Abstract
Disruption of the O-mannosylation pathway involved in functional glycosylation of α-dystroglycan gives rise to congenital muscular dystrophies. Protein O-linked mannose β-1,4-N-acetylglucosaminyltransferase 2 (POMGNT2) catalyzes the first step toward the functional matriglycan structure on α-dystroglycan that is responsible for binding extracellular matrix proteins and certain arenaviruses. Alternatively, protein O-linked mannose β-1,2-N-acetylglucosaminyltransferase 1 (POMGNT1) catalyzes the first step toward other various glycan structures present on α-dystroglycan of unknown function. Here, we demonstrate that POMGNT1 is promiscuous for O-mannosylated peptides, whereas POMGNT2 displays significant primary amino acid selectivity near the site of O-mannosylation. We define a POMGNT2 acceptor motif, conserved among 59 vertebrate species, in α-dystroglycan that when engineered into a POMGNT1-only site is sufficient to convert the O-mannosylated peptide to a substrate for POMGNT2. Additionally, an acceptor glycopeptide is a less efficient substrate for POMGNT2 when two of the conserved amino acids are replaced. These findings begin to define the selectivity of POMGNT2 and suggest that this enzyme functions as a gatekeeper enzyme to prevent the vast majority of O-mannosylated sites on proteins from becoming modified with glycan structures functional for binding laminin globular domain-containing proteins.
Keywords: dystroglycan, enzyme kinetics, glycobiology, glycoprotein, glycosyltransferase
Introduction
Congenital muscular dystrophy (CMD)3 describes a family of genetic, degenerative diseases characterized by contractures, myopathy, and in some cases central nervous system abnormalities. Many CMDs are caused by defects in the formation of a functional dystrophin glycoprotein complex that links the actin cytoskeleton to the extracellular matrix (ECM). α-Dystroglycan (α-DG), encoded by the DAG1 gene, provides the physical link to laminin globular (LG) domain-containing proteins in the ECM (1); however, there are only a few known mutations in the DAG1 coding sequence that lead to CMD (2). A subset of CMDs, termed secondary dystroglycanopathies, is caused by mutations in genes encoding enzymes responsible for glycosylating α-DG in its mucin-like domain (residues 313–489). These secondary dystroglycanopathies range in severity from mild limb-girdle muscular dystrophy to the more severe Walker-Warburg syndrome (3–5). The causal genes for secondary dystroglycanopathies have been identified as encoding enzymes in the pathway associated with the biosynthesis of the O-mannosyl (O-Man) glycans (6, 7).
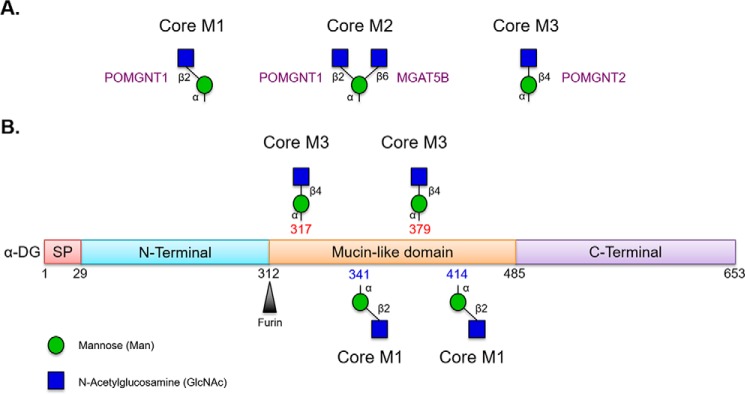
The O-mannosylation pathway begins in the endoplasmic reticulum (ER) where a complex of POMT1 and POMT2 catalyzes the transfer of mannose from dolicholphosphomannose to serine and threonine residues in an α-linkage to α-DG (8) and presumably a handful of other proteins (9). Bifurcation of the pathway then occurs by the addition of an N-acetylglucosamine (GlcNAc) in either a β2 or a β4 linkage (see Fig. 1). Two enzymes, POMGNT1 and POMGNT2, respectively, mediate these additions. In most cases on α-DG, a β-1,2-linked GlcNAc residue can be added to the initial mannose residue by POMGNT1 in the cis-Golgi (10). This core M1 structure can be branched by another GlcNAc addition to give rise to the core M2 glycan structure (11). Much more rarely on α-DG, POMGNT2 will add a β-1,4-linked GlcNAc to the initial mannose residue in the ER, leading to the formation of the core M3 glycan structure (see Fig. 1).
FIGURE 1.
Core O-Man structures on α-dystroglycan. A, POMGNT1 is responsible for generating the M1 core glycan structure that can be branched by MGAT5B to generate the M2 core, whereas POMGNT2 is responsible for generating the M3 core glycan structure. B, schematic of known O-mannosylated sites on α-dystroglycan addressed in this study. Thr-317 and Thr-379 are elaborated with the M3 core glycan structure, whereas Thr-341 and Thr-414 are elaborated with M1 core glycan structures that can be further elaborated to core M2 glycan structures. Glycan symbols follow guidelines outlined in Ref. 38. SP, signal peptide.
After POMGNT2-mediated β-1,4-GlcNAc addition, the glycan is subjected to further extension with a β-1,3-linked N-acetylgalactosamine by B3GALNT2 and phosphorylation of the reducing end mannose at the 6-position by POMK to give rise to the phosphotrisaccharide core M3 glycan structure while still in the ER (12–14). From here, it has been recently demonstrated that FKTN and FKRP appear to be responsible for extending the core M3 phosphotrisaccharide in the Golgi by addition of two ribitol phosphate units in phosphodiester linkages (15). TMEM5 then apparently adds a xylose to the distal ribitol that is followed by B4GAT1-catalyzed addition of glucuronic acid in a β-1,4 linkage to the xylose (16, 17). This primer permits LARGE1 to catalyze the addition of a repeating disaccharide (α-1,3-linked xylose-β-1,3-linked glucuronic acid) that is the functional component, termed matriglycan, responsible for binding to LG domains of ECM proteins (2, 18, 19).
Human α-DG has at least 25 O-mannosylation sites (20–22). The majority of the O-mannosylation sites on α-DG are populated by core M1 and M2 glycan structures via the action of POMGNT1 (M1) followed by MGAT5B (M2) (20–23). Site mapping studies have identified only two positions, Thr-317 and Thr-379, on α-DG with M3 core structures, although some evidence suggests 319 and 381 may also be sites of M3 modification (Fig. 1) (12, 18, 20, 21). Paradoxically, from a spatial-temporal perspective, O-Man-modified α-DG encounters POMGNT2 in the ER before POMGNT1 in the cis-Golgi but is preferentially modified by POMGNT1. This led us to hypothesize that POMGNT2 must demonstrate substrate selectivity beyond simply an O-Man-modified amino acid.
Here, we explore the specificity of POMGNT2 and compare it with POMGNT1. We synthesized multiple O-mannosylated peptides derived from known M1- and M3-modified sites of α-DG and tested their ability to be acceptor substrates for the two enzymes. POMGNT2 displays selectivity based on the primary amino acid sequence in proximity to the site of O-mannosylation, whereas POMGNT1 is promiscuous. We identified a sequence motif, highly conserved in vertebrates, in α-DG that appears to modulate POMGNT2 substrate specificity in vitro. We demonstrated sufficiency of the extended motif by engineering the sequence into a typical M1 O-mannosylated peptide that resulted in it being a POMGNT2 acceptor. We also demonstrate that replacement of conserved amino acids compromises an M3 peptide for extension by POMGNT2. Intriguingly, a conservative degenerate sequence based on our identified motif is present in several human membrane/secreted proteins.
Results
Acceptor Selectivity of POMGNT1 and POMGNT2 Using Synthetic α-DG Glycopeptides
To identify primary amino acid determinants of POMGNT2 selectivity, we used solid-phase peptide synthesis to generate synthetic glycopeptides whose sequences are those from known O-mannosylated regions of human α-DG (Table 1). Direct physical evidence for core M3 extension at position 379 in α-DG has been shown previously (24), whereas the threonine at position 341 in α-DG has been demonstrated as a POMT1/POMT2 acceptor that does not carry an M3 core (25). We selected these two sites (379 and 341) because we predicted that their extensions differ in core glycan structure whereas their immediate primary amino acid sequences share a similar Thr(-O-Man)-Pro-Thr (TPT) motif. The synthetic glycopeptides were designed to be 21 amino acids in length with the mannosylated threonine as the central residue (residue 11) to evaluate nearby C-terminal and N-terminal amino acid determinants (Table 1).
TABLE 1.
Comparison of POMGNT1 and POMGNT2 kinetics with various M1 and M3 synthetic glycopeptide acceptors
An asterisk in the acceptor sequence indicates the mannosylated threonine residue. Kinetic parameters of Man341 and Man414 with POMGNT2 were not measurable as indicated by the dash.
| Acceptor | Acceptor sequence | Core glycan structure | Km | kcat | kcat/Km |
|---|---|---|---|---|---|
| mm | s−1 | m−1 s−1 | |||
| POMGNT1 | |||||
| Man317 | PKRVRRQIHAT*PTPVTAIGPP | M3 | 1.2 ± 0.1 | 13 ± 0.5 | 10 × 103 |
| Man379 | TIRTRGAIIQT*PTLGPIQPTR | M3 | 2.6 ± 0.4 | 32 ± 3.3 | 12 × 103 |
| Man379-ETP | TIETPGAIIQT*PTLGPIQPTR | Modified M3 | 0.9 ± 0.1 | 11 ± 0.4 | 12 × 103 |
| Man341 | IQEPPSRIVPT*PTSPAIAPPT | M1 | 0.1 ± 0.04 | 1.0 ± 0.1 | 10 × 103 |
| Man341-RPR | IQRPRSRIVPT*PTSPAIAPPT | Modified M1 | 3.2 ± 0.7 | 13 ± 1.5 | 4.0 × 103 |
| Man414 | YVEPT*AV | M1 | 11 ± 3.0 | 16 ± 3.3 | 1.5 × 103 |
| POMGNT2 | |||||
| Man317 | PKRVRRQIHAT*PTPVTAIGPP | M3 | 2.2 ± 0.2 | 12 ± 0.5 | 5.5 × 103 |
| Man379 | TIRTRGAIIQT*PTLGPIQPTR | M3 | 0.8 ± .04 | 16 ± 1.4 | 20 × 103 |
| Man379-ETP | TIETPGAIIQT*PTLGPIQPTR | Modified M3 | 2.9 ± 0.7 | 11 ± 1.4 | 3.8 × 103 |
| Man341 | IQEPPSRIVPT*PTSPAIAPPT | M1 | — | — | — |
| Man341-RPR | IQRPRSRIVPT*PTSPAIAPPT | Modified M1 | 6.0 ± 1.1 | 3.3 ± 1.0 | 0.6 × 103 |
| Man414 | YVEPT*AV | M1 | — | — | — |
| ShortMan379 | GAIIQT*PTLGPIQPTR | Modified M3 | 0.8 ± 0.3 | 10 ± 1.4 | 12 × 103 |
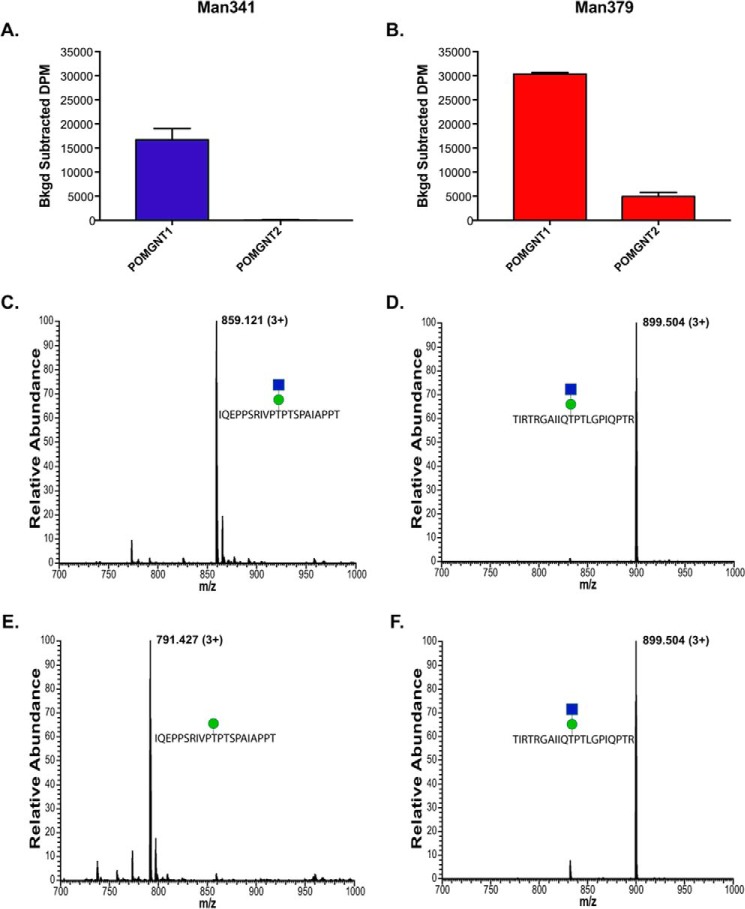
To establish whether the synthetic glycopeptides were substrates for POMGNT1 and POMGNT2, we performed overnight radioactive transfer assays. Recombinant human POMGNT1 catalyzed GlcNAc transfer to both the Man341 and Man379 synthetic glycopeptides (Fig. 2, A and B). To confirm the composition of POMGNT1 reaction products, parallel transfer assays using non-radiolabeled UDP-GlcNAc were performed using the Man341 and Man379 glycopeptides as acceptor substrates, and the reaction products were analyzed by mass spectrometry (MS) (Fig. 2, C and D). The observed peaks at 859.121 and 899.504 m/z in the full FTMS correspond to the addition of a N-acetylhexosamine residue (+203) to the Man341 and Man379 glycopeptides, respectively. Thus, POMGNT1 will extend the mannose in synthetic glycopeptides at positions 341 and 379 of the α-DG sequence in vitro. These results clearly demonstrate that POMGNT1 exhibits minimal acceptor selectivity between core M1 and M3 sites on these two α-DG-derived glycopeptides.
FIGURE 2.
Unlike POMGNT1, POMGNT2 exhibits acceptor selectivity. A and B, radioactive assay of POMGNT1 and POMGNT2 activity with Man341 (A), an M1 acceptor, and Man379 (B), an M3 acceptor. Transfer is measured in background-corrected dpm. Error bars represent S.E. of three replicates. C and D, FTMS spectra verifying POMGNT1-extended Man341 (1.16-ppm mass accuracy) (C) and POMGNT1-extended Man379 (2.53-ppm mass accuracy) (D). The green circle represents a mannose, and the blue square represents an N-acetylglucosamine (38). E and F, FTMS spectra verifying POMGNT2 Man341 product (1.11-ppm mass accuracy) (E) and POMGNT2-extended Man379 (1.11-ppm mass accuracy) (F). The green circle represents a mannose, and the blue square represents an N-acetylglucosamine (38).
In comparison, POMGNT2 showed preferential acceptor selectivity for the known M3 site in the α-DG sequence. Radiolabel transfer assays showed no detectable transfer of the sugar to the acceptor Man341 glycopeptide by POMGNT2 (Fig. 2A) but did show transfer of GlcNAc to the Man379 synthetic glycopeptide (Fig. 2B). Parallel non-radioactive transfer assays followed by MS analysis identified the composition of the POMGNT2 reaction products and verified the transfer results observed in the radioactive assays (Fig. 2, E and F). The predominant peak at 791.427 m/z in the full FTMS corresponds to the unmodified Man341 glycopeptide with a single mannose (Fig. 2E). In contrast to the results seen in Fig. 2E, the observed peak at 899.504 m/z in the full FTMS in Fig. 2F corresponds to the addition of an N-acetylhexosamine residue (+203) to the Man379 glycopeptide. These results suggest that POMGNT2 preferentially modifies specific sites on α-DG intended for core M3 glycan elaboration.
Kinetic Parameters of POMGNT1 and POMGNT2 with Synthetic α-DG Glycopeptides
To further characterize the substrate specificities of POMGNT1 and POMGNT2, additional core M1 and core M3 synthetic glycopeptides were generated. Man414 and Man317 are known O-mannosylated regions of α-DG (18, 21). Evidence for core M3 extension at position 317 in α-DG has been shown previously (12, 18). Man317 is 21 amino acids in length, contains the TPT motif, and has the mannosylated threonine as the central residue (residue 11), similar to Man379 and Man341 (Table 1). Man414 is only seven amino acids in length and lacks the TPT motif (Table 1). However, the kinetics of Man414 with POMGNT1 have been studied previously (26), and the homologous residue in rabbit (Oryctolagus cuniculus) has been site-mapped with mannose (21), making it a useful predicted core M1 glycopeptide for this study.
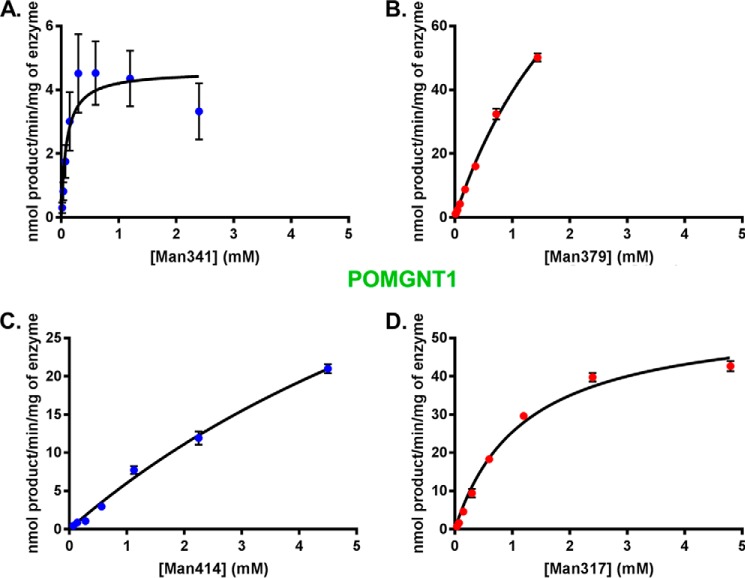
Glycosyltransferase reaction kinetics for POMGNT1 with the four synthesized glycopeptides were investigated by UDP-GloTM assays. The α-DG sequences in the four synthetic glycopeptides were all utilized by POMGNT1 as acceptors (Fig. 3, A–D). Inspection of the Km values derived from non-linear regression analyses of the experimentally obtained values reveals that the affinity of POMGNT1 for synthetic acceptor glycopeptides containing a TPT motif (Man317, Man379, and Man341) is greater than the affinity for the ShortMan414 synthetic glycopeptide lacking a TPT motif (Table 1). POMGNT1 has the fastest turnover (kcat) with the Man341 synthetic glycopeptide, but catalytic efficiency (kcat/Km) is an order of magnitude greater for the core M3 synthetic glycopeptides Man317 and Man379 (Table 1).
FIGURE 3.
POMGNT1 transfers to both M1 and M3 acceptors. POMGNT1 kinetics with Man341 (A), Man379 (B), Man414 (C), or Man317 (D) acceptor glycopeptide measured by UDP-Glo assay are shown. Error bars represent S.E. from three experiments. See Table 1 for a list of kinetic parameters.
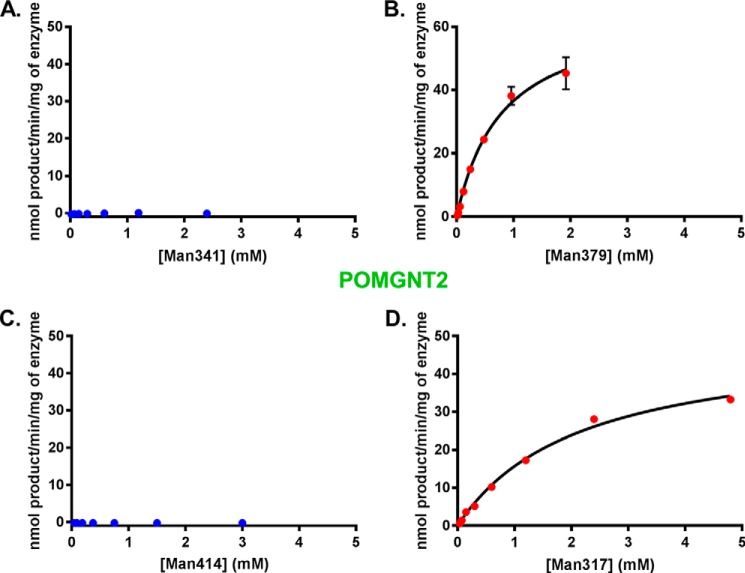
To validate the acceptor selectivity of POMGNT2, we also performed UDP-Glo assays with the four synthesized glycopeptides to investigate glycosyltransferase reaction kinetics. Transfer of GlcNAc to Man341 and Man414 by POMGNT2 (Fig. 4, A and C) was below the level of detection, whereas Man379 and Man317 (Fig. 4, B and D) are clearly acceptor substrates for POMGNT2 activity. The measured Km, kcat, and kcat/Km for Man317 and Man379 synthetic glycopeptides with POMGNT2 are similar (Table 1). These data are consistent with the results obtained in our initial transfer assays and support the proposal that the features of the primary amino acid sequence in the region of the TPT sequence are determinants of POMGNT2 selectivity.
FIGURE 4.
POMGNT2 only transfers to M3 acceptors. POMGNT2 kinetics with Man341 (A), Man379 (B), Man414 (C), or Man317 (D) acceptor glycopeptide measured by UDP-Glo assay are shown. Error bars represent S.E. from three experiments. See Table 1 for a list of kinetic parameters.
A Primary Amino Acid Motif in α-DG Is Favorable for POMGNT2 Activity
It was suggested previously that all O-mannosylated sites on α-DG have a conserved TPT motif at the mannosylated threonine (18, 25), although site mapping studies have demonstrated that only a subset of mapped sites follow this pattern (20–22). Indeed, the primary amino acid sequences around mapped O-mannose sites on α-DG (excluding sites Thr-317 and Thr-379) are heterogeneous (Fig. 5A). To identify primary sequence elements that govern the observed preferences on POMGNT2 acceptor substrate selectivity, α-DG amino acid sequences surrounding sites Thr-317 and Thr-379 from 59 vertebrate species with orthologues of human DAG1, POMGNT1 or POMGNT2, and FKTN or B4GAT1 were aligned using WebLogo (27) (Fig. 5B). The previously identified TPT motif was evident in our alignment, but other conserved amino acids were observed that are not present around Thr-341. Interestingly, our alignment of 317/379 M3 sites across species demonstrated that arginines at −6 and −8 and an Ile at −3 were conserved in addition to the Pro at +1 and the Thr at +2. Thus, the RXRXXIXXTPT motif is a proposed conserved sequence for M3 extension (Fig. 5B).
FIGURE 5.
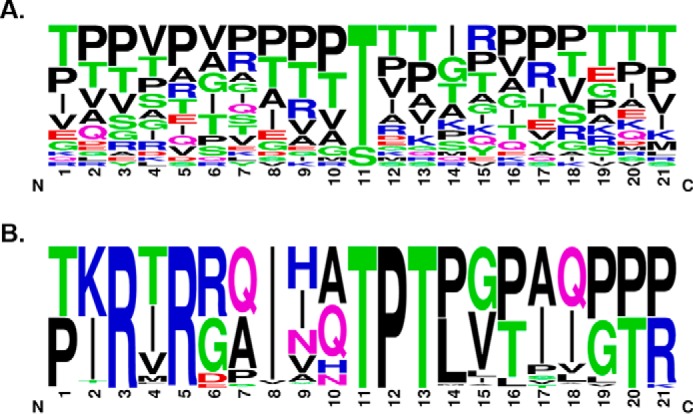
A conserved consensus sequence for POMGNT2 activity. A, sequence alignment of 21-mer sequences of α-DG centered on known O-mannose sites from human and rabbit (20, 21) excluding Thr-317 and Thr-379. The logo was made using Berkeley's WebLogo program. B, sequence alignment of 21-mer sequences of α-DG centered on human sites Thr-317 and Thr-379 from all Ensembl vertebrates with orthologues of DAG1, POMGNT1 or POMGNT2, and FKTN or B4GAT1 (total of 59 species; see “Experimental Procedures” for a complete list). The logo was made using Berkeley's WebLogo program.
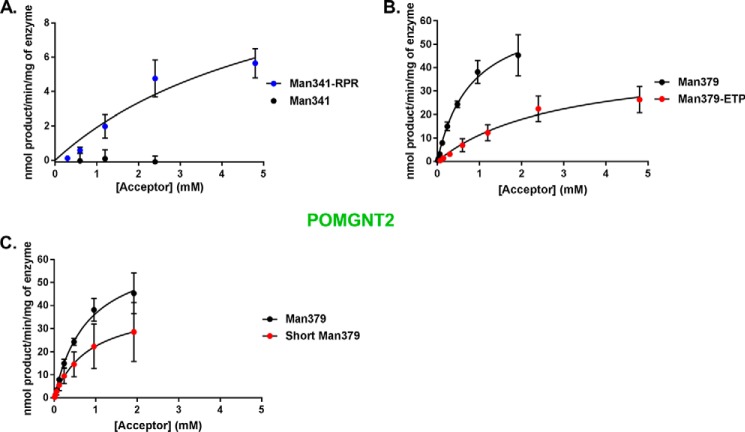
Because this motif is only present at known core M3 sites and not at core M1 sites, we hypothesized that the RXR portion of the primary amino acid sequence motif of α-DG might confer extension by POMGNT2. To test this, we synthesized a modified version of the Man341 peptide that already contains the TPT sequence and an Ile at −3 that we refer to as Man341-RPR. In this glycopeptide, we replaced the two divergent amino acids at −6 and −8 with arginines to introduce the conserved RXR motif. Glycosyltransferase reaction kinetics of POMGNT2 with this modified glycopeptide were investigated by UDP-Glo assay. In contrast to the undetectable reaction with Man341, POMGNT2 transferred GlcNAc to Man341-RPR (Fig. 6A and Table 1). Thus, we successfully converted a core M1 non-acceptor peptide into a core M3 acceptor for POMGNT2 in vitro by the addition of our identified motif.
FIGURE 6.
A primary amino acid motif in α-DG is permissible for POMGNT2 activity. POMGNT2 kinetics with Man341-RPR and Man341 (A), Man379-ETP and Man379 (B), and ShortMan379 and Man379 (C) acceptor glycopeptides were measured by UDP-Glo assay. Error bars represent S.E. from three experiments. See Table 1 for a list of kinetic parameters.
To further test that the RXR portion of the primary amino acid sequence motif of α-DG is important for glycan extension by POMGNT2, we synthesized a new glycopeptide based on the sequence at a known core M3 site (379) but with the two N-terminal arginine residues altered to the divergent residues of a core M1 acceptor (Man341). We have designated this modified core M3 glycopeptide as Man379-ETP. Glycosyltransferase reaction kinetics of POMGNT2 with this modified glycopeptide were investigated by UDP-Glo assays. In comparison with the kinetics of POMGNT2 with Man379, POMGNT2 has a lower affinity and a greater than 5-fold reduction in catalytic efficiency for Man379-ETP (Fig. 6B and Table 1). The replacement of our identified RXR motif in a core M3 acceptor with divergent residues reduced but did not eliminate POMGNT2 activity.
Lastly, to test the necessity of the RXR portion of the primary amino acid sequence motif of α-DG for POMGNT2 activity, we synthesized a truncated version of the Man379 M3 glycopeptide that we refer to as ShortMan379. The N terminus of this glycopeptide begins immediately following the RXR motif. Glycosyltransferase reaction kinetics of POMGNT2 with this truncated glycopeptide were investigated by the UDP-Glo assay. POMGNT2 utilized ShortMan379 as an acceptor substrate with an affinity similar to Man379 but with a less than 1-fold reduction in catalytic efficiency (Fig. 6C and Table 1). Thus, the RXR portion of the motif appears to not be essential in the context of a synthetic peptide for POMGNT2 activity.
Discussion
Although POMGNT2 is poised to modify α-DG in the ER before it encounters POMGNT1 in the cis-Golgi, only two M3 sites have been identified on α-DG. Thus, it seems likely that POMGNT2 demonstrates acceptor substrate preferences beyond simply an O-Man-modified residue. We tested this hypothesis regarding specificity by examining the impact of local primary amino acid sequence around O-Man sites on synthetic peptides as acceptor substrates for POMGNT1 and POMGNT2.
Using a set of O-Man glycopeptide substrates, we have shown that POMGNT2 has a preference for acceptors with mannosylated residues at positions Thr-317 and Thr-379, whereas POMGNT1 has no significant acceptor substrate preferences among the various synthetic glycopeptides tested (Figs. 2–4 and Table 1). Analysis of the sites that are POMGNT2-dependent demonstrate a RXRXXIXXTPT motif that is conserved among vertebrate α-DGs (Fig. 5B). We also observed that this sequence is not found in any of the mapped sites from other O-mannosylated proteins (9), consistent with α-DG being the only demonstrated protein to contain M3 glycans (Fig. 5A). We found that replacement of a divergent sequence on a POMGNT1 acceptor that was only missing the conserved RXR motif converted it to a POMGNT2 acceptor, demonstrating that replacing the two amino acids was sufficient to confer activity (Fig. 6A). Likewise, replacement of the arginines in the Man379 peptide with amino acids found in the M1 peptide of Man341 reduced the efficiency of POMGNT2 to catalyze the addition of GlcNAc to the O-Man peptide more than 5-fold (Fig. 6B). However, although the addition of the RXR motif to a core M1 acceptor is sufficient to make it a substrate for POMGNT2, the complete removal of the RXR motif on a core M3 acceptor does not abolish POMGNT2 activity (Fig. 6C). Taken together, these results suggest that the RXR motif allows for extension by POMGNT2 at core M3 sites but is not essential for a short synthetic O-Man peptide. These results support a case of sufficiency in the absence of necessity that deviates from the normal necessary and sufficient or necessary but not sufficient arguments. We would rationalize that when there is sequence upstream of the site of action, such as that actually found in the full-length α-dystroglycan protein, non-basic amino acids replacing the RXR portion of the motif generate steric or electrostatic clashes that prevent proper binding of the substrate protein.
Interestingly, for Man317, the identified RXR motif is upstream of the known furin cleavage site. However, as POMGNT2 is an ER-resident glycosyltransferase and furin is located in the Golgi, POMGNT2 acts first and thus has the capability to interact with residues upstream of the furin cleavage site, and this may at least partially explain the requirement for the N terminus for synthesis of functionally glycosylated mature α-DG (19).
Our current model, based on the data presented here, is that POMGNT2 selectivity determines which sites on α-DG become modified with the core M3 glycan structure. In turn, only the core M3 glycan structure can be extended by B3GALNT2, phosphorylated by POMK, and further elaborated to become the functional matriglycan for α-DG (2, 13). Functional glycosylation of α-DG and, in particular, matriglycan synthesis stemming from the POMGNT2-dependent core M3 glycan structure are required for binding to ECM proteins with LG domains and maintaining overall ECM integrity (11). Thus, POMGNT2 acts a gatekeeper enzyme for functional glycosylation of α-DG.
The strict RXRXXIXXTPT motif that we have presented here is not present on any other secreted or membrane-associated protein in humans except for α-DG at Thr-317/319 and Thr-379/381 (2, 13). Relaxing the sequence constraints to allow for conservative replacements generates a motif of (R/K)X(R/K)XX(I/L/V)XX(T/S)P(T/S). This motif is found on a handful of membrane/secreted human proteins including SRPX, CLEC18C, FREM2, MANBA, SEMA3E, SPACA7, and TMEM182. However, if we examine conservation of the motif in these proteins across 59 vertebrate species, as we did for α-DG, we see poor conservation (data not shown). This lends further support to the working model that only α-DG contains sequences that are substrates for POMGNT2 that go on to become functionally glycosylated with matriglycan (2).
We have identified and partially characterized a primary amino acid sequence motif governing acceptor specificity for POMGNT2 toward O-mannosylated substrates. Additional studies are required to fully characterize the functional roles of individual amino acids in this motif. Structural analyses of POMGNT2 in complex with various acceptor substrates would greatly assist in defining the molecular details of the POMGNT2 gatekeeping mechanism that we have established here. Furthermore, future in vivo studies testing the role of the RXRXXIXXTPT motif in POMGNT2 acceptor selectivity will be invaluable to complement our in vitro findings presented here.
Experimental Procedures
Cell Culture and Protein Purification
The catalytic domains of human POMGNT1 (amino acid residues 60–660; UniProt Q8WZA1) and POMGNT2 (amino acid residues 25–580; UniProt Q8NAT1) were expressed as soluble, secreted fusion proteins by transient transfection of HEK293 suspension cultures (28). The coding regions were amplified from Mammalian Gene Collection (29) clones using primers that appended a tobacco etch virus protease cleavage site (30) to the N-terminal end of the coding region and attL1 and attL2 Gateway adaptor sites to the 5′- and 3′-terminal ends of the amplimer products. The amplimers were recombined via BP clonase reaction into the pDONR221 vector, and the DNA sequences were confirmed. The pDONR221 clones were then recombined via LR clonase reaction into a custom Gateway adapted version of the pGEn2 mammalian expression vector (28, 31) to assemble a recombinant coding region comprising a 25-amino acid N-terminal signal sequence from the Trypanosoma cruzi lysosomal α-mannosidase (32) followed by a His8 tag, 17-amino acid AviTag (33), “superfolder” GFP (34), the nine-amino acid sequence encoded by attB1 recombination site followed by the tobacco etch virus protease cleavage site and the respective glycosyltransferase catalytic domain coding region. Suspension culture HEK293f cells (Life Technologies) were transfected as described previously (28), and the culture supernatant was subjected to nickel-nitrilotriacetic acid superflow chromatography (Qiagen). Enzyme preparations eluted with 300 mm imidazole were concentrated to ∼1 mg/ml using an ultrafiltration pressure cell membrane (Millipore) with a 10-kDa molecular mass cutoff.
Glycopeptide Synthesis
The glycopeptide synthesis here extends earlier work describing synthesis of O-Man-Ser and -Thr peptide synthesis building blocks as well as O-Man glycopeptides (35). The glycopeptides were prepared as C-terminal carboxamides and acetylated at the N terminus to emulate the situation in the native protein. For this work, all couplings except those for glycosylated residues were carried out on an automated microwave-assisted solid-phase peptide synthesizer (CEM Corp. Liberty microwave synthesizer) using standard protocols in the instrument software on Rink amide resin (∼0.5 meq/g; Novabiochem) via an NR-N-(9-fluorenyl)methoxycarbonyl (Fmoc)-based approach with N,N-dimethylformamide (DMF) as the primary solvent. 20% 4-methylpiperidine in DMF was used for Fmoc removal. 2-(1H-Benzotriazole-1-yl)-oxy-1,1,3,3-tetramethyluronium hexafluorophosphate/1-hydroxybenzotriazole in the presence of N,N-diisopropylethylamine (DIPEA) were used as the coupling reagents for standard amino acids. For the coupling of the glycosylated amino acid Fmoc-Thr(α-d-Man(Ac)4)-OH (Sussex Research), the peptide resin was removed from the synthesizer, and coupling was performed manually using a CEM Corp. Discover microwave apparatus. 2-(7-Aza-1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate and 1-hydroxy-7-azabenzotriazole in the presence of DIPEA were the activating reagents. Typically two couplings at ∼1.5-fold excess of glycosylated amino acid to the resin loading were done for this amino acid to conserve reagent. Upon completion of the manual coupling reaction, as determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), glycopeptide resins were returned to the automatic synthesizer to complete assembly. After final N-deprotection, the glycopeptides were manually N-acetylated by treatment with DMF/acetic anhydride/DIPEA (85:10:5, v/v) for ∼30 min, and O-acetyl protection on the mannosyl residues was subsequently removed by two successive treatments with hydrazine/MeOH (70:20, v/v) for an hour each. Glycopeptides were then cleaved from the resin as C-terminal carboxamides with simultaneous removal of remaining amino acid side chain protection through treatment with TFA/triisopropylsilane/H2O (95:2.5:2.5) for ∼4 h. The resin was filtered off, and the TFA solution was concentrated on a rotary evaporator to a few milliliters. The remaining concentrate was added dropwise to cold ether from which the crude glycopeptides precipitated. After centrifugation and removal of the ether supernatant, the glycopeptides were redissolved and purified via HPLC over an Ultra II 250 × 10.0-mm 5-μm C18 column (Restek Corp.) with a 0.1% TFA in water, 0.1% TFA in acetonitrile solvent gradient. Purity was verified by analytical HPLC and MALDI-TOF MS (see supplemental Fig. 1). Yields were in the range of 30–50%.
Radiolabel Transfer Assays
The radiometric assays were carried out in reactions containing 100 mm MES (pH 6.5), 10 mm MnCl2, 2 mm UDP-GlcNAc mixed with 10 nCi of 3H-labeled UDP-GlcNAc and 1 mm glycopeptide acceptor. Reactions were incubated for 21 h at 37 °C, then quenched by addition of 5 μl of 1% TFA, and boiled at 100 °C for 5 min. Reaction products were purified by reverse-phase separation using C18 Sep-Pak microspin columns (The Nest Group) by loading and washing with 0.1% formic acid and elution with 80% acetonitrile with 0.1% formic acid. Disintegrations per minute (dpm) were counted using a liquid scintillation counter (Beckman) to determine the amount of [3H]GlcNAc incorporated into the glycopeptides. The data presented represent the average of at least three independent experiments.
Mass Spectrometry
Cold glycosyltransferase reactions used for analysis by mass spectrometry were carried out identically to the radioactive transfer assays but without radioactive UDP-GlcNAc. After reverse phase separation, the product was vacuumed to dryness and resuspended in 100 μl of 0.1% formic acid. Samples were filtered using a 0.2-μm Nanosep microcentrifuge filter (Pall Life Sciences) and transferred to an autosampler vial with glass insert (Thermo ScientificTM). The samples were run on a Thermo Scientific Orbitrap FusionTM LumosTM mass spectrometer. Full Fourier transform MS Spectra were analyzed using Xcalibur Qual Browser software, and MS/MS scans were analyzed using ByonicTM version 2.6.46 (Protein Metrics Inc.) using a precursor mass tolerance of 10 ppm and a fragmentation mass tolerance of 0.3 daltons followed by manual interpretation.
UDP-Glo Glycosyltransferase Assays
UDP-Glo glycosyltransferase assays (Promega) were performed using 50 mm Tris-HCl (pH 7.5), 5 mm MnCl2, 100 μm UDP-GlcNAc, 40 ng of enzyme, and varying amounts of glycopeptide acceptor substrates at 37 °C for 2 h in a white, flat bottom, 384-well plate. After the glycosyltransferase reaction, an equal volume of UDP detection reagent was added to simultaneously convert the UDP product to ATP and generate light in a luciferase reaction. The light generated was detected using a GloMax-Multi+ luminometer (Promega). Luminescence was correlated to UDP concentration by using a UDP standard curve. Kinetic parameters were extracted from the data after fitting to the Michaelis-Menten equation using the non-linear regression fit in GraphPad Prism version 7.1. The data presented represent the average of at least three independent experiments.
WebLogo Consensus Sequence Alignment
All vertebrate species available on the Ensembl genome browser (release 85) (36) with orthologues of human DAG1, POMGNT1 or POMGNT2, and FKTN or B4GAT1 (Xenopus tropicalis, Latimeria chalumnae, Anolis carolinensis, Choloepus hoffmanni, Tursiops truncatus, Pelodiscus sinensis, Meleagris gallopavo, Gallus gallus, Anas platyrhynchos, Taeniopygia guttata, Ficedula albicollis, Dipodomys ordii, Echinops telfairi, Ochotona princeps, Loxodonta africana, Procavia capensis, Myotis lucifugus, Cavia porcellus, Rattus norvegicus, Mus musculus, Erinaceus europaeus, Dasypus novemcinctus, Otolemur garnettii, Ictidomys tridecemlineatus, Bos taurus, Ovis aries, O. cuniculus, Mustela putorius furo, Canis lupus familiaris, Felis catus, Ailuropoda melanoleuca, Tarsius syrichta, Tupaia belangeri, Pteropus vampyrus, Sus scrofa, Equus caballus, Callithrix jacchus, Papio anubis, Macaca mulatta, Chlorocebus sabaeus, Nomascus leucogenys, Gorilla gorilla, Pan troglodytes, Homo sapiens, Ornithorhynchus anatinus, Macropus eugenii, Monodelphis domestica, Sarcophilus harrisii, Lepisosteus oculatus, Danio rerio, Astyanax mexicanus, Tetraodon nigroviridis, Takifugu rubripes, Oryzias latipes, Xiphophorus maculatus, Poecilia formosa, Gadus morhua, Gasterosteus aculeatus, and Oreochromis niloticus) were aligned to human DAG1 using Clustal Omega (37). The 10 amino acids upstream and downstream of the threonine at position 317 and 379 in human DAG1 for all species were extracted from the alignment and used for analysis in Berkeley's WebLogo program (version 3) (27).
Author Contributions
L. W., D. L., and K. W. M. conceived and coordinated the study. S. M. H. and L. W. wrote the paper. S. M. H., S. P., D. S., and L. W. designed, performed, and analyzed all experiments shown. S. W. and K. W. M. designed and constructed vectors for expression of proteins. S. M. H., S. P., and S. W. expressed and purified proteins. M. E. and S. M. H. synthesized the glycopeptides under the direction of D. L. and G.-J. B. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank M. Osman Sheikh for assistance in the preparation of the manuscript and Robert Bridger and Jeremy L. Praissman for technical assistance with mass spectrometry. We also thank all members of the Wells laboratory for thoughtful discussions.
This work was supported in part by National Institutes of Health Grants R01GM111939 (to D. L. and L. W.), P01GM107012 (to G.-J. B., K. W. M., and L. W.), P41GM103490 (to K. W. M. and L. W.), P41GM103390 (to K. W. M. and D. L.), and S10RR027155 for purchase of the peptide synthesizer (to D. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Fig. 1.
- CMD
- congenital muscular dystrophy
- POMGNT2
- protein O-linked mannose β-1,4-N-acetylglucosaminyltransferase 2
- POMGNT1
- protein O-linked mannose β-1,2-N-acetylglucosaminyltransferase 1
- ECM
- extracellular matrix
- α-DG
- α-dystroglycan
- LG
- laminin globular
- O-Man
- O-mannosyl
- ER
- endoplasmic reticulum
- GlcNAc
- N-acetylglucosamine
- MS
- mass spectrometry
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- DMF
- N,N-dimethylformamide
- DIPEA
- N,N-diisopropylethylamine
- MALDI-TOF
- matrix-assisted laser desorption/ionization time-of-flight.
References
- 1. Ervasti J. M., and Campbell K. P. (1993) A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol. 122, 809–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yoshida-Moriguchi T., and Campbell K. P. (2015) Matriglycan: a novel polysaccharide that links dystroglycan to the basement membrane. Glycobiology 25, 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Live D., Wells L., and Boons G.-J. (2013) Dissecting the molecular basis of the role of the O-mannosylation pathway in disease: α-dystroglycan and forms of muscular dystrophy. ChemBioChem 14, 2392–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Godfrey C., Foley A. R., Clement E., and Muntoni F. (2011) Dystroglycanopathies: coming into focus. Curr. Opin. Genet. Dev. 21, 278–285 [DOI] [PubMed] [Google Scholar]
- 5. Voglmeir J., Kaloo S., Laurent N., Meloni M. M., Bohlmann L., Wilson I. B., and Flitsch S. L. (2011) Biochemical correlation of activity of the α-dystroglycan-modifying glycosyltransferase POMGnT1 with mutations in muscle-eye-brain disease. Biochem. J. 436, 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jae L. T., Raaben M., Riemersma M., van Beusekom E., Blomen V. A., Velds A., Kerkhoven R. M., Carette J. E., Topaloglu H., Meinecke P., Wessels M. W., Lefeber D. J., Whelan S. P., van Bokhoven H., and Brummelkamp T. R. (2013) Deciphering the glycosylome of dystroglycanopathies using haploid screens for Lassa virus entry. Science 340, 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wells L. (2013) The O-Mannosylation pathway: glycosyltransferases and proteins implicated in congenital muscular dystrophy. J. Biol. Chem. 288, 6930–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manya H., Chiba A., Yoshida A., Wang X., Chiba Y., Jigami Y., Margolis R. U., and Endo T. (2004) Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc. Natl. Acad. Sci. U.S.A. 101, 500–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vester-Christensen M. B., Halim A., Joshi H. J., Steentoft C., Bennett E. P., Levery S. B., Vakhrushev S. Y., and Clausen H. (2013) Mining the O-mannose glycoproteome reveals cadherins as major O-mannosylated glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 110, 21018–21023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takahashi S., Sasaki T., Manya H., Chiba Y., Yoshida A., Mizuno M., Ishida H., Ito F., Inazu T., Kotani N., Takasaki S., Takeuchi M., and Endo T. (2001) A new β-1,2-N-acetylglucosaminyltransferase that may play a role in the biosynthesis of mammalian O-mannosyl glycans. Glycobiology 11, 37–45 [DOI] [PubMed] [Google Scholar]
- 11. Praissman J. L., and Wells L. (2014) Mammalian O-mannosylation pathway: glycan structures, enzymes, and protein substrates. Biochemistry 53, 3066–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yagi H., Nakagawa N., Saito T., Kiyonari H., Abe T., Toda T., Wu S.-W., Khoo K.-H., Oka S., and Kato K. (2013) AGO61-dependent GlcNAc modification primes the formation of functional glycans on α-dystroglycan. Sci. Rep. 3, 3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoshida-Moriguchi T., Willer T., Anderson M. E., Venzke D., Whyte T., Muntoni F., Lee H., Nelson S. F., Yu L., and Campbell K. P. (2013) SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science 341, 896–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ogawa M., Nakamura N., Nakayama Y., Kurosaka A., Manya H., Kanagawa M., Endo T., Furukawa K., and Okajima T. (2013) GTDC2 modifies O-mannosylated α-dystroglycan in the endoplasmic reticulum to generate N-acetyl glucosamine epitopes reactive with CTD110.6 antibody. Biochem. Biophys. Res. Commun. 440, 88–93 [DOI] [PubMed] [Google Scholar]
- 15. Kanagawa M., Kobayashi K., Tajiri M., Manya H., Kuga A., Yamaguchi Y., Akasaka-Manya K., Furukawa J., Mizuno M., Kawakami H., Shinohara Y., Wada Y., Endo T., and Toda T. (2016) Identification of a post-translational modification with ribitol-phosphate and its defect in muscular dystrophy. Cell Rep. 14, 2209–2223 [DOI] [PubMed] [Google Scholar]
- 16. Praissman J. L., Willer T., Sheikh M. O., Toi A., Chitayat D., Lin Y. Y., Lee H., Stalnaker S. H., Wang S., Prabhakar P. K., Nelson S. F., Stemple D. L., Moore S. A., Moremen K. W., Campbell K. P., and Wells L. (2016) The functional O-mannose glycan on α-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife 5, e14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Praissman J. L., Live D. H., Wang S., Ramiah A., Chinoy Z. S., Boons G.-J., Moremen K. W., and Wells L. (2014) B4GAT1 is the priming enzyme for the LARGE-dependent functional glycosylation of α-dystroglycan. eLife 3, e03943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hara Y., Kanagawa M., Kunz S., Yoshida-Moriguchi T., Satz J. S., Kobayashi Y. M., Zhu Z., Burden S. J., Oldstone M. B., and Campbell K. P. (2011) Like-acetylglucosaminyltransferase (LARGE)-dependent modification of dystroglycan at Thr-317/319 is required for laminin binding and arenavirus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 17426–17431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanagawa M., Saito F., Kunz S., Yoshida-Moriguchi T., Barresi R., Kobayashi Y. M., Muschler J., Dumanski J. P., Michele D. E., Oldstone M. B., and Campbell K. P. (2004) Molecular recognition by LARGE is essential for expression of functional dystroglycan. Cell 117, 953–964 [DOI] [PubMed] [Google Scholar]
- 20. Nilsson J., Nilsson J., Larson G., and Grahn A. (2010) Characterization of site-specific O-glycan structures within the mucin-like domain of α-dystroglycan from human skeletal muscle. Glycobiology 20, 1160–1169 [DOI] [PubMed] [Google Scholar]
- 21. Stalnaker S. H., Hashmi S., Lim J.-M., Aoki K., Porterfield M., Gutierrez-Sanchez G., Wheeler J., Ervasti J. M., Bergmann C., Tiemeyer M., and Wells L. (2010) Site mapping and characterization of O-glycan structures on α-dystroglycan isolated from rabbit skeletal muscle. J. Biol. Chem. 285, 24882–24891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harrison R., Hitchen P. G., Panico M., Morris H. R., Mekhaiel D., Pleass R. J., Dell A., Hewitt J. E., and Haslam S. M. (2012) Glycoproteomic characterization of recombinant mouse α-dystroglycan. Glycobiology 22, 662–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee J. K., Matthews R. T., Lim J. M., Swanier K., Wells L., and Pierce J. M. (2012) Developmental expression of the neuron-specific N-acetylglucosaminyltransferase Vb (GnT-Vb/IX) and identification of its in vivo glycan products in comparison with those of its paralog, GnT-V. J. Biol. Chem. 287, 28526–28536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshida-Moriguchi T., Yu L., Stalnaker S. H., Davis S., Kunz S., Madson M., Oldstone M. B., Schachter H., Wells L., and Campbell K. P. (2010) O-Mannosyl phosphorylation of α-dystroglycan is required for laminin binding. Science 327, 88–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Manya H., Suzuki T., Akasaka-Manya K., Ishida H.-K., Mizuno M., Suzuki Y., Inazu T., Dohmae N., and Endo T. (2007) Regulation of mammalian protein O-mannosylation: preferential amino acid sequence for o-mannose modification. J. Biol. Chem. 282, 20200–20206 [DOI] [PubMed] [Google Scholar]
- 26. Mo K.-F., Fang T., Stalnaker S. H., Kirby P. S., Liu M., Wells L., Pierce M., Live D. H., and Boons G.-J. (2011) Synthetic, structural, and biosynthetic studies of an unusual phospho-glycopeptide derived from α-dystroglycan. J. Am. Chem. Soc. 133, 14418–14430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meng L., Forouhar F., Thieker D., Gao Z., Ramiah A., Moniz H., Xiang Y., Seetharaman J., Milaninia S., Su M., Bridger R., Veillon L., Azadi P., Kornhaber G., Wells L., et al. (2013) Enzymatic basis for N-glycan sialylation: structure of rat α2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J. Biol. Chem. 288, 34680–34698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gerhard D. S., Wagner L., Feingold E. A., Shenmen C. M., Grouse L. H., Schuler G., Klein S. L., Old S., Rasooly R., Good P., Guyer M., Peck A. M., Derge J. G., Lipman D., Collins F. S., et al. (2004) The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC). Genome Res 14, 2121–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Phan J., Zdanov A., Evdokimov A. G., Tropea J. E., Peters H. K. 3rd, Kapust R. B., Li M., Wlodawer A., and Waugh D. S. (2002) Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 277, 50564–50572 [DOI] [PubMed] [Google Scholar]
- 31. Barb A. W., Meng L., Gao Z., Johnson R. W., Moremen K. W., and Prestegard J. H. (2012) NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry 51, 4618–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vandersall-Nairn A. S., Merkle R. K., O'Brien K., Oeltmann T. N., and Moremen K. W. (1998) Cloning, expression, purification, and characterization of the acid α-mannosidase from Trypanosoma cruzi. Glycobiology 8, 1183–1194 [DOI] [PubMed] [Google Scholar]
- 33. Beckett D., Kovaleva E., and Schatz P. J. (1999) A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pédelacq J. D., Cabantous S., Tran T., Terwilliger T. C., and Waldo G. S. (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 [DOI] [PubMed] [Google Scholar]
- 35. Liu M., Borgert A., Barany G., and Live D. (2008) Conformational consequences of protein glycosylation: preparation of O-mannosyl serine and threonine building blocks, and their incorporation into glycopeptide sequences derived from α-dystroglycan. Biopolymers 90, 358–368 [DOI] [PubMed] [Google Scholar]
- 36. Yates A., Akanni W., Amode M. R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L., Girón C. G., Gordon L., Hourlier T., Hunt S. E., Janacek S. H., et al. (2016) Ensembl 2016. Nucleic Acids Res. 44, D710–D716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., et al. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.