Abstract
Type 1 diabetes (T1D) is an autoimmune disease that is clinically silent until the majority of β cells are destroyed. There is an unmet need for reliable and cost-effective biomarkers to predict and diagnose diabetes at an early stage. A number of stable microRNAs (miRNAs) have been reported in serum and plasma and are now being investigated as biomarkers of different diseases. We measured the levels of 745 miRNAs in sera of children with recent-onset T1D and age-matched controls using locked nucleic acid–enhanced (LNA-enhanced) quantitative PCR profiling. Thirty-five miRNAs were significantly different between the groups, and 27 miRNAs were elevated in T1D. Good discriminating power was obtained for 6 miRNAs (miR-454-3p, miR-222-3p, miR-144-5p, miR-345-5p, miR-24-3p, and miR-140-5p), which were not elevated at later stages of diabetes. In silico pathway analysis, based on inferred miRNA target genes, associated glycosaminoglycan biosynthesis as well as PI3K/Akt, MAPK, and Wnt signaling pathways with early stages of T1D. Among the 27 upregulated miRNAs in T1D, 2 miRNAs significantly correlated with hemoglobin A1c (HbA1c), as did 5 of 8 downregulated miRNAs. A total of 134 miRNAs significantly correlated with HbA1c when stratifying hyperglycemia-induced miRNAs from T1D-specific miRNAs. In conclusion, we have identified a serum miRNA pattern of recent-onset T1D and signaling pathways that may be involved in its pathogenesis.
A panel of circulating serum miRNAs is identified in children with recent-onset type 1 diabetes.
Introduction
Type 1 diabetes (T1D) arises from autoimmune destruction of insulin-producing β cells, resulting in insufficient insulin secretion and elevated blood glucose levels. T1D accounts for 5% to 10% of all diabetes worldwide (1) and generally presents in children or young adults. Currently, T1D cannot be diagnosed or easily predicted until the majority of β cells have been destroyed and symptoms appear. At the time of diagnosis, β cell mass is typically reduced by 70%–80% (2) and intensive treatment can preserve residual β cell function (3). Earlier detection of T1D may therefore be expected to improve clinical outcomes by allowing for treatment to be initiated when a greater residual β cell mass remains.
MicroRNAs (miRNAs), a family of endogenously produced single-stranded RNA molecules of approximately 22 nucleotides in length, have emerged in the last decade as key regulators of gene expression and cell function (4). Their differential expression within tissues has been detected in various disorders, including cancer, cardiovascular disease, and T2D (5). Surprisingly perhaps, stable miRNAs can be found at high concentrations in body fluids and can be readily detected and measured by quantitative PCR (qPCR) using sequence-specific primers (6), making them potentially useful as biomarkers. In particular, circulating miRNA patterns are currently being investigated as potential biomarkers for metabolic diseases and cancers as a means of providing noninvasive, rapid, sensitive, and specific diagnostic measures. Moreover, miRNA signatures may also be correlated with clinical staging of disease and used as prognostic markers or indicators of disease progression.
Numerous studies have reported that patients with T2D have dysregulated miRNA profiles in solid tissue samples and biological fluids (7, 8). However, as each study used different profiling methods and platforms, their results are inconsistent, and a definitive T2D profile has not yet been established. A recent meta-analysis (9) of studies investigating blood and solid tissue samples from patients with T2D or in animal models of diabetes found dysregulation of 40 miRNAs in various tissues and blood, potentially identifying biomarkers of T2D. In contrast, there is little information about the miRNA profiles of patients with T1D. Unbiased miRNA profiling of various blood cell types from patients with T1D and controls (10–12) has provided insights into the miRNAs related to the autoimmunity component of T1D. Profiling of circulating miRNAs may offer new biomarkers for prediction and early detection of T1D and may also provide insights into mechanisms of pathogenesis or tissue damage. In the present study, we examined the serum miRNA profiles of 10 children with recent onset of T1D (within 42 days after diagnosis), as compared with healthy children without T1D, and then validated our findings in a second larger cohort to identify miRNAs that may serve as early biomarkers of T1D.
Results
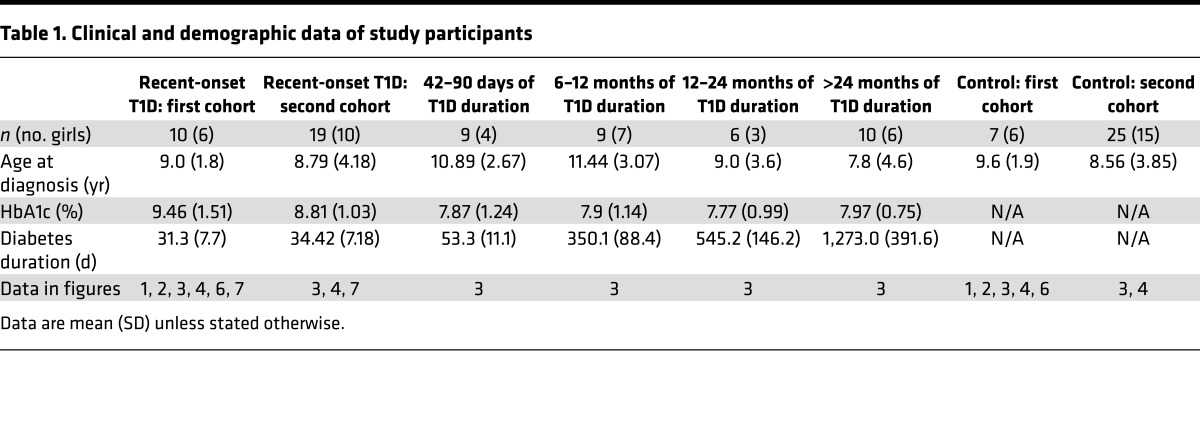
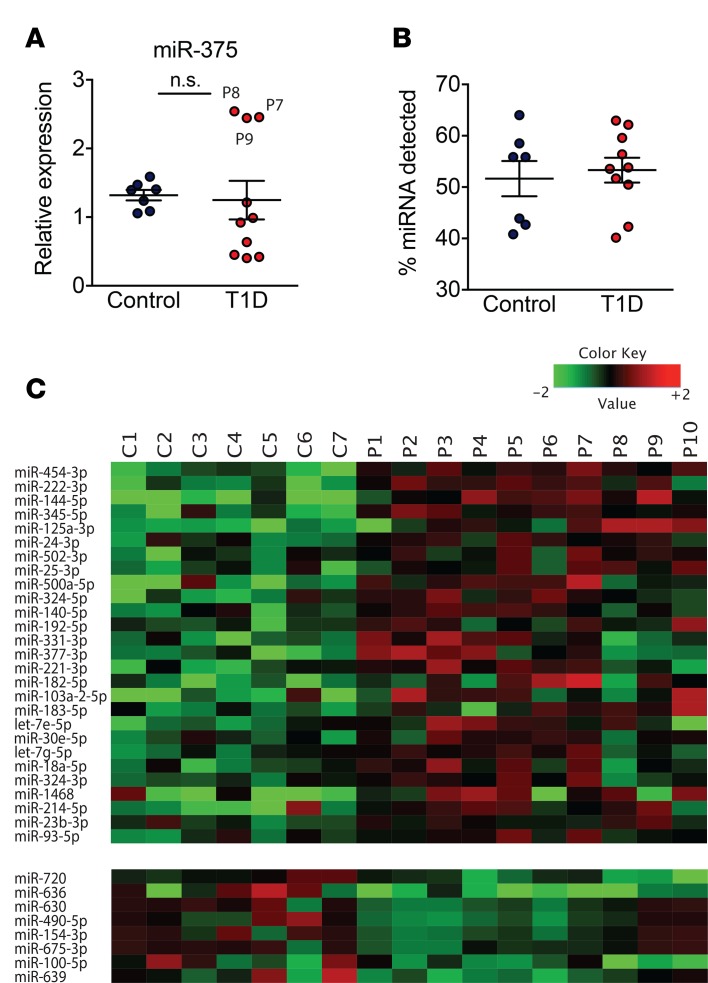
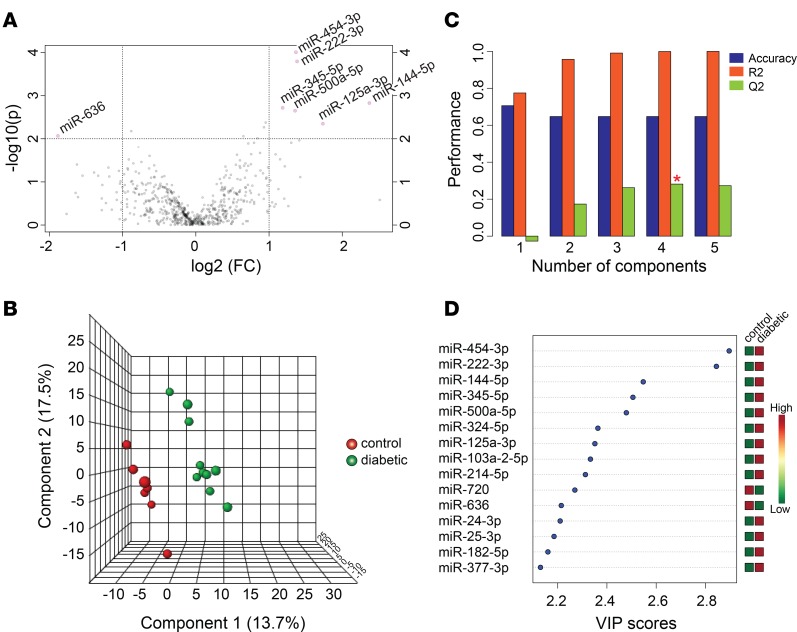
To identify early miRNA expression signatures of T1D, RNA was isolated from the sera of 10 children (aged 6 years to 12 years, mean 9.0 ± 1.8 years) with recent onset of diabetes ranging from 21 to 42 days (mean 31.3 ± 7.7 days) since day of diagnosis and their age-matched controls (aged 6 years to 12 years, mean 9.6 ± 1.9 years; Table 1). The control group was chosen from children who visited the British Columbia Children’s Hospital endocrinology division and were found to be healthy, with normal variations in growth and puberty and no evidence of either autoimmune disease or diabetes following comprehensive outpatient assessment. We first sought to investigate whether miR-375, a miRNA that we reported to be increased in the circulation of prediabetic mice (6) and a miRNA reported to be lower in the sera of children with newly diagnosed T1D (13), was altered in the sera of children with recent onset of diabetes. Measurement of miR-375 levels with qRT-PCR indicated that miR-375 was not significantly different in the sera of children with recent onset of diabetes (Figure 1A), prompting us to perform an unbiased miRNA screen. The cDNA from children with recent-onset diabetes and controls was profiled using a human miRNA qPCR platform with locked nucleic acid–enhanced (LNA-enhanced) primers (14). There was no difference in the number of miRNAs detected in the T1D samples compared with controls (Figure 1B). The miRNA detection rate ranged from 41% to 64% in the control samples and 40% to 63% in the T1D samples. Of the 745 miRNAs analyzed, 15% were not present in any of the samples tested. miRNA levels in each individual were global mean normalized and subjected to heatmap analysis (Figure 1C). Of all the circulating miRNAs detected in this study, 35 were significantly different (P ≤ 0.05) between the 2 groups, with 27 being elevated in the T1D samples (Table 2).
Table 1. Clinical and demographic data of study participants.
Figure 1. Candidate miRNA biomarkers of type 1 diabetes.
(A) miR-375 levels from the sera of 7 control and 10 children with recent onset of type 1 diabetes (T1D) were measured using qRT-PCR. (B and C) miRNA levels from the sera of 7 control and 10 children with recent onset of T1D were quantified using a locked nucleic acid–enhanced (LNA-enhanced) qPCR profiling platform. (B) Percentage of total miRNAs with Ct values <38 in each sample. (C) Heatmap showing significantly different expression levels of miRNAs among control (C1–C7) and T1D patients (P1–P10). P ≤ 0.05 by Student’s t test. (A and B) Data were analyzed by Student’s t test and are expressed as the mean ± SEM.
Table 2. Differentially expressed circulating miRNAs.
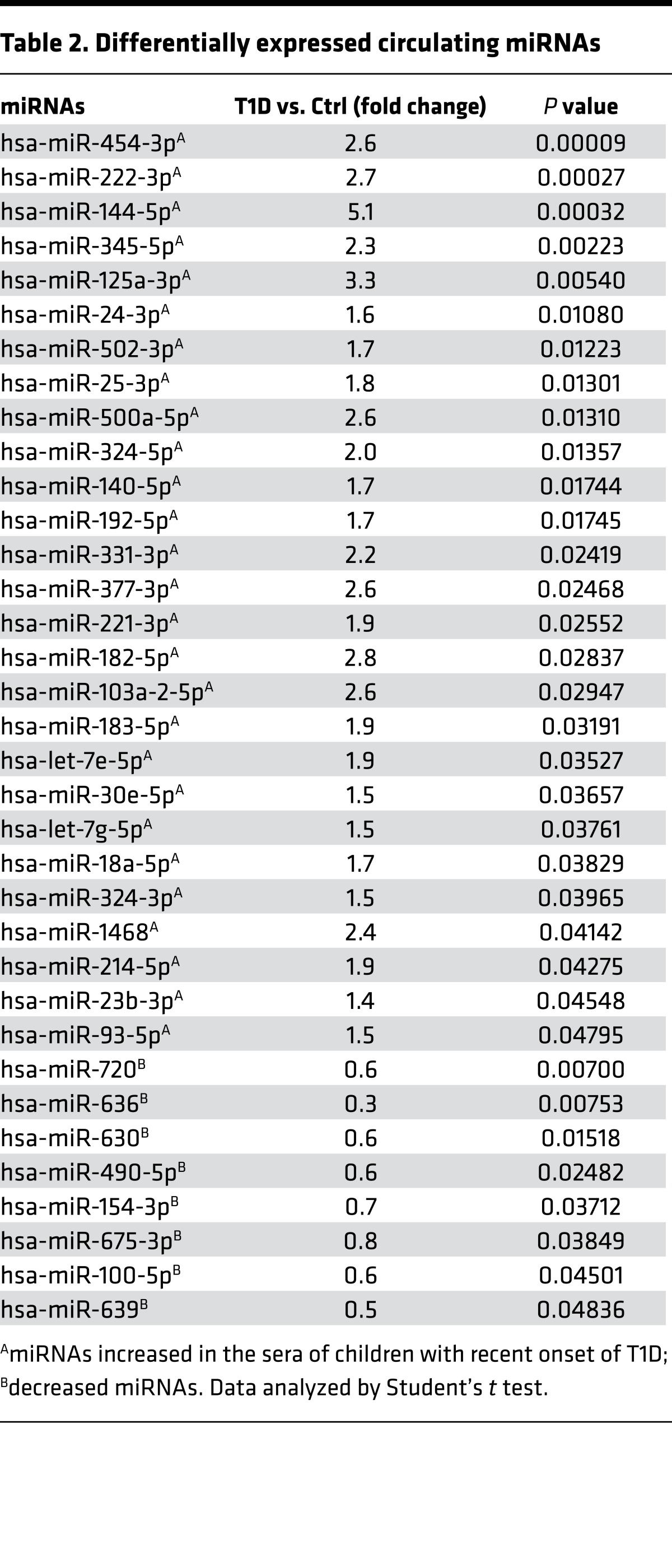
To identify the miRNAs with the greatest fold differences as well as high statistical significance between the 2 groups, volcano plots were constructed. This analysis indicated that miR-454-3p, miR-222-3p, miR-345-5p, miR-500a-5p, miR-125a-3p, miR-144-5p, and miR-636 had the greatest change with the highest significance (Figure 2A). Next, we performed multivariate analysis using the partial least squares supervised classification method (PLS-DA; Figure 2B). We analyzed 3 performance measures: prediction accuracy, R2 (the sum of the squares captured by the model), and Q2 (cross-validated R2) using a different number of components. Using Q2 as a measure, we determined that 4 miRNAs were optimal to define the pattern of recent-onset T1D (Figure 2C). Using PLS-DA, we also analyzed the importance of each miRNA in explaining the variance between the groups and ranked each miRNA based on variable importance in projection scores (Figure 2D). The top 4 miRNAs with the highest scores were miR-454-3p, miR-222-3p, miR-144-5p, and miR-345-5p (Figure 2D).
Figure 2. Multivariate analysis using partial least squares–discriminant analysis.
(A) Volcano plots showing changes in miRNA levels between children with recent-onset type 1 diabetes (T1D) and controls. The log fold change is depicted on the x axis, and log10 of the P value is shown on the y axis. Data analyzed by Student’s t test. (B) Partial least squares–discriminant analysis 3D plot. Green circles: children with T1D; red circles: control group. (C) The three performance measures (prediction accuracy, R2, and Q2) using a different number of components. The red asterisk indicates the best values of the currently selected measures (Q2). (D) The top 15 miRNAs ranked by variable of importance (VIP) scores.
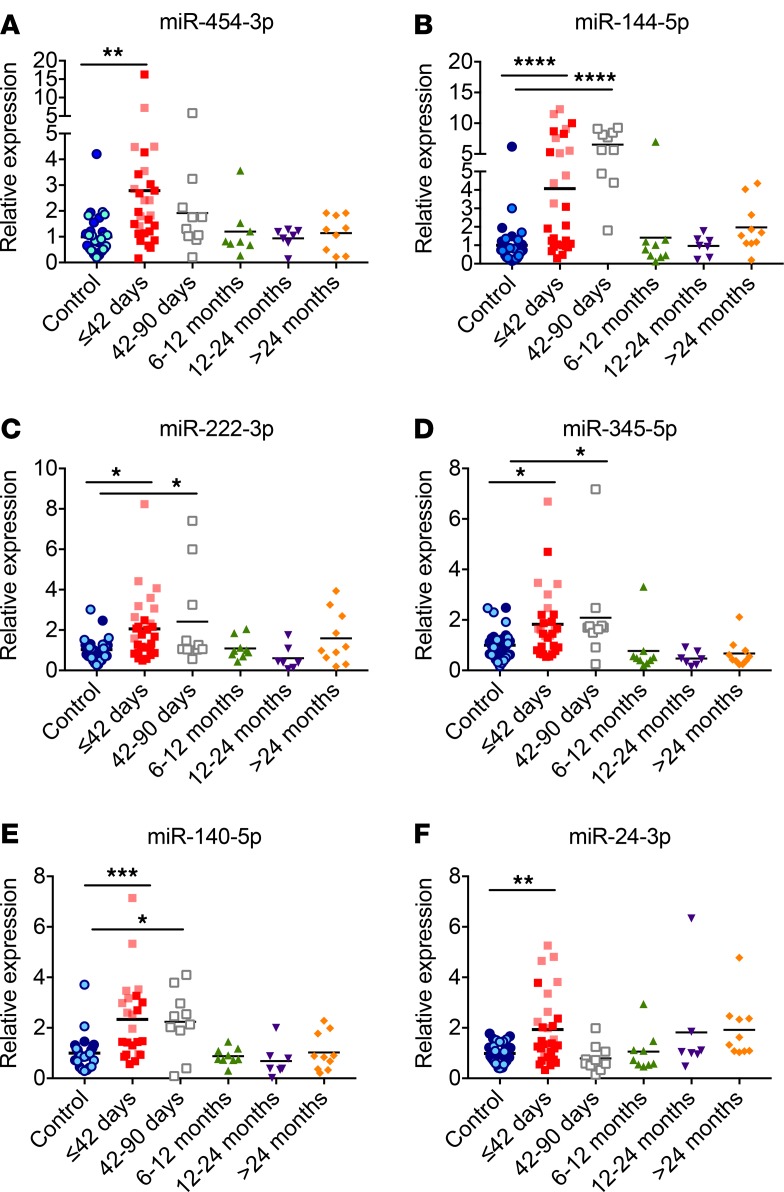
We next investigated the levels of these 4 miRNAs in samples from a larger cohort of patients with recent-onset T1D. We also sought to determine whether any of these miRNAs could be used for assessment of disease staging in children with T1D. We obtained additional serum samples from controls and from children with different durations of T1D and grouped them by diabetes duration. We isolated RNA from the sera and measured miRNA levels by qRT-PCR using LNA-enhanced primers. Levels of miR-454-3p, miR-222-3p, miR-144-5p, and miR-345-5p were significantly elevated in the sera of children with recent-onset T1D samples compared with nondiabetic controls (Figure 3, A–D), confirming the results obtained from the profiling platform. miR-454-3p levels were specifically increased during the first 42 days following diabetes diagnosis and were not higher at later stages of the disease. miR-144-5p, miR-222-3p, and miR-345 levels were significantly higher up until 90 days after diabetes diagnosis but were comparable to control levels beyond 6 months after diagnosis. We next measured the levels of miR-24-3p and miR-140-5p, 2 miRNAs randomly selected from the list of miRNAs that showed differences in the qPCR platform (Table 2). miR-140-5p levels were significantly higher up until 90 days after diabetes diagnosis but were comparable to control levels beyond 6 months after diagnosis (Figure 3E). miR-24-3p levels were increased in recent-onset T1D samples but were comparable to controls at later stages (Figure 3F). We also measured the levels of miR-144-3p, a miRNA that was unchanged between the control and T1D group in the profiling experiment, and our analysis indicated that this miRNA was not different between the controls and children with recent-onset T1D (data not shown). The validation between the profiling and subsequent individual qPCR assays in a larger cohort of samples supports the identified miRNAs as markers of early stages of T1D.
Figure 3. Circulating miRNA levels at different stages of type 1 diabetes.
RNA was isolated from the sera of children at different stages after diabetes diagnosis, and levels of (A) miR-454-3p, (B) miR-144-5p, (C) miR-222-3p, (D) miR-345-5p, (E) miR-140-5p, and (F) miR-24-3p were quantified using qPCR with locked nucleic acid–enhanced primers. miRNA levels from the sera of the first cohort of children indentified with diabetes are indicated with lighter shades of blue (controls) and lighter shades of red (≤42 days after diagnosis). Mean values are indicated with a line. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 after 1-way ANOVA with Dunnett’s post-hoc test. n = 7–32.
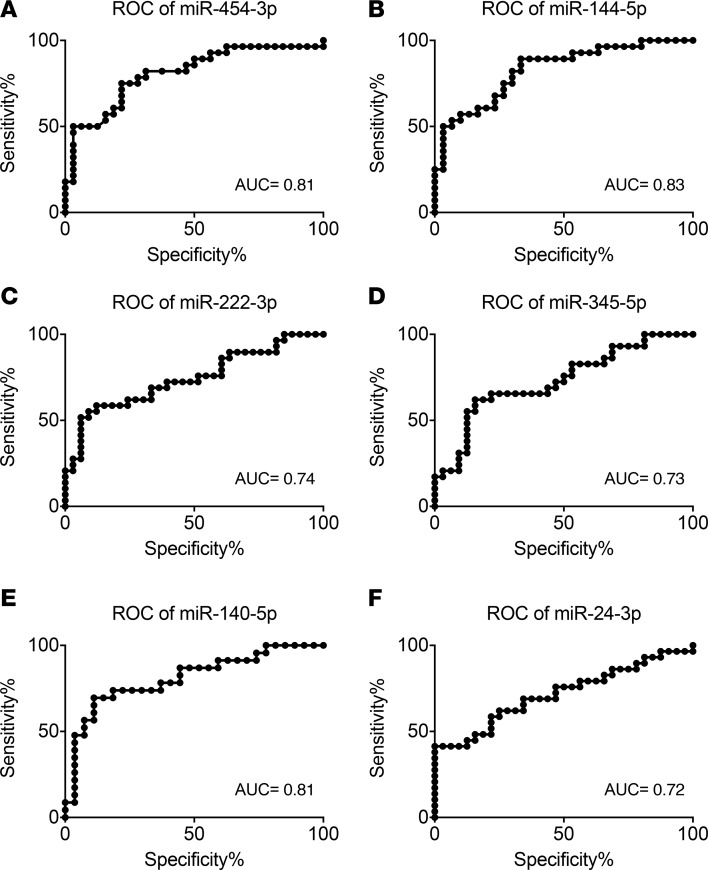
We next analyzed the sensitivity and specificity of 6 miRNAs by generating receiver-operating characteristic (ROC) curves. ROC curves plot the sensitivity against the specificity for the different possible cutpoints of a diagnostic test, and the area under the ROC curves shows the accuracy of the test, separating groups tested into those with and without the disease. An area of 1 indicates that the test is perfectly accurate to distinguish between those with and without the disease, whereas an area of 0.5 indicates that the test fails to distinguish between the groups. Areas under the ROC curve for miR-454-3p, miR-222-3p, miR-144-5p, miR-345-5p, miR-24-3p, and miR-140-5p were 0.81, 0.74, 0.83, 0.73, 0.72, and 0.81, respectively (Figure 4), suggesting that levels of miR-454-3p, miR-144-5p, and miR-140-5p particularly can distinguish between recent-onset diabetes and controls with high sensitivity and specificity.
Figure 4. Receiver-operating characteristic curve analysis for detecting children with recent-onset type 1 diabetes.
True positive rate (sensitivity) is plotted as a function of the false positive rate (indicated in the figure as 100% specificity) for different miRNA levels. The AUC is a measure of how well a quantitative test can distinguish between subjects with and without a disease. Recent-onset type 1 diabetes (n = 29); controls (n = 32). ROC, receiver-operating characteristic.
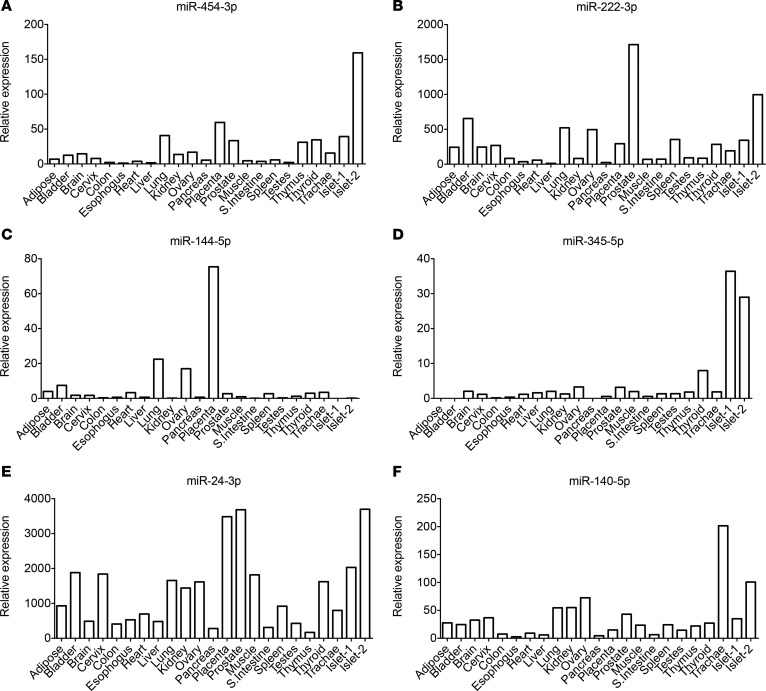
Little is known about the origin or role of circulating miRNAs. While some studies suggest that they can mediate cell-cell communication (15, 16), others propose that they are by-products of dying cells (17). To test whether miR-454-3p, miR-222-3p, miR-144-5p, miR-345-5p, miR-24-3p, and miR-140-5p exhibit tissue-specific patterns and thereby potentially identify their source, we isolated RNA from various human tissues and analyzed miRNA expression by qRT-PCR. miR-345-5p had a low expression in the pancreas but was enriched in the pancreatic islets, suggesting that it may have an important role in regulating β cell function (Figure 5). Interestingly, miR-144-5p was expressed at very low levels in both pancreas and islets. Other tested miRNAs were rather ubiquitously expressed and did not show any apparent tissue specificity.
Figure 5. The abundance of miRNAs in different human tissues.
An equal amount of RNA from various human tissues (each a pool of 3 tissue donors) and 2 different human islet preparations was reverse transcribed, and miRNA expression was determined for (A) miR-454-3p, (B) miR-222-3p, (C) miR-144-5p, (D) miR-345-5p, (E) miR-24-3p, and (F) miR-140-5p by quantitative real-time PCR. Cycle 33 (Ct = 30) was arbitrarily set as 1.
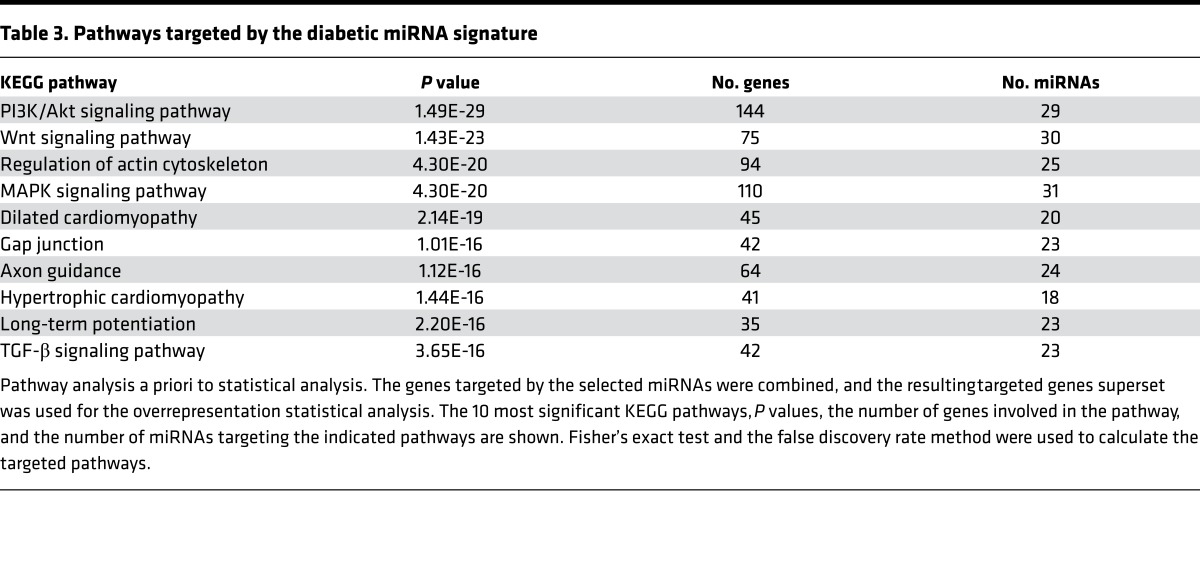
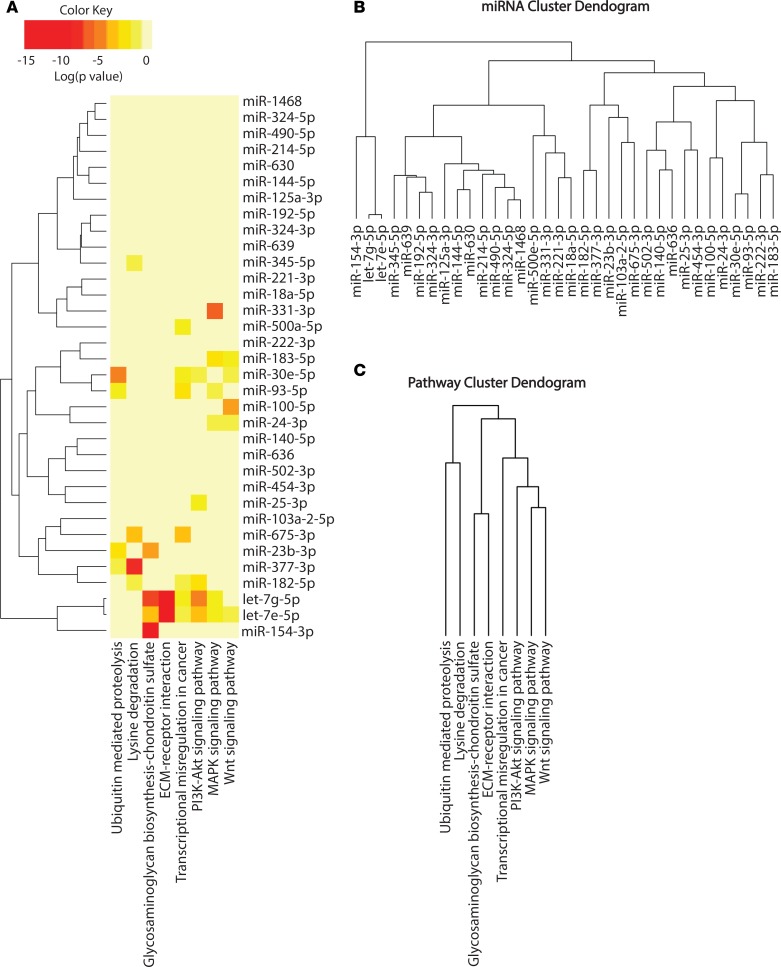
To identify potential biological functions affected by the miRNA signature of recent-onset diabetes, we performed in silico pathway analysis based on the inferred miRNA target genes, using the DIANA-miRPath v2.0 web server. An enrichment analysis of miRNA target genes in KEGG pathways was performed by uniting the genes targeted by all the miRNAs found to be different in the qPCR platform prior to statistical calculation. A conservative Fisher’s exact test and the false discovery rate method were used to calculate the targeted pathways. Table 3 lists the pathways targeted by the diabetic miRNA signature. PI3K/Akt, WNT, regulation of actin cytoskeleton, MAPK, and dilated cardiomyopathy are the top 5 targeted significant pathways. We also analyzed the pathways affected by these miRNAs a posteriori of the statistical analysis. Briefly, significance levels between all possible miRNA-pathway pairs were calculated, and a merged P value was obtained for each pathway by applying Fisher’s combined probability method (Fisher’s method). All results were corrected for multiple hypothesis testing by applying Benjamini and Hochberg’s false discovery rate. In addition, in this analysis, PI3K/Akt, MAPK, and WNT signaling pathways were among the significantly targeted pathways (Figure 6A), and let-7g-5p and let-7e-5p were the closely related miRNAs targeting similar pathways. Interestingly, glycosaminoglycan biosynthesis was the most statistically significant targeted pathway. Clustering the miRNAs based on their influence on molecular pathways (Figure 6B) indicated that miR-125a-3p, miR-144-5p, miR-630, miR-214-5p, miR-490-5p, miR-324-5p, and miR-1468 appear to target similar pathways. By taking into account the significance levels of the interactions, we next clustered the pathways targeted by similar lists of miRNAs (Figure 6C). MAPK and WNT signaling pathways were predicted to be targeted by a specific cluster of miRNAs, as were glycosaminoglycan biosynthesis and extracellular matrix (ECM) receptor interaction pathways.
Table 3. Pathways targeted by the diabetic miRNA signature.
Figure 6. miRNA pathway analysis.
(A) Clustering is based on significance levels. Darker colors represent higher significance. On the miRNA axis, miRNAs displaying similar pathway targeting patterns are clustered together. (B) Hierarchical clustering of miRNAs based on the significance level of interaction. (C) Pathways are clustered based on the subset of miRNAs that target each pathway.
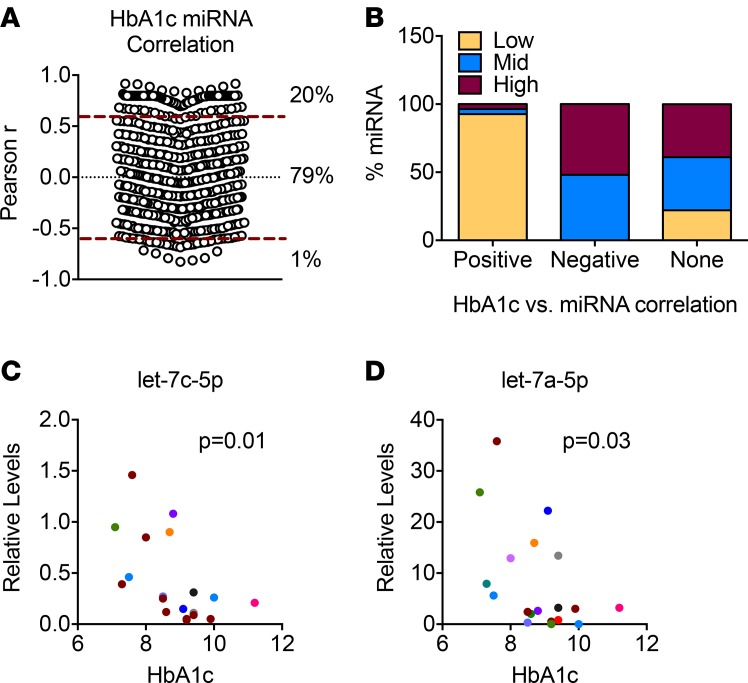
Hemoglobin A1c (HbA1c) levels of children with recent onset of diabetes, whose miRNA levels were analyzed with qPCR profiling, varied from 7.7% to 12.4% (9.46% ± 1.51%, mean ± SD; Table 1). Of 634 detected miRNAs in the sera, we found 134 miRNAs (21%) were significantly correlated with HbA1c levels (Figure 7A), with 20% having a positive correlation and 1% having a negative correlation. We next analyzed the abundance of miRNAs correlating with HbA1c levels. We classified miRNAs into 3 equal ranks, low, mid, or high, based on their Ct values on the qPCR platform. Interestingly, 93% of the miRNAs correlating positively with HbA1c levels had low abundance in the sera from patients with diabetes (Figure 7B), whereas negatively correlating miRNAs were similarly distributed in the mid and high ranks. miRNAs having no correlation with HbA1c levels were distributed relatively evenly between low, mid, and high ranks, at 22.1%, 39%, and 38.8%, respectively. We also analyzed the relation between HbA1c levels and miRNAs that were differentially expressed in the sera of T1D patients. Of the 27 miRNAs that were increased in the T1D samples, 2 miRNAs, miR-324-5p and let-7e-5p, showed significant negative correlation (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/jci.insight.89656DS1). A positive correlation with HbA1c levels was seen in 5 of the 8 miRNAs that were decreased in the T1D samples (Supplemental Table 1). miRNAs showing significant correlation with HbA1c levels, regardless of their association with T1D, are shown in Supplemental Table 1. To assess the reproducibility of these findings, we analyzed the levels of let-7c-5p and let-7a-5p in samples from a second cohort of recent-onset T1D. Let-7c-5p and let-7a-5p were abundantly present in the sera and were the most significantly correlated miRNAs with HbA1c levels (Supplemental Table 1). Similar to the results obtained from the profiling experiments, let-7c-5p and let-7a-5p levels showed strong negative correlation with HbA1c levels (P = 0.01 and P = 0.03, respectively) in the second cohort (Figure 7, C and D).
Figure 7. Correlation between miRNAs and hemoglobin A1c levels.
(A) A Pearson’s correlation was performed to determine the relationship between 634 miRNA levels and hemoglobin A1c (HbA1c) levels in 10 serum samples from children with type 1 diabetes (T1D). Twenty percent of all miRNAs correlated positively with HbA1c levels, whereas 1% displayed negative correlation. P ≤ 0.05 was considered significant. (B) miRNAs were classified into 3 groups based on their circulating levels: low, mid, and high. The percentage of miRNAs from each class contributing to positive, negative, or no miRNA-HbA1c association is shown. (C and D) Let-7c-5p and let-7a-5p levels were quantified using quantitative PCR (qPCR) with locked nucleic acid–enhanced primers in serum samples from a second cohort of patients with T1D, and correlation with HbA1c levels is shown. Cycle 30 (Ct = 30) was arbitrarily set as 1. Data was analyzed using Pearson`s correlation and 2-tailed P values are indicated.
Discussion
We used a LNA-based qPCR profiling platform to identify miRNAs with abnormal levels in sera from children with recent-onset T1D. We discovered a set of miRNAs that discriminated between 10 diabetic and 7 nondiabetic controls with high sensitivity and specificity. Expression patterns of 6 of 6 miRNAs were confirmed using individual qRT-PCR assays in samples from a total of 29 patients with recent-onset T1D and 32 controls. Levels of these miRNAs were investigated at different stages of diabetes and were found to be specifically increased in the samples collected from patients with early stages of diabetes (90 days), as the levels in the sera of children with diabetes longer than 6 months were comparable to controls. Tissue expression analysis indicated a ubiquitous expression profile for 5 of 6 miRNAs analyzed, and 1 miRNA was enriched in pancreatic islets. Functional annotation of genes that are potentially regulated by the miRNAs implicated by the profiling experiments imply that PI3K/Akt, MAPK, and WNT signaling pathways may be involved in the early pathogenesis of T1D. In addition, a set of miRNAs significantly correlated with HbA1c levels, suggesting that circulating miRNA levels are altered in hyperglycemia.
Nielsen et al. (18) profiled pooled serum samples of two T1D cohorts and an age-matched control group using Solexa sequencing and discovered 47 differentially expressed miRNAs. Although the aims of their study were similar to ours, the approaches differed in multiple ways, including the discovery platform used as well as the profiled sample size. RNA sequencing is a powerful technique for discovery-phase research, based on high sensitivity, specificity, and reproducibility, yet qPCR is superior to many miRNA quantification methods, including microarray and RNA sequencing (19, 20). A recent study compared 12 commercially available miRNA profiling platforms (14) and found that LNA-based miRNA platforms offer the best combination of sensitivity and specificity. We had 100% reproducibility when validating the expression level of 6 miRNAs using LNA-enhanced primers, supporting the reliability of the LNA-based qRT-PCR discovery platform. We profiled individual sera from 10 patients and 7 controls and selected miRNAs with statistical significance of P < 0.05 as differentially regulated miRNAs. Nielsen et al. focused on miRNAs with a greater than 2-fold difference between the pooled T1D and control groups. Although they validated some of their findings in larger cohorts, the significance of the discovered miRNAs may be compromised due to profiling of pooled samples. Despite the methodological differences, 4 of the 24 miRNAs reported to be elevated by Nielsen et al. in the sera of T1D children (miR-24-3p, miR-25-3p, miR-222-3p, and miR-103) were also increased in our measurements, pointing to some common miRNA biomarkers identified in both studies.
We measured the levels of 6 miRNAs identified in our profiling experiments in a larger cohort of children with recent onset of T1D and found that all 6 miRNAs were increased in the sera of subjects with recent onset of diabetes compared with controls. It will also be important to examine the levels of miRNAs identified in this study in adults, as the rate of β cell destruction is variable: rapid in infants and children and slower in adults (21). A caveat of our study is that we grouped our subjects based on the duration of their diabetes after diagnosis; however, asymptomatic, clinically “silent,” diabetes is variable in length. It is possible that early diabetes onset can be detected with miRNAs other than those identified in this study, if the samples were staged differently based on C-peptide levels, genetics, autoantibodies, and metabolic markers of the disease. Regardless, as we have examined the levels of miRNAs after clinical presentation of T1D and because T1D does not present until 80% to 90% of the β cells are destroyed, we believe the signature we have identified reflects early symptomatic diabetes onset. Peak insulitis occurs at or near the onset of T1D (22). It will be interesting to investigate whether the miRNAs identified in this study could play a causal role in insulitis.
To gain insights into the potential tissue source of the 6 miRNAs identified in this study and specifically test whether these miRNAs are enriched in the pancreatic islets, we measured the miRNA expression levels in various tissues from healthy adult humans, including pancreas and islets. miR-345-5p was enriched in the pancreatic islets, suggesting that it may have an important role in regulating β cell function. It may also serve as a biomarker of pancreatic islet dysfunction. Expression levels of miR-454-3p, miR-144-5p, and miR-222-3p were rather ubiquitous, implying that their elevations in recent onset of T1D could reflect a systemic or secondary response to β cell injury. Alternatively, it is possible that levels of these miRNAs are altered in the islets of prediabetic patients and reflect β cell injury. Although a link between diabetes and miR-144-5p or miR-345-5p has not been reported, the level of miR-454-3p was increased in the peripheral blood mononuclear cells of individuals with T1D (10), inferring a role for this miRNA in the cellular immune response in T1D. miR-140-5p and miR-222-3p were increased in sera of patients with T2D (23) and in the sera of obese individuals (24). miR-222-3p has been described to induce endothelial dysfunction (25). As high glucose concentration modifies vascular endothelial cell ultrastructure in T1D, increased miR-222-3p levels may reflect changes in endothelial cell structure/function and might be a useful biomarker to monitor the effect of hyperglycemia on vascular endothelium. miR-24-3p and miR-25-3p were elevated in the sera of T1D children (18), and overexpression of miR-24-3p has been shown to inhibit β cell proliferation and insulin secretion (26), while miR-25-3p overexpression inhibits insulin biosynthesis (27), implying a role for both of these miRNAs in modulating β cell function in T1D.
The miRNAs identified from the profiling experiments may have important roles in T1D, either by promoting disease onset or alleviating the symptoms. They may also be reflective of the activated signaling cascades in diabetes. To gain insight into pathways affected by the miRNAs identified in the profiling experiment, KEGG pathway prediction analysis was performed. PI3K/Akt, MAPK, WNT, transcriptional misregulation, ECM receptor interaction, glycosaminoglycan biosynthesis-chondroitin sulfate, lysine degradation, and ubiquitin-mediated proteolysis pathways were predicted to be affected by the miRNAs identified in this study. Some of these pathways have been proposed to have a role in diabetes by affecting β cell function. For example, 2 SNPs in the transcription factor TCF7L2, a protein involved in Wnt signaling, are the most highly correlated polymorphisms linked to T2D (28, 29), and stimulation of Wnt signaling increases β cell proliferation (30). Similarly, AKT signaling increases β cell proliferation (30), regulates the development and function of innate immune cells, and is involved in regulating the inflammatory response (31). Dysregulated Wnt and Akt signaling may be a significant contributor to T1D pathogenesis, either by affecting β cell proliferation and/or modulating innate immune cell function; the contribution of the identified miRNAs remains to be investigated. Recent studies underline the importance of islet and lymphoid tissue ECM composition in promoting immune cell activation, migration, and destruction of islet β cells in T1D. For example, heparan sulfate, a glycosaminoglycan that interacts with collagen IV and laminin to provide mechanical support and a barrier to cell migration, accumulates in healthy β cells but is decreased in T1D (32). Treatment of female NOD mice with a heparin sulfate mimetic reduced the percentage of islets with destructive insulitis and prevented diabetes in 50% of T1D-susceptible mice (32). Hyaluronan, another ubiquitous component of the cell surface and ECM, plays a role in controlling T cell movement (33). Injections of hyaluronidase 1 hour before cell transfer of diabetogenic splenocytes prevented the development of diabetes (34). Hence, our findings suggest that aberrant miRNA levels may impair expression of structural cell components that in turn enhance immune cell infiltration and invasion.
Hyperglycemia is common to both T1D and T2D, and HbA1c levels provide an indication of blood glucose levels in the recent preceding months. We analyzed the correlation between all profiled miRNAs and HbA1c levels. Of all the detected miRNAs, 20% showed a positive correlation and 1% showed a negative correlation with hyperglycemia. Among the 35 dysregulated miRNAs we identified in profiling experiments, 28 miRNAs were not correlated with HbA1c levels, indicating specificity of these miRNAs to T1D rather than hyperglycemia itself. Let-7c-5p and let-7a-5p, 2 let-7 family members abundantly present in the serum, were negatively correlated with HbA1c levels in 2 separate cohorts. The promoter of let-7a-3 is reported to be hypermethylated in diabetic nephropathy, leading to its downregulation (35). Of all positively correlated miRNAs, 93% were present at low levels in the serum. Hyperglycemia causes a variety of pathological changes in small vessels, arteries, and peripheral nerves, and it induces tissue damage in endothelial cells by increasing reactive oxygen species (36). Endothelial dysfunction develops within the first decade after T1D onset (37), and alterations in peripheral vascular dynamics occur within the first 5 years after diagnosis (38). We speculate that the miRNAs that positively correlated with HbA1c levels and whose expression was low in the circulation may be released from the endothelial cells and may reflect the early tissue damage induced by hyperglycemia. If correct, there may be an increase in the levels of these low-abundance serum miRNAs in other endothelial dysfunction–related pathologies, including hypertension or atherosclerosis.
In conclusion, our study has identified a set of 35 circulating miRNAs differentially expressed in the sera of children with T1D, 6 of which were validated in a larger cohort and were increased in early stages of T1D onset but not beyond 90 days after diagnosis. Future efforts will be aimed at addressing the value of these serum miRNAs for T1D prediction using natural history repository samples.
Methods
Study population.
An observational cohort of children with T1D was recruited through the Type 1 Diabetes Clinic at BC Children’s Hospital. The control age-matched children were selected from a pool of children referred to the BC Children’s Hospital Endocrinology Clinic for assessment and were investigated for benign conditions, such as early puberty. All the controls used in this study were found to be in the normal range for growth and puberty following comprehensive outpatient assessment at the BC Children’s Hospital Endocrinology Clinic and had no endocrine abnormality or autoimmune conditions. Peripheral blood was collected in serum clot tubes and allowed to clot at room temperature for 30 minutes. Tubes were centrifuged at 1,200 g for 10 minutes at room temperature, and the serum supernatant was aliquoted and stored at –80°C.
RNA isolation and qRT-PCR.
Human serum samples were stored at –80°C. After thawing, they were centrifuged at 3,000 g for 5 minutes at 4°C to remove cryoprecipitates and cell debris. Total RNA was isolated from 200 μl human serum in the presence of a MS2 carrier RNA (Roche) using a miRNAeasy kit (Qiagen), including a DNAse digestion step. Purified RNA was kept at –80°C before being used for reverse transcription. RNA was reverse transcribed with a Universal cDNA synthesis kit (Exiqon). qRT-PCR was performed using SYBR Green master mix (Exiqon) with LNA-based miRNA primers (Exiqon). The product numbers of each primer are as follows: hsa-miR-454-3p: 205663, hsa-miR-144-5p: 204670, hsa-miR-222-3p:204551, hsa-miR-345-5p:206006, hsa-miR-140-5p: 204540, hsa-miR-24-3p:204260. Relative values were calculated with the ΔCT method. Samples from different cohorts of recent-onset subjects were measured at different times and were graphed together after normalizing miRNA levels to respective control levels that were measured at the same time. A human RNA panel was purchased from Life Technologies, and RNA from each tissue was a pool from 3 subjects. Human islets were obtained from Ziliang Ao and Garth L. Warnock from the Irving K. Barber Human Islet Isolation Laboratory (Vancouver, British Columbia, Canada).
miRNA profiling by qRT-PCR.
RNA isolated from the serum was sent to Roswell Park Cancer Institute for miRNA profiling. Briefly, 8.8 μl RNA was reverse transcribed in a total volume of 44 μl, in duplicates, using the Universal cDNA synthesis kit (Exiqon). Total cDNA was mixed with SYBR Green Master Mix (Exiqon) and ROX and dispensed into 2 miRNA Ready-to-Use PCR, Human panel I+II, V2.M (Exiqon) plates using an electronic multichannel pipettor. Plates were run on the ABI 7900 qPCR machine with the exact same cycling conditions and parameters for every plate. Three wells on each plate were assigned for inter-plate calibration, eliminating run-to-run variation when comparing multiple plates. All Ct values above 38 were set to 38 as the maximum value and regarded as nonamplification. Data were global mean normalized.
Statistics.
Statistical analyses were performed using GraphPad Prism software 7, and data are presented as the mean ± SEM, unless stated otherwise. miRNAs with Ct values lower than 38 from the profiling platform were considered as present, and levels of each miRNA were global mean normalized. Significant miRNAs between the control and T1D group were calculated using 2-tailed Student’s t test, and P ≤ 0.05 was considered significant. Levels of miRNAs from different duration of diabetes samples were compared using 1-way ANOVA with Dunnett’s post-hoc test.
Identification of significant features and multivariate analysis were performed using the open-access web-based software MetaboAnalyst (39). Data were global mean normalized, the pooled average from the control group was formed, and values were log transformed. Data were mean centered and divided by the SD of each variable. Volcano plots were generated showing significance versus fold changes. P values of less than 0.05 were considered statistically significant. Multivariate statistical analysis was performed using partial least squares–discriminant analysis (PLS-DA). Supervised clustering was performed for a maximum of 5 components, and prediction accuracy was determined by assessing the prediction accuracy based on cross-validation (Q2). Variable importance in projection was calculated by weighing the sum of squares of the PLS loadings, taking into account the amount of explained variance of each component.
miRNA pathway analysis was performed using the open-access web-server DIANA-miRPath v2.0 (40). The in silico target prediction of differentially expressed miRNAs was computed using the DIANA-microT-CDS (41), a broadly used miRNA target prediction algorithm exhibiting the highest sensitivity and specificity compared with other analyses. An enrichment analysis of miRNA target genes in KEGG pathways was performed in 2 ways, both a priori and a posteriori of the statistical analysis. In the first test, gene targets of the differentially expressed miRNAs were combined into a common set and incorporated into an enrichment analysis. A conservative Fisher’s exact test and the false discovery rate method were used to calculate the significantly targeted KEGG pathways. As for the a posteriori analysis, significance levels between all possible miRNA-pathway pairs were calculated, and by applying Fisher’s combined probability method and combining the significance values, a merged P value for each pathway was obtained.
Hierarchical clustering of targeted pathways and miRNAs was performed using DIANA-miRPath v2.0 (40). Clustering was based on a complete linkage clustering method, where squared Euclidian distances were calculated as distance measures, utilizing absolute P values in all calculations and accounting for the significance levels of the interactions.
Study approval.
The study protocol was reviewed and approved by the University of British Columbia Clinical Research Ethics Boards. All parents provided written consent, and children provided assent when capable.
Author contributions
SE and TJK designed, directed, and interpreted experiments. SE performed experiments. TJK is the guarantor of this work, had full access to all the data, and takes full responsibility for the integrity of data and the accuracy of data analysis. CP, RT, and AM were responsible for the recruitment, specimen handling and storage, and related clinical data collection of the T1D and healthy control pediatric subjects. SE wrote the manuscript, and all authors contributed to editing the manuscript.
Supplementary Material
Acknowledgments
TJK gratefully acknowledges funding from the JDRF for this research. SE is a recipient of a JDRF postdoctoral fellowship. We thank Maria Glavas and Jennifer E. Bruin for critical reading of the manuscript. We would also like to thank Ziliang Ao and Garth L. Warnock from the Irving K. Barber Human Islet Isolation Laboratory (Vancouver, British Columbia, Canada) for providing human islets. CP and RT gratefully acknowledge funding from the Canadian Institutes for Health Research for the SLE/D Team for Childhood Autoimmunity.
Footnotes
AM’s present address is: Division of Clinical and Metabolic Genetics, The Hospital for Sick Children, University of Toronto, Toronto, Ontario, Canada.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2017;2(4):e89656. https://doi.org/10.1172/jci.insight.89656.
Contributor Information
Suheda Erener, Email: serener@mail.ubc.ca.
Ashish Marwaha, Email: akm103ox@gmail.com.
Rusung Tan, Email: rusung.tan@ubc.ca.
Constadina Panagiotopoulos, Email: dpanagiotopoulos@cw.bc.ca.
References
- 1.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2005;28(suppl 1):S37–S42. doi: 10.2337/diacare.28.suppl_1.s37. [DOI] [PubMed] [Google Scholar]
- 2.Cnop M, Welsh N, Jonas JC, Jörns A, Lenzen S, Eizirik DL. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 2005;54(suppl 2):S97–S107. doi: 10.2337/diabetes.54.suppl_2.s97. [DOI] [PubMed] [Google Scholar]
- 3.Keymeulen B, et al. Insulin needs after CD3-antibody therapy in new-onset type 1 diabetes. N Engl J Med. 2005;352(25):2598–2608. doi: 10.1056/NEJMoa043980. [DOI] [PubMed] [Google Scholar]
- 4.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108(12):3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha TY. MicroRNAs in human diseases: from cancer to cardiovascular disease. Immune Netw. 2011;11(3):135–154. doi: 10.4110/in.2011.11.3.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erener S, Mojibian M, Fox JK, Denroche HC, Kieffer TJ. Circulating miR-375 as a biomarker of β-cell death and diabetes in mice. Endocrinology. 2013;154(2):603–608. doi: 10.1210/en.2012-1744. [DOI] [PubMed] [Google Scholar]
- 7.Zampetaki A, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107(6):810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 8.Pescador N, Pérez-Barba M, Ibarra JM, Corbatón A, Martínez-Larrad MT, Serrano-Ríos M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS One. 2013;8(10):e77251. doi: 10.1371/journal.pone.0077251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu H, Leung SW. Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia. 2015;58(5):900–911. doi: 10.1007/s00125-015-3510-2. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi P, et al. MicroRNA expression profiling and functional annotation analysis of their targets in patients with type 1 diabetes mellitus. Gene. 2014;539(2):213–223. doi: 10.1016/j.gene.2014.01.075. [DOI] [PubMed] [Google Scholar]
- 11.Sebastiani G, Grieco FA, Spagnuolo I, Galleri L, Cataldo D, Dotta F. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes Metab Res Rev. 2011;27(8):862–866. doi: 10.1002/dmrr.1262. [DOI] [PubMed] [Google Scholar]
- 12.Hezova R, et al. microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol. 2010;260(2):70–74. doi: 10.1016/j.cellimm.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Marchand L, et al. miRNA-375 a sensor of glucotoxicity is altered in the serum of children with newly diagnosed type 1 diabetes. J Diabetes Res. 2016;2016:1869082. doi: 10.1155/2016/1869082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mestdagh P, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11(8):809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 15.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen LB, et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp Diabetes Res. 2012;2012:896362. doi: 10.1155/2012/896362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang K, Peng X, Luo J, Gou D. Identification of circulating miRNA biomarkers based on global quantitative real-time PCR profiling. J Anim Sci Biotechnol. 2012;3(1):4. doi: 10.1186/2049-1891-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.American Diabetes Association. (2) Classification and diagnosis of diabetes. Diabetes Care. 2015;38(suppl):S8–S16. doi: 10.2337/dc15-S005. [DOI] [PubMed] [Google Scholar]
- 22.In’t Veld P. Insulitis in human type 1 diabetes: The quest for an elusive lesion. Islets. 2011;3(4):131–138. doi: 10.4161/isl.3.4.15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ortega FJ, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37(5):1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 24.Ortega FJ, et al. Targeting the circulating microRNA signature of obesity. Clin Chem. 2013;59(5):781–792. doi: 10.1373/clinchem.2012.195776. [DOI] [PubMed] [Google Scholar]
- 25.Xue Y, et al. MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1α in the progression of atherosclerosis. Atherosclerosis. 2015;241(2):671–681. doi: 10.1016/j.atherosclerosis.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, et al. MicroRNA-24/MODY gene regulatory pathway mediates pancreatic β-cell dysfunction. Diabetes. 2013;62(9):3194–3206. doi: 10.2337/db13-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Setyowati Karolina D, Sepramaniam S, Tan HZ, Armugam A, Jeyaseelan K. miR-25 and miR-92a regulate insulin I biosynthesis in rats. RNA Biol. 2013;10(8):1365–1378. doi: 10.4161/rna.25557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant SF, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 29.Florez JC, et al. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med. 2006;355(3):241–250. doi: 10.1056/NEJMoa062418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fatrai S, et al. Akt induces β-cell proliferation by regulating cyclin D1, cyclin D2, and p21 levels and cyclin-dependent kinase-4 activity. Diabetes. 2006;55(2):318–325. doi: 10.2337/diabetes.55.02.06.db05-0757. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Kinase AKT controls innate immune cell development and function. Immunology. 2013;140(2):143–152. doi: 10.1111/imm.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziolkowski AF, Popp SK, Freeman C, Parish CR, Simeonovic CJ. Heparan sulfate and heparanase play key roles in mouse β cell survival and autoimmune diabetes. J Clin Invest. 2012;122(1):132–141. doi: 10.1172/JCI46177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evanko SP, Potter-Perigo S, Bollyky PL, Nepom GT, Wight TN. Hyaluronan and versican in the control of human T-lymphocyte adhesion and migration. Matrix Biol. 2012;31(2):90–100. doi: 10.1016/j.matbio.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss L, et al. Induction of resistance to diabetes in non-obese diabetic mice by targeting CD44 with a specific monoclonal antibody. Proc Natl Acad Sci U S A. 2000;97(1):285–290. doi: 10.1073/pnas.97.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng R, et al. Promoter hypermethylation of let-7a-3 is relevant to its down-expression in diabetic nephropathy by targeting UHRF1. Gene. 2015;570(1):57–63. doi: 10.1016/j.gene.2015.05.073. [DOI] [PubMed] [Google Scholar]
- 36.Nishikawa T, et al. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404(6779):787–790. doi: 10.1038/35008121. [DOI] [PubMed] [Google Scholar]
- 37.Järvisalo MJ, et al. Endothelial dysfunction and increased arterial intima-media thickness in children with type 1 diabetes. Circulation. 2004;109(14):1750–1755. doi: 10.1161/01.CIR.0000124725.46165.2C. [DOI] [PubMed] [Google Scholar]
- 38.Ladeia AM, Ladeia-Frota C, Pinho L, Stefanelli E, Adan L. Endothelial dysfunction is correlated with microalbuminuria in children with short-duration type 1 diabetes. Diabetes Care. 2005;28(8):2048–2050. doi: 10.2337/diacare.28.8.2048. [DOI] [PubMed] [Google Scholar]
- 39.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6(6):743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 40.Vlachos IS, et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40(Web Server issue):W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28(6):771–776. doi: 10.1093/bioinformatics/bts043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.