Abstract
Prozymes are catalytically inactive enzyme paralogs that dramatically stimulate the function of weakly active enzymes through complex formation. The two prozymes described to date reside in the polyamine biosynthesis pathway of the human parasite Trypanosoma brucei, an early branching eukaryote that lacks transcriptional regulation and regulates its proteome through posttranscriptional and posttranslational means. Arginine methylation is a common posttranslational modification in eukaryotes catalyzed by protein arginine methyltransferases (PRMTs) that are typically thought to function as homodimers. We demonstrate that a major T. brucei PRMT, TbPRMT1, functions as a heterotetrameric enzyme-prozyme pair. The inactive PRMT paralog, TbPRMT1PRO, is essential for catalytic activity of the TbPRMT1ENZ subunit. Mutational analysis definitively demonstrates that TbPRMT1ENZ is the cofactor-binding subunit and carries all catalytic activity of the complex. Our results are the first demonstration of an obligate heteromeric PRMT, and they suggest that enzyme-prozyme organization is expanded in trypanosomes as a posttranslational means of enzyme regulation.
Keywords: allosteric regulation, post-translational modification (PTM), protein methylation, Trypanosoma brucei, trypanosome, PRMT, arginine methylation, prozyme
Introduction
Trypanosoma brucei, the causative agent of human African trypanosomiasis, poses a severe health risk in Sub-Saharan Africa. An estimated 70 million people are at risk of the infection, and the World Health Organization estimates about 20,000 new cases per year (1). In the search for new treatments, understanding the basic biology of the parasite is a cornerstone on the path to discovery of novel biological processes that could potentially serve as drug targets (2). Despite the eukaryotic nature of T. brucei, the organism has been shown to harbor many unique biological processes or, in some cases, to extensively utilize mechanisms that are rarely used in higher eukaryotes (3–5). The exceptional biology of trypanosomes is likely a consequence of their long separate evolutionary history and numerous adaptations to the parasitic lifestyle (6).
Trypanosomes undergo significant morphological and physiological changes during their life cycle, which includes transitions between mammalian and insect vector hosts that provide distinct environmental conditions and temperatures (7). Strikingly, these organisms accomplish adaptations to changing environments and differentiation to several distinct life cycle forms all in the absence of transcriptional control of their genome (3). Rather, gene expression is regulated at posttranscriptional and posttranslational levels. Posttranslational modification of proteins is one such mechanism that allows rapid responses to internal and external cues (8). Our proteome-wide studies revealed that about 15% of the proteome of T. brucei insect vector procyclic forms (PFs)2 harbors arginine methyl marks (9, 10).3 Thus, arginine methylation is poised to play a crucial role in regulating T. brucei biology. In support of this notion, we showed that TbPRMT1-catalyzed arginine methylation of the essential RNA-binding protein DRBD18 acts as a switch that controls the RNA-stabilizing and -degrading activity of this major transcriptome regulator as well as the composition of DRBD18-containing ribonucleoproteins (11).
An enzyme family containing three major types of protein arginine methyltransferases (PRMTs) catalyzes arginine methylation (12). All PRMTs catalyze formation of ω-NG-monomethylarginine (MMA), type I PRMTs catalyze ω-NG,NG-asymmetric dimethylarginine (ADMA), and type II PRMTs create ω-NG,N′G-symmetric dimethylarginine (SDMA). Humans possess nine PRMTs (12). T. brucei apparently contains just four PRMTs, and we showed that together these enzymes have the capacity to catalyze MMA, ADMA, and SDMA formation (13–16). TbPRMT7 was the first characterized PRMT to unquestionably create only MMA, making it a type III PRMT (13). TbPRMT5, despite its evolutionary divergence, exhibits a type II activity (15), and TbPRMT6 and TbPRMT1 have both been shown to create ADMA, therefore belonging to the type I PRMT group (14, 16). Like human PRMT1, TbPRMT1 is responsible for the majority of ADMA formation in vivo (14, 17). However, perplexingly, in vitro, TbPRMT1 exhibits a very narrow substrate specificity and weak activity (14). We recently showed that TbPRMT1 protein stability is mutually dependent on a previously uncharacterized T. brucei PRMT paralog tentatively named TbPRMT3 based on its homology to human PRMT3 (18). Typically, this type of mutual stability dependence is observed in proteins belonging to the same complex (19, 20). TbPRMT3 has not exhibited any in vitro activity in our hands, and its primary sequence harbors mutations in conserved PRMT motifs; thus, we suspected it to be an inactive PRMT paralog. The observed in vivo dependence of a very weak enzyme and a catalytically dead paralog strikingly resembles a scenario that has been described in the T. brucei polyamine synthesis pathway: the prozyme paradigm (21). Here, the functional forms of two enzymes involved in polyamine biosynthesis consist of a barely active enzyme in complex with an inactive enzyme paralog, termed prozyme. The functional complex comprises two proteins in which the prozyme allosterically activates the enzyme, and the activity of the complex exceeds that of the “active” subunit alone on average by 2000-fold (19, 21).
In this study, we show that TbPRMT1 functions as a heterotetrameric complex formed by the enzymatic subunit TbPRMT1ENZ (previously TbPRMT1) and the inactive PRMT paralog TbPRMT1PRO (previously TbPRMT3). Our results demonstrate a novel PRMT organization and represent the first expansion of the trypanosome prozyme paradigm outside the polyamine synthesis pathway. These findings suggest that allosteric enzyme activation by catalytically inactive paralogs may be a more widespread mechanism for posttranslational regulation in trypanosomes than previously appreciated. Furthermore, our results suggest the presence of novel PRMT regulatory mechanisms that could also function in higher organisms under specific conditions.
Results
TbPRMT1PRO Is Missing Key Catalytic Residues
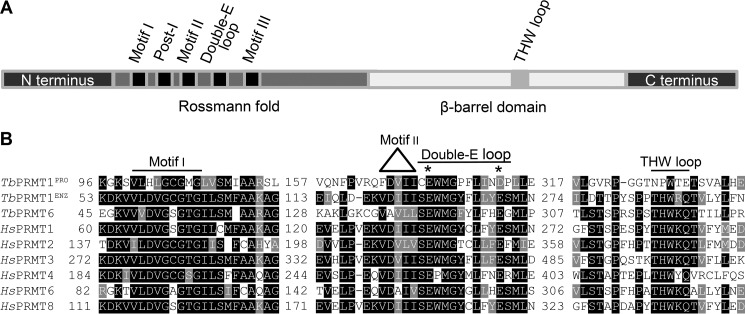
The T. brucei genome encodes five proteins with high homology to human PRMTs, four of which have been characterized previously (13–16). Pairwise BLAST comparisons with human PRMTs indicated that the remaining putative TbPRMT (Tb927.10.3560) has the highest sequence similarity to human PRMT3, and therefore this enzyme has been formerly referred to as TbPRMT3 (18, 22). In light of the functional results presented in this work, we have renamed Tb927.10.3560 as TbPRMT1PRO. Our studies also have led us to rename the former TbPRMT1 (Tb927.1.4690) as TbPRMT1ENZ, and these names will be used hereafter. To begin to understand the function of TbPRMT1PRO, we first examined its amino acid sequence. In general, type I PRMTs comprise a Rossmann fold that harbors conserved motifs I, post-I, II, and III and the double-E loop as well as a β-barrel domain containing a THW loop (Fig. 1A). The TbPRMT1PRO amino acid sequence reveals conserved motifs I, post-I, II, and III. However, this protein harbors a Glu to Asp mutation within its double-E loop (Fig. 1B). Although conservative, the analogous mutation in rat PRMT1 was shown previously to decrease in vitro methylation activity to 0.03% compared with wild type enzyme (23). Furthermore, based on phylogenetic analysis, TbPRMT1PRO clusters with type I PRMTs (24), and as such, it is expected to contain a THW loop. Strikingly, neither the threonine nor the histidine residue is conserved in the region of TbPRMT1PRO corresponding to the THW loop (Fig. 1B). These observations suggested that TbPRMT1PRO could be an inactive PRMT paralog.
FIGURE 1.
TbPRMT1PRO lacks motifs important for PRMT activity. A, schematic representation of PRMT motifs (not to scale). B, selected motifs from Clustal Omega alignment of T. brucei and Homo sapiens type I PRMTs. * marks conserved residues in the double-E loop.
TbPRMT1PRO Forms a Complex with TbPRMT1ENZ
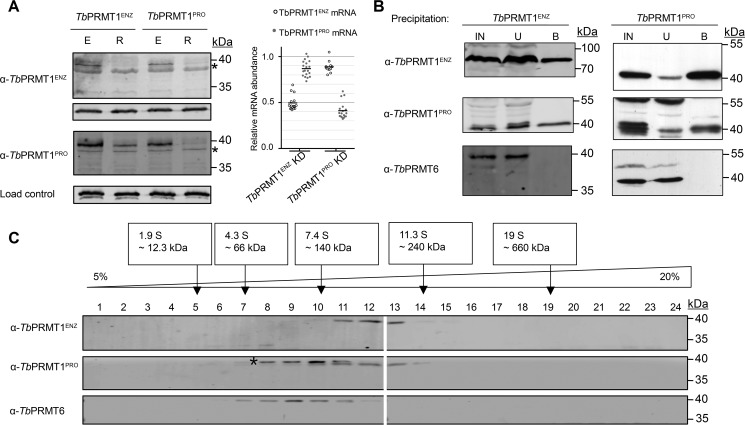
TbPRMT1ENZ has been described previously as an active enzyme that is responsible for the majority of ADMA formation in vivo (14). Surprisingly, in vitro we observed a narrow substrate specificity and very low activity compared with rat PRMT1 (14). Attempts to detect TbPRMT1PRO activity have been unsuccessful, reinforcing the idea that it possibly is a catalytically inactive PRMT paralog. While investigating the interplay of TbPRMTs in T. brucei PF, we noticed that TbPRMT1ENZ and TbPRMT1PRO share mutual protein stability dependence (18). Because this phenomenon is commonly associated with proteins that form a complex, we explored the possibility that TbPRMT1ENZ and TbPRMT1PRO form a PRMT heteromeric complex. First, we asked whether the mutual stability dependence of TbPRMT1ENZ and TbPRMT1PRO is conserved in both culturable T. brucei life cycle stages. We induced RNAi of either TbPRMT1ENZ or TbPRMT1PRO in the clinically relevant bloodstream form (BF) T. brucei and used Western blotting to examine levels of both proteins (Fig. 2A, left). We observed a decrease in the protein levels of both proteins regardless of which PRMT was repressed. To rule out the possibility that our antibodies simply do not discriminate between TbPRMT1ENZ and TbPRMT1PRO, the antibodies were tested against Escherichia coli-expressed recombinant proteins. Fig. 3B, lanes a and c, clearly show the specificity of each antibody to its intended target. Furthermore, to ensure that our RNAi constructs were specific to only a single PRMT, we measured TbPRMT1ENZ or TbPRMT1PRO mRNA levels in both knockdown cell lines. In either case, we observed no significant change in the mRNA level of the non-target PRMT upon RNAi induction (Fig. 2A, right) as reported previously in the PF life cycle stage (18). Thus, the mutual stability dependence of TbPRMT1ENZ and TbPRMT1PRO occurs on the protein level in both life cycle stages.
FIGURE 2.
TbPRMT1ENZ and TbPRMT1PRO form a complex. A, left, the stabilities of TbPRMT1ENZ and TbPRMT1PRO proteins are mutually dependent. Lysed BF T. brucei cells with the indicated TbPRMT either expressed (E) or repressed (R) were probed with antibodies against specific TbPRMTs. α-p22 antibody was used as a load control. * marks a nonspecific band. The image is representative of three biological replicates. A, right, RNAi-mediated depletion of either TbPRMT1PRO or TbPRMT1ENZ mRNA does not significantly affect the mRNA level of their counterpart. mRNA levels of each TbPRMT1 subunit were determined using qRT-PCR. Data were plotted as induced/uninduced RNAi cell mRNA levels. Data points represent technical replicates within two biological replicates. The horizontal line represents the mean. B, TbPRMT1ENZ-Myc-BirA* and TbPRMT1PRO-LSH were expressed from an exogenous locus in separate cell lines. Input (IN), unbound (U), and bound (B) proteins from precipitations of tagged TbPRMTs were probed with α-TbPRMT antibodies. α-TbPRMT6 served as a specificity control. The image is representative of two biological replicates. C, glycerol gradient sedimentation patterns of TbPRMT1ENZ and TbPRMT1PRO overlap in a range suggesting a tetramer formation. Wild type 29-13 procyclic form T. brucei cell lysate was fractionated on a 5–20% glycerol gradient. Fractions were probed with α-TbPRMT antibodies. Size standards were run on a parallel gradient. * marks a contaminating species recognized by the TbPRMT1PRO antibody. The image is representative of two biological replicates.
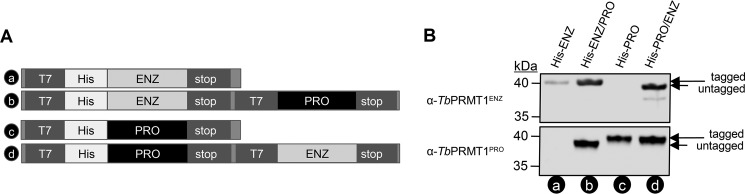
FIGURE 3.
A, schematic of pETDuet-1 constructs used to express His-TbPRMT1ENZ (a), His-TbPRMT1ENZ/PRO (b), His-TbPRMT1PRO (c), and His-TbPRMT1PRO/ENZ (d). B, TbPRMT1ENZ and TbPRMT1PRO interact directly. Constructs a–d shown in A were expressed in E. coli. His-tagged protein was purified, and the elutions were probed with the indicated α-TbPRMT antibodies. The image is representative of two biological replicates.
Next, we investigated whether TbPRMT1ENZ and TbPRMT1PRO are a part of the same complex in vivo. Because the stability data suggested the same mechanism in both life cycle stages, we performed all subsequent experiments in T. brucei PF, which grows to higher densities and therefore is more suitable for proteomic experiments. We expressed N-terminally tagged TbPRMT1ENZ-Myc-BirA* and C-terminally tagged TbPRMT1PRO-linker-Strep-His (LSH) in separate cell lines and precipitated the tagged proteins using α-Myc antibody or metal affinity resin, respectively. Western blots containing input, unbound, and bound samples were then probed with α-TbPRMT1ENZ, α-TbPRMT1PRO, and α-TbPRMT6 antibodies (Fig. 2B). We observed that TbPRMT1ENZ and TbPRMT1PRO were efficiently precipitated in both reactions, whereas TbPRMT6 was not present in either. These data are in agreement with our hypothesis that TbPRMT1ENZ and TbPRMT1PRO form a heteromer. To confirm this observation, we utilized a less direct but more native approach to investigate the same question. We lysed wild type PF T. brucei cells, separated native complexes on a 5–20% glycerol gradient, and probed the gradient fractions with α-TbPRMT antibodies (Fig. 2C). TbPRMT1ENZ (39 kDa) and TbPRMT1PRO (42 kDa) co-sedimented in fractions that, according to the size standards that were run on a parallel gradient, roughly correspond to a size of a tetramer (160 kDa). In multiple experiments, we noticed that α-TbPRMT1PRO antibody detected two bands at ∼40 kDa; however, upon induction of RNAi in PF cells, only the lower band was visibly diminished, leading us to conclude that the upper band is likely nonspecific (data not shown). In contrast to TbPRMT1ENZ and TbPRMT1PRO, TbPRMT6 (41 kDa) peaked in a fraction corresponding to a dimer. Together, these data indicate that TbPRMT1ENZ and TbPRMT1PRO are part of the same complex in vivo.
As our in vivo studies did not allow us to determine whether TbPRMT1ENZ and TbPRMT1PRO interact directly, we used an in vitro approach to answer this question. To this end, we utilized a pETDuet bacterial expression vector that allows for co-expression of two proteins under separate T7 promoters (Fig. 3A). The two TbPRMT1 subunits were first cloned separately to permit expression of a single histidine (His)-tagged PRMT (Fig. 3A, constructs “a” and “c”). For simplicity, these will be referred to as His-ENZ and His-PRO. In separate plasmids, the remaining TbPRMT1 ORF was cloned into the second site, which resulted in two additional constructs containing His-TbPRMT1ENZ with untagged TbPRMT1PRO or His-TbPRMT1PRO with untagged TbPRMT1ENZ. These constructs will be hereafter referred to as His-ENZ/PRO and His-PRO/ENZ (Fig. 3A, constructs “b” and “d”). Each of the four constructs was separately expressed in E. coli, and metal affinity resin was used to purify the His-tagged protein under stringent washing conditions (1 m NaCl). Next, we probed Western blots containing the eluted proteins with α-TbPRMT1ENZ and α-TbPRMT1PRO antibodies (Fig. 3B). We observed that, in both cases, the untagged subunit was purified together with the His-tagged protein, clearly demonstrating a strong, direct non-ionic interaction between TbPRMT1ENZ and TbPRMT1PRO.
TbPRMT1ENZ/TbPRMT1PRO Heteromer Is the Functional Unit of the Major Type I PRMT in T. brucei
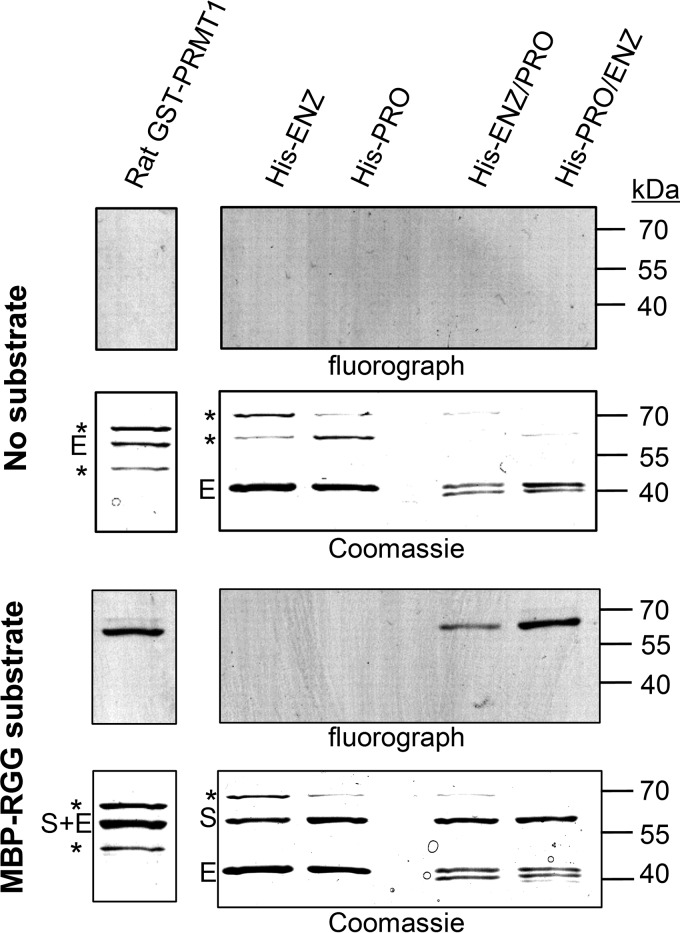
Having confirmed His-TbPRMT1ENZ/TbPRMT1PRO heteromer formation in vivo and in vitro, we next wanted to investigate the in vitro activity of the heteromer. Because our previous TbPRMT1ENZ characterization showed that TbPRMT1ENZ down-regulation almost completely abolishes ADMA formation in T. brucei (14), we expected the heteromer to exhibit activity comparable with the major mammalian type I PRMT, PRMT1. To test the PRMT activities of TbPRMT1ENZ, TbPRMT1PRO, and the heteromer containing both proteins and compare their activities with mammalian PRMT1, we incubated rat PRMT1, His-ENZ, His-PRO, His-ENZ/PRO, or His-PRO/ENZ with S-adenosyl-l-[methyl-3H]methionine ([3H]AdoMet) in the absence or presence of an RGG-rich substrate (Fig. 4). Following the reactions, proteins were resolved by SDS-PAGE and Coomassie-stained, and the radiolabeled signal was visualized by fluorography. We observed that under our experimental conditions both heteromers, regardless of which subunit was fused to the N-terminal His tag, exhibited activity, whereas neither subunit alone produced any signal. Furthermore, the His-PRO/ENZ complex showed activity comparable with that of the rat PRMT1. From these data, we conclude that the TbPRMT1ENZ/TbPRMT1PRO heteromer is the active form of TbPRMT1.
FIGURE 4.
TbPRMT1ENZ/PRO heteromer is the active form of a major T. brucei PRMT. For the in vitro methylation assay, MBP-RGG substrate was incubated with TbPRMTs in the presence of [3H]AdoMet. Proteins were separated by SDS-PAGE and visualized by fluorography and Coomassie staining. TbPRMT1ENZ (His-ENZ) and TbPRMT1PRO (His-PRO) do not exhibit any activity under the selected experimental conditions. The heteromer containing His-TbPRMT1PRO/ENZ exhibits activity comparable with rat GST-PRMT1. Reactions where MBP-RGG was excluded were used as a control (no substrate). S, substrate; E, enzyme; *, contaminant. The image is representative of two technical replicates.
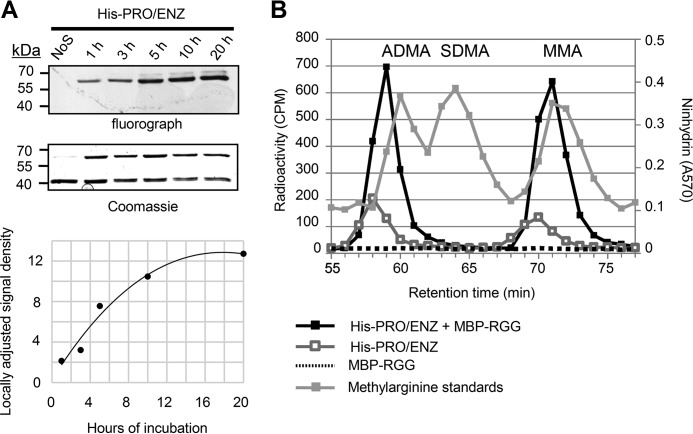
We next wanted to determine the type of methylarginine produced by TbPRMT1. To this end, we performed a methylation reaction containing TbPRMT1 heteromer, MBP-RGG substrate, and [3H]AdoMet. The reaction was allowed to proceed for 2 h based on a time course experiment showing that at this time point the reaction was progressing within the linear range (Fig. 5A). Subsequently, the proteins were TCA-precipitated, hydrolyzed into amino acids, and analyzed on a cation exchange chromatography column together with unlabeled MMA, ADMA, and SDMA standards. Reactions omitting either substrate or enzyme served as controls (Fig. 5B). We observed a significant amount of ADMA and MMA present in the experimental sample, whereas no trace of SDMA was recorded. This led us to conclude that the TbPRMT1 heteromer is a type I PRMT. We also observed a low level of both ADMA and MMA in the PRMT-only control. To determine whether this can be attributed to automethylation of the PRMT, we developed the gel presented in Fig. 4 for an additional month and concluded that the methylated species likely arise primarily from methylation of bacterial proteins that were present at low levels as contaminants in our TbPRMT1 preparation (data not shown). Analysis of either TbPRMT1ENZ or TbPRMT1PRO alone by cation exchange chromatography revealed no activity as expected (data not shown), supporting the classification of TbPRMT1ENZ/TbPRMT1PRO heteromer as the major T. brucei type I PRMT.
FIGURE 5.
A, under our experimental conditions, His-PRO/ENZ complex activity progresses in a linear range at the 2-h reaction time. Five identical methylation reactions were set up. At each time point, a single reaction was quenched. After separation by SDS-PAGE, the gel was incubated with EN3HANCE solution for 1 h, dried, and exposed to film at −80 °C overnight. The density of each signal was measured and plotted (lower panel). NoS, no substrate was added to reaction. The image is representative of two technical replicates. B, high resolution ion exchange chromatography analysis of methylarginine derivatives catalyzed by His-TbPRMT1PRO/ENZ. His-TbPRMT1PRO/ENZ was incubated with substrate and TCA-precipitated. The resulting protein mixture was digested by acid hydrolysis. Amino acids were analyzed by cation exchange chromatography in the presence of unlabeled ADMA, SDMA, and MMA standards. The experiment was performed once.
TbPRMT1 Forms a Heterotetramer in Solution
Having determined that the TbPRMT1 heteromer is a functional PRMT, we were intrigued by the possibility that this heteromer could be larger than the canonical PRMT dimer. The glycerol gradient sedimentation of native TbPRMT1 complex suggested this enzyme could function as a tetramer (Fig. 2C). PRMTs can reportedly exist in large complexes in vivo (25, 26) and form homo-oligomers in vitro (27–30). Furthermore, the recent human PRMT8 crystal structure revealed a homotetrameric PRMT architecture, suggesting the possibility of heterotetrameric intermember interactions between two PRMT dimers (31). To investigate the size of the TbPRMT1 heteromer we used multiangle light scattering (MALS). These studies showed that TbPRMT1 adopts exclusively a tetrameric structure (∼160 kDa) in solution (Fig. 6). In contrast to the heteromer, TbPRMT1PRO exists as a dimer in solution (Fig. 6). TbPRMT1ENZ displays a high degree of aggregation when expressed alone, which precluded MALS analysis of this protein. Together, our data indicate that TbPRMT1 requires both TbPRMT1ENZ and TbPRMT1PRO to adopt the active heterotetrameric structure.
FIGURE 6.
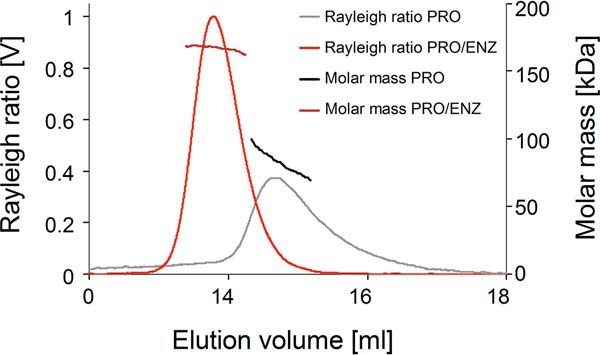
Molecular mass determination and Rayleigh ratio of TbPRMT1ENZ/PRO and of TbPRMT1PRO by MALS coupled to size exclusion chromatography (Superdex 200 10/300 column). The light scattering Rayleigh ratio (left ordinate; TbPRMT1ENZ/PRO, light red; TbPRMT1PRO, gray) and the molar mass distribution (right ordinate; TbPRMT1ENZ/PRO, dark red; TbPRMT1PRO, black) versus the elution volume are shown. TbPRMT1PRO forms a heterotetramer, whereas TbPRMT1PRO is dimeric. TbPRMT1ENZ on its own is unstable and could therefore not be analyzed by MALS. The image is representative of two biological replicates.
TbPRMT1ENZ Is the Catalytic Subunit of the TbPRMT1 Heteromer
The primary sequence of TbPRMT1PRO suggested that it might be a catalytically inactive PRMT paralog. Furthermore, TbPRMT1ENZ, despite its homology with human PRMT1, failed to show signs of catalytic activity in this study and only weak activity in our previous study (14). Together, these facts are reminiscent of a previously described prozyme paradigm typified by two different enzymes in the T. brucei polyamine biosynthesis pathway that function as heteromers in which an inactive enzyme paralog allosterically activates a catalytically active subunit (19, 21, 32, 33). With this paradigm and our data in mind, we asked whether TbPRMT1ENZ is the sole subunit in the TbPRMT1 complex that carries catalytic function. To investigate this possibility, we turned to a mutagenesis approach. We hypothesized that an introduction of mutations that are known to inactivate PRMTs into TbPRMT1ENZ would lead to a total loss of heteromer activity, whereas the same mutations introduced into TbPRMT1PRO sequence should leave the activity intact. Previous studies indicated that a mutation in conserved motif I residues leads to loss of PRMT activity, likely due to disruption of AdoMet binding (34, 35). Thus, we introduced a Gly to Arg mutation in the second Gly of the GXGXG in motif I of either TbPRMT1ENZ or TbPRMT1PRO in the context of the heteromeric enzyme. This mutation has been shown to completely abolish activity of a yeast PRMT1 homolog (35). To diversify our approach, we also introduced a mutation that would inactivate a PRMT by a different mechanism. We mutated the second Glu in the double-E loop to Gln (E135Q), a mutation previously shown to abolish PRMT activity without affecting AdoMet binding (23, 36). As noted above, TbPRMT1PRO carries an Asp at this position (D180Q). Studies of mammalian PRMT1 showed that, although Glu to Asp mutation seriously hinders activity, Gln at this position completely abolished measurable PRMT activity (23).
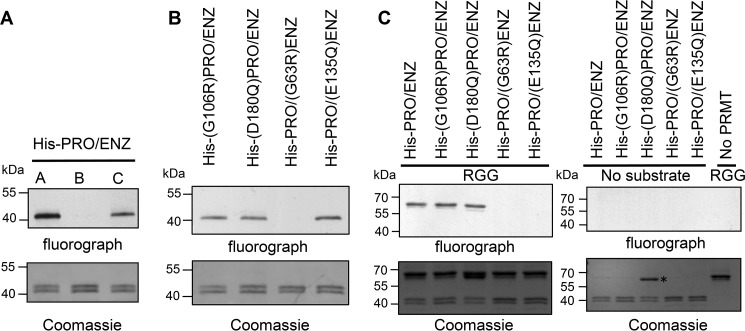
The mutations described above were separately introduced into a pETDuet construct encoding for His-PRO and untagged ENZ (Fig. 3A, construct d), and the proteins were purified via the His tag. The presence of both proteins in these purifications was verified by Western blotting (data not shown), and the equimolar representation of each subunit was confirmed by visual examination of the Coomassie staining (Fig. 7).
FIGURE 7.
TbPRMT1ENZ is the catalytic subunit of TbPRMT1ENZ/PRO complex. A, AdoMet is specifically cross-linked to His-ENZ/PRO complex. His-ENZ/PRO was incubated with [3H]AdoMet (A). After 10 min of incubation, a 50-fold excess of unlabeled AdoMet (B) or dATP (C) was added. Samples were then incubated at 4 °C for ∼18 h and UV cross-linked. After separation of proteins by SDS-PAGE, the gel was incubated with EN3HANCE solution for 1 h, dried, and exposed to film. The image is representative of two biological replicates. B, AdoMet binding capacity of TbPRMT1ENZ/PRO. TbPRMTs were incubated with [3H]AdoMet and UV cross-linked. Proteins were separated by SDS-PAGE and visualized by fluorography and Coomassie staining. Gly → Arg mutation targets AdoMet binding motif I, whereas Glu → Gln (or Asp → Gln in the case of TbPRMT1PRO) emulates a mutation shown previously to abolish rat PRMT1 activity without affecting AdoMet binding capacity of the enzyme. The experiment was performed once. C, in vitro methylation assays of His-TbPRMT1PRO/ENZ mutants. TbPRMT1 wild type (His-PRO/ENZ) or mutant heteromers were incubated with MBP-RGG substrate in the presence of [3H]AdoMet (left panel). Proteins were separated by SDS-PAGE and visualized by fluorography and Coomassie staining. Either MBP-RGG substrate or PRMT was omitted in the control reactions (right panel). The image is representative of three technical replicates.
First, we analyzed the AdoMet binding properties of wild type and mutant TbPRMT1 heterotetramers to determine whether both subunits directly participate in AdoMet binding. Even in homodimeric yeast and mammalian PRMTs, it is not clear whether AdoMet binds to one or both monomers. Because deletion of the dimerization arm of rat PRMT1 leads to a complete loss of AdoMet binding, it is generally accepted that dimerization is a necessary prerequisite to bind AdoMet (23). Similarly, mutation of the dimerization arm was recently shown to abolish dimerization and catalytic activity in TbPRMT7 (36). However, crystal structures of the conserved PRMT core show that the active site is likely formed within a single monomer rather than on the interface of the two subunits, leaving both subunits with equal potential to bind the cofactor (23, 36, 37). Interestingly, human PRMT8, which has been recently found to multimerize in solution, reportedly contains a single AdoMet molecule per dimer (31), although this finding has been challenged (38). In our case, despite the natural mutations in TbPRMT1PRO that suggest inactivity, its motif I, which is thought to play an important role in AdoMet binding, is well conserved. Therefore, we could not rule out direct involvement of TbPRMT1PRO subunit in AdoMet binding. To determine the roles of TbPRMT1ENZ and TbPRMT1PRO in AdoMet binding within the active heteromer, we UV cross-linked [3H]AdoMet to the His-PRO/ENZ heteromer, resolved the proteins by SDS-PAGE, and visualized the radioactive signal by fluorography. We observed a single radioactive band whose signal was fully outcompeted by a 50-fold excess of unlabeled AdoMet but not by excess dATP, thereby demonstrating the specificity of the cross-linking (Fig. 7A). We then performed the same assay using the heteromers in which one of the subunits carried an inactivating mutation (Fig. 7B). We observed that both double-E loop mutants, His-(D180Q)PRO/ENZ and His-PRO/(E135Q)ENZ, were capable of AdoMet binding, which was in accord with mutational studies on mammalian PRMT1 showing that, although this residue in the double-E loop is crucial for catalysis, it contributes very little to AdoMet binding (23). The retained ability to bind AdoMet also reassured us that the introduced mutations do not distort protein folding. We next analyzed AdoMet binding in heteromers with motif I mutations in each of the subunits. Remarkably, although AdoMet binding of the heteromer was abolished when motif I of TbPRMT1ENZ was mutated (Fig. 7B, His-PRO/(G63R)ENZ), the heteromer harboring mutation of motif I in the TbPRMT1PRO subunit was perfectly capable of AdoMet binding (Fig. 7B, His-(G106R)PRO/ENZ). This led us to conclude that TbPRMT1ENZ is the sole AdoMet-binding subunit of the complex.
To investigate the contribution of the TbPRMT1ENZ and TbPRMT1PRO subunits to the catalytic activity of the heteromeric TbPRMT1 complex, we next performed a gel-based in vitro methylation assay. TbPRMT1, [3H]AdoMet, and substrate were mixed and incubated for 18 h at 22 °C, and methylated products were visualized by fluorography (Fig. 7C). Heteromers containing mutations in either double-E loop (His-(D180Q)PRO/ENZ) or motif I (His-(G106R)PRO/ENZ) of TbPRMT1PRO exhibited activity comparable with wild type complex. In stark contrast, complexes that carried mutations in either the double-E loop or motif I of TbPRMT1ENZ were catalytically dead (Fig. 7C, His-PRO/(G63R)ENZ and His-PRO/(E15Q)ENZ). Therefore, we conclude that TbPRMT1ENZ is the catalytic subunit of the TbPRMT1 heterotetramer.
Discussion
In this report, we describe a novel mode of PRMT organization and function. TbPRMT1, the major type I PRMT in the early branching eukaryote T. brucei (14), is a functional heterotetramer comprising two subunits. The previously reported TbPRMT1 protein (14, 22) (here renamed TbPRMT1ENZ) bears a striking sequence identity to human PRMT1. By itself, TbPRMT1ENZ exhibits no detectable activity under the conditions tested here despite retaining all critical PRMT motifs and displaying over 50% amino acid identity with human PRMT1. Nevertheless, TbPRMT1ENZ constitutes the catalytically active subunit of the tetramer. The second subunit of the tetramer previously referred to as TbPRMT3 (18, 22) is renamed here as TbPRMT1PRO. TbPRMT1PRO is catalytically inactive but is essential for the allosteric activation of TbPRMT1ENZ. This unique PRMT organization represents the first reported obligate heteromeric PRMT in any organism.
Two other T. brucei enzymes, deoxyhypusine synthase and AdoMetDC, both within the polyamine synthesis pathway, have been shown to function in a similar manner (19, 21, 32, 33, 39). In both cases, a catalytically inactive enzyme paralog termed prozyme dramatically stimulates the function of the true enzyme. This mode of organization and activation was coined the “prozyme paradigm” by Phillips and co-workers (21), thus leading to the TbPRMT1 naming convention described above. Inactive enzyme paralogs, called pseudoenzymes, are common in eukaryotes. By some estimates, 10% of human enzyme domains are predicted to be catalytically inactive, and this estimate is even higher for worms and flies at 15% (40). The importance of these proteins is becoming increasingly apparent as pseudoenzymes are assigned biological functions. For example, iRhoms, inactive enzymes resembling rhomboid proteases, act in the endoplasmic reticulum to promote degradation of specific proteins or maturation of others (41). In another example, ornithine decarboxylase homologs regulate activity of ornithine decarboxylases by countering their inhibitory antienzymes (42). Nevertheless, the degree of activation observed in trypanosome enzyme-prozyme pairs is exceptional. It has been postulated that the dramatic control of enzyme activities via the enzyme-prozyme mechanism was expanded in trypanosomes in response to their lack of transcriptional control and the almost exclusive reliance on posttranscriptional and posttranslational mechanisms of gene regulation (43). The present study is the first report of an enzyme-prozyme complex in trypanosomes outside the polyamine synthesis pathway. Our data suggest that this type of control mechanism may indeed be amplified in these organisms and that additional examples of enzyme-prozyme pairs likely await discovery in trypanosomes.
Novel means of enzyme regulation stemming from the enzyme-prozyme mechanism have been reported. For example, the AdoMetDC prozyme is present in limiting amounts, and it is rapidly up-regulated when AdoMetDC activity is chemically inhibited, allowing increased flux through the pathway (33). This regulation takes place at the level of prozyme translation, apparently triggered by decarboxylated AdoMet (44). TbPRMT1 is unlikely to be controlled by an analogous mechanism because the stabilities of TbPRMT1PRO and TbPRMT1ENZ proteins are mutually exclusive, similar to deoxyhypusine synthase and its prozyme (19). Intriguingly, however, TbPRMT1PRO, but not TbPRMT1ENZ, is reportedly phosphorylated (8, 45), bound to mRNA (46), and trafficked to stress granules in starved trypanosomes (47). This suggests that TbPRMT1PRO could be, in certain situations, operating independently of TbPRMT1ENZ, possibly as a homodimer (Fig. 6). An interesting observation made during our work was that, although TbPRMT1PRO can be readily purified from E. coli in the absence of the enzyme, TbPRMT1ENZ is notably unstable by itself and aggregates. This led us to postulate that TbPRMT1PRO may act as a chaperone toward TbPRMT1ENZ, and the mutual protein stability effect we observed could reflect different protein degradation mechanisms for the two subunits. In that case, sequestration of TbPRMT1PRO into stress granules would efficiently abolish TbPRMT1 activity for the duration of the stress and allow for a quick recovery once the cells enter a less hostile environment. Further research will focus on mechanisms of TbPRMT1 regulation, allowing us to conclusively determine the advantages T. brucei gains from utilizing a heteromeric PRMT1.
The discovery of a heterotetrameric, prozyme-activated PRMT may also have relevance to methyltransferase organization and regulation in higher eukaryotes. The most striking parallel with the prozyme paradigm emerged from the work on the human RNA methyltransferase complex METTL3-METTL14. In this complex, METTL3 constitutes the catalytic core and binds AdoMet but requires allosteric activation and stabilization by METTL14 (48, 49). Although METTL14 has been reported to exhibit weak in vitro methylation activity (50), the phylogenetic analysis suggests that the METTL14 catalytic core has lost its function (51). In another example, mammalian PRMT7 and PRMT9 both harbor two catalytic modules in tandem, forming a pseudodimer. The data suggest that in both cases only the N-terminal PRMT module contains conserved residues and binds AdoMet, although the inactive module is necessary for the enzyme activity (52–55), which is somewhat reminiscent of activation of TbPRMT1ENZ by TbPRMT1PRO. In regard to possible PRMT multimerization, we show here that TbPRMT1 forms a tetramer in vitro and sediments at a size corresponding to a tetramer in wild type cell lysate separated on a glycerol gradient. Based primarily on crystallographic studies, types I and III PRMTs are considered to function predominantly as homodimers with the exception of yeast PRMT1 (HMT1) (23, 27, 36, 37, 56, 57). However, two recent structural studies of human PRMT8 revealed a tetrameric enzyme bound to a single molecule of AdoMet per canonical dimer (31) or a possible octameric helical assembly (38). Furthermore, larger oligomers of type I PRMTs are often observed by both size exclusion chromatography and glycerol gradient fractionation, and some evidence suggests that the oligomerization may be necessary for PRMT activity (23, 28, 29, 38, 58, 59). Together, these findings imply that PRMT oligomerization may be more common than previously thought. Moreover, not only have PRMTs been shown to homooligomerize, but some studies support the possibility that PRMTs in mammals and plants have the ability to form heteromers. For example, mammalian PRMT2 can interact with PRMT1 both in vivo and in vitro, although the extent to which this occurs in vivo is unknown (60). Interestingly, recombinant PRMT2 enhanced the activity of recombinant PRMT1 up to 15-fold even when PRMT2 harbored inactivating mutations, suggesting a possible regulatory role for PRMT2. Human PRMT3 was first identified in a yeast two-hybrid screen using PRMT1 as bait (29), and the interaction of human PRMT1 and PRMT8 has been documented by several high throughput studies and directed experiments (61–64). In Arabidopsis, two CARM1 (PRMT4) homologs interact in vitro and in vivo, although homodimeric CARM1 complexes are also present (65). The functional significance of these heteromeric PRMT interactions remains to be explored. However, one possibility is that, although trypanosomes evolved to utilize heteromeric PRMT1 as their main arginine methylation enzyme, the same mechanism may be used in mammals and plants, albeit on a much smaller scale and under specific conditions, to provide an additional level of enzyme regulation.
Experimental Procedures
T. brucei Cell Culture and Generation of Cell Lines and Antibodies
TbPRMT1ENZ and TbPRMT1PRO doxycycline-inducible RNAi cell lines were generated as described (18). Briefly, plasmids were linearized using the NotI restriction site and electroporated into a BF single marker T. brucei strain (66) with the Amaxa NucleofactorTM system. Cells were cultured in HMI-11 medium supplemented with 10% FBS (67). Transformants were selected with 2.5 μg/ml phleomycin, and clones were obtained by limiting dilution. RNAi was induced for 4 days with 2.5 μg/ml doxycycline.
pLEW100-Myc-BirA* vector (68) was a kind gift from Graham Warren (Max F. Perutz Laboratories, University of Vienna and Medical University of Vienna, Vienna, Austria). The complete open reading frame for TbPRMT1ENZ was amplified using TbPRMT1ENZ 5′ AflII and TbPRMT1ENZ BamHI 3′ STOP primers (primers listed in Table 1) and cloned into the AflII and BamHI sites of pLEW100-Myc-BirA*. To facilitate readthrough of PRMT1 in these sites, the stop codon of Myc-BirA* was deleted using the primers BirA STOPdel 5′ and BirA STOPdel 3′.
TABLE 1.
List of primers
| Primer name | Primer sequence |
|---|---|
| 4A | AGCTTCAATTGCTCGAGGGATCCATGTGGAGCC |
| 4B | CCACATGGATCCCTCGAGCAATTGA |
| 5A | ATCCGCAGTTCGAGAAGCATCATCACCATCA |
| 5B | GGTGATGATGCTTCTCGAACTGCGGATGGCT |
| 6A | CCACCATCACTCTAGATAGA |
| 6B | GATCTCTATCTAGAGTGATGGTGGTGAT |
| 13A | TCGAGGGATCCGGTGGCTCCGGAGGTAGTA |
| 13B | GATCTACTACCTCCGGAGCCACCGGATCCC |
| qPCR1 TbPRMT1ENZ 5′ | CGTTCTCACTGCTTTGTTTG |
| qPCR1 TbPRMT1ENZ 3′ | TTTCCGAAGAAGTGGAAGAG |
| qPCR1 TbPRMT1PRO 5′ | GCGTGTATGGGGGTATTTAG |
| qPCR1 TbPRMT1PRO 3′ | AACATGTTCTTGCACACGAC |
| qPCR2 TbPRMT1ENZ 5′ | GGTTTAAGGGCACAATTCGC |
| qPCR2 TbPRMT1ENZ 3′ | ACACCTTTTGTTTCCCGAGG |
| qPCR2 TbPRMT1PRO 5′ | GAAGCAATGTCCTCACAGACT |
| qPCR2 TbPRMT1PRO 3′ | AAGCAGACAAACAGACGCT |
| TbPRMT1ENZ 5′ AflII | GACTTAAGATGACGGTGGACGCAAATGCCGCC |
| TbPRMT1ENZ BamHI 3′ STOP | GAGGATCCCTACCGCAGCCGAAAATCCTGG |
| BirA STOPdel 5′ | CGCAGAGAAGCTCGAGCTTAAGATGACGGTG |
| BirA STOPdel 3′ | CACCGTCATCTTAAGCTCGAGCTTCTCTGCG |
| pETDuet BamH1 frame fix 5′ | CATCATCACCACAGCGCAGGATCCATG |
| pETDuet BamH1 frame fix 3′ | CATGGATCCTGCGCTGTGGTGATGATG |
| TbPRMT1ENZ-ORF-BamHI-5′ | GTAGGATCCATGACGGTGGACGCAAATG |
| TbPRMT1ENZ -ORF-HindIII-3′ | GTAAAGCTTCTACCGCAGCCGAAAATCCT |
| TbPRMT1ENZ -ORF-NdeI-5′ | GTACATATGATGACGGTGGACGCAAATG |
| TbPRMT1ENZ -ORF-XhoI-3′ | GTACTCGAGCTACCGCAGCCGAAAATCCT |
| TbPRMT1PRO-ORF-BamHI-5′ | GTAGGATCCATGTCACCGAAGAAAAACTCGG |
| TbPRMT1PRO ORF-HindIII-3′ | GTAAAGCTTTCAATACCTTTGGTAGTTGTACGTG |
| TbPRMT1PRO-ORF-NdeI-5′ | GTACATATGATGTCACCGAAGAAAAACTCGG |
| TbPRMT1PRO-ORF-XhoI-3′ | GTACTCGAGTCAATACCTTTGGTAGTTGTACGTG |
| G63R TbPRMT1ENZ 5′ | CTTGATGTTGGTTGCAGGACGGGAATCCTTTC |
| G63R TbPRMT1ENZ 3′ | GAAAGGATTCCCGTCCTGCAACCAACATCAAG |
| G106R TbPRMT1PRO 5′ | GCATTTGGGGTGCAGAATGGGGTTGG |
| G106R TbPRMT1PRO 3′ | CCAACCCCATTCTGCACCCCAAATGC |
| E135Q TbPRMT1ENZ 5′ | TCCTACTCTATCAGTCTATGTTAAACACCG |
| E135Q TbPRMT1ENZ 3′ | CGGTGTTTAACATAGACTGATAGAGTAGGA |
| D180Q TbPRMT1PRO 5′ | GACCATTCTTGATCAATCAGCCACTGCTAGAAGAGGC |
| D180Q TbPRMT1PRO 3′ | GCCTCTTCTAGCAGTGGCTGATTGATCAAGAATGGTC |
The plasmid pLEW100-LSH with a C-terminal linker followed by the Strep(II) tag and eight repeats of His was constructed as follows. First, Strep(II)-His8 tag was cloned into pLEW100 (66) digested with HindIII/BamHI. 15 pmol of each primer used in the reaction described below was phosphorylated with T4 polynucleotide kinase (ThermoFisher) and ATP in manufacturer supplied buffer A for 30 min at 37 °C followed by inactivation for 10 min at 75 °C. Primer pairs were pooled and incubated at 94 °C for 1 min, 50 °C for 1 min, and 24 °C for 1 min to facilitate annealing. Primers 4A, 4B, 5A, 5B, 6A, and 6B were then pooled; the primer mixture (0.75 pmol of each primer) was ligated with 200 ng of restricted pLEW100 plasmid with T4 DNA ligase; and ligated plasmids were transformed into DH5-α E. coli. Linker was then added to the resulting plasmid by digesting pLEW100-Strep(II)-His8 with XhoI/BamHI and ligating in phosphorylated primers 13A and 13B as above. Both pLEW100-Myc-BirA*-PRMT1ENZ and pLEW100-PRMTPRO-LSH were linearized with NotI restriction enzyme and transfected into T. brucei strain 29-13 PF cells. Resultant cell lines were selected with 2.5 μg/ml phleomycin or 1 μg/ml puromycin, respectively. TbPRMT1ENZ, TbPRMT1PRO, TbPRMT6, and p22 antibodies were described previously (16, 18, 69).
Co-immunoprecipitation of PRMT1 Isoforms
The immunoprecipitation of TbPRMT1ENZ with the tagged TbPRMT1PRO-LSH was performed following a 4-day induction of TbPRMT1PRO-LSH using 2.5 μg/ml tetracycline. Cells were pelleted and resuspended in 6.5 ml of PBS with 0.1% Nonidet P-40, an EDTA-free protease inhibitor tablet (Roche Applied Science; one tablet/50 ml of lysis buffer), 50 μg/ml DNase I, and 1 mm CaCl2. Cells were lysed for 20 min at 4 °C followed by centrifugation at 14,400 × g for 15 min. The recovered supernatant was bound to TALON® cobalt beads (Clontech) for 1 h at 4 °C. The flow-through was collected followed by washing with 50 ml of PBS. Bound TbPRMT1PRO was then eluted with 150 mm imidazole. Eluted fractions were analyzed by Western blotting.
The reciprocal immunoprecipitation was performed using PF cells expressing Myc-BirA*-PRMT1ENZ. Cells were induced for 4 days using 2.5 μg/ml doxycycline and collected by centrifugation. The cell pellet was resuspended in 7.4 ml of PBS with 0.1% Nonidet P-40, an EDTA-free protease inhibitor tablet (one tablet/50 ml of lysis buffer), and 1 mm DTT. Cells were lysed for 20 min at 4 °C, and then the supernatant was collected by centrifugation at 14,400 × g for 15 min. The supernatant was then bound to 100 μl of anti-Myc-Sepharose beads (ICL, Inc.) for 3 h at 4 °C. The flow-through was collected, and the beads were rinsed with 20 ml of PBS with 0.1% Tween 20. Bound TbPRMT1ENZ was eluted with 100 mm glycine (pH 2.5) directly into 1 m Tris (pH 8.7) to neutralize the elutions. Eluted fractions were analyzed by Western blotting.
Recombinant Protein Cloning, Mutagenesis, and Expression
TbPRMT1 was expressed in BL21 E. coli strain using pETDuet-1 vector that allows for co-expression of two genes under separate T7 promoters. First, we inserted a single nucleotide into pETDuet-1 sequence between the His tag and BamHI site to correct a frameshift that would arise from using said site (primers pETDuet BamH1 frame fix 5′ and pETDuet BamH1 frame fix 3′). The TbPRMT1ENZ or TbPRMT1PRO full open reading frame, excluding the stop codon, was then amplified using TbPRMT1ENZ-ORF-BamHI-5′ and TbPRMT1ENZ-ORF-HindIII-3′ or TbPRMT1PRO-ORF-BamHI-5′ and TbPRMT1PRO ORF-HindIII-3′ primers and cloned into the first multiple cloning site (MCS) using BamHI/HindIII restriction sites (Fig. 3A, constructs a and c) to allow for expression of a single N-terminally His-tagged protein. To generate constructs b and d (Fig. 3A), the full open reading frame of the second TbPRMT1 subunit, including the stop codon, was amplified with TbPRMT1ENZ-ORF-NdeI-5′ and TbPRMT1ENZ-ORF-XhoI-3′ or TbPRMT1PRO-ORF-NdeI-5′ and TbPRMT1PRO-ORF-XhoI-3′ primers and cloned into the NdeI/XhoI site. Construct d was then used to generate catalytically inactive mutants. Overlapping primers containing the desired mutation were extended by KOD Hot Start DNA polymerase (Novagen) and digested overnight by DpnI prior to transformation.
Recombinant protein expression was induced in BL21 E. coli strain by adding 0.1 mm isopropyl 1-thio-β-d-galactopyranoside and subsequent growth at 18 °C overnight. Bacterial pellets were resuspended and incubated for 20 min in PBS (pH 7.4) supplemented with 0.1 mm PMSF, 50 μg/ml DNase I (Sigma), 1 mm CaCl2, and 50 μg/ml lysozyme and then sonicated. NaCl was then added at 1 m followed by three more rounds of sonication. The lysate was centrifuged for 20 min at 25,000 × g, and the supernatant was incubated for 3.5 h with TALON metal affinity resin (Clontech). The resin was washed, and proteins were eluted with PBS containing 200 mm imidazole. Samples were then dialyzed against PBS and flash frozen. The purification protocol of mutant protein included 10 μg/ml RNase A during lysis and 15 mm imidazole throughout. Rat GST-PRMT1 and MBP-RGG substrate were expressed and purified as described previously (14, 70).
The protein used for light scattering was expressed from a pETDuet-1 vector with TbPRMT1PRO in MCS1 (restriction sites NcoI and NotI) and TbPRMT1ENZ in MCS2 (NdeI and XhoI). The vector contained a PreScission protease (GE Healthcare) cleavage site for His tag removal from TbPRMT1PRO. Protein expression followed the same protocol as described previously for TbPRMT7 (36). Briefly, protein purification encompassed affinity chromatography on a nickel-nitrilotriacetic acid column (Qiagen), ion exchange chromatography on a HiTrap Heparin HP 5-ml column (GE Healthcare), and gel filtration on a HiLoad Superdex 200 16/60 column (GE Healthcare).
In Vitro Methylation Assays and Methylated Species Identification
In vitro methylation assays were performed as described previously (14). Briefly, 1.2 μm total PRMT was mixed with 0.2 or 2 μm substrate and 0.7 μm [3H]AdoMet (55–85 Ci (2.03–3.15 TBq)/mmol; PerkinElmer Life Sciences) in PBS (pH 7.4) containing 1 mm PMSF in a final volume of 50 μl. Reactions we carried out for ∼18 h at 22 °C. After separation by SDS-PAGE, the gel was incubated with EN3HANCE solution (PerkinElmer Life Sciences) for 1 h, dried, and exposed to film at −80 °C overnight. The samples shown in Fig. 7C also contained 2 mm DTT, which led to substantial reduction of necessary exposure time from ∼18 to 1 h.
Time Course of His-PRO/ENZ Complex Activity
Under our experimental conditions, we showed that product formation from the His-PRO/ENZ complex was linearly dependent upon time for at least the 2-hour reaction time used for chromatographic analysis in Fig. 5B. Five PBS-based reactions containing 60 pmol of His-PRO/ENZ, 10 pmol of MBP-RGG substrate, and 1 mm PMSF were initiated by addition of 2 μCi of [3H]AdoMet (55–85 Ci (2.03–3.15 TBq)/mmol). The total reaction volume was 50 μl. In vitro methylation was allowed to progress for 1, 3, 5, 10, and 20 h; reactions were stopped by the addition of SDS sample buffer. After separation by SDS-PAGE, the gel was incubated with EN3HANCE solution for 1 h, dried, and exposed to film at −80 °C overnight.
Identification of methylarginine species was performed as described previously (36). Briefly, His-TbPRMT1PRO/ENZ was incubated with substrate and TCA-precipitated. The resulting protein mixture was digested by acid hydrolysis. Amino acids were analyzed by cation exchange chromatography in the presence of unlabeled ADMA, SDMA, and MMA standards.
AdoMet-PRMT UV Cross-linking
To cross-link PRMTs to [3H]AdoMet, 1.6 μm total PRMT was mixed with 1.4 μm [3H]AdoMet (78 Ci/mmol) in 50 mm phosphate buffer (pH 7.4) in the presence of 5 mm DTT in a final volume of 50 μl and incubated at 4 °C for ∼18 h. Samples were then UV cross-linked for 10 min on ice 1 cm from the UV lamp using a UV Stratalinker 2400 (Stratagene). After separation of proteins by SDS-PAGE, the gel was Coomassie-stained, incubated with EN3HANCE solution for 1 h, dried, and exposed to film at −80 °C for 1 week.
To determine the specificity of UV cross-linking assay to AdoMet, 1.6 μm total PRMT was mixed with 1.4 μm [3H]AdoMet in 50 mm phosphate buffer (pH 7.4) in the presence of 5 mm DTT and incubated at 22 °C for ∼10 min (50-μl reaction volume). Following initial incubation, a 50× excess of dATP or unlabeled AdoMet was added to control reactions. Samples were then incubated at 4 °C for ∼18 h. Samples were UV cross-linked for 10 min on ice 1 cm from the UV lamp using a UV Stratalinker 2400. After separation of proteins by SDS-PAGE, the gel was Coomassie-stained, incubated with EN3HANCE solution for 1 h, dried, and exposed to film at −80 °C for 1 week.
Multiangle Light Scattering
Untagged TbPRMT1ENZ/PRO and TbPRMT1PRO were characterized by multiangle light scattering following size exclusion chromatography. The identity of the proteins used for light scattering analysis was confirmed by mass spectrometry analysis. Protein at 50 μm (TbPRMT1ENZ/PRO) and 130 μm (TbPRMT1PRO) was injected onto a Superdex 200 10/300 GL size exclusion chromatography column equilibrated in a buffer containing 20 mm HEPES (pH 7.5), 200 mm NaCl, and 0.5 mm tris(2-carboxyethyl)phosphine. The chromatography system was connected in series with an 18-angle light scattering detector (DAWN HELEOS) and refractive index detector (OptilabrEX) (Wyatt Technology). Data were collected every second at a flow rate of 0.15 ml/min at 25 °C. Data analysis was carried out using the program ASTRA, yielding the molar mass and mass distribution (polydispersity) of the sample.
Quantitative RT-PCR
TbPRMT1ENZ and TbPRMT1PRO RNAi cell lines were grown for 4 days in the absence or presence of 2.5 (replicate 1) or 4 μg/ml (replicate 2) doxycycline. 250 ml of each cell culture was then harvested and resuspended in 1 ml of TRIzol reagent (Ambion). RNA was isolated according to the manufacturer's instructions and subsequently re-extracted by an acidic phenol RNA purification procedure. 10 μg of RNA/sample was DNase-treated using an Ambion DNA-free kit (Invitrogen) and re-extracted with acidic phenol. cDNA was synthesized using random hexamer primers with an iScript Select cDNA Synthesis kit (Bio-Rad) according to the manufacturer's instructions. Levels of each PRMT mRNA were then assayed by qRT-PCR using qPCR1 TbPRMT1ENZ and qPCR1 TbPRMT1PRO primer pairs in the first replicate. Second replicate qPCR used primer pairs qPCR2 TbPRMT1ENZ and qPCR2 TbPRMT1PRO. Between samples, mRNA levels were normalized to the levels of β-tubulin mRNA.
Glycerol Gradient Sedimentation
5–20% glycerol gradients were prepared as follows. 5.5 ml of buffer A (20 mm HEPES (pH 7.9), 10 mm MgCl2, 50 mm KCl, 1 mm EDTA, and 20% glycerol) was poured into an ultracentrifugation tube and frozen at −80 °C. 5.5 ml of buffer B (20 mm HEPES (pH 7.9), 10 mm MgCl2, 50 mm KCl, 1 mm EDTA, and 5% glycerol) was layered on top of the frozen buffer A. The tubes then underwent four freeze/thaw cycles at −80 °C to create a linear glycerol gradient. 1 × 109 wild type 29-13 procyclic form T. brucei cells per sample were harvested. The cell pellet was resuspended in 0.5 ml of lysis buffer (10 mm Tris (pH 8.0), 150 mm NaCl, 0.1% Nonidet P-40, an EDTA-free protease inhibitor tablet (one tablet/50 ml of lysis buffer), 50 μg/ml DNase I, and 1 mm CaCl2). Cells were lysed for 20 min at 4 °C by addition of Triton X-100 to a final concentration of 1% (v/v). Lysates were cleared by centrifugation for 15 min at 15,000 × g. Cleared lysate was loaded on top of a 5–20% gradient and centrifuged at 32,000 rpm for 16 h in an SW41 Beckman rotor. Size standards consisting of proteins with known sedimentation coefficients were run on a parallel gradient (cytochrome c (1.9S), BSA (4.3S), yeast alcohol dehydrogenase (7.4S), catalase (11.3S), and thyroglobulin (19S)). 0.5-ml fractions were collected and probed with α-TbPRMT antibodies. Size standard fraction contents were visualized by Coomassie staining.
Author Contributions
L. K. R., L. K., S. G. C., and E. W. D. conceived the project. L. K., E. W. D., J. C. F., and K. J. carried out the experiments. All authors contributed to interpretation of the data. L. K. and L. K. R. wrote the manuscript, and S. G. C. and E. W. D. provided valuable support with the writing.
Acknowledgments
We thank Kyle Smith (University at Buffalo) and Doris Berman (Rockefeller University) for technical assistance, Günter Blobel (Rockefeller University) for support, and David King (University of California, Berkeley) for mass spectrometry analysis.
This work was supported by National Institutes of Health Grants R01 AI060260 (to L. K. R.) and R01 GM026020 (to S. G. C.), National Institutes of Health Ruth L. Kirschstein National Service Award GM007185 to (K. J.), and American Heart Association Predoctoral Fellowship 15PRE24480155 (to L. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
K. Lott and L. Read, unpublished results.
- PF
- procyclic form
- PRMT
- protein arginine methyltransferase
- Tb
- T. brucei
- MMA
- ω-NG-monomethylarginine
- ADMA
- ω-NG,NG-asymmetric dimethylarginine
- SDMA
- ω-NG,N′G-symmetric dimethylarginine
- BF
- bloodstream form
- LSH
- linker-Strep-His
- AdoMet
- S-adenosyl-l-methionine
- MBP
- maltose-binding protein
- MALS
- multiangle light scattering
- AdoMetDC
- AdoMet decarboxylase
- MCS
- multiple cloning site
- qRT-PCR
- quantitative RT-PCR
- qPCR
- quantitative PCR.
References
- 1. Keating J., Yukich J. O., Sutherland C. S., Woods G., and Tediosi F. (2015) Human African trypanosomiasis prevention, treatment and control costs: a systematic review. Acta Trop. 150, 4–13 [DOI] [PubMed] [Google Scholar]
- 2. Bilbe G. (2015) Infectious diseases. Overcoming neglect of kinetoplastid diseases. Science 348, 974–976 [DOI] [PubMed] [Google Scholar]
- 3. Daniels J. P., Gull K., and Wickstead B. (2010) Cell biology of the trypanosome genome. Microbiol. Mol. Biol. Rev. 74, 552–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferguson M. A. (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112, 2799–2809 [DOI] [PubMed] [Google Scholar]
- 5. Read L. K., Lukeš J., and Hashimi H. (2016) Trypanosome RNA editing: the complexity of getting U in and taking U out. Wiley Interdiscip. Rev. RNA 7, 33–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cavalier-Smith T. (2010) Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol. Lett. 6, 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vickerman K. (1985) Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull. 41, 105–114 [DOI] [PubMed] [Google Scholar]
- 8. Urbaniak M. D., Martin D. M., and Ferguson M. A. (2013) Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J. Proteome Res. 12, 2233–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lott K., Li J., Fisk J. C., Wang H., Aletta J. M., Qu J., and Read L. K. (2013) Global proteomic analysis in trypanosomes reveals unique proteins and conserved cellular processes impacted by arginine methylation. J. Proteomics 91, 210–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fisk J. C., Li J., Wang H., Aletta J. M., Qu J., and Read L. K. (2013) Proteomic analysis reveals diverse classes of arginine methylproteins in mitochondria of trypanosomes. Mol. Cell. Proteomics 12, 302–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lott K., Mukhopadhyay S., Li J., Wang J., Yao J., Sun Y., Qu J., and Read L. K. (2015) Arginine methylation of DRBD18 differentially impacts its opposing effects on the trypanosome transcriptome. Nucleic Acids Res. 43, 5501–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fisk J. C., Sayegh J., Zurita-Lopez C., Menon S., Presnyak V., Clarke S. G., and Read L. K. (2009) A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei. J. Biol. Chem. 284, 11590–11600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pelletier M., Pasternack D. A., and Read L. K. (2005) In vitro and in vivo analysis of the major type I protein arginine methyltransferase from Trypanosoma brucei. Mol. Biochem. Parasitol. 144, 206–217 [DOI] [PubMed] [Google Scholar]
- 15. Pasternack D. A., Sayegh J., Clarke S., and Read L. K. (2007) Evolutionarily divergent type II protein arginine methyltransferase in Trypanosoma brucei. Eukaryot. Cell 6, 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fisk J. C., Zurita-Lopez C., Sayegh J., Tomasello D. L., Clarke S. G., and Read L. K. (2010) TbPRMT6 is a type I protein arginine methyltransferase that contributes to cytokinesis in Trypanosoma brucei. Eukaryot. Cell 9, 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang J., Frankel A., Cook R. J., Kim S., Paik W. K., Williams K. R., Clarke S., and Herschman H. R. (2000) PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 275, 7723–7730 [DOI] [PubMed] [Google Scholar]
- 18. Lott K., Zhu L., Fisk J. C., Tomasello D. L., and Read L. K. (2014) Functional interplay between protein arginine methyltransferases in Trypanosoma brucei. Microbiologyopen 3, 595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nguyen S., Jones D. C., Wyllie S., Fairlamb A. H., and Phillips M. A. (2013) Allosteric activation of trypanosomatid deoxyhypusine synthase by a catalytically dead paralog. J. Biol. Chem. 288, 15256–15267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hashimi H., Cicová Z., Novotná L., Wen Y. Z., and Lukes J. (2009) Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA 15, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Willert E. K., Fitzpatrick R., and Phillips M. A. (2007) Allosteric regulation of an essential trypanosome polyamine biosynthetic enzyme by a catalytically dead homolog. Proc. Natl. Acad. Sci. U.S.A. 104, 8275–8280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fisk J. C., and Read L. K. (2011) Protein arginine methylation in parasitic protozoa. Eukaryot. Cell 10, 1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X., and Cheng X. (2003) Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 11, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y. C., Wang J. D., Chen C. H., Chen Y. W., and Li C. (2015) A novel BLAST-based relative distance (BBRD) method can effectively group members of protein arginine methyltransferases and suggest their evolutionary relationship. Mol. Phylogenet. Evol. 84, 101–111 [DOI] [PubMed] [Google Scholar]
- 25. Gary J. D., and Clarke S. (1998) RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid. Res. Mol. Biol. 61, 65–131 [DOI] [PubMed] [Google Scholar]
- 26. Hung C. J., Chen D. H., Shen Y. T., Li Y. C., Lin Y. W., Hsieh M., and Li C. (2007) Characterization of protein arginine methyltransferases in porcine brain. J. Biochem. Mol. Biol. 40, 617–624 [DOI] [PubMed] [Google Scholar]
- 27. Troffer-Charlier N., Cura V., Hassenboehler P., Moras D., and Cavarelli J. (2007) Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. EMBO J. 26, 4391–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin W. J., Gary J. D., Yang M. C., Clarke S., and Herschman H. R. (1996) The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 271, 15034–15044 [DOI] [PubMed] [Google Scholar]
- 29. Tang J., Gary J. D., Clarke S., and Herschman H. R. (1998) PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J. Biol. Chem. 273, 16935–16945 [DOI] [PubMed] [Google Scholar]
- 30. Rho J., Choi S., Seong Y. R., Cho W. K., Kim S. H., and Im D. S. (2001) Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J. Biol. Chem. 276, 11393–11401 [DOI] [PubMed] [Google Scholar]
- 31. Lee W. C., Lin W. L., Matsui T., Chen E. S., Wei T. Y., Lin W. H., Hu H., Zheng Y. G., Tsai M. D., and Ho M. C. (2015) Protein arginine methyltransferase 8: tetrameric structure and protein substrate specificity. Biochemistry 54, 7514–7523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Velez N., Brautigam C. A., and Phillips M. A. (2013) Trypanosoma brucei S-adenosylmethionine decarboxylase N terminus is essential for allosteric activation by the regulatory subunit prozyme. J. Biol. Chem. 288, 5232–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Willert E. K., and Phillips M. A. (2008) Regulated expression of an essential allosteric activator of polyamine biosynthesis in African trypanosomes. PLoS Pathog. 4, e1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neault M., Mallette F. A., Vogel G., Michaud-Levesque J., and Richard S. (2012) Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 40, 9513–9521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McBride A. E., Weiss V. H., Kim H. K., Hogle J. M., and Silver P. A. (2000) Analysis of the yeast arginine methyltransferase Hmt1p/Rmt1p and its in vivo function. Cofactor binding and substrate interactions. J. Biol. Chem. 275, 3128–3136 [DOI] [PubMed] [Google Scholar]
- 36. Debler E. W., Jain K., Warmack R. A., Feng Y., Clarke S. G., Blobel G., and Stavropoulos P. (2016) A glutamate/aspartate switch controls product specificity in a protein arginine methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 113, 2068–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang X., Zhou L., and Cheng X. (2000) Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 19, 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Toma-Fukai S., Kim J. D., Park K. E., Kuwabara N., Shimizu N., Krayukhina E., Uchiyama S., Fukamizu A., and Shimizu T. (2016) Novel helical assembly in arginine methyltransferase 8. J. Mol. Biol. 428, 1197–1208 [DOI] [PubMed] [Google Scholar]
- 39. Willert E. K., and Phillips M. A. (2009) Cross-species activation of trypanosome S-adenosylmethionine decarboxylase by the regulatory subunit prozyme. Mol. Biochem. Parasitol. 168, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pils B., and Schultz J. (2004) Inactive enzyme-homologues find new function in regulatory processes. J. Mol. Biol. 340, 399–404 [DOI] [PubMed] [Google Scholar]
- 41. Adrain C., and Freeman M. (2012) New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat. Rev. Mol. Cell Biol. 13, 489–498 [DOI] [PubMed] [Google Scholar]
- 42. Ivanov I. P., Firth A. E., and Atkins J. F. (2010) Recurrent emergence of catalytically inactive ornithine decarboxylase homologous forms that likely have regulatory function. J. Mol. Evol. 70, 289–302 [DOI] [PubMed] [Google Scholar]
- 43. Nguyen S., Leija C., Kinch L., Regmi S., Li Q., Grishin N. V., and Phillips M. A. (2015) Deoxyhypusine modification of eukaryotic translation initiation factor 5A (eIF5A) is essential for Trypanosoma brucei growth and for expression of polyprolyl-containing proteins. J. Biol. Chem. 290, 19987–19998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiao Y., Nguyen S., Kim S. H., Volkov O. A., Tu B. P., and Phillips M. A. (2013) Product feedback regulation implicated in translational control of the Trypanosoma brucei S-adenosylmethionine decarboxylase regulatory subunit prozyme. Mol. Microbiol. 88, 846–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nett I. R., Martin D. M., Miranda-Saavedra D., Lamont D., Barber J. D., Mehlert A., and Ferguson M. A. (2009) The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol. Cell. Proteomics 8, 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lueong S., Merce C., Fischer B., Hoheisel J. D., and Erben E. D. (2016) Gene expression regulatory networks in Trypanosoma brucei: insights into the role of the mRNA-binding proteome. Mol. Microbiol. 100, 457–471 [DOI] [PubMed] [Google Scholar]
- 47. Fritz M., Vanselow J., Sauer N., Lamer S., Goos C., Siegel T. N., Subota I., Schlosser A., Carrington M., and Kramer S. (2015) Novel insights into RNP granules by employing the trypanosome's microtubule skeleton as a molecular sieve. Nucleic Acids Res. 43, 8013–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang P., Doxtader K. A., and Nam Y. (2016) Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., Gong Z., Wang Q., Huang J., Tang C., Zou T., and Yin P. (2016) Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578 [DOI] [PubMed] [Google Scholar]
- 50. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., Dai Q., Chen W., and He C. (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Iyer L. M., Zhang D., and Aravind L. (2016) Adenine methylation in eukaryotes: apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays 38, 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miranda T. B., Miranda M., Frankel A., and Clarke S. (2004) PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J. Biol. Chem. 279, 22902–22907 [DOI] [PubMed] [Google Scholar]
- 53. Hasegawa M., Toma-Fukai S., Kim J. D., Fukamizu A., and Shimizu T. (2014) Protein arginine methyltransferase 7 has a novel homodimer-like structure formed by tandem repeats. FEBS Lett. 588, 1942–1948 [DOI] [PubMed] [Google Scholar]
- 54. Hadjikyriacou A., Yang Y., Espejo A., Bedford M. T., and Clarke S. G. (2015) Unique features of human protein arginine methyltransferase 9 (PRMT9) and its substrate RNA splicing factor SF3B2. J. Biol. Chem. 290, 16723–16743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang Y., Hadjikyriacou A., Xia Z., Gayatri S., Kim D., Zurita-Lopez C., Kelly R., Guo A., Li W., Clarke S. G., and Bedford M. T. (2015) PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. 6, 6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weiss V. H., McBride A. E., Soriano M. A., Filman D. J., Silver P. A., and Hogle J. M. (2000) The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat. Struct. Biol. 7, 1165–1171 [DOI] [PubMed] [Google Scholar]
- 57. Wang C., Zhu Y., Chen J., Li X., Peng J., Chen J., Zou Y., Zhang Z., Jin H., Yang P., Wu J., Niu L., Gong Q., Teng M., and Shi Y. (2014) Crystal structure of arginine methyltransferase 6 from Trypanosoma brucei. PLoS One 9, e87267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Herrmann F., Pably P., Eckerich C., Bedford M. T., and Fackelmayer F. O. (2009) Human protein arginine methyltransferases in vivo—distinct properties of eight canonical members of the PRMT family. J. Cell Sci. 122, 667–677 [DOI] [PubMed] [Google Scholar]
- 59. Lim Y., Kwon Y. H., Won N. H., Min B. H., Park I. S., Paik W. K., and Kim S. (2005) Multimerization of expressed protein-arginine methyltransferases during the growth and differentiation of rat liver. Biochim. Biophys. Acta 1723, 240–247 [DOI] [PubMed] [Google Scholar]
- 60. Pak M. L., Lakowski T. M., Thomas D., Vhuiyan M. I., Hüsecken K., and Frankel A. (2011) A protein arginine N-methyltransferase 1 (PRMT1) and 2 heteromeric interaction increases PRMT1 enzymatic activity. Biochemistry 50, 8226–8240 [DOI] [PubMed] [Google Scholar]
- 61. Weimann M., Grossmann A., Woodsmith J., Özkan Z., Birth P., Meierhofer D., Benlasfer N., Valovka T., Timmermann B., Wanker E. E., Sauer S., and Stelzl U. (2013) A Y2H-seq approach defines the human protein methyltransferase interactome. Nat. Methods 10, 339–342 [DOI] [PubMed] [Google Scholar]
- 62. Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., et al. (2015) The BioPlex Network: a systematic exploration of the human interactome. Cell 162, 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee J., Sayegh J., Daniel J., Clarke S., and Bedford M. T. (2005) PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 280, 32890–32896 [DOI] [PubMed] [Google Scholar]
- 64. Rolland T., Taşan M., Charloteaux B., Pevzner S. J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R., Kamburov A., Ghiassian S. D., Yang X., Ghamsari L., Balcha D., et al. (2014) A proteome-scale map of the human interactome network. Cell 159, 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Niu L., Zhang Y., Pei Y., Liu C., and Cao X. (2008) Redundant requirement for a pair of PROTEIN ARGININE METHYLTRANSFERASE4 homologs for the proper regulation of Arabidopsis flowering time. Plant Physiol. 148, 490–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wirtz E., Leal S., Ochatt C., and Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 67. Baltz T., Baltz D., Giroud C., and Crockett J. (1985) Cultivation in a semi-defined medium of animal infective forms of Trypanosoma brucei, T. equiperdum, T. evansi, T. rhodesiense and T. gambiense. EMBO J. 4, 1273–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morriswood B., Havlicek K., Demmel L., Yavuz S., Sealey-Cardona M., Vidilaseris K., Anrather D., Kostan J., Djinovic-Carugo K., Roux K. J., and Warren G. (2013) Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot. Cell 12, 356–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hayman M. L., Miller M. M., Chandler D. M., Goulah C. C., and Read L. K. (2001) The trypanosome homolog of human p32 interacts with RBP16 and stimulates its gRNA binding activity. Nucleic Acids Res. 29, 5216–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pelletier M., Xu Y., Wang X., Zahariev S., Pongor S., Aletta J. M., and Read L. K. (2001) Arginine methylation of a mitochondrial guide RNA binding protein from Trypanosoma brucei. Mol. Biochem. Parasitol. 118, 49–59 [DOI] [PubMed] [Google Scholar]