Abstract
Adult neurogenesis, the process of generating mature neurons from neuronal progenitor cells, makes critical contributions to neural circuitry and brain function in both healthy and disease states. Neurogenesis is a highly regulated process in which diverse environmental and physiological stimuli are relayed to resident neural stem cell populations to control the transcription of genes involved in self-renewal and differentiation. Understanding the molecular mechanisms governing neurogenesis is necessary for the development of translational strategies to harness this process for neuronal repair. Here we report that the Ras-related GTPase RIT1 serves to control the sequential proliferation and differentiation of adult hippocampal neural progenitor cells, with in vivo expression of active RIT1 driving robust adult neurogenesis. Gene expression profiling analysis demonstrates increased expression of a specific set of transcription factors known to govern adult neurogenesis in response to active RIT1 expression in the hippocampus, including sex-determining region Y-related HMG box 2 (Sox2), a well established regulator of stem cell self-renewal and neurogenesis. In adult hippocampal neuronal precursor cells, RIT1 controls an Akt-dependent signaling cascade, resulting in the stabilization and transcriptional activation of phosphorylated Sox2. This study supports a role for RIT1 in relaying niche-derived signals to neural/stem progenitor cells to control transcription of genes involved in self-renewal and differentiation.
Keywords: Akt, PKB, GTPase, hippocampus, neurogenesis, neuron, Ras protein, RIT1, Sox2, dentate gyrus
Introduction
The adult brain harbors germinal cell niches in the subventricular zone of the lateral ventricles and the subgranular zone in the dentate gyrus of the hippocampus (1–3). The activation of these relatively quiescent neural progenitor cell (NPC)2 populations and their capacity to differentiate into specialized cells is under rigorous cellular control (4). Transduction of a variety of extracellular niche stimuli results in the activation of intracellular regulatory mechanisms within NPCs, signaling cascades that include transcription factors, and epigenetic regulators that serve to finely coordinate gene expression during neurogenesis (5). The transcription factor sex-determining region Y-related HMG box 2 (Sox2) is a member of the SOXB1 family of transcription factors with established roles in maintaining stem cell/progenitor cell properties in diverse cellular populations (6, 7). Genetic deletion of Sox2 causes neurodegeneration and impaired neurogenesis in the adult mouse brain, whereas human Sox2 mutations are associated with anophthalmia, a disorder characterized by cognitive disabilities and defects in hippocampal development (8, 9). Although the role of Sox2 in stem cell maintenance within the neurogenic niche has been described previously (10, 11), the molecular mechanisms that control Sox2 activation in response to appropriate neurogenic cues remain poorly characterized.
ES cell self-renewal and pluripotency are regulated by a core group of transcription factors, including Sox2 (12–14). Although Sox2 is not highly expressed in ES cells, its protein levels are under stringent control. For example, moderate increases in Sox2 lead to differentiation of ES cells primarily into neural ectodermal cells (15), whereas reduced levels of Sox2 trigger differentiation toward the trophectoderm cell fate (16). Furthermore, Sox2 has a critical role in lineage specification (17), and Sox2 proteins levels are differentially regulated in distinct cell lineages during early development. Although Sox2 expression is under rigid transcriptional control (14), additional posttranscriptional mechanisms have recently been reported. In embryonic stem cells, Sox2 stabilization and transcriptional activation are controlled by a balance of site-specific methylation and phosphorylation (18, 19). However, it is unclear whether a similar regulatory cascade operates in NPCs, and the molecular mechanisms that regulate Sox2 activity in the neurogenic niche remain to be identified.
RIT1 is member of the Ras-related family of small GTP-binding proteins, a group of structurally related and evolutionarily conserved proteins that share the ability to undergo guanine nucleotide-dependent conformational change (20, 21). Functioning with their allied regulatory and effector protein networks, Ras-related GTPases serve as critical cellular biotimers, coupling diverse cellular stimuli to the spatial and temporal regulation of signal transduction pathways that contribute to almost every aspect of cellular physiology. RIT1 is widely expressed, including throughout the human and mouse brain (22–24). At the molecular level, we have previously described roles for RIT1 in the regulation of axonal and dendritic growth (23), activation of Akt (25–27), and control of cAMP response element-binding protein transcriptional activity (27). More recently, we identified a role for RIT1 in the survival of adult-born hippocampal neurons following traumatic brain injury (28). Following cortical contusion, RIT1 deficiency resulted in a significant delay in injury-induced hippocampal neurogenesis, suggesting that RIT1 might be an integral component of a signaling pathway involved in neural progenitor activation (28). To generate a deeper understanding RIT1 function in the CNS, we developed a conditional mouse model allowing doxycycline-regulated expression of activated RIT1. Here we report that active RIT1 expression drives robust hippocampal neurogenesis through activation of a pro-neural transcriptional program. RIT1 signaling controls the transcriptional activity of Sox2 in neural progenitor cells, supporting a key role for RIT1 in the dynamic regulation of adult neurogenesis.
Results
RIT1 Is Expressed in the Dentate Gyrus
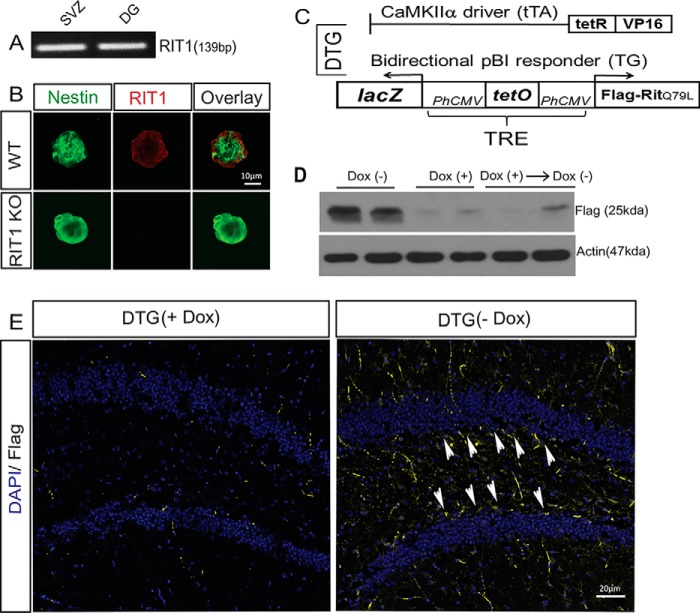
RIT1 loss sensitizes immature hippocampal neurons to stress-dependent apoptosis and blunts hippocampal neural progenitor cell activation following traumatic brain injury (23). Consistent with a role for RIT1 in regulating adult neurogenesis, semiquantitative RT-PCR analysis confirmed RIT1 expression in both neurogenic niches of the CNS (Fig. 1A), the subgranular zone in the dentate gyrus of the hippocampus, and the subventricular zone of the lateral ventricles. This was validated using confocal laser-scanning imaging of RIT1 protein expression in WT and RIT1−/− subgranular zone neurospheres (Fig. 1B). To model RIT1 activation and examine its functional effect on neurogenesis, we generated a line of constitutively active RIT1-overexpressing transgenic mice using a neuron-specific binary tetracycline/doxycycline (Dox)-regulated system in which double transgenic (DTG) mice express FLAG-tagged RIT1Q79L when Dox is removed from the diet (29) (Fig. 1C). Western blotting confirmed Dox-regulated expression of the transgene (Fig. 1D). Moderate in vivo overexpression of active RIT1 was observed in the dentate gyrus of young adult DTG mice 3 weeks after removal of the Dox diet (Fig. 1E).
FIGURE 1.
Generation of the conditional RIT1 mouse model. A, semiquantitative RT-PCR demonstrates RIT1 expression in the adult mouse subventricular zone (SVZ) and DG (n = 4). B, representative confocal images of RIT1 protein expression in WT and RIT1−/− cultured neurospheres. C, schematic representing the binary transgenic system regulated by Dox to inducibly overexpress RIT1. D, representative Western blotting analysis showing FLAG-RIT1Q79L protein expression in brain extracts from 4-month-old DTG mice ± Dox versus WT control. E, representative immunohistochemistry for FLAG-RIT1Q79L (yellow) in the dentate gyrus of 4-month-old DTG mice on Dox (+) or 3 weeks after removal of the Dox diet (−). Arrowheads, FLAG-expressing cells. Nuclei (DAPI) are shown in blue (magnification ×20).
RIT1 Signaling Induces Pro-neural Gene Expression
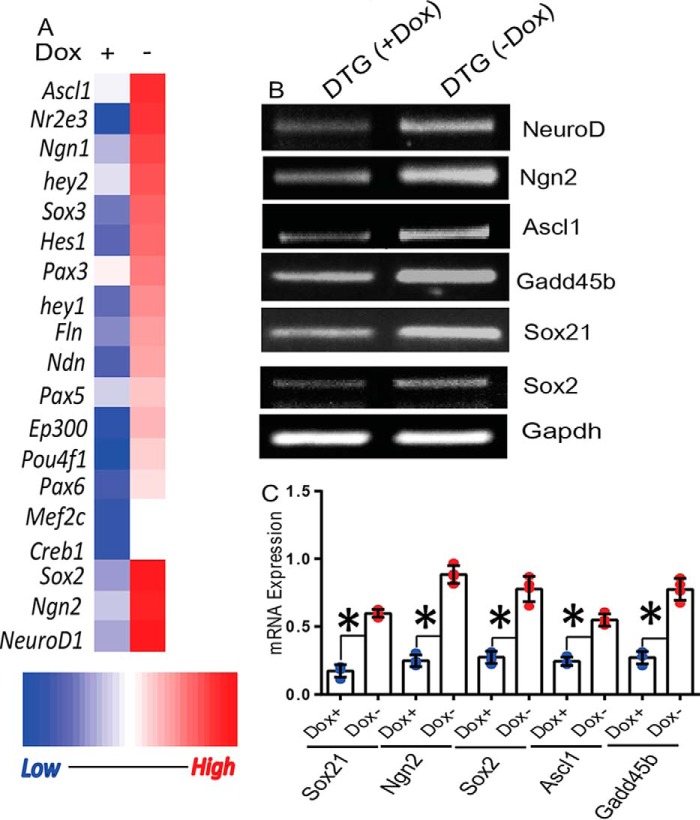
Cell cycle regulators, transcription factors, and epigenetic control proteins are key regulators of adult neurogenesis (30). Because RIT1 is known to control a variety of transcription factors (21, 27, 31), we performed a pathway-focused PCR array analysis of the dentate gyrus from DTG mice 3 weeks after removal of doxycycline from the diet to investigate the expression of genes known to regulate neurogenesis and neural stem cell differentiation. As seen in Fig. 2A, active RIT1 expression stimulates the expression of Sox2 (p < 0.05) along with a collection of pro-neural genes, including Ngn2 (p < 0.01), Ascl1 (p < 0.05), and NeuroD1 (p < 0.05). These results were independently verified using semiquantitative RT-PCR (Fig. 2, B and C). Because Sox2 has been shown to directly bind the promoters of NeuroD1 and Ngn2 to enable activation of the neuronal differentiation program in response to appropriate neurogenic stimuli (32), we also examined the expression of more widely known targets of Sox2 transcription, e.g. Sox21 and Gadd45b. As seen in Fig. 2C, both Sox21 and Gadd45b levels increased in RIT1Q79L-overexpressing DTG mice. Taken together, these data suggest that constitutively active RIT1 signaling leads to activation of Sox2 and expression of pro-neural genes in the dentate gyrus.
FIGURE 2.
Pro-neural gene expression profile associated with RIT1 expression. A, total RNA prepared from the dentate gyrus of DTG mice ± Dox diet was subjected to PCR array analysis for neurogenic transcription factors, and the data were plotted as a heat map. B, semiquantitative RT-PCR validation of the array data and downstream targets of Sox2 (Sox21 and Gadd45b). C, quantification of RT-PCR data. Results are presented as mean ± S.E. calculated from three separate experiments. *, p < 0.05.
Active RIT1 Stabilizes Sox2 Protein Levels in Vivo and in Vitro
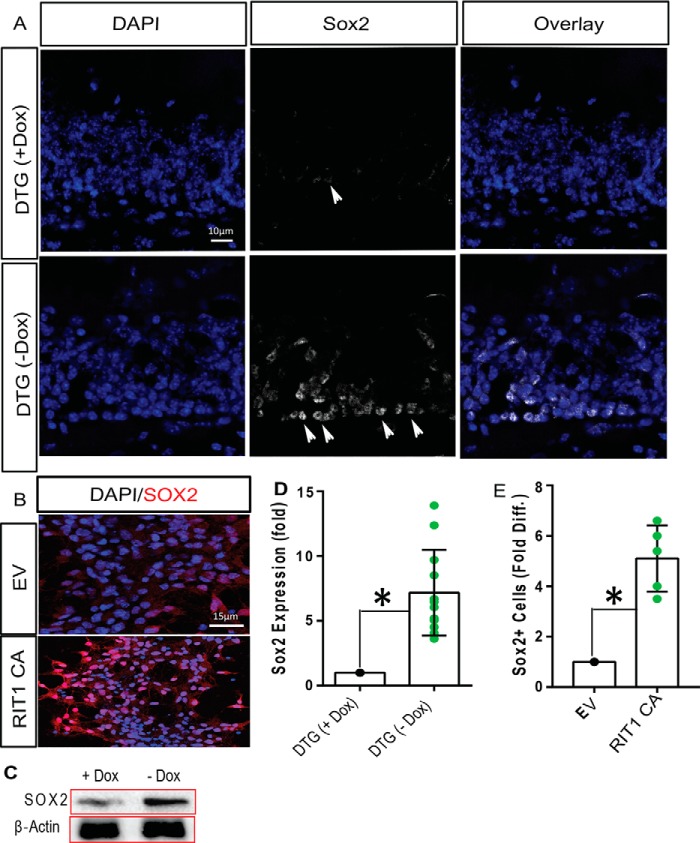
To determine whether RIT1-dependent pro-neural gene induction involves Sox2 activation, we next examined whether RIT1 signaling regulates Sox2 protein levels in vivo and in vitro. Immunohistochemical analysis using laser-scanning confocal microscopy showed a prominent increase in Sox2 protein levels (p < 0.01) in the dentate gyrus of DTG mice following Dox withdrawal (Fig. 3, A and D). This result was confirmed by immunoblotting (Fig. 3C) Transient transfection of primary hippocampal neural progenitor cells (HNPCs) with a vector expressing active RIT1 also resulted in elevated Sox2 levels relative to empty vector (p < 0.05) (Fig. 3, B and E). Together with the gene expression analysis, these data suggest that RIT1 regulates Sox2 function in the hippocampus.
FIGURE 3.
RIT1 overexpression increases Sox2. A, immunofluorescence for Sox2 in white (white arrowheads, magnification ×20), showing that protein expression is greatly increased in the DG of DTG mice after Dox removal (n = 6/group). Nuclei are shown in blue. B, immunofluorescence of Sox2 (red) in HNPCs following transfection with FLAG-RIT1Q79L (CA) or EV control. Nuclei are shown in blue (n = 3). C, immunoblot analysis of endogenous Sox2 in the DG of DTG mice before and after Dox removal (n = 3/group). Note that Sox2 levels increase upon expression of active RIT1. D and E, quantitation of Sox2 expression and Sox2-expressing cells. Results are presented as mean ± S.E. calculated from three separate experiments. *, p < 0.05.
Active RIT1 Promotes HNPC Expansion
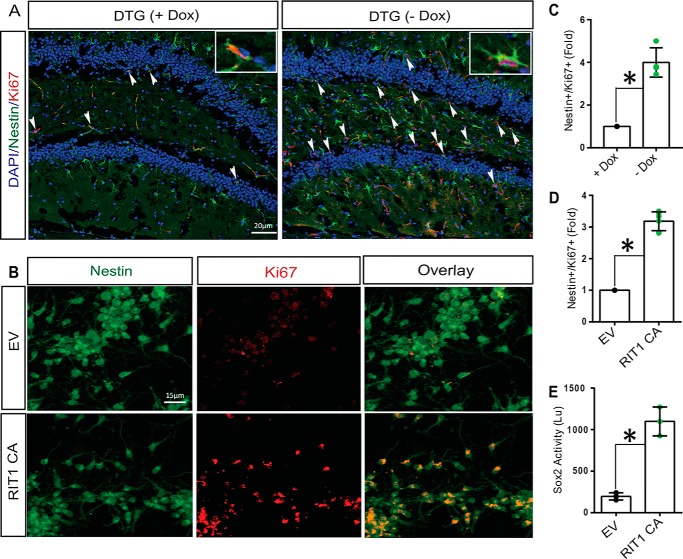
Sox2 plays important roles in maintaining neural stem cell/progenitor cell properties, including their capacity to proliferate and self-renewal (10, 11). Because RIT1 signaling was capable of directing a pro-neural transcriptional program, including Sox2 activation, we reasoned that it might also regulate HNPC proliferation. We performed immunostaining to identify proliferating (Nestin+/Ki67+) hippocampal neuronal stem cells. As seen in Fig. 4, A and C, expression of activated RIT1 leads to robust amplification of Nestin+/Ki67+ cells within the dentate gyrus. These results were further confirmed by transfecting HNPCs with a vector expressing active RIT1 and resulted in increased proliferation, as monitored by the number of Nestin+/Ki67+ cells (Fig. 4, B, and D; p < 0.05). RIT1-dependent HNPC expansion was accompanied by increased Sox2 transcriptional activity, monitored using a luciferase reporter assay in transfected HNPCs (Fig. 4E, p < 0.01). Cumulatively, these data demonstrate that RIT1 regulates HNPC proliferation in parallel with Sox2 transcriptional activation.
FIGURE 4.
Neuronal RIT1 expression increases HNPC proliferation and Sox2 transcriptional activity. A and C, representative immunofluorescent images (A) and quantification (C) of the DG from DTG mice immunostained for Nestin (green) and Ki67 (red) to label proliferating neuronal stem cells. Nuclei (DAPI) are shown in blue. B, HNPCs were transfected with Myc-tagged RIT1Q79L (CA) or EV, and proliferation was assessed by immunohistochemical detection of Nestin (green) and Ki67 (red) co-labeled cells. D, quantification of HNPC proliferation (Nestin+/Ki67+) as mean ± S.E. from three independent experiments. E, HNPCs were transfected as in A in the presence of a Sox2 luciferase reporter construct. Luciferase activity was evaluated 48 h post-transfection as described under “Experimental Procedures.” Results are presented as mean ± S.E. for three independent experiments repeated in triplicate. *, p < 0.05.
RIT1-Sox2 Signaling Regulates Adult Hippocampal Neurogenesis
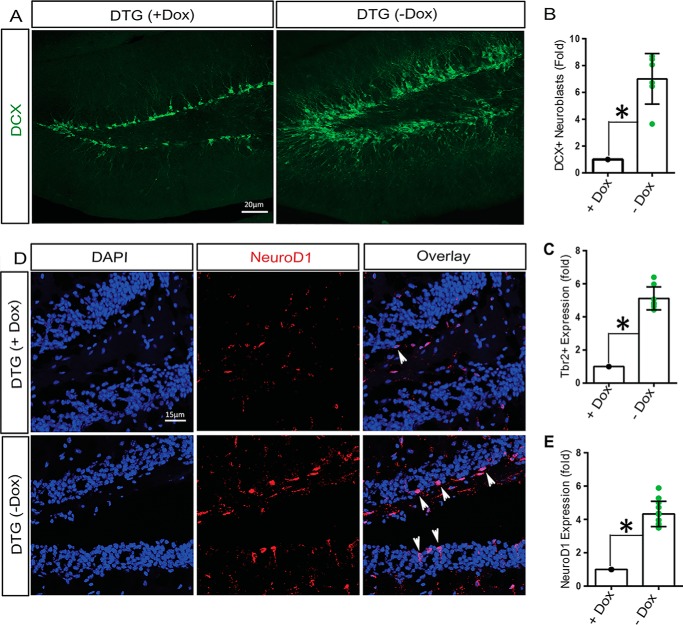
To examine the in vivo effect of active RIT1 expression on hippocampal neurogenesis, 3-month-old DTG mice were shifted to a Dox-free diet for 3 weeks, and the number of immature doublecortin-positive (DCX+) neuroblasts was assessed by immunostaining. As seen in Fig. 5A, expression of activated RIT1 results in a marked increase in DCX+ neuroblasts compared with DTG mice under Dox suppression (p < 0.01) (Fig. 5, A and B). Consistent with an ability of Rit to promote adult hippocampal neurogenesis, there were significant increases in both Tbr2+ (Fig. 5C, p < 0.05) and NeuroD1+ intermediate neural progenitors (Fig. 5, D and E; p < 0.05) in DTG mice following Dox withdrawal. These data strongly suggest that RIT1 signaling regulates hippocampal neurogenesis with an accompanying expansion of immediate neural precursor cells.
FIGURE 5.
RIT1 induces hippocampal neurogenesis. A, immunofluorescence detection of DCX (green) expression in the DG of DTG mice (n = 6/group). B, quantification of DCX+ neuroblasts presented as mean ± S.E. *, p < 0.05. C, quantification of Trb2+ intermediate neural precursors in the DG of DTG ± Dox diet mice for 3 weeks (n = 6/group). *, p < 0.05. D, immunofluorescence detection of NeuroD1 (red, arrowheads), showing that expression is greatly increased in the DG of DTG mice 3 weeks after removal from the Dox diet (n = 6/group) compared with littermates remaining under Dox suppression. Nuclei are shown in blue (magnification ×20). E, quantification of NeuroD1+ neuroblasts (n = 6/treatment group) as mean ± S.E. *, p < 0.05.
RIT1-mediated Sox2 Activation Involves Akt Signaling
Our previous studies have shown that RIT1 activates a p38/mTORC2/Akt signaling cascade to promote cell survival in response to oxidative stress (26). Because Akt is known to phosphorylate Sox2 at Thr118, increasing both protein stability and transcriptional activity (18), we next asked whether RIT1 controls an Akt/Sox2 signaling cascade in HNPCs. Expression of activated RIT1 increased the number of HNPCs expressing activated Akt (p < 0.01), monitored by anti-phospho-Akt Ser473 immunostaining (Fig. 6, A and B). In agreement with the increase in active Akt, there was an 8-fold increase in the number of phospho-Sox2-Thr118+ nuclei in HNPCs transfected with active RIT1 compared with empty vector (EV) controls (Fig. 7, A and C; p < 0.01). Pharmacological inhibitor studies suggest that RIT1-dependent Sox2 activation requires Akt signaling because treatment with the Akt inhibitor triciribine resulted in a marked decrease in the number of phospho-Sox2-Thr118+ nuclei in RIT1Q79L-overexpressing HNPCs (Fig. 7, A and C). Moreover, Akt inhibition blocked the RIT1Q79L-mediated increase in Tuj1-immunoreactive neurons in transfected HNPCs (Fig. 7, B and D; p < 0.05). Taken together, these data suggest that the RIT1-mediated increase in hippocampal neurogenesis requires Akt-dependent Sox2 activation.
FIGURE 6.
RIT1 regulates Akt in HNPCs. A, HNPCs were transfected with either EV or FLAG-tagged RIT1Q97L (CA), and immunohistochemistry was used to detect Nestin (red) and active Akt (green, phospho-Akt (Ser473)) (magnification ×20). B, quantification of HNPC proliferation as mean ± S.E. from three independent experiments. *, p < 0.05.
FIGURE 7.
RIT1-mediated Sox2 activation requires Akt. A, HNPCs were transfected with either EV or activated RIT1 (CA) and treated with or without Akt inhibitor (triciribine, 10 μm) for 12 h, and immunofluorescence was used to detect Nestin (red) and cellular levels of Sox2-Thr118 phosphorylation (green). B, HNPCs expressing FLAG-tagged RIT1Q79L were treated with Akt inhibitor and grown in complete neuronal differentiation medium for 6 days. Neuronal differentiation was assessed by immunofluorescence detection of Tuj1+ neurons (magnification ×20). C and D, quantification of phospho-Sox2+ and Tuji1+ neurons, respectively, as mean ± S.E. from three independent experiments. *, p < 0.05.
Discussion
NSC proliferation and differentiation are regulated by a variety of extracellular niche signals (33). Transcriptional cascades play fundamental roles in NSC regulation and are dynamically regulated by a large number of synergistic and antagonistic niche signals to ensure a ready supply of progenitors to meet the demand for new neurons, oligodendrocytes, and astrocytes (34). Although the core transcription factor Sox2 is a master regulator of neural stem cell biology (7), playing a critical role in neurogenesis within the adult brain (32), the molecular mechanisms that control Sox2-dependent neuronal differentiation remain incompletely characterized.
Here, using a conditional mouse overexpression model (DTG mice), we identify a role for the RIT1 GTPase in Sox2 regulation and the control of neural progenitor/stem cells. We observed prominent expression of a set of pro-neural transcription factors, including Sox2, in the dentate gyrus of mice expressing active RIT1 (Fig. 2). Because Sox2 has recently been shown to bind to the promoters of poised pro-neural genes in NPCs to enable an appropriate neuronal differentiation (32, 35) program, including the neurogenic genes Ngn2 and NeuroD1, we reasoned that RIT1-dependent neurogenic gene expression might rely on Sox2 activation. Indeed, active RIT1 expression both increases Sox2 protein levels (Fig. 3) and stimulates Sox2 transcriptional activation (Fig. 4). Presumably, RIT1-dependent Sox2 regulation provides a novel molecular mechanism for the control of hippocampal neurogenesis in response to select neurogenic stimuli. In keeping with this hypothesis, active RIT1 expression drives robust NPC expansion within the dentate gyrus (Fig. 4). Moreover, although RIT1 deficiency permits normal levels of basal neurogenesis, we have previously noted significant defects in hippocampal neurogenesis following traumatic brain injury (28). Sox2-deficient HNPCs display increased cell death during neuronal differentiation (32), and Sox2-null mice have deficits in hippocampal neurogenesis and augmented neurodegeneration (36, 37), suggesting that RIT1 may be needed for injury-mediated Sox2 activation. Studies are underway to test this hypothesis.
In embryonic stem cells, Sox2 is regulated by competing posttranslational modifications (18, 19). Site-specific methylation promotes Sox2 ubiquitination and degradation. In contrast, Akt phosphorylates Sox2 at Thr118 (18, 38), antagonizing methylation, to stabilize Sox2 levels and induce transcription. As RIT1 was shown to regulate an mTORC2-Akt signaling cascade to promote cellular oxidative stress survival (26), we explored whether RIT1-mediated Sox 2 activation might involve Akt signaling. Active RIT1 signaling results in Akt activation in HNPCs (Fig. 6) and increased levels of total and Sox2-Thr118p+ nuclei (Fig. 7), suggesting that RIT1 controls an Akt-Sox2 cascade to promote neurogenesis. In support of this mechanism, pharmacological Akt inhibition blocked active RIT1-dependent Sox2-Thr118 phosphorylation and the generation of Tuj1+ neurons in transfected HNPCs (Fig. 7).
The self-renewal and pluripotency of embryonic stem cells is regulated by a core set of transcription factors, including Oct3/Oct4, Sox2, and Nanog (12–14). Recent advances in cell reprogramming have fueled intense interest in the regulation of Sox2 and its role in neural fate determination. Expression of Sox2 alone or as part of a core set of transcription factors has been shown to drive somatic cell reprogramming (39–44). In the adult CNS, retroviral delivery of Sox2 has been found to directly or indirectly reprogram resident astrocytes into induced neurons in the injured adult cerebral cortex (45–47). Importantly, there is a cellular progression for Sox2-mediated conversion of adult astrocytes to neurons, involving the neural commitment of progenitors (46). These induced adult neuroblasts are capable of in vivo proliferation and generate mature neurons when supplied with neurotrophic factors or following small-molecule treatment (46, 47), which may provide a novel strategy for neuronal regenerative therapy. Because RIT1 controls neurogenesis and stimulates a Sox2/pro-neurogenic transcriptional program, it will be important in future to examine whether active RIT1 might be used for resident astroglial cell reprogramming.
In summary, these studies extend our understanding of Sox2 regulation, identifying RIT1-Akt-Sox2 cascade signaling as a mechanism governing NPC proliferation and neurogenesis. Because RIT1 is known to couple diverse neuronal mitogens and cellular stress stimuli to transcriptional activation, an intriguing possibility is that RIT1-mediated Sox2 activation plays a role in neurogenic niche sensing, serving to link select environmental or physiological stimuli to neural stem cell self-renewal and differentiation. Growing literature indicates that Sox2-dependent reprogramming strategies allow the conversion of glial cells into neurons, allowing the possibility of regenerative cell therapy for repair of the damaged brain (45–49). Identification of the cellular stimuli that activate RIT1 or inducible delivery of active RIT1 (50) might provide a novel therapeutic repair strategy, especially during states of crisis such as the aftermath of traumatic brain injury.
Experimental Procedures
Materials
The following materials were used: DMEM and F12 nutrient mixture (Gibco), triciribine (Millipore), Jetprime transfection reagent with buffer (Polyplus), doxycycline, fetal bovine serum (Gibco), N2 supplement (Thermo Fisher), B27 supplement (Thermo Fisher), RT-PCR buffer, RT Aid enzyme and ribonuclease inhibitor (Thermo Fisher), RT2 Profiler neurogenesis PCR array system (SA Biosciences), and SYBR Green with reference control (Bio-Rad).
Animals
All experimental procedures were approved by the University of Kentucky Institutional Animal Care and Use Committee in accordance with guidelines established by the National Institutes of Health in the Guide for the Care and Use of Laboratory Animals. Animals were housed up to 5 mice/cage in the University of Kentucky Medical Center vivarium with a 14:10-h light/dark photoperiod and were provided food and water ad libitum. DTG mice were produced by transgenesis to overexpress RIT1Q79L in a subset of CNS cells using a regulated binary system based on the tetracycline transactivator protein and the Tet operator (pBI Tet responder, Clontech). Active RIT1 was engineered to contain three copies of the FLAG tag peptide at the N terminus so that it could be distinguished from the endogenous protein. A fragment from the pBI-FLAG-RIT1Q79L vector was released and microinjected into the pronuclei of oocytes (University of Cincinnati Gene Targeting Core, Cincinnati, OH) from C57BL/6 mice. Mice expressing the tTA gene under the control of the Ca2+-calmodulin-dependent kinase II α promoter, CaMKII-tTA (Tet-Off), were obtained from The Jackson Laboratory (003010) (51), permitting co-expression of LacZ and FLAG-tagged RIT1Q79L within the CNS, including the dentate gyrus (29). From the prenatal period to the time of study initiation, DTG transgenic mice (and their dams) were given dietary supplementation of Dox, an analog of tetracycline, in their chow (200 mg/kg) to suppress FLAG-tagged RIT1Q79L expression. tTA littermates also received Dox-supplemented food. Selective neuronal FLAG-RIT1 expression was achieved by switching mice to a normal chow diet (−Dox) for 3 weeks.
Isolation and Passage of HNPCs
HNPCs were isolated from wild-type mice as described previously (52). Briefly, mice were euthanized and immediately sterilized using 70% ethanol. The brain was quickly dissected and immersed in dissection buffer (HBSS (1×) with no Ca2+ or Mg2+ and 1× antibiotic solution (1× antibiotic-antimycotic, Gibco)). Using a stereomicroscope, the dentate gyrus (DG) was dissected and placed in dissection buffer on ice. Typically, four to five hippocampi were pooled. After several washes with HBSS (1×), the tissue was incubated with enzymatic digestion solution (0.25% trypsin in 1× HBSS with activated papain) at 37 °C for 30–45 min with frequent shaking. Following digestion, trypsin activity was quenched by repeated washing with 5–10 ml of DMEM, and prewarmed culture medium (N2 containing DMEM/F12 (1:1) with antibiotics, EGF (20 ng/ml), and FGF2 (10 ng/ml)) was added. The tissues were triturated using fire polished Pasteur pipettes three to four times to release hippocampal NPCs. Approximately, 50 × 104 cells were plated in 12-well plates for 4–5 days. Neurospheres are evident from day 3 onward. For passaging purposes, the neurospheres from the respective groups were pooled and mechanically dissociated before replating for further culture. All cells used in this study were passaged at least twice. Neurospheres were counted using a grid on a microscope.
Cell Transfections and Treatments
For HNPC transfections, fresh neurospheres were passaged, and ∼105 cells were plated on poly-d-lysine-coated 12/18-mm coverglasses after appropriate sterilization using nitric acid and repeated autoclaving (53). Cells were transfected 72 h using Jetprime transfection reagent after the initial plating, which is necessary for HNPCs to regain contact. All DNA constructs used in this study have been described previously (27, 28, 54, 55). Jetprime transfection reagent has been shown previously to mediate efficient transfection of murine neuronal stem cells (53). Briefly, pCMV10-Myc RIT1Q79L or empty pCMV10 (1 μg) vector was mixed with 2 μl/μg Jetprime reagent according to the protocol of the manufacturer. For transfection, 200 μl of culture medium was collected and mixed with the transfection mixture and distributed in a dropwise method with frequent stirring. The cells were incubated for 6–7 h and washed with Dulbecco's phosphate-buffered saline (1×, Corning), and fresh complete medium was added. Cells were allowed to recover for 48 h. To monitor the transfection efficiency, we performed RT-PCR for RIT1 and confocal laser-scanning microscopy for the tag protein Myc in these cultures. We could always get >75% transfection efficiency in our experiments. For neuronal differentiation assays, cells after treatments were differentiated by addition of 1% FBS and retinoic acid (1 μm) for 4–6 days.
Tissue Collection and Processing
Animals received an overdose of Fatal-plus (65 mg/kg intraperitoneal sodium pentobarbital) and were perfused with 0.98% saline followed by 4% paraformaldehyde (56). Brains were removed from the skull immediately and post-fixed in 4% paraformaldehyde at 4 °C for 2 days. Brains were then washed extensively to remove the excess paraformaldehyde and incubated in increasing concentrations of 10–30% sucrose solution overnight for 3 days at room temperature. Finally, fully immersed brains were cryosectioned. The tissue blocks were embedded in optimal cutting compound (OCT) and snap-frozen. All blocks were allowed to stand at −80 °C for at least 2 days before sectioning using a cryostat.
Immunohistochemistry
Coronal brain sections, 15 and 40 μm, were cut and mounted on positive fixed microfrosted glass slides (Fisher Scientific). General antigen retrieval was performed in citrate buffer (pH 6) (57). BrdU antigen retrieval was performed using warm trypsin (0.25%) containing 2 n HCl. Sections were washed in PBS and incubated in blocking and permeabilizing buffer (1% serum (matching the host of the secondary antibody that was used) and 1% Triton X-100 in PBS) for 10 min at room temperature, followed by extensive washing (in PBS (pH 7.4)). Primary antibodies against RIT1 (14G7) (Santa Cruz Biotechnology), FLAG, Ki67 (rabbit), BrdU (Sigma), Doublecortin (DCX) (Millipore), Sox2 (Abcam), NeuroD1 (donkey) (Santa Cruz), Nestin (Covance), Akt, phospho-AktS473, βIII tubulin, ERK1/2, anti-phospho-ERK1/2 (Cell Signaling), and phospho-Sox2T118 (ECM Biosciences) were diluted in blocking serum, incubated overnight with sections at 4 °C, followed by extensive washing with 1× PBS (room temperature). Then, secondary antibodies, either conjugated with Alexa 488, Alexa 568, Alexa 594, or phycoerythrin, were applied to the sections for 2 h in the dark, followed by extensive washing with 1× PBS (room temperature). The sections were air-dried, mounted with DAPI-containing medium, and imaged 2–3 days later for coverslips to settle. For immunocytochemistry, cells were fixed in 4% paraformaldehyde for 15 min at room temperature and permeabilized and blocked as described for tissue sections. Imaging was performed using either a Nikon A1 or C2 confocal microscope. All images were acquired using the NIS Elements software package.
PCR Array
The mouse neurogenesis RT2 Profiler PCR array (Qiagen) was used for neurogenesis-specific transcription factor screening. Briefly, mice were sacrificed, and total RNA was prepared (Promega SV total RNA kit) from the dentate gyrus of DTG mice (lifetime doxycycline diet (control) or 3 weeks after removal of the Dox diet (−Dox)) and immediately frozen at −70 °C. Aliquots of RNA (500 ng from each group) were reverse-transcribed using a Bio-Rad RT kit. The cDNA was diluted 1:3 in ultrapure water for the final PCR. The cDNA was mixed with SYBR Green into the array plates, and cycling was performed according to the instructions of the manufacturer. A melt curve analysis was performed to check for product integrity. The values were obtained using the ΔCT method and plotted as heat maps.
Semiquantitative Real-time PCR
Briefly, total RNA was prepared from the dissected hippocampi and subventricular zones separately under aseptic conditions from each mouse (56) using a Promega SV total RNA isolation kit. RNA was stored immediately at −70 °C in aliquots. RNA (100 ng/sample) was reverse-transcribed using the iScript first strand synthesis kit (Bio-Rad) with the following gene-specific primers: Sox2, 5′AAGCCATGAATGCAGAGGAGGACT3′ (forward) and 5′AGCTGCAGG CAGCCGGCGACC3′ (reverse); Ascl1, 5′CCCCCAACTACTCCAACGAC3′ (forward) and 5′GTCCAGCAGCTCTTGTTCCT3′ (reverse); Sox215, ′CTCATCCTTCCTCCCTCCCG3′ (forward) and 5′CCAAGCCAGCGGACTCAGAGAC3′ (reverse); Gadd45b, 5′CCTGGCCATAGACGAAGAAG3′ (forward) and 5′AGCCTCTGCATGCCTGATAC3′ (reverse); BDNF, NeuroD1, 5′AAGCCATGAATGCAG AGGAGGACT3′ (forward) and 5′AGCTGCAGGCAGCCGGCGACC3′ (reverse); Neurog2, 5′TCACGAAG ATCGAGACGCTG3′ (forward) and 5′CTCCAGGAGGAAGGTGGAGA3′ (reverse); and GAPDH, 5′TGCACCACCAACTGCTTAGC3′ (forward) and 5′GGCATGGACTGTGGTCATGAG3′ (reverse). PCR products were amplified using DreamTaq Green (Thermo Fisher), resolved on 1–2% agarose gels, and imaged with Gel Logic 112 (Fisher Biotech).
Luciferase Gene Reporter Assays
The Cignal Sox2 reporter (Qiagen, CCS-0038L) contains repeats of the Sox2 promoter and Sox2 binding sites driving firefly luciferase expression. HNPCs were allowed to adhere for 72 h after replating and then transfected with EV or RIT1 Q79L (RIT1 CA). Approximately 72 h post-transfection, cells were washed with PBS and lysed for 30 min in passive lysis buffer (Promega luciferase kit). The lysate was centrifuged at 12,000 × g for 30 s at room temperature and put on ice for immediate use or frozen at −80 °C. A 10- to 15-μl aliquot of the lysis supernatant was mixed with 100 μl of detection reagent, and luminescence was recorded using a luminometer with a 10-s time interval. The readings were noted and averaged for statistical purposes.
Statistical Analysis
All data are represented as mean ± S.E. Statistical analysis was carried out by either non-parametric unpaired one-tailed t test or one-way analysis of variance combined with post hoc analysis using Tukey-Kramer multiple comparisons. Comparisons with p < 0.05 were considered significant.
Author Contributions
S. M. conducted most of the experiments, analyzed the results, and wrote most of the paper. W. C. generated the DTG transgenic mouse and performed studies examining how active RIT1 alters neurogenesis. D. A. A. conceived the idea for the project, helped to generate the transgenic model, and wrote the manuscript with S. M. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Wang Chi and Yu Chin (Markey Cancer Center, University of Kentucky) for help with the generation of heat maps. We also acknowledge Linda Simmerman (Spinal Cord and Brain Injury Center) and Carole Moncman (Department of Molecular and Cellular Biochemistry) for help with confocal microscopy.
This work was supported in part by NINDS, National Institutes of Health Grant R01 NS045103, Kentucky Spinal Cord and Head Injury Research Trust Grant 12-1A, and a Kentucky Lung Cancer research grant. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- NPC
- neural progenitor/stem cell
- Dox
- doxycycline
- DTG
- double transgenic
- HNPC
- hippocampal neural progenitor cell
- DCX
- doublecortin
- EV
- empty vector
- DG
- dentate gyrus
- CA
- constitutively active
- HBSS
- Hank's Balanced Salt Solution.
References
- 1. Aimone J. B., Li Y., Lee S. W., Clemenson G. D., Deng W., and Gage F. H. (2014) Regulation and function of adult neurogenesis: from genes to cognition. Physiol. Rev. 94, 991–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bond A. M., Ming G. L., and Song H. (2015) Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell 17, 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gotz M., Nakafuku M., and Petrik D. (2016) Neurogenesis in the developing and adult brain: similarities and key differences. Cold Spring Harb. Perspect. Biol. 8, 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alunni A., and Bally-Cuif L. (2016) A comparative view of regenerative neurogenesis in vertebrates. Development 143, 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ming G. L., and Song H. (2011) Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu K., Lin B., Zhao M., Yang X., Chen M., Gao A., Liu F., Que J., and Lan X. (2013) The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell. Signal. 25, 1264–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sarkar A., and Hochedlinger K. (2013) The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12, 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fantes J., Ragge N. K., Lynch S. A., McGill N. I., Collin J. R., Howard-Peebles P. N., Hayward C., Vivian A. J., Williamson K., van Heyningen V., and FitzPatrick D. R. (2003) Mutations in SOX2 cause anophthalmia. Nat. Genet. 33, 461–463 [DOI] [PubMed] [Google Scholar]
- 9. Kelberman D., Rizzoti K., Avilion A., Bitner-Glindzicz M., Cianfarani S., Collins J., Chong W. K., Kirk J. M., Achermann J. C., Ross R., Carmignac D., Lovell-Badge R., Robinson I. C., and Dattani M. T. (2006) Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J. Clin. Invest. 116, 2442–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Archer T. C., Jin J., and Casey E. S. (2011) Interaction of Sox1, Sox2, Sox3 and Oct4 during primary neurogenesis. Dev. Biol. 350, 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyagi S., Kato H., and Okuda A. (2009) Role of SoxB1 transcription factors in development. Cell Mol. Life Sci. 66, 3675–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., and Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heng J. C., and Ng H. H. (2010) Transcriptional regulation in embryonic stem cells. Adv. Exp. Med. Biol. 695, 76–91 [DOI] [PubMed] [Google Scholar]
- 14. Young R. A. (2011) Control of the embryonic stem cell state. Cell 144, 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kopp J. L., Ormsbee B. D., Desler M., and Rizzino A. (2008) Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 26, 903–911 [DOI] [PubMed] [Google Scholar]
- 16. Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A. A., Ko M. S., and Niwa H. (2007) Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9, 625–635 [DOI] [PubMed] [Google Scholar]
- 17. Wang Z., Oron E., Nelson B., Razis S., and Ivanova N. (2012) Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10, 440–454 [DOI] [PubMed] [Google Scholar]
- 18. Fang L., Zhang L., Wei W., Jin X., Wang P., Tong Y., Li J., Du J. X., and Wong J. (2014) A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell 55, 537–551 [DOI] [PubMed] [Google Scholar]
- 19. Wang J., Zhang Y., Hou J., Qian X., Zhang H., Zhang Z., Li M., Wang R., Liao K., Wang Y., Li Z., Zhong D., Wan P., Dong L., Liu F., et al. (2016) Ube2s regulates Sox2 stability and mouse ES cell maintenance. Cell Death Differ. 23, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Colicelli J. (2004) Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi G. X., Cai W., and Andres D. A. (2013) Rit subfamily small GTPases: regulators in neuronal differentiation and survival. Cell. Signal. 25, 2060–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee C. H., Della N. G., Chew C. E., and Zack D. J. (1996) Rin, a neuron-specific and calmodulin-binding small G-protein, and Rit define a novel subfamily of ras proteins. J. Neurosci. 16, 6784–6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lein P. J., Guo X., Shi G. X., Moholt-Siebert M., Bruun D., and Andres D. A. (2007) The novel GTPase Rit differentially regulates axonal and dendritic growth. J. Neurosci. 27, 4725–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wes P. D., Yu M., and Montell C. (1996) RIC, a calmodulin-binding Ras-like GTPase. EMBO J. 15, 5839–5848 [PMC free article] [PubMed] [Google Scholar]
- 25. Berger A. H., Imielinski M., Duke F., Wala J., Kaplan N., Shi G. X., Andres D. A., and Meyerson M. (2014) Oncogenic RIT1 mutations in lung adenocarcinoma. Oncogene 33, 4418–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cai W., and Andres D. A. (2014) mTORC2 is required for rit-mediated oxidative stress resistance. PLoS ONE 9, e115602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi G. X., Cai W., and Andres D. A. (2012) Rit-mediated stress resistance involves a p38-mitogen- and stress-activated protein kinase 1 (MSK1)-dependent cAMP response element-binding protein (CREB) activation cascade. J. Biol. Chem. 287, 39859–39868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cai W., Carlson S. W., Brelsfoard J. M., Mannon C. E., Moncman C. L., Saatman K. E., and Andres D. A. (2012) Rit GTPase signaling promotes immature hippocampal neuronal survival. J. Neurosci. 32, 9887–9897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakagawa S., Kim J. E., Lee R., Malberg J. E., Chen J., Steffen C., Zhang Y. J., Nestler E. J., and Duman R. S. (2002) Regulation of neurogenesis in adult mouse hippocampus by cAMP and the cAMP response element-binding protein. J. Neurosci. 22, 3673–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao C., Deng W., and Gage F. H. (2008) Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660 [DOI] [PubMed] [Google Scholar]
- 31. Rusyn E. V., Reynolds E. R., Shao H., Grana T. M., Chan T. O., Andres D. A., and Cox A. D. (2000) Rit, a non-lipid-modified Ras-related protein, transforms NIH3T3 cells without activating the ERK, JNK, p38 MAPK or PI3K/Akt pathways. Oncogene 19, 4685–4694 [DOI] [PubMed] [Google Scholar]
- 32. Amador-Arjona A., Cimadamore F., Huang C. T., Wright R., Lewis S., Gage F. H., and Terskikh A. V. (2015) SOX2 primes the epigenetic landscape in neural precursors enabling proper gene activation during hippocampal neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 112, E1936–E1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Telley L., Govindan S., Prados J., Stevant I., Nef S., Dermitzakis E., Dayer A., and Jabaudon D. (2016) Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351, 1443–1446 [DOI] [PubMed] [Google Scholar]
- 34. Conover J. C., and Notti R. Q. (2008) The neural stem cell niche. Cell Tissue Res. 331, 211–224 [DOI] [PubMed] [Google Scholar]
- 35. Cimadamore F., Amador-Arjona A., Chen C., Huang C. T., and Terskikh A. V. (2013) SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc. Natl. Acad. Sci. U.S.A. 110, E3017–E3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Episkopou V. (2005) SOX2 functions in adult neural stem cells. Trends Neurosci. 28, 219–221 [DOI] [PubMed] [Google Scholar]
- 37. Ferri A. L., Cavallaro M., Braida D., Di Cristofano A., Canta A., Vezzani A., Ottolenghi S., Pandolfi P. P., Sala M., DeBiasi S., and Nicolis S. K. (2004) Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131, 3805–3819 [DOI] [PubMed] [Google Scholar]
- 38. Jeong C. H., Cho Y. Y., Kim M. O., Kim S. H., Cho E. J., Lee S. Y., Jeon Y. J., Lee K. Y., Yao K., Keum Y. S., Bode A. M., and Dong Z. (2010) Phosphorylation of Sox2 cooperates in reprogramming to pluripotent stem cells. Stem Cells 28, 2141–2150 [DOI] [PubMed] [Google Scholar]
- 39. Han D. W., Tapia N., Hermann A., Hemmer K., Höing S., Araúzo-Bravo M. J., Zaehres H., Wu G., Frank S., Moritz S., Greber B., Yang J. H., Lee H. T., Schwamborn J. C., Storch A., and Schöler H. R. (2012) Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 10, 465–472 [DOI] [PubMed] [Google Scholar]
- 40. Kim J., Efe J. A., Zhu S., Talantova M., Yuan X., Wang S., Lipton S. A., Zhang K., and Ding S. (2011) Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 108, 7838–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lujan E., Chanda S., Ahlenius H., Südhof T. C., and Wernig M. (2012) Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. U.S.A. 109, 2527–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ring K. L., Tong L. M., Balestra M. E., Javier R., Andrews-Zwilling Y., Li G., Walker D., Zhang W. R., Kreitzer A. C., and Huang Y. (2012) Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 11, 100–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takahashi K., and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 44. Thier M., Wörsdörfer P., Lakes Y. B., Gorris R., Herms S., Opitz T., Seiferling D., Quandel T., Hoffmann P., Nöthen M. M., Brüstle O., and Edenhofer F. (2012) Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10, 473–479 [DOI] [PubMed] [Google Scholar]
- 45. Heinrich C., Bergami M., Gascón S., Lepier A., Viganò F., Dimou L., Sutor B., Berninger B., and Götz M. (2014) Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem Cell Reports 3, 1000–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Niu W., Zang T., Smith D. K., Vue T. Y., Zou Y., Bachoo R., Johnson J. E., and Zhang C. L. (2015) SOX2 reprograms resident astrocytes into neural progenitors in the adult brain. Stem Cell Reports 4, 780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Niu W., Zang T., Zou Y., Fang S., Smith D. K., Bachoo R., and Zhang C. L. (2013) In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 15, 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao X., Wang X., Xiong W., and Chen J. (2016) In vivo reprogramming reactive glia into iPSCs to produce new neurons in the cortex following traumatic brain injury. Sci. Rep. 6, 22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yi X., Jin G., Zhang X., Mao W., Li H., Qin J., Shi J., Dai K., and Zhang F. (2013) Cortical endogenic neural regeneration of adult rat after traumatic brain injury. PLoS ONE 8, e70306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Poulou M., Mandalos N. P., Karnavas T., Saridaki M., McKay R. D., and Remboutsika E. (2016) A “hit and run” approach to inducible direct reprogramming of astrocytes to neural stem cells. Front. Physiol. 7, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mayford M., Bach M. E., Huang Y. Y., Wang L., Hawkins R. D., and Kandel E. R. (1996) Control of memory formation through regulated expression of a CaMKII transgene. Science 274, 1678–1683 [DOI] [PubMed] [Google Scholar]
- 52. Guo W., Patzlaff N. E., Jobe E. M., and Zhao X. (2012) Isolation of multipotent neural stem or progenitor cells from both the dentate gyrus and subventricular zone of a single adult mouse. Nat. Protoc. 7, 2005–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Park D. H., Hong S. J., Salinas R. D., Liu S. J., Sun S. W., Sgualdino J., Testa G., Matzuk M. M., Iwamori N., and Lim D. A. (2014) Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 8, 1290–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi G. X., and Andres D. A. (2005) Rit contributes to nerve growth factor-induced neuronal differentiation via activation of B-Raf-extracellular signal-regulated kinase and p38 mitogen-activated protein kinase cascades. Mol. Cell. Biol. 25, 830–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shi G. X., Jin L., and Andres D. A. (2011) A rit GTPase-p38 mitogen-activated protein kinase survival pathway confers resistance to cellular stress. Mol. Cell. Biol. 31, 1938–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mir S., Sen T., and Sen N. (2014) Cytokine-induced GAPDH sulfhydration affects PSD95 degradation and memory. Mol. Cell 56, 786–795 [DOI] [PubMed] [Google Scholar]
- 57. Johnson J. O., Pioro E. P., Boehringer A., Chia R., Feit H., Renton A. E., Pliner H. A., Abramzon Y., Marangi G., Winborn B. J., Gibbs J. R., Nalls M. A., Morgan S., Shoai M., Hardy J., et al. (2014) Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 17, 664–666 [DOI] [PMC free article] [PubMed] [Google Scholar]