in mouse SN injected with indicated virus. Scale bar = 500 mm. (F) Stereological assessment of TH+ and Nissl positive neurons in the SN pars compacta (SNpc) of indicated injection groups (n = 5 GFP, n = 6 PARIS, n = 5 PARIS/PINK1, n = 4 PARIS-SA-DM, and n = 4 PARIS-SA-DM/PINK1 injected hemispheres). Quantified data (Figures 6D and 6F) expressed as mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, analysis of variance (ANOVA) test followed by Tukey HSD post-hoc analysis. β-actin was used as a loading control. See also Figure S6.
PINK1 knockdown in an adult mouse brain leads to PARIS-dependent DA neuronal loss
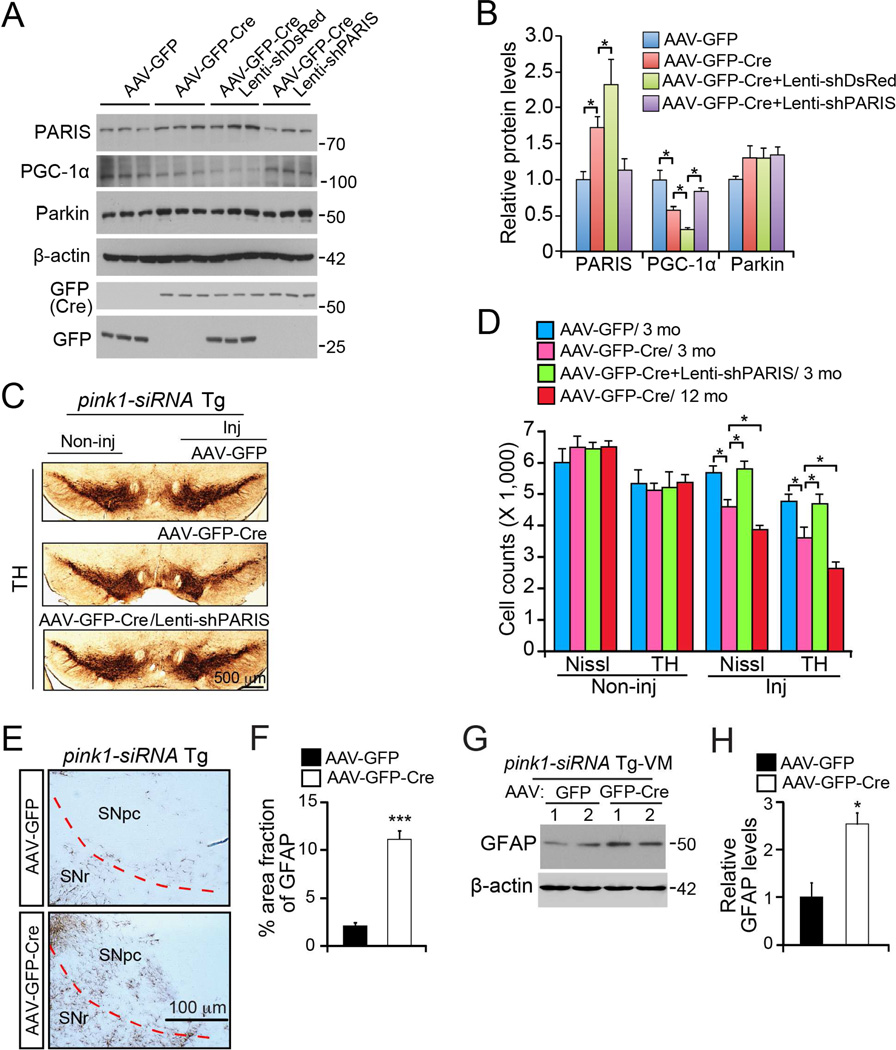
Knockout of parkin in adult mice leads to degeneration of DA neurons that is PARIS dependent (Shin et al., 2011; Stevens et al., 2015). To investigate whether adult knockdown of PINK1 leads to DA neuron degeneration, we used conditional pink1-RNAi transgenic mice (Zhou et al., 2007). Mice were stereotaxically injected with AAV-GFP-Cre, which efficiently transduces TH+ DA neurons (Figure S6A and S6B) leading to >80% knockdown of PINK1 mRNA (Figure S6C), and reduced PARIS S613 phosphorylation despite increased PARIS (Figure S6D and S6E). PINK1 knockdown leads to a ~30% reduction in PGC-1α mRNA (Figure S6C). PINK1 knockdown leads to a two-fold increase in PARIS protein and reduction of PGC-1α protein which are rescued by PARIS knockdown (Figure 7A and 7B).
Figure 7. PARIS is required for loss of dopaminergic neurons and reduction of PGC-1 a in conditional PINK1 ablation.
(A) Western blot analysis of PARIS, PGC-1α, parkin, and β-actin in the VM of pink1-siRNA Tg mice 4 weeks after SN injection with the indicated virus. Two month old mice were used for injection. (B) Quantification of band intensities of PARIS, PGC-1α, and parkin normalized to β-actin (n = 3). (C) Representative TH immunohistochemistry of DA neurons in the SN 3 months after injection of AAV-GFP or AAV-GFP-Cre viruses and lenti-shPARIS as indicated. Scale bar = 500 mm. (D) Stereological counting of TH+ and Nissl stained neurons 3 months and 12 months after AAV-GFP-Cre injection of pink1-siRNA transgenic mice with and without coinjection of lenti-shRNA-PARIS. (n = 4 at 3 months; n = 5 GFP-Cre injected, 3 non-injected hemispheres at 12 months). (E) Representative GFAP immunohistochemistry of pink1-siRNA mouse VM injected with AAV-GFP or AAV-GFP-Cre viruses. SNpc, SN pars compacta. SNr, SN reticularis. Scale bar = 100 mm. (F) Percentage of GFAP positive area in the SNpc of AAV-GFP or AAV-GFP-Cre injected pink1-siRNA transgenic mice (n = 10 sections from 3 mice per group). (G) Representative western blots of GFAP in AAV-GFP or AAV-GFP-Cre injected pink1-siRNA mouse midbrain. (H) Relative quantification of GFAP in (G) normalized to β-actin (n = 4). Quantified data expressed as mean ± s.e.m., *P < 0.05, ***P < 0.001, a two-tailed Student’s t test (Figures 7F and 7H). Analysis of variance (ANOVA) test followed by Tukey HSD post-hoc analysis (Figures 7B and 7D). See also Figures S7.
To address potential off-target effects of shRNA to PINK1 and PARIS, AAV that express human PINK1 (AAV-hPINK1) or a different shRNA to mouse PARIS (AAV-shPARIS#2-RFP) (Figure S7). AAV-shPARIS#2, efficiently transduced GFP-Cre positive DA neurons (Figure S7A). PGC-1α repression was evaluated at the cellular level by probing pink1-siRNA mouse VM with antibodies to TH, PGC-1α, and GFP after injection with AAV-GFP or AAV-GFP-Cre and AAV-hPINK1 viruses (Figure S7B and S7C). TH+ neurons transduced with GFP-Cre, had reduced PGC-1α signal intensity compared to AAV-GFP control (Figure S7B, and S7C) consistent with the western blot analysis (Figure 7A and 7B). Co-injection of AAV-hPINK1 virus prevented the PGC-1α repression (Figure S7B and S7C). We also examined whether PGC-1α repression by PINK1 depletion is PARIS dependent in DA neurons by laser capture microdissection. TH+ and AAV virus transduced (GFP+) neurons from pink1-siRNA transgenic mice stereotaxically injected with AAV-GFP, AAV-GFP-Cre, and AAV-GFP-Cre/shRNA-PARIS in the VM (VM) were collected for RNA extraction (Figure S7D and S7E). DA neurons transduced with both AAV-GFP-Cre and shPARIS, were protected from PGC-1α downregulation following PINK1 ablation (Figure S7D and S7E) consistent with the western blot analysis (Figure 7A and 7B).
Three months after AAV-GFP-Cre injection of pink1-RNAi mouse VM the number of DA neurons was assessed via stereologic analysis. AAV-GFP-Cre mediated expression of shRNA-PINK1 in adult animals leads to a 30% reduction in TH+ neurons that is prevented when PARIS is also knocked down (Figure 7C and 7D). This DA neuron degeneration is progressive since at 12 months post-injection there is about 45% loss of TH+ neurons (Figure 7D). PINK1 depleted mice show astrogliosis in the substantia nigra (SN) by GFAP immunoreactivity and western blot (Figure 7E–7H).
Along with PGC-1α restoration, human PINK1 expression in PINK1 depleted mice reversed DA neuron loss as determined by stereologic counting of TH+ neurons (Figure S7F and S7G). Co-injection of AAV-shPARIS #2-RFP also provided nearly complete protection from DA neuronal toxicity due to PINK1 depletion (Figure S7F and S7G). These results taken together confirm that in vivo knockdown of PINK1 in adult mice leads to loss of DA neurons that is PARIS dependent.
DISCUSSION
Using gain and loss of function studies we show here that PINK1 acts as a priming kinase for parkin-mediated ubiquitination of PARIS, which controls the steady state levels of PARIS. Substrate phosphorylation is a common regulatory mechanism by which E3 ligases recognize their cognate substrate (Hunter, 2007). Phosphorylation of PARIS by PINK1 occurs under basal conditions both in vitro and in vivo and the absence of PINK1 leads to reduced levels of phosphorylated PARIS, which are not efficiently recognized by parkin for ubiquitination and proteasomal degradation. PARIS accumulates in the absence of PINK1 where it transcriptionally represses PGC-1α leading to the degeneration of DA neurons.
Parkin and PINK1 are key players in multiple domains of mitochondrial health and quality control, including fission/fusion, transport, mitophagy and biogenesis (Corti and Brice, 2013; Pickrell and Youle, 2015; Scarffe et al., 2014; Voigt et al., 2016; Winklhofer, 2014). In addition to their role at the mitochondria, both parkin and PINK1 have roles outside the mitochondria. For instance, parkin is active in multiple cellular compartments including the cytosol (von Coelln et al., 2004a; West et al., 2007), synaptic terminals (Fallon et al., 2006; Helton et al., 2008), mitochondria (Geisler et al., 2010), endosomes (Song et al., 2016) and nucleus (da Costa et al., 2009; Lee et al., 2015). PINK1 is also found outside the mitochondria where different processed forms occur in the cytosol under basal conditions (Fedorowicz et al., 2014; Haque et al., 2008; Lin and Kang, 2008, 2010; Murata et al., 2011; Sha et al., 2010; Weihofen et al., 2008). These different forms of PINK1 may not simply be intermediates that are targeted for proteasomal degradation as there is evidence that cytosolic PINK1 may regulate dendritic complexity (Dagda et al., 2014), vesicular release of dopamine, calcium-activated potassium channels, long-term potentiation (Kitada et al., 2007; Shan et al., 2009) and mitochondrial transport by phosphorylating Milton or Miro (Liu et al., 2012; Matenia et al., 2012). Thus, it could be possible that PARIS phosphorylation under physiological conditions is mediated by cytosolic PINK1. Consistent with this notion we observe basal levels of phosphorylation of cytosolic ubiquitin and PARIS under physiological conditions. PINK1 activity and phosphorylation of parkin (Fiesel et al., 2015) and ubiquitin under basal conditions have also been reported (Fiesel et al., 2015; Koyano et al., 2014; Ordureau et al., 2014). Other mitochondrial proteins such as fumarase, apoptosis inducing factor (AIF), aconitase, NAD(P)H dehydrogenase among others and have both mitochondrial and extramitochondrial localizations due to a variety of mechanisms (Yogev and Pines, 2011). Thus, it will be important in future studies to determine the mechanisms accounting for the non-mitochondrial presence of PINK1 and phosphorylation of cytosolic substrates. In addition to cytosolic PINK1, mitochondrial PINK1 can come into contact with proteins in the cytoplasm, since processed PINK1 is targeted to the outer mitochondrial membrane with its kinase domain facing the cytosol (Zhou et al., 2008), enabling PINK1 to have full access to cytosolic substrates. Thus, PINK1 through potential different mechanisms is capable of phosphorylating cytosolic substrates.
Structural analysis of parkin indicates that it exists in an autoinhibited state and thus requires relief of this autoinhibition to become active (Trempe et al., 2013; Wauer and Komander, 2013). During mitochondrial membrane depolarization parkin is activated by PINK1 phosphorylation of parkin and ubiquitin (Kane et al., 2014; Kazlauskaite et al., 2014; Kazlauskaite et al., 2015; Koyano et al., 2014; Kumar et al., 2015; Ordureau et al., 2014; Sauve et al., 2015; Wauer et al., 2015a; Wauer et al., 2015b; Yamano et al., 2015). What accounts for parkin activation under physiological conditions in the cytosol and non-mitochondrial organelles is not known and requires further study. However, our observation that phospho-ubiquitin is present in the cytosol and is PINK1 dependent suggests that phosphorylation of ubiquitin by PINK1 may contribute to activation of cytosolic parkin. In addition, parkin is likely to be regulated by its association with its E2s (Fiesel et al., 2014), CHIP and substrates (Byrd and Weissman, 2013; Imai et al., 2002; Shin et al., 2011).
Dopamine neurons do not degenerate in germ line parkin knockout mice (Goldberg et al., 2003; Itier et al., 2003; Von Coelln et al., 2004b). Crossing germ line parkin knockout mice to mitochondrial mutator mice causes loss of DA neurons and elevation of phosphorylated ubiquitin suggesting that PINK1 is activated and that defects in mitophagy could be contributing to loss of DA neurons (Pickrell et al., 2015). On the other hand, mitophagy defects were not found in parkin knockouts in response to mitochondrial stressors (Kageyama et al., 2014; Kageyama et al., 2012; Sterky et al., 2011). Moreover, in adult conditional parkin knockouts, which exhibit progressive degeneration of DA neurons, mitochondrial mass was reduced and defects in mitophagy were not observed (Stevens et al., 2015). PARIS up-regulation and PGC-1α suppression seem to be the primary driver of DA neuron loss due to the absence or inactivation of parkin (Shin et al., 2011; Stevens et al., 2015) and PINK1 in PD since loss of DA neurons is PARIS dependent. Since PARIS regulates mitochondrial biogenesis through its actions on PGC-1α under normal conditions (i.e. active PINK1 and parkin) there is likely a homeostatic mechanism that controls the content of mitochondria in a cell through degradation (mitophagy) and synthesis (biogenesis). Consistent with this notion is our observation that following induction of mitophagy through mitochondrial depolarization via CCCP there is enhanced PARIS phosphorylation and reduction in its levels, which would relieve its repression on PGC-1α and augment biogenesis. Moreover, it has been reported that PGC-1α is involved in the regulation of mitochondrial autophagy and degradation as well as biogenesis (Baldelli et al., 2014; Vainshtein et al., 2015a; Vainshtein et al., 2015b). Further studies are required to investigate this possibility.
In conclusion, since the PARIS/PGC-1α cell death pathway appears to play a role in sporadic PD (Clark et al., 2011; Eschbach et al., 2015; Shin et al., 2011; Su et al., 2015; Zheng et al., 2010), therapeutic strategies aimed at inhibiting PARIS or maintaining PGC-1α function could be beneficial in both sporadic PD and PD due to mutations in parkin or PINK1.
EXPERIMENTAL PROCEDURES
Full experimental details can be found in the Supplemental Experimental Procedures.
Antibodies
Antibodies specific to PARIS that is phosphorylated at serine 613 (anti-pS613-PARIS antibodies) were raised by immunizing a rabbit with a phospho-peptide (CDPFK-(phosphorylated)S-PASKGPLASTD, synthesized in the Johns Hopkins Sequencing Facility) and purified in a column conjugated with the phosphorylated PARIS peptide (SulfoLink Immobilization Kit for Peptides, Thermo Scientific).
Plasmids
TetP-PARIS-FLAG was constructed by cloning PCR amplified C-terminal FLAG tagged human PARIS into the SalI site of pTRE-Dual2 vector (Clontech). Serine 322 and serine 613 of human PARIS were mutagenized into alanine by using site directed mutagenesis following the manufacturer’s instruction to generate S322A, S613A, and double phosphorylation mutant PARIS (QuikChange® II XL Site-Directed Mutagenesis Kit, Stratagene).
Subcellular fractionation
SH-SY5Y cells and mouse brain tissues were subjected to subcellular fractionation using the Qproteome Mitochondria Isolation Kit (Qiagen) following the manufacturer instructions. Cytosolic fractions were concentrated with acetone precipitation.
Tet-off mediated pulse chase study
SH-SY5Y cells were transfected with pCMV-tTA and TetP-PARIS-FLAG and indicated DNA constructs. One day later, doxycycline (200 ng per ml, Invitrogen) was added to culture media and PARIS-FLAG was measured by western blot.
In vitro ubiquitination/phosphorylation assay
Recombinant PARIS WT or phosphorylation SA-DM mutant proteins were incubated with ubiquitin S65A (10 µg), UBE1 (250 ng), UbcH7 (500 ng), TcPINK1 and rat Parkin (full length, aa 219–465, 2 µg) in ubiquitin conjugation reaction buffer (Boston Biochem) supplemented with 2 mM ATP at 37° C for 40 minutes.
Virally induced conditional PINK1 knockdown mouse model
PINK1-siRNA transgenic mice were generously provided by Dr. Xugang Xia (Thomas Jefferson University). In this mouse strain, expression of shRNA to mouse PINK1 is blocked by a floxed STOP sequence in between the U6 promoter and shPINK1 sequence (Zhou et al., 2007). A recombinant adeno-associated virus expressing GFP-fused Cre recombinase (AAV-GFP-Cre) was stereotaxically introduced into two-month-old mice SN at coordinates (1.3 mm lateral, 3.2mm caudal, 4.3 mm ventral relative to Bregma).
Chromatin immunoprecipitation
SH-SY5Y cells were fixed, glycine quenched, and lysed in 500 ml of SDS buffer containing protease inhibitors. Chromatin was digested with micrococcal nuclease and sonicated to 100–250bp DNA/Protein complex using a Diagenode UCD-300. Pre-cleared samples were incubated with mouse anti-PARIS antibodies (Neuromab) overnight. Elutes were subjected to reverse cross-linking and DNA was recovered by phenol-chloroform-ethanol purification.
Laser capture microdissection
The brains were rapidly removed and frozen on dry ice with OCT tissue freezing medium for cryostat sectioning (Tissue-Tek). An RNase inhibitor and sterile PBS were used during immunofluorescence labeling for tyrosine hydroxylase (TH) and GFP. Double positive neurons were collected by LCM (Leica P.A.L.M. laser microdissection) and used for RNA extraction.
Statistics
Quantitative data are presented as the mean ± s.e.m. Statistical significance was assessed either via an unpaired two-tailed Student’s t-test or nonparametric Mann-Whitney U-test for two-group comparison or an ANOVA test with Tukey’s HSD post hoc analysis for comparison of more than three groups.
Supplementary Material
Highlights.
PARIS is a PINK1 Substrate
PINK1 phosphorylation of PARIS primes it for parkin ubiquitination and degradation
PINK1 controls PARIS regulated PGC-1α levels and dopamine neuron survival
Conditional knockdown of PINK1 leads to PARIS-dependent loss of dopamine neurons
Acknowledgments
This work was supported by grants from the NIH/NINDS NS38377, the JPB Foundation, and Samsung Biomedical Research Institute. The authors acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation and the Diana Helis Henry Medical Research Foundation through their direct engagement in the continuous active conduct of medical research in conjunction with The Johns Hopkins Hospital and the Johns Hopkins University School of Medicine and the Foundation’s Parkinson’s Disease Programs M-1, M-2, H-2014. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases. W.S. was supported by NIH/NINDS R01 #NS085070, the Michael J. Fox Foundation for Parkinson’s Research and the Foundation for Mitochondrial Medicine, Mayo Clinic Foundation, the Centers for Individualized and Regenerative Medicine, the Marriott Family Foundation, and a Gerstner Family Career Development Award. F.C.F. is the recipient of a fellowship from the American Parkinson Disease Foundation (APDA). This research was also supported by grants from the NRF (NRF-2016R1A2B4008271, 2015R1C1A1A01052708) funded by the Korea Ministry of Science, ICT & Future Planning and was also supported by a Samsung Biomedical Research Institute grant (SMX1161351 and SMX1161191). Dr. T. and V. Dawson are founders of Valted, LLC and hold an ownership equity interest in the company. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, one table, and Supplemental References and can be found with this article online at http://www.cell.com/cell-reports/home
AUTHOR CONTRIBUTIONS
Conceptualization, V.L.D. and T.M.D.; Methodology, Y.L., D.A.S., S.-U.K., V.L.D., J-H.S., and T.M.D.; Formal Analysis, S-U.K. and J-H.S.; Investigation, Y.L., D.A.S., S.-U.K., H.J., Y.-I.L., L.A.S., G.E.U., H.K., S.H., K.A., S.N., S.P.Y.; Writing – Original Draft, Y.L., D.A.S., V.L.D. and T.M.D. ; Writing – Review and Editing, Y.L., D.A.S., S.-U.K., H.J., Y.-I.L., H.S.K., L.A.S., G.E.U., H.K., S.H., T.-I.K., K.A., S.B., J.W.K., S.N., S.P.Y., F.C.F., W.S., V.L.D., J.-H.S., T.M.D.; Funding Acquisition, V.L.D., J.-H.S., T.M.D.; Resources, H.S.K., J.K., T-I.K., S.B., F.C.F. and W.S.; Supervision, V.L.D., J.-H.S., T.M.D.
REFERENCES
- Baldelli S, Aquilano K, Ciriolo MR. PGC-1αlpha buffers ROS-mediated removal of mitochondria during myogenesis. Cell death & disease. 2014;5:e1515. doi: 10.1038/cddis.2014.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd RA, Weissman AM. Compact Parkin only: insights into the structure of an autoinhibited ubiquitin ligase. EMBO J. 2013;32:2087–2089. doi: 10.1038/emboj.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciron C, Zheng L, Bobela W, Knott GW, Leone TC, Kelly DP, Schneider BL. PGC-1αlpha activity in nigral dopamine neurons determines vulnerability to alpha-synuclein. Acta Neuropathol Commun. 2015;3:16. doi: 10.1186/s40478-015-0200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Clark J, Reddy S, Zheng K, Betensky RA, Simon DK. Association of PGC-1αlpha polymorphisms with age of onset and risk of Parkinson’s disease. BMC Med Genet. 2011;12:69. doi: 10.1186/1471-2350-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti O, Brice A. Mitochondrial quality control turns out to be the principal suspect in parkin and PINK1-related autosomal recessive Parkinson’s disease. Curr Opin Neurobiol. 2013;23:100–108. doi: 10.1016/j.conb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson’s disease. Physiol Rev. 2011;91:1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- da Costa CA, Sunyach C, Giaime E, West A, Corti O, Brice A, Safe S, Abou-Sleiman PM, Wood NW, Takahashi H, et al. Transcriptional repression of p53 by parkin and impairment by mutations associated with autosomal recessive juvenile Parkinson’s disease. Nat Cell Biol. 2009;11:1370–1375. doi: 10.1038/ncb1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Pien I, Wang R, Zhu J, Wang KZ, Callio J, Banerjee TD, Dagda RY, Chu CT. Beyond the mitochondrion: cytosolic PINK1 remodels dendrites through protein kinase A. J Neurochem. 2014;128:864–877. doi: 10.1111/jnc.12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschbach J, von Einem B, Muller K, Bayer H, Scheffold A, Morrison BE, Rudolph KL, Thal DR, Witting A, Weydt P, et al. Mutual exacerbation of peroxisome proliferator-activated receptor gamma coactivator 1alpha deregulation and alpha-synuclein oligomerization. Ann Neurol. 2015;77:15–32. doi: 10.1002/ana.24294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol. 2006;8:834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- Fedorowicz MA, de Vries-Schneider RL, Rub C, Becker D, Huang Y, Zhou C, Alessi Wolken DM, Voos W, Liu Y, Przedborski S. Cytosolic cleaved PINK1 represses Parkin translocation to mitochondria and mitophagy. EMBO Rep. 2014;15:86–93. doi: 10.1002/embr.201337294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel FC, Ando M, Hudec R, Hill AR, Castanedes-Casey M, Caulfield TR, Moussaud-Lamodiere EL, Stankowski JN, Bauer PO, Lorenzo-Betancor O, et al. (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep. 2015;16:1114–1130. doi: 10.15252/embr.201540514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel FC, Moussaud-Lamodiere EL, Ando M, Springer W. A specific subset of E2 ubiquitin-conjugating enzymes regulate Parkin activation and mitophagy differently. J Cell Sci. 2014;127:3488–3504. doi: 10.1242/jcs.147520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, Meloni EG, Wu N, Ackerson LC, Klapstein GJ, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- Haque ME, Thomas KJ, D’Souza C, Callaghan S, Kitada T, Slack RS, Fraser P, Cookson MR, Tandon A, Park DS. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc Natl Acad Sci U S A. 2008;105:1716–1721. doi: 10.1073/pnas.0705363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helton TD, Otsuka T, Lee MC, Mu Y, Ehlers MD. Pruning and loss of excitatory synapses by the parkin ubiquitin ligase. Proc Natl Acad Sci U S A. 2008;105:19492–19497. doi: 10.1073/pnas.0802280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, Takahashi R. CHIP is associated with Parkin, a gene responsible for familial Parkinson’s disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- Itier JM, Ibanez P, Mena MA, Abbas N, Cohen-Salmon C, Bohme GA, Laville M, Pratt J, Corti O, Pradier L, et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet. 2003;12:2277–2291. doi: 10.1093/hmg/ddg239. [DOI] [PubMed] [Google Scholar]
- Jiang H, Kang SU, Zhang S, Karuppagounder S, Xu J, Lee YK, Kang BG, Lee Y, Zhang J, Pletnikova O, et al. Adult Conditional Knockout of PGC-1αlpha Leads to Loss of Dopamine Neurons. eNeuro. 2016;3 doi: 10.1523/ENEURO.0183-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Hoke A, Dawson VL, Dawson TM, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33:2798–2813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J Cell Biol. 2012;197:535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Martinez-Torres RJ, Wilkie S, Kumar A, Peltier J, Gonzalez A, Johnson C, Zhang J, Hope AG, Peggie M, et al. Binding to serine 65-phosphorylated ubiquitin primes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep. 2015;16:939–954. doi: 10.15252/embr.201540352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kitada T, Pisani A, Porter DR, Yamaguchi H, Tscherter A, Martella G, Bonsi P, Zhang C, Pothos EN, Shen J. Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proc Natl Acad Sci U S A. 2007;104:11441–11446. doi: 10.1073/pnas.0702717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- Kumar A, Aguirre JD, Condos TE, Martinez-Torres RJ, Chaugule VK, Toth R, Sundaramoorthy R, Mercier P, Knebel A, Spratt DE, et al. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J. 2015;34:2506–2521. doi: 10.15252/embj.201592337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kim JJ, Nam HJ, Gao B, Yin P, Qin B, Yi SY, Ham H, Evans D, Kim SH, et al. Parkin Regulates Mitosis and Genomic Stability through Cdc20/Cdh1. Mol Cell. 2015;60:21–34. doi: 10.1016/j.molcel.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Kang UJ. Characterization of PINK1 processing, stability, and subcellular localization. J Neurochem. 2008;106:464–474. doi: 10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Kang UJ. Structural determinants of PINK1 topology and dual subcellular distribution. BMC Cell Biol. 2010;11:90. doi: 10.1186/1471-2121-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, Millan I, Shen A, Saxton W, Kanao T, et al. Parkinson’s disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 2012;8:e1002537. doi: 10.1371/journal.pgen.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I, Dawson VL, Dawson TM. Recent advances in the genetics of Parkinson’s disease. Annual review of genomics and human genetics. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matenia D, Hempp C, Timm T, Eikhof A, Mandelkow EM. Microtubule affinity-regulating kinase 2 (MARK2) turns on phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1) at Thr-313, a mutation site in Parkinson disease: effects on mitochondrial transport. J Biol Chem. 2012;287:8174–8186. doi: 10.1074/jbc.M111.262287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudo G, Makela J, Di Liberto V, Tselykh TV, Olivieri M, Piepponen P, Eriksson O, Malkia A, Bonomo A, Kairisalo M, et al. Transgenic expression and activation of PGC-1αlpha protect dopaminergic neurons in the MPTP mouse model of Parkinson’s disease. Cell Mol Life Sci. 2012;69:1153–1165. doi: 10.1007/s00018-011-0850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata H, Sakaguchi M, Jin Y, Sakaguchi Y, Futami J, Yamada H, Kataoka K, Huh NH. A new cytosolic pathway from a Parkinson disease-associated kinase, BRPK/PINK1: activation of AKT via mTORC2. J Biol Chem. 2011;286:7182–7189. doi: 10.1074/jbc.M110.179390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, Olszewski JL, Koerber JT, Xie T, Beausoleil SA, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Pickrell AM, Huang CH, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, Harper JW, Youle RJ. Endogenous Parkin Preserves Dopaminergic Substantia Nigral Neurons following Mitochondrial DNA Mutagenic Stress. Neuron. 2015;87:371–381. doi: 10.1016/j.neuron.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochet JC, Hay BA, Guo M. Molecular insights into Parkinson’s disease. Prog Mol Biol Transl Sci. 2012;107:125–188. doi: 10.1016/B978-0-12-385883-2.00011-4. [DOI] [PubMed] [Google Scholar]
- Sauve V, Lilov A, Seirafi M, Vranas M, Rasool S, Kozlov G, Sprules T, Wang J, Trempe JF, Gehring K. A Ubl/ubiquitin switch in the activation of Parkin. EMBO J. 2015;34:2492–2505. doi: 10.15252/embj.201592237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarffe LA, Stevens DA, Dawson VL, Dawson TM. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 2014;37:315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha D, Chin LS, Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum Mol Genet. 2010;19:352–363. doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, Liu B, Li L, Chang N, Li L, Wang H, Wang D, Feng H, Cheung C, Liao M, et al. Regulation of PINK1 by NR2B–containing NMDA receptors in ischemic neuronal injury. J Neurochem. 2009;111:1149–1160. doi: 10.1111/j.1471-4159.2009.06398.x. [DOI] [PubMed] [Google Scholar]
- Shiba K, Arai T, Sato S, Kubo S, Ohba Y, Mizuno Y, Hattori N. Parkin stabilizes PINK1 through direct interaction. Biochem Biophys Res Commun. 2009;383:331–335. doi: 10.1016/j.bbrc.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) repression of PGC-1αlpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A, Bhaumik D, Chinta SJ, Rane A, Rajagopalan S, Lieu CA, Lithgow GJ, Andersen JK. Mitochondrial Quality Control via the PGC1alpha-TFEB Signaling Pathway Is Compromised by Parkin Q311X Mutation But Independently Restored by Rapamycin. J Neurosci. 2015;35:12833–12844. doi: 10.1523/JNEUROSCI.0109-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A, Rane A, Rajagopalan S, Chinta SJ, Andersen JK. Detrimental effects of oxidative losses in parkin activity in a model of sporadic Parkinson’s disease are attenuated by restoration of PGC1alpha. Neurobiol Dis. 2016;93:115–120. doi: 10.1016/j.nbd.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Song P, Trajkovic K, Tsunemi T, Krainc D. Parkin Modulates Endosomal Organization and Function of the Endo-Lysosomal Pathway. J Neurosci. 2016;36:2425–2437. doi: 10.1523/JNEUROSCI.2569-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky FH, Lee S, Wibom R, Olson L, Larsson NG. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc Natl Acad Sci U S A. 2011;108:12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens DA, Lee Y, Kang HC, Lee BD, Lee YI, Bower A, Jiang H, Kang SU, Andrabi SA, Dawson VL, et al. Parkin loss leads to PARIS-dependent declines in mitochondrial mass and respiration. Proc Natl Acad Sci U S A. 2015;112:11696–11701. doi: 10.1073/pnas.1500624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Chu Y, Kordower JH, Li B, Cao H, Huang L, Nishida M, Song L, Wang D, Federoff HJ. PGC-1αlpha Promoter Methylation in Parkinson’s Disease. PLoS One. 2015;10:e0134087. doi: 10.1371/journal.pone.0134087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- Vainshtein A, Desjardins EM, Armani A, Sandri M, Hood DA. PGC-1αlpha modulates denervation-induced mitophagy in skeletal muscle. Skeletal muscle. 2015a;5:9. doi: 10.1186/s13395-015-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainshtein A, Tryon LD, Pauly M, Hood DA. Role of PGC-1αlpha during acute exercise-induced autophagy and mitophagy in skeletal muscle. American journal of physiology Cell physiology. 2015b;308:C710–C719. doi: 10.1152/ajpcell.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Berlemann LA, Winklhofer KF. The mitochondrial kinase PINK1: functions beyond mitophagy. J Neurochem. 2016 doi: 10.1111/jnc.13655. [DOI] [PubMed] [Google Scholar]
- von Coelln R, Dawson VL, Dawson TM. Parkin-associated Parkinson’s disease. Cell Tissue Res. 2004a;318:175–184. doi: 10.1007/s00441-004-0924-4. [DOI] [PubMed] [Google Scholar]
- Von Coelln R, Thomas B, Savitt JM, Lim KL, Sasaki M, Hess EJ, Dawson VL, Dawson TM. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci U S A. 2004b;101:10744–10749. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 2015a;524:370–374. doi: 10.1038/nature14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Swatek KN, Wagstaff JL, Gladkova C, Pruneda JN, Michel MA, Gersch M, Johnson CM, Freund SM, Komander D. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015b;34:307–325. doi: 10.15252/embj.201489847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen A, Ostaszewski B, Minami Y, Selkoe DJ. Pink1 Parkinson mutations, the Cdc37/Hsp90 chaperones and Parkin all influence the maturation or subcellular distribution of Pink1. Hum Mol Genet. 2008;17:602–616. doi: 10.1093/hmg/ddm334. [DOI] [PubMed] [Google Scholar]
- West AB, Dawson VL, Dawson TM. The Role of PARKIN in Parkinson’s Disease. In: Dawson TM, editor. Parkinson’s Disease: Genetics and Pathogenesis. Informa Healthcare USA; 2007. pp. 199–218. [Google Scholar]
- Winklhofer KF. Parkin and mitochondrial quality control: toward assembling the puzzle. Trends Cell Biol. 2014;24:332–341. doi: 10.1016/j.tcb.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Woodroof HI, Pogson JH, Begley M, Cantley LC, Deak M, Campbell DG, van Aalten DM, Whitworth AJ, Alessi DR, Muqit MM. Discovery of catalytically active orthologues of the Parkinson’s disease kinase PINK1: analysis of substrate specificity and impact of mutations. Open Biol. 2011;1:110012. doi: 10.1098/rsob.110012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Queliconi BB, Koyano F, Saeki Y, Hirokawa T, Tanaka K, Matsuda N. Site-specific Interaction Mapping of Phosphorylated Ubiquitin to Uncover Parkin Activation. J Biol Chem. 2015;290:25199–25211. doi: 10.1074/jbc.M115.671446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O, Pines O. Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim Biophys Acta. 2011;1808:1012–1020. doi: 10.1016/j.bbamem.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, et al. PGC-1αlpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3001059. 52ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Falkenburger BH, Schulz JB, Tieu K, Xu Z, Xia XG. Silencing of the Pink1 gene expression by conditional RNAi does not induce dopaminergic neuron death in mice. Int J Biol Sci. 2007;3:242–250. doi: 10.7150/ijbs.3.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.