Abstract
Histone acetylation has a regulatory role in gene expression and is necessary for proper tissue development. To investigate the specific roles of histone deacetylases (HDACs) in rod differentiation in neonatal mouse retinas, we used a pharmacological approach that showed that inhibition of class I but not class IIa HDACs caused the same phenotypic changes seen with broad spectrum HDAC inhibitors, most notably a block in the differentiation of rod photoreceptors. Inhibition of HDAC1 resulted in increase of acetylation of lysine 9 of histone 3 (H3K9) and lysine 12 of histone 4 (H4K12) but not lysine 27 of histone 3 (H3K27) and led to maintained expression of progenitor-specific genes such as Vsx2 and Hes1 with concomitant block of expression of rod-specific genes. ChiP experiments confirmed these changes in the promoters of a group of progenitor genes. Based on our results, we suggest that HDAC1-specific inhibition prevents progenitor cells of the retina from exiting the cell cycle and differentiating. HDAC1 may be an essential epigenetic regulator of the transition from progenitor cells to terminally differentiated photoreceptors.
Keywords: development, epigenetics, histone deacetylase inhibitor (HDAC inhibitor) (HDI), photoreceptor, retina, rhodopsin, H3K9 acetylation, H4K12 acetylation, explant culture, recoverin
Introduction
Histone modifications regulate both chromatin structure and gene expression, and changing patterns of such modifications are an integral part of normal tissue development (1). In particular, a series of acetylated derivatives of histone 3 (H3)4 and histone 4 (H4) are thought to be sensitive indicators, and possibly predictors, of gene expression, but the complexity of this histone code is far from understood (2, 3). The positively charged lysines in the N-terminal tails of histones H3 and H4 bind to negatively charged phosphates of DNA and restrict DNA accessibility, promoting chromatin condensation and repressing gene transcription. Acetylation of lysines 9, 14, 18, and 27 in H3 and lysines 8, 12, and 16 in H4 histones inhibits DNA-nucleosome interaction, opening chromatin for transcription and activating gene expression (1, 4–8).
Acetylation of histones is regulated by two antagonist enzyme groups: histone acetyltransferases and histone deacetylases (HDACs) (9). The family of mammalian HDACs is composed of four classes based on their homologies to yeast proteins and enzymatic function (10, 11). HDAC1, HDAC2, HDAC3, and HDAC8 belong to class I (12, 13). Because of their higher levels of expression, nuclear localization, and high enzymatic activity toward histone substrates, class I HDACs are thought to play a central role in cell differentiation and tissue development, especially in neurogenesis (11, 14). Class I HDACs exert their functions as subunits in chromatin complexes such as Sin3, nucleosome remodeling and histone deacetylation (NuRD), corepressor for element-1-silencing transcription factor (CoREST) and nuclear receptor co-repressor/silencing mediator for retinoid or thyroid hormone receptors (NCoR/SMRT). Class II HDACs have several additional protein domains, demonstrate low enzymatic activity in vitro, and are found in cytoplasm as well as in nuclei. They are subdivided into class IIa (HDAC4, HDAC5, HDAC7, and HDAC9) and class IIb (HDAC6 and HDAC10). Class II HDACs are expressed at lower levels, show more restricted patterns of expression, and shuttle between cytoplasm and nuclei. They are thought to function by recruiting class I HDACs or other transcriptional repressors to chromatin complexes (10, 15).
In the central nervous system (CNS), HDACs regulate differentiation of both neuronal and glial lineages (16, 17). Several studies of histone acetylation in neural development using pan-HDAC inhibitors or knockdown or knock-out models have given somewhat contradictory results (for a review, see Ref. 18), suggesting that spatial-temporal differences in expression of different HDACs are crucial for their function. So far, there have been few studies of the actions of individual HDACs.
Given the importance of these enzymes in various cellular functions, they have become targets for treatment of cancer, neurological disorders, and other diseases. It has been shown by protein crystallography that HDAC inhibitors interact directly with the zinc-binding site and that this interaction is required for inhibitory activity, which can be reversible or irreversible (19). The HDAC inhibitors are classified by structure and can be divided into four major classes: short-chain fatty acids such as butyrate (CAS 156-54-7) and valproic acid (CAS 99-66-1); cyclic tetrapeptides such as trapoxin A (CAS 133155-89-2), romidepsin (CAS 128517-07-7), and apicidin (CAS 183506–66-3); hydroxamic acids such as vorinostat (CAS 149647-78-9) and trichostatin A (TSA) (CAS 58880-19-6); and benzamides such as entinostat (CAS 209783-80-2); and 4-(dimethylamino)-N-[6-(hydroxyamino)-6-oxohexyl]benzamide (CAS 193551-00-7).
Sodium butyrate was first discovered by Riggs et al. (20) to cause acetylated histones to accumulate in HeLa and Friend erythroleukemia cells, and further studies by Candido et al. (21) showed that the effect was due to the inhibition of histone deacetylases in a reversible manner. In a deacetylase activity assay with recombinant human HDACs (rhHDACs), entinostat exhibits EC50 in the 100 nm range for rhHDAC1. No inhibition was found for rhHDACs 4, 6, 7 and 8 and an EC50 in the low μm range was described for rhHDACs 2, 3, and 9 (22). The same assay showed that apicidin inhibits rhHDACs 2 and 3 in the nanomolar range (120 and 8 nm, respectively) with no inhibitory effect on rhHDAC1 and class II rhHDACs (22). CAS 193551-00-7 is a benzamide derived from entinostat with selective inhibitory activity for HDAC1 in the range of 100 nm (23). An in vitro enzyme inhibition assay has shown the selectivity for only HDAC1 with an effect on cell proliferation in murine erythroleukemia cells (23). In primary cell cultures, 1 μm CAS 193551-00-7 had the same effect as specific HDAC1 siRNA (24). At a concentration of 1 μm, CAS 193551-00-7 does not inhibit HDAC3 (25).
Romidepsin is produced by Chromobacterium violaceum and in an assay using HDACs prepared from 293T cells showed that the IC50 for HDAC1 and HDAC2 is in the 40 nm range, whereas inhibition for class II HDAC4 is 500 nm and for HDAC6 is 14,000 nm (26–28). The class IIa inhibitor MC-1568 was shown to be selective for this class of HDACs with an IC50 of 220 nm. No inhibitory activity was found for HDAC1, whereas 55% of HDAC4 activity was inhibited in these enzymes immunoprecipitated from human breast cancer ZR-75.1 cell lysates (29).
We have used the mouse retina as a model to investigate the role of HDACs in terminal differentiation of a specific neuronal cell type, the rod photoreceptor. All retinal cell types originate from a multipotent retinal progenitor cell in a time-dependent manner with horizontal, cone, amacrine, and ganglion cells born embryonically and the great majority of rod, bipolar, and Müller cells being born postnatally (for a review, see Ref. 30). After postnatal day 1 (PN1), rod photoreceptors, which represent the majority of cells in the retina, begin differentiation and start expressing a number of cell type-specific molecules, including the visual pigment protein rhodopsin. Previous studies have linked non-selective inhibition of HDACs by TSA or sodium butyrate to a complete blockage of rod photoreceptor development and an induction of apoptosis in mice (31, 32). These studies, however, could not define whether all or only some of the specific HDACs were important in retinal development. Here we have used a pharmacological approach to show that inhibition of class I HDACs, particularly HDAC1, leads to the same phenotypic changes seen with broad spectrum inhibitors. Under specific inhibition of HDAC1, acetylation of H3K9 and H4K12 but not H3K27 is increased, and expression of progenitor-specific genes is maintained with concomitant block of expression of rod-specific genes. HDAC1 appears to be key enzyme controlling the passage of cells from a progenitor to a terminally differentiated state.
Results
Inhibition of Class I HDAC Selectively Increases H4K12ac and H3K9ac Acetylation and Decreases Rhodopsin Expression
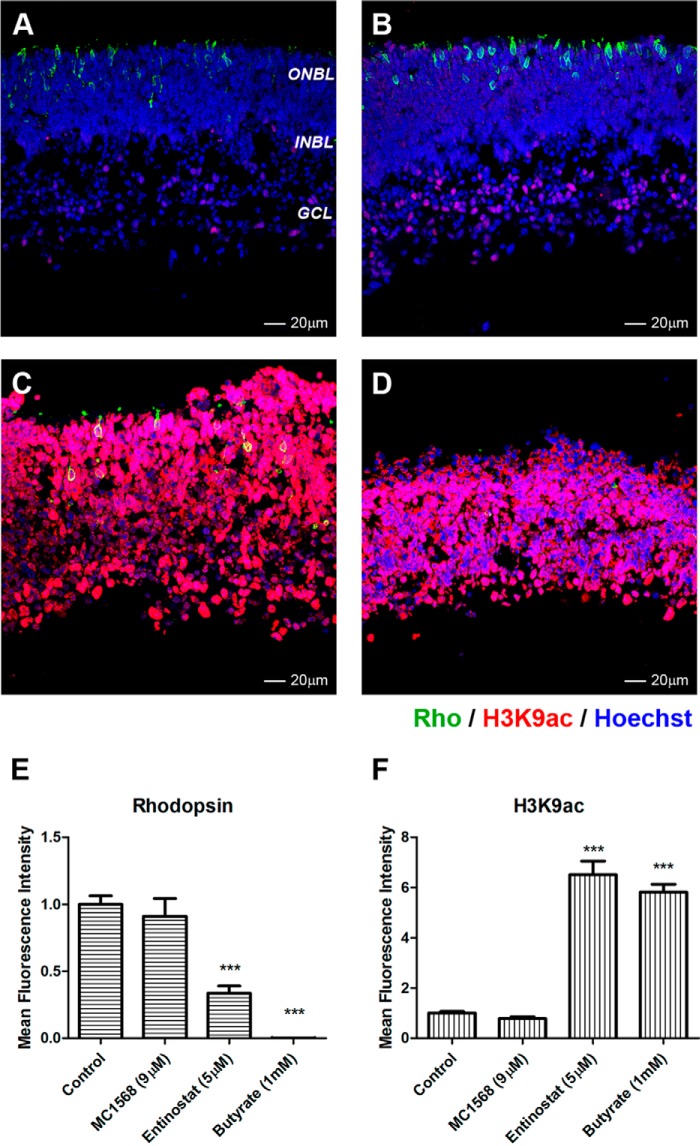
To analyze HDAC expression in the retina, total RNA was isolated from retinas at embryonic day 16 (E16), PN1, PN7, and PN21 and compared with expression in the body and in brain. Most HDACs are expressed in retina, with the exception of HDAC7 (HDAC8 was not tested), and show varying levels throughout development (Fig. 1). General inhibition of HDACs by nonselective inhibitor TSA or sodium butyrate blocks rod photoreceptor development in mouse retina (31). To test whether there was selectivity in the inhibition of rod differentiation by HDACs, we compared the effects of inhibitors of two classes. In retina explants cultured from PN1–PN4, we tested a panel of inhibitors for class I and class IIa HDACs (Table 1) and followed rhodopsin expression as a marker of rod photoreceptor differentiation. No difference in rhodopsin staining was observed in retinas treated for 96 h with class IIa HDAC inhibitor MC-1568 when compared with control (Fig. 2, A, B, and E). In contrast, inhibition of class I HDACs by entinostat for 96 h showed a 60% decrease in rhodopsin labeling, a decrease comparable with rhodopsin down-regulation by sodium butyrate (Fig. 2, C, D, and E). Co-staining with antibodies recognizing one histone acetylation site, H3K9ac, showed that the level of acetylation was dramatically increased under entinostat and sodium butyrate treatment but not under MC-1568 treatment (Fig. 2, C, D, and F).
FIGURE 1.
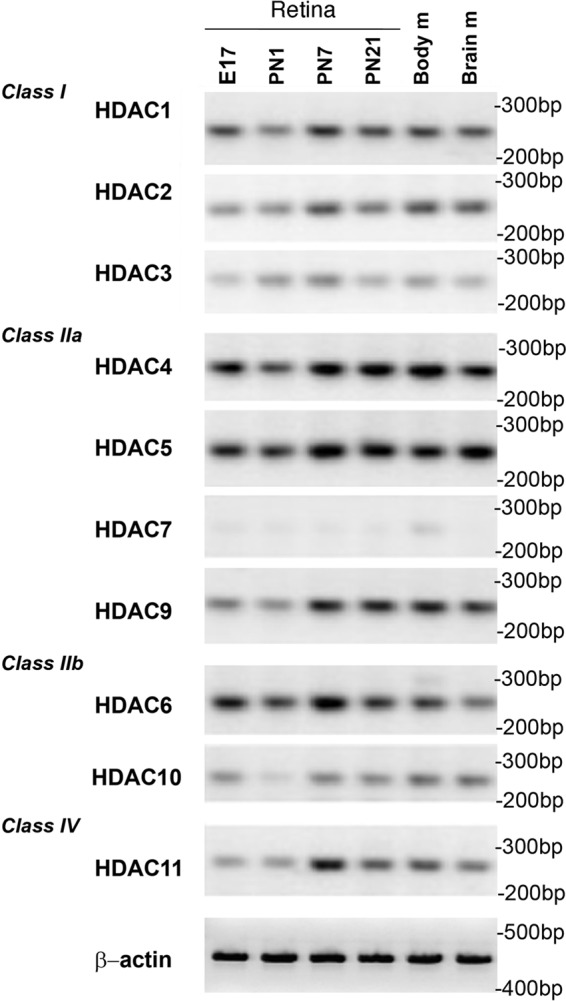
HDAC expression in the mouse retina during development from E16 to PN21measured by RT-PCR. RNA was isolated from mouse retina at specific developmental times, and samples from brain and body mixed tissue (adult) were used as controls. β-Actin RT-PCR was used as a loading control.
TABLE 1.
HDAC inhibitors used in this work
| HDACs | Inhibitor | IC50 | Refs. |
|---|---|---|---|
| HDAC1 | CAS 193551-00-7 | 100 (HDAC1); >1,000 nm (HDAC3) | 23, 25 |
| HDAC3 | Apicidin | 8 nm (HDAC3); 120 nm (HDAC 2); > 10,000 nm (HDACs 1, 4, 6, 7, and 9) | 22, 33, 34 |
| HDACs 1 and 2 | Romidepsin | 36 nm (HDAC1); 47 nm (HDAC2); 510 nm (HDAC4); >10,000 nm (HDAC6) | 26–28 |
| HDACs 1, 2, 3, and 9 | Entinostat (MS-275) | 181 nm (HDAC1); 1100 nm (HDAC2); 2300 nm (HDAC3); 505 nm (HDAC9); >10,000 nm (HDACs 4, 6, 7, and 8) | 22, 35–37 |
| Class IIa | MC-1568 | 220 nm (HDACs 4, 5, 7, and 9) | 29, 38, 39 |
| Global | Sodium butyrate | 0.8 mm | 20, 21, 40, 41 |
FIGURE 2.
Inhibition of class I HDAC affects rhodopsin expression. A–D, immunofluorescence microscopy of PN1 retina explants cultured for 96 h with DMSO (control; A); 9 μm MC-1568 (HDAC class IIa inhibitor; B), 5 μm entinostat (HDAC class I inhibitor; C), and 1 mm sodium butyrate (pan-HDAC inhibitor; D). Cryosections were stained with anti-RHO (green) and anti-H3K9ac (red) antibodies, and nuclei were counterstained with Hoechst 33358 (blue). ONBL, outer neuroblast layer; INBL, inner neuroblast layer; GCL, ganglion cell layer. Image quantification of immunofluorescence intensity was performed for three biological replicates with three technical replicates for each sample and normalized to control ±S.E. E, rhodopsin expression. F, H3K9ac levels. ***, p < 0.001. Error bars represent S.E.
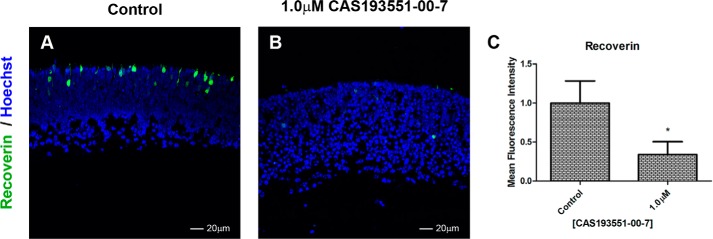
Entinostat inhibits all class I HDACs. Because selective inhibitors for HDAC1 and HDAC3 are available, we wanted to determine whether inhibition of one or both of these enzymes was responsible for the results seen with entinostat. We incubated retinas with either CAS 193551-00-7, an HDAC1 selective inhibitor, or apicidin, an HDAC3 selective inhibitor. Inhibition of HDAC1 using CAS 193551-00-7 resulted in a dose-dependent decrease in rhodopsin (Fig. 3, A–G and I) and an increase in labeling by antibodies recognizing H4K12ac (Fig. 3, A–D and J) and H3K9ac (Fig. 3, F and K) but no change in labeling with anti-H3K27ac (Fig. 3, G, H, and L). At the highest concentration of CAS 193551-00-7, there was a clear loss of retinal structure. To confirm that HDAC1 inhibition was not selectively impacting rhodopsin expression but had a more general effect on photoreceptors generation, we examined another photoreceptor specific protein, recoverin, and found that levels were decreased under specific HDAC1 inhibition similarly to rhodopsin (Fig. 4).
FIGURE 3.
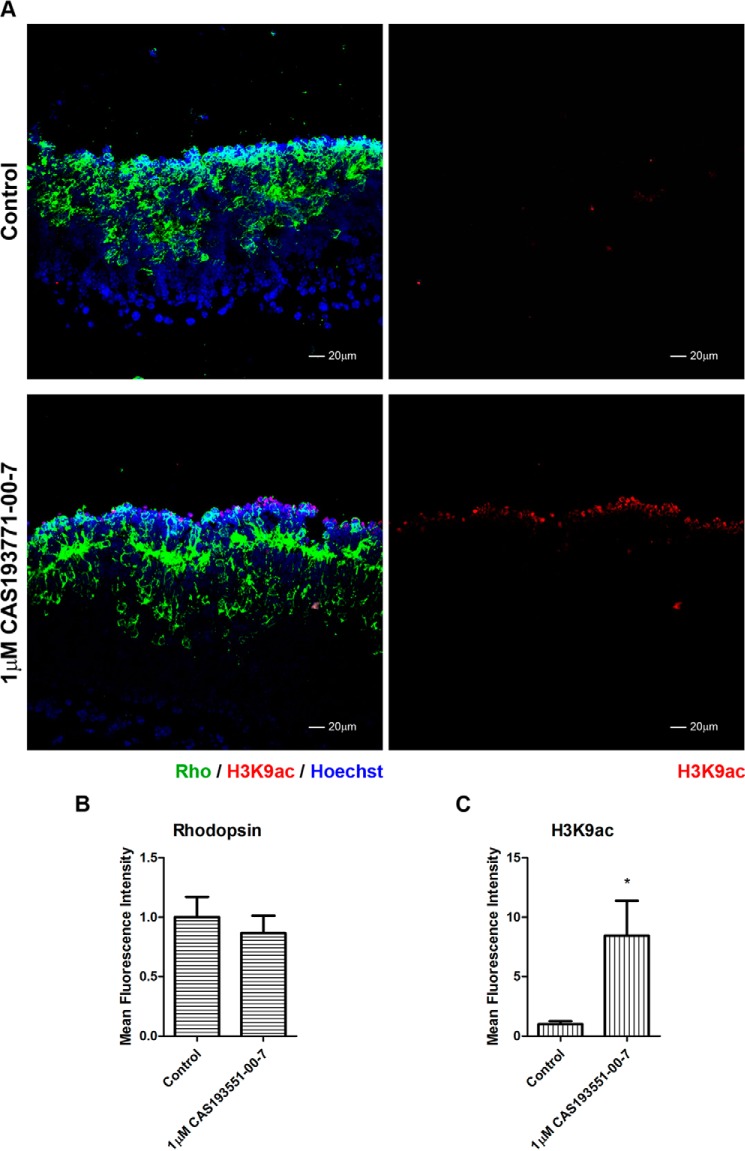
Specific inhibition of HDAC1 blocks rhodopsin expression. A–H, immunofluorescence microscopy of PN1 retina explants cultured for 96 h with DMSO (control; A, E, and G) or treated with different concentrations of CAS 193551-00-7 (HDAC1i): 0.1 (B), 0.5 (C), and 1.0 μm (D, F, and H). Cryosections were stained with anti-RHO (green) (A–H), anti-H4K12ac (red) (A–D), anti-H3K9ac (red) (E and F), and anti-H3K27ac (red) (G and H) antibodies, and nuclei were counterstained with Hoechst 33358 (blue). Image quantification of immunofluorescence intensity was performed for three biological replicates with three technical replicates for each sample and normalized to control ±S.E. I, rhodopsin expression. J, H4K12ac levels. K, H3K9ac levels. L, H3K27ac levels. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars represent S.E.
FIGURE 4.
Specific inhibition of HDAC1 affects recoverin expression. A and B, immunofluorescence microscopy of PN1 retina explants cultured for 96 h with DMSO (control; A) or treated with 1.0 μm CAS 193551-00-7 (HDAC1i; B). Cryosections were stained with anti-recoverin (green), and nuclei were counterstained with Hoechst 33358 (blue). Image quantification of immunofluorescence intensity was performed for three biological replicates with three technical replicates for each sample and normalized to control ±S.E. C, recoverin expression. *, p < 0.05. Error bars represent S.E.
Inhibition of HDAC3 by apicidin (Fig. 5, A–C) resulted in a modest decrease (25%) in rhodopsin staining (Fig. 5D) and no visible change in anti-H4K12ac (Fig. 5E) or H3K9ac labeling (Fig. 5F). There is no HDAC2-specific inhibitor, and we were unable to assess HDAC2-specific regulation of retina development, but we could not rule out a possible role of HDAC2 in this process. From these results, we conclude that class I HDACs are important for rod differentiation as defined by the appearance of rhodopsin- or recoverin-expressing cells in developing retina and that within this class HDAC1 is the most important for this process.
FIGURE 5.
Effect of HDAC3 inhibition on rhodopsin expression. A–F, immunofluorescence microscopy of PN1 retina explants cultured for 96 h with DMSO (control; A and D) or treated with different concentrations of apicidin (HDAC3i): 8 (B and E) and 16 nm (C and F). Cryosections were stained with anti-RHO (green), anti-H4K12ac (red) (A, B, and C), and anti-H3K9ac (D, E, and F) antibodies, and nuclei were counterstained with Hoechst 33358 (blue). Image quantification of immunofluorescence intensity was performed for three biological replicates with three technical replicates for each sample and normalized to control ±S.E. G, rhodopsin expression. H, H4K12ac levels. I, H3K9ac levels. *, p < 0.05; **, p < 0.01. Error bars represent S.E.
To confirm the effects of the different HDAC inhibitors (HDACis) on the global pattern of histone acetylation in the retina shown by immunofluorescence staining, we performed Western blotting using antibodies against H3K9ac, H3K27ac, and H4K12ac marks. PN1 explants were incubated in the presence of six different HDACis for 96 h. As shown in Fig. 6, inhibition of all HDACs with sodium butyrate increased the acetylation level of all three histone modifications. Selective inhibition of HDACs 1 and 2 by romidepsin or class I HDACs by entinostat also increased the levels of the three acetylated histones. Selective inhibition of HDAC1 by CAS 193551-00-7 showed an increased acetylation for H4K12ac and H3K9ac but not for H3K27ac, matching the immunofluorescence results. In contrast, inhibition of HDAC3 by apicidin resulted in little to no change in the acetylation levels when compared with the control, confirming the lack of change in immunocytochemical labeling of H4K12ac and H3K9ac described above. Similarly, inhibition of class IIa HDACs by MC-1568 had no effect on the acetylation levels. From these results, we conclude that HDAC1 has a more widespread effect on histone acetylation in PN1–PN4 retina explants than either HDAC3 or class IIa HDACs.
FIGURE 6.

Selective inhibition of HDAC1 increases global level of H3K9ac and H4K12ac but not H3K27ac. Western blots of samples isolated from mouse PN1 retina explants cultured for 96 h with DMSO (control) or treated with 1.0 μm CAS 193551-00-7 (HDAC1i), 16 nm apicidin (HDAC3i), 54 nm romidepsin (HDAC1,2i), 5 μm entinostat (HDAC class I inhibitor), 36 μm MC-1568 (HDAC class IIa inhibitor), or 5 μm sodium butyrate (pan-HDAC inhibitor) are shown. Blots were probed with antibodies against H4K12ac (A), H3K9ac (B), and H3K27ac (C). Coomassie staining of core histones was used as a loading control (D). Band intensity quantification was performed for three biological replicates for each histone acetylation mark and normalized to control ±S.E. E, the dashed rectangle in Coomassie staining of core histones indicates the position where Western blots were spliced. **, p < 0.01. Error bars represent S.E.
HDAC1 Inhibition Results in Less Cell Proliferation and More Cell Death
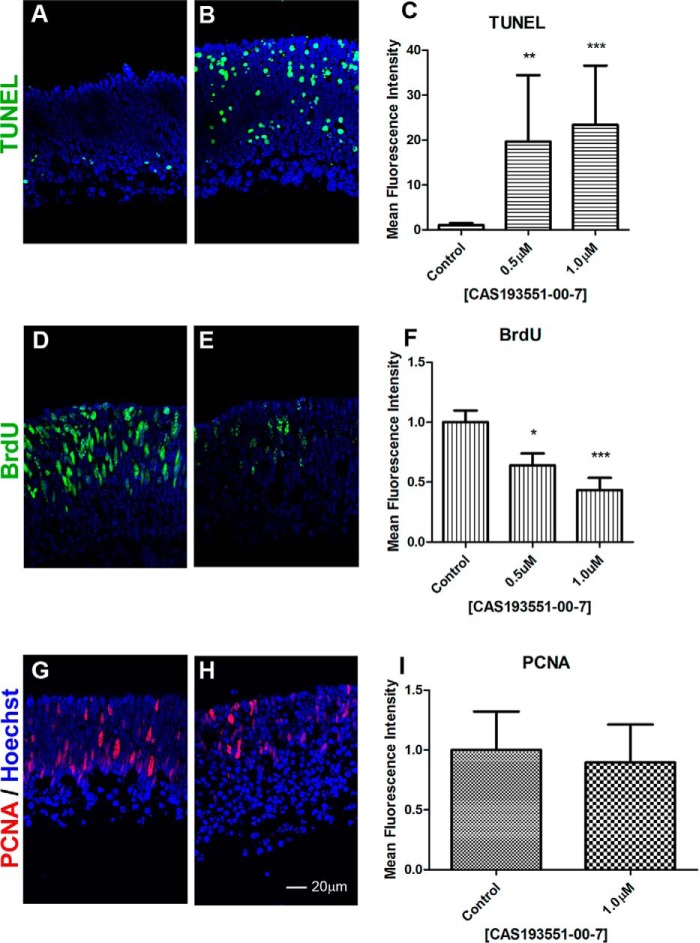
Pan-HDAC inhibition in retina explants by TSA resulted in increased cell death and reduction in proliferation (31). We tested whether an HDAC1-specific inhibitor had the same effect by assaying cell death by TUNEL and cell proliferation by BrdU incorporation and proliferating cell nuclear antigen (PCNA) staining. Retina explants treated for 48 h with CAS 193551-00-7 (HDAC1i) showed an increase in the number of apoptotic cells by TUNEL assay (Fig. 7, A–C) and reduction in BrdU incorporation (Fig. 7, D–F). For comparison, explants treated with apicidin (HDAC3i) had no difference in proliferation as judged by BrdU incorporation (data not shown). Retinal explants treated for 96 h with HDAC1i showed a similar number of cells positive for PCNA when compared with the control (Fig. 7, G–I).
FIGURE 7.
HDAC1 inhibition reduces BrdU incorporation and increases apoptosis. Immunofluorescence microscopy of PN1 retina explants cultured for 48 h (A–F) or 96 h (G–I) with DMSO (control; A, D, and G) or treated with 1.0 μm CAS 193551-00-7 (HDAC1i; B, E, and H) was performed. Cryosections were stained with TUNEL labeling (A and B), (green), anti-BrdU antibodies for BrdU incorporation (green) (D and E), or anti-PCNA antibodies (red) (G and H), and nuclei were counterstained with Hoechst 33358 (blue). Image quantification of immunofluorescence intensity was performed for three biological replicates with three technical replicates for each sample and normalized to control ±S.E. C, TUNEL labeling. F, BrdU incorporation. I, PCNA level. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars represent S.E.
HDAC1 Inhibition Effect on Retina Is Reversible and Stage-specific
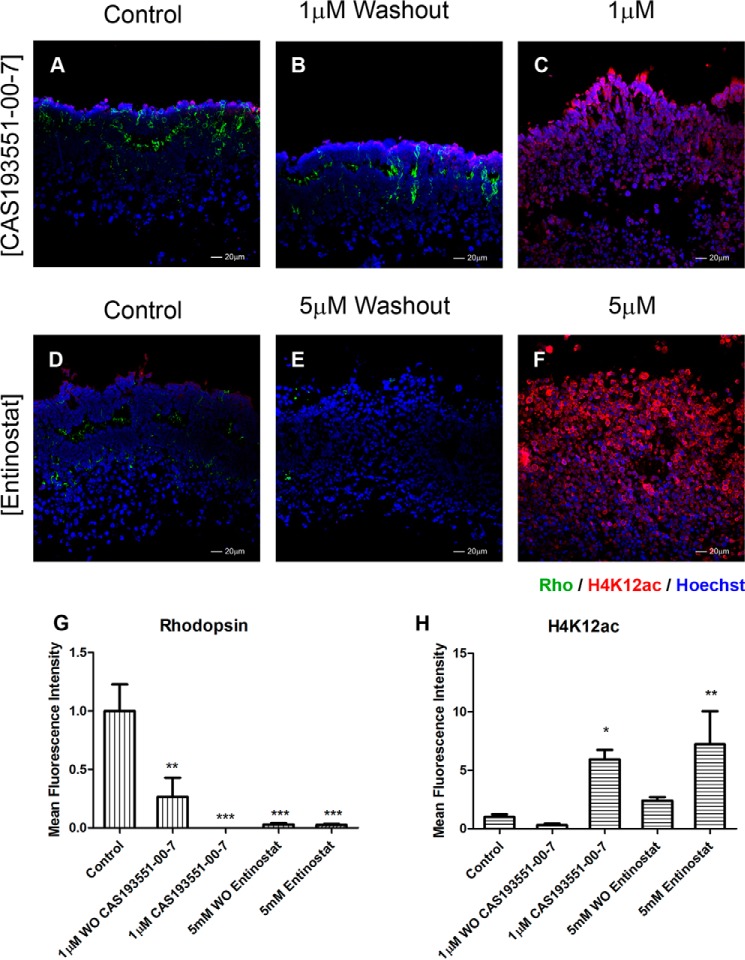
To determine whether HDAC1 inhibition merely paused development or permanently altered the retinal cells, we studied whether CAS 193551-00-7 treatment is reversible. Retina explants were incubated for 96 h in the presence of CAS 193551-00-7 (Fig. 8, A–C) or entinostat for comparison (Fig. 8, D–F), then inhibitors were washed out, and retinas were cultured for another 96 h without the inhibitors. After HDAC1i washout, rod photoreceptor development resumed with an increase in rhodopsin levels compared with explants maintained with inhibitor (Fig. 8G). H4K12 acetylation levels following washout were reduced (Fig. 8H). Both results suggest that the effects of HDAC1 inhibition on rod development are reversible. Although washout of entinostat partially decreased H4K12ac level, it failed to promote rhodopsin expression.
FIGURE 8.
HDAC1 inhibition is reversible. Immunofluorescence microscopy of PN1 retina explants cultured for 8 days with DMSO (control; A and D), 1.0 μm CAS 193551-00-7 (HDAC1i; C), or 5 μm entinostat (HDAC class I inhibitor; F) was performed. For washout (WO) conditions, explants were treated for 96 h with inhibitor and then cultured without inhibitors for an additional 96 h: CAS 193551-00-7 (B) and entinostat (E). Cryosections were stained with anti-RHO (green) (A–F) and anti-H4K12ac (red) (A–F) antibodies, and nuclei were counterstained with Hoechst 33358 (blue). Image quantification of immunofluorescence intensity was performed for three biological replicates with three technical replicates for each sample ±S.E. G, rhodopsin expression. H, H4K12ac levels. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars represent S.E.
We next investigated whether the effect of selective HDAC1 inhibition is stage-specific and treated explant cultures from PN7 retinas for 48 h with CAS 193551-00-7 (Fig. 9A). We did not observe any changes in retina structure or rhodopsin level compared with control (Fig. 9, A, left panels, and B) despite prominent up-regulation of acetylation of H3K9 (Fig. 9, A and C). This suggests that HDAC1 acts at a distinct developmental window during the first postnatal week of mouse retina maturation when precursors differentiate into rod photoreceptors.
FIGURE 9.
HDAC1 inhibition of rhodopsin is stage-specific. A, immunofluorescence microscopy of PN7 retina explants cultured for 48 h with DMSO (control; upper panels) or 1.0 μm CAS 193551-00-7 (HDAC1i; lower panels). Cryosections were stained with anti-RHO (green) (left panels) and anti-H3K9ac (red) antibodies, and nuclei were counterstained with Hoechst 33358 (blue). Image quantification of immunofluorescence intensity was performed for three biological replicates with three technical replicates for each sample ±S.E. B, rhodopsin expression. C, H3K9ac levels. *, p < 0.05. Error bars represent S.E.
Inhibition of HDAC1 Prevents the Increased Expression of a Specific Subset of Photoreceptor Genes while Maintaining the Levels of Expression of Genes Associated with Retinal Progenitors
To determine whether the effect of HDAC1 inhibition was on rod photoreceptor development in general or selectively on expression of rod-specific genes such as rhodopsin or recoverin, we tested RNA levels of a panel of 31 genes that represent different groups of genes known to be down-regulated or up-regulated during retina maturation. RNA was collected from retinal explants treated for 96 h with 0.5 or 1.0 μm CAS 193551-00-7, and genes were considered up- or down-regulated if the expression was significantly different from controls with a p value <0.05.
These data revealed a number of important results (Fig. 10A). First, all nine rod-specific genes tested were expressed at lower levels in both treated groups with Rho, Pde6b, Samd11, Sag, Rom1, and Nrl showing more than 10-fold lower expression, demonstrating that HDAC1 inhibition blocks the program of rod photoreceptor differentiation rather than affecting any individual gene.
FIGURE 10.
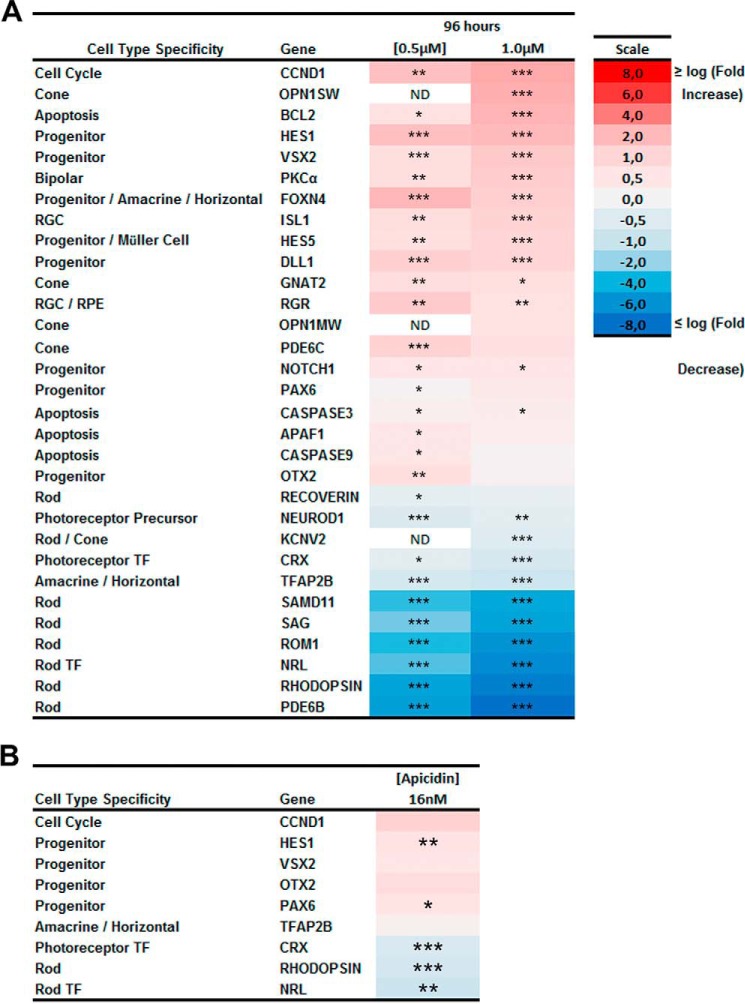
Inhibition of HDAC1 blocks expression of rod-specific genes while promoting expression of progenitor genes. A, heat map of expression of 31 genes measure by RT-PCR for retinas treated for 96 h with 0.5 or 1.0 μm CAS 193551-00-7 (HDAC1i) compared with control with only DMSO with the log -fold increase in red and decrease in blue. B, heat map of expression of nine genes measure by RT-PCR for retinas treated for 96 h with 16 nm apicidin (HDAC3i) compared with control with the log2 -fold increase in red and decrease in blue. All samples were performed for three biological replicates with three technical replicates each. The relative expression level for each gene was calculated by the 2−ΔΔCt method and normalized to GAPDH. *, p < 0.05; **, p < 0.01; ***, p < 0.001. ND, not determined; RPE, retinal pigmented epithelium; RGC, retinal ganglion cell; TF, transcription factor.
Second, seven genes of 13 that were up-regulated in both treated samples were progenitor genes, Otx2, Dll1, Notch1, Vsx2, Foxn4, and Hes1, or a cell cycle regulator, Ccnd1, with 2–5 times higher expression in treated compared with untreated explants. This suggests that a second major effect of HDAC1 inhibition is the maintenance of expression of the group of key progenitor genes that are normally turned off as the retina develops and could be achieved by keeping histone acetylation at promoters of these progenitor genes.
Third, several genes specific for non-rod retinal cell types were also up-regulated under HDAC1 inhibition. Expression levels were increased for a marker of bipolar cells (42), Pkcα (2.6-fold); a marker of Müller glia cells (43), Hes5 (2.1-fold); a marker of retinal ganglion cells (44), Isl (2.2-fold); and four markers of cone photoreceptors (45), Pde6c, Opn1sw, Opn1mw, and Gnat2 (>1.5 times) but not Kcnv2 gene that is expressed in both cone and rod photoreceptors. Bipolar and Müller cells differentiate postnatally, and our results are in accordance to previous studies and might indicate that bipolar and Müller cell fate may be driven by a loss of HDAC activity (31). We conclude from this that HDAC1 is essential and specific for rod photoreceptor differentiation. Fourth, in agreement with results of a decrease in cell proliferation and an increase in cell death under HDAC1 inhibition (Fig. 7), expression of apoptotic genes (46) Bcl2, Apaf1, Casp3, and Casp9 was up-regulated.
For comparison, we tested the expression pattern of a panel of nine genes selected from the earlier panel to represent genes that showed significant changes in expression in retinal explants treated with HDAC3 inhibitor apicidin as a decrease in rhodopsin staining was also observed in retinas treated with this inhibitor. Transcription factors Crx and Nrl and rhodopsin expression were modestly down-regulated (<2-fold) in the treated retinas as compared with control (Fig. 10B). Conversely, Pax6 and Hes1 expression was up-regulated slightly (< 1.5-fold) as compared with control.
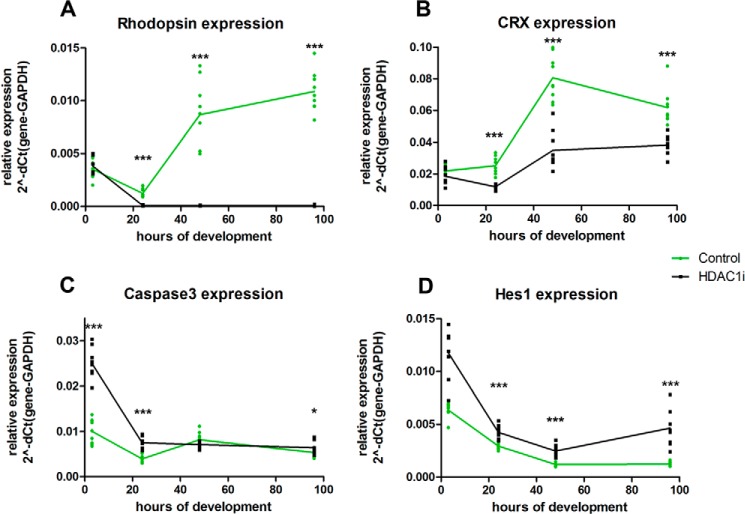
We then evaluated the time course of changes in expression level under HDAC1i for several key genes. At 3-h incubation of retina explant culture, there were no changes in Rho level with HDAC1i, but after 3 h its expression increased in control samples but markedly declined in HDAC1i-treated explants (Fig. 11A). Crx expression had a different time course compared with rhodopsin; at 3-h incubation with HDAC1i, its level was not significantly different from that in control explants, but then its level fell and remained significantly below control levels even at 96 h of incubation (Fig. 11B). We used caspase3 expression as a proxy to assess the process of apoptosis under HDAC1i. The level of Casp3 was already up-regulated at 3 h under HDAC1i, but it returned to the level in the control explant after 48 h (Fig. 11C). Levels of Hes1 decreased in control samples up to 48 h and then plateaued, whereas under HDAC1i expression the level of Hes1 was higher compared with control at all time points, and it began to increase after 48 h (Fig. 11D). We conclude from these experiments that there is a burst of apoptosis in retina explants under HDAC1i that starts earlier than down-regulation of Rho and that low RHO levels are due to combination of apoptosis of progenitors and maintained expression of the suppressor HES1.
FIGURE 11.
Time course of expression level for several key genes under HDAC1 inhibition. Expression of four genes was measured by RT-PCR for retinas treated for 3, 24, 48, and 96 h with 1.0 μm CAS 193551-00-7 (HDAC1i) compared with control with only DMSO. The relative expression level for each gene was calculated by the ΔCt to GAPDH. Data were obtained for three biological replicates with three technical replicates each. p values were as follows for comparison between control and HDAC1i-treated samples: *, p < 0.05; ***, p < 0.001.
HDAC1 Inhibition Leads to Maintenance of High Histone Acetylation Levels on Promoters of Progenitor Genes
As described in the previous section, HDAC1 inhibition led to maintenance of expression of a group of key regulatory progenitor genes. To test whether this was achieved by keeping histones acetylated at the promoters of these genes, we examined the promoters of progenitor gene Vsx2 (Fig. 12A) and rod-specific gene Rho (Fig. 12B) by chromatin immunoprecipitation (ChIP) and RT-PCR. We first analyzed existing retina developmental genome-wide data sets to understand how chromatin organization changed during development for these two genes. We mapped ChIP-sequencing data for inhibitory mark trimethylation of lysine 27 of histone 3 (H3K27me3) (E17, PN1, PN7, and PN15); active mark dimethylation of lysine 4 of histone 3 (H3K4me2) (E17, PN1, PN7, and PN15) (47); binding sites for photoreceptor-specific transcription factors CRX (PN56) (48) and NRL (PN28) (49); polymerase (Pol) II-binding sites (PN2 and PN25) (50); and DNase-hypersensitive sites (DHS) (51), which correspond to open chromatin regions (PN1, PN7, and PN56) (Fig. 12, A and B). The Vsx2 gene showed chromatin features characteristic for developmental down-regulation (Fig. 12A) as H3K4me2 marks and DHS decreased during retina maturation, but H3K27me3 accumulated around the promoter. There are no Pol II- or CRX-binding sites around the promoter of Vsx2. The Rho promoter showed the opposite patterns of chromatin features (Fig. 12B) with essentially no H2K27me3, increasing amounts of the active mark H3K4me2, and increased DHS as well as Pol II-, NRL-, and CRX-binding sites in mature retina.
FIGURE 12.
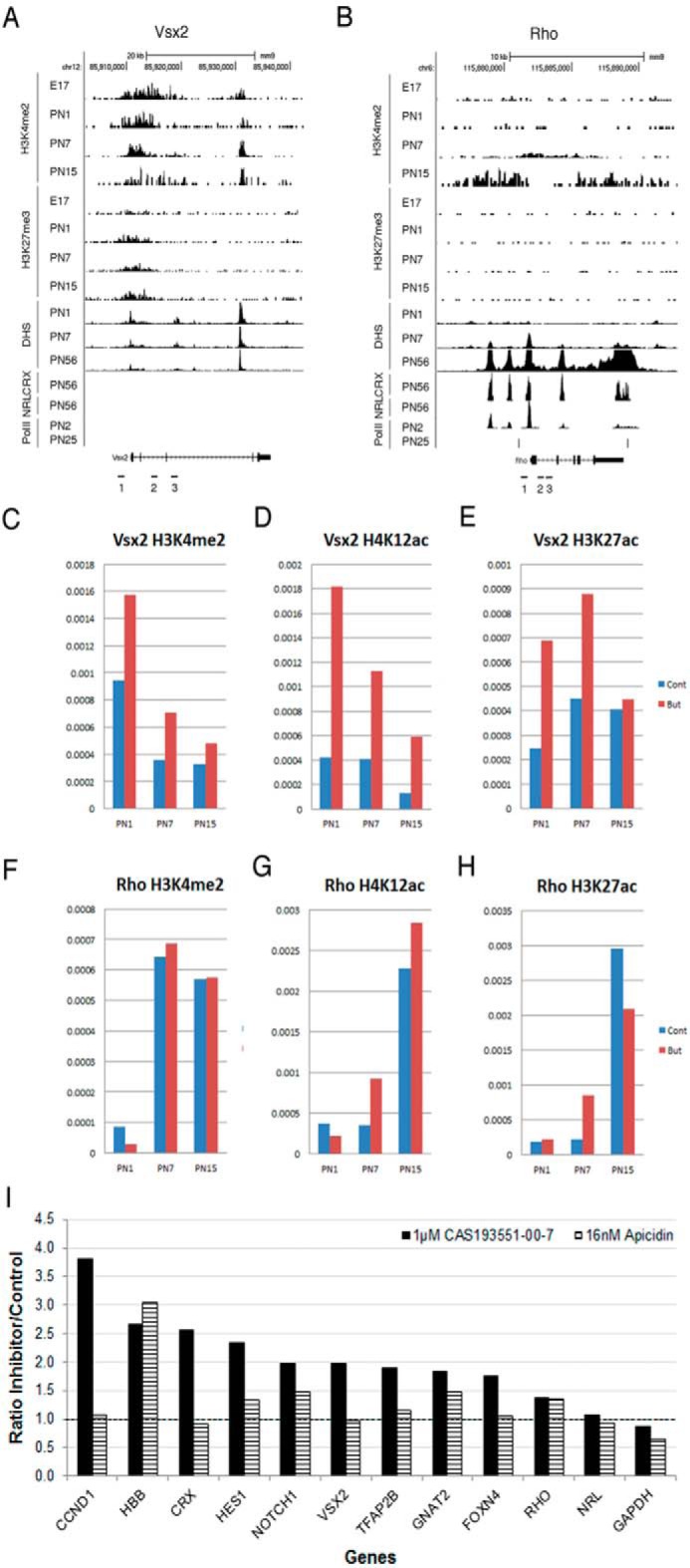
HDAC1 inhibition maintains levels of histone acetylation on promoters of progenitor genes. A and B, combined genome-wide tracks of mouse retina samples for H3K4me2 and H3K27me3 ChIP-sequencing analysis, DHS, and NRL-CRX- and Pol II-binding sites for mouse Vsx2 (A) and Rho (B) gene loci. C–H, ChIP analysis of the developmental changes in accumulation of H3K4me2 (C and F), H4K12ac (D and G), H3K27ac (E and H) at Vsx2 (C–E) and Rho (F–H) genes on samples from mouse retina at PN1, PN7, and PN15. ChIP experiments were carried with 5 mm sodium butyrate (red) or without (blue) in two biological replicates; quantitative RT-PCRs were done with three sets of primers for the area around TSS and promoter of the gene in three technical replicates. Primers pair locations are depicted as numbered black bars underneath genome maps of the genes (A and B). y axis, fraction of input represented as average accumulation of histone modification for three sets of primers. I, comparison of H4K12ac accumulation on gene promoters in retina explant cultured for 96 h with either 1.0 μm CAS 193551-00-7(HDAC1i), 16 nm apicidin (HDAC3i), or medium only. ChIP experiments were done in triplicate for CAS 193551-00-7 and in duplicate for apicidin; quantitative RT-PCRs were done with primers for the area around TSS and promoter of the gene in three technical replicates.
We next assessed levels of acetylation marks H3K27ac and H4K12ac and compared these with H3K4me2 on the Vsx2 and Rho promoters and transcription start site (TSS) by ChIP-RT-PCR at three stages of retina development, PN1, PN7, and PN15, using three pairs of genomic primers. Levels of H4K12ac (Fig. 12D) at the Vsx2 gene locus were decreased during retina development, similar to those of H3K4me2 (Fig. 12C), whereas H3K27ac levels did not change (Fig. 12E). Addition of the pan-HDAC inhibitor sodium butyrate during the ChIP procedure substantially increased the level of all three active histone modification marks at Vsx2 locus, especially H4K12ac at all developmental stages. In comparison, Rho locus was characterized by increased accumulation of all three active histone modifications during development (Fig. 12, F–H), but addition of sodium butyrate had no effect, or only a small effect, on this accumulation.
Because we saw an effect of histone modification marks at the promoter of Vsx2 using the pan-HDAC inhibitor sodium butyrate, we decided to verify the effects of HDAC1 inhibitor CAS 193551-00-7 and HDAC3 inhibitor apicidin on the histone acetylation mark H4K12ac around the promoter and TSS of this and several other progenitor genes that were up-regulated under HDAC inhibition as well as several rod-specific genes that were down-regulated under HDAC inhibition. PCRs used primers specific for promoter regions in 11 genes that showed statistical difference in the expression levels when treated with both inhibitors (Fig. 12I). Treatment with HDAC1i increased the H4K12ac level in most of the genes analyzed, especially Ccnd1, Crx, Hes1, Notch1, and Vsx2 (>2-fold change). Only Nrl, Rho, and Gapdh showed no difference in H4K12ac between treatment and control. In contrast, we observed no increase in the H4K12ac mark in retinas treated with HDAC3i on any of the 11 genes (Fig. 12I). Interestingly, Hbb, a gene not expressed in the retina, showed substantial increases in H4K12ac after treatment with either HDAC1 or HDAC3 inhibitor.
Discussion
Histone acetylation is generally thought to be associated with the opening of chromatin and the activation of transcription. The phenotypic effect of modulating histone acetylation is, however, more complex. Inhibiting HDACs can induce differentiation in both embryonic and adult CNS progenitors (16, 52, 53), but following conditional deletion of HDAC1 and HDAC2 progenitors were unable to undergo differentiation into mature neurons and underwent cell death (17).
Some of the differences in reported roles of HDACs may well be due to the different temporal and spatial distribution of the various isoforms. We found members of most classes of HDACs in the retina, each with its own developmental profile, corroborating data from others (31, 54–56). Previous studies have shown that non-selective inhibition of HDACs in the developing retina blocked formation of rod photoreceptors (31, 32), raising the question of which class of HDACs might be involved in regulating rod development. Using a variety of HDAC inhibitors, we found that HDAC1 was key in allowing the differentiation of rod photoreceptors. Although this was originally defined using expression of opsin and recoverin as markers of rods, a group of other rod-specific genes showed similar blocked expression, indicating that the whole program of rod development was inhibited. Interestingly, genes for other retinal cell types showed increased expression. This was true for the other late developing retinal cells (bipolar cells and Müller glial cells) as well as early developing retinal cells (ganglion cells and cone photoreceptors). It is possible that when the progenitor pool is prevented from moving into a rod differentiation pathway the cells take on other fates.
HDAC1-null mice are lethal, and the animals die before E10.5 (57). Lethal defects in neuronal elements are seen also in zebrafish deleted for HDAC1 (58–60). In mice lacking HDAC1 in the central nervous system, neuronal progenitors do not differentiate and undergo cell death (17). Similarly, we found an increase in cell death and a decrease in cell proliferation when retinal explants were incubated with an HDAC1 inhibitor. A decrease in BrdU incorporation has been observed in the retinas of zebrafish HDAC1 mutants (60). One of the genes whose expression is increased when HDAC1 is inhibited is Ccnd1. The up-regulation of Ccnd1 in the treated retinas was unexpected because a decrease in BrdU incorporation was observed. At this point in development, photoreceptor progenitor cells become postmitotic and begin to differentiate. It is likely that because cells fail to leave the cell cycle cyclin D1 accumulates and induces apoptosis (61). This is with agreement with our observed up-regulation of the apoptotic genes Apaf1, Bcl2, and Casp3.
Although BrdU incorporation decreased in the presence of HDAC1 inhibitors, PCNA labeling remained the same. This is probably due to the different distribution of these two proliferation markers during the cell cycle. PCNA starts to increase through G1, peaking at the G1/S phase, whereas BrdU is only incorporated in the S phase. This would suggest that inhibiting HDAC1 prevents cells from moving into S phase, leading to an increase in cell death and movement into different differentiation pathways. Not all the rod progenitors die because washout of the HDAC1 inhibitor led to resumption of rod generation, indicating survival of a pool of rod progenitors.
Although we have not tested all HDAC isoforms, the only other enzyme regulating rod formation was HDAC3. The effect of HDAC3 was less prominent as determined both by reduction of rhodopsin expression and by changes in histone acetylation assayed by both immunocytochemistry and Western blotting. More modest changes in expression of particular genes were also observed. In particular, levels of RNA for the transcription factor Crx were reduced almost 2-fold, and this may have been sufficient to account for the reduced rhodopsin expression.
Acetylation of non-histone proteins can affect a number of cellular functions because it influences mRNA localization and stability as well as protein signaling, transcription, and degradation (for a review, see Ref. 62). Because HDAC1 lacks a nuclear export signal, this enzyme is primary localized in the nucleus; therefore, the effect of its inhibition is probably due to its action on histone proteins and not on non-histone targets. In contrast, HDAC3 has a nuclear export signal. It has been shown that HDAC3 can influence STAT3 phosphorylation at serine 727 through the interaction of phosphatase 2A protein (63). The persistent activation of STAT3 was shown to suppress rod cell fate by decreasing Crx and rhodopsin expression and increasing the precursor markers Hes1 and OTX2 (64–66). This effect on non-histone HDAC substrates could explain our results for HDAC3 inhibition because the mRNA levels for Crx and rhodopsin were down-regulated and Hes1 was up-regulated in retinas treated with apicidin as compared with control.
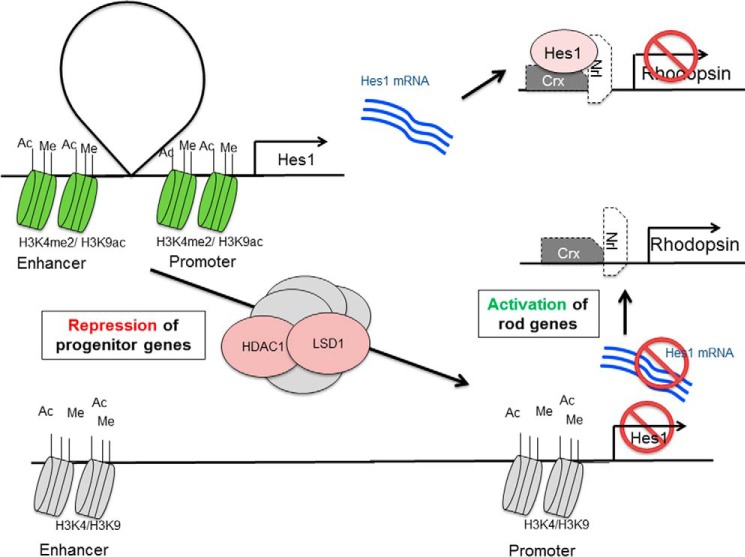
In previous experiments, we showed that inhibiting histone demethylation by lysine demethylase 1 (LSD1) also blocked differentiation and maintains expression of progenitor genes that are normally down-regulated as the tissue matures (67). Our conclusion in that study was that among the genes whose expression was maintained were transcriptional repressors such as Hes1 that are known to block rod differentiation. Similarly, inhibition of HDAC1 caused maintained expression of Hes1. From these data, we suggest that the changes of gene expression seen with HDAC1 inhibition may be due to its direct action on the chromatin of progenitor genes. Higher levels of histone acetylation lead to an open chromatin conformation around the promoter and TSS and prevent the normal developmental switching off of the gene with subsequent effects on cell proliferation and further cell differentiation. It has been shown that when HDAC1 and LSD1 deacetylate histones and demethylate H3K4me2/1, respectively, as parts of complexes CoREST and NuRD this causes repression of both neuronal and non-neuronal genes (68–72). In the course of retina development, the two histone-modifying enzymes HDAC1 and LSD1 may work in a common complex at the point of transition from late progenitor to differentiated rod photoreceptor (schematized in Fig. 13). Because neither HDAC1 nor LSD1 have DNA binding properties, the temporal and spatial control of the actions of these enzymes during development must reside in other components of the complexes. In retinoblastoma cells, HDAC1 is found together with LSD1 in CoREST complex-repressed genes where the complex is specifically bound to target gene promoters through the nuclear receptor NR2E1 (TLX) (73).
FIGURE 13.
Schematic presentation of proposed function of HDAC1 and LSD1 complex during retina maturation. Top, in progenitors, the repressor gene Hes1 is expressed, and it blocks the differentiation of rods. Bottom, by the action of protein complexes containing HDAC1 and LSD1, histone modifications at the Hes1 enhancer and promoter are changed, and the gene is no longer expressed. Lack of HES1 protein allows activation of rod genes and rod terminal differentiation.
In summary, we have shown that inhibition of HDAC1, but not HDAC3, can prevent the developmental decrease in expression of progenitor genes and the developmental increase in expression of genes characteristic of terminally differentiated rod photoreceptors. The actions of HDAC1 affect both H3K9 and H4K12 but not H3K27, indicating targeting of this enzyme to specific chromatin sites. The similarities in phenotype observed by inhibiting HDAC1 and LSD1 and their co-expression in chromatin-modifying complexes suggest a coordinated interaction between acetylation and methylation in the regulation of the transition from progenitor to differentiated photoreceptor in the mammalian retina and possibly other tissues. Thus, our results demonstrate that, in addition to important roles of transcriptional factors during development and cell type specification, epigenetic regulation is essential for proper retina maturation. An interesting application for epigenetic modifiers such as inhibitors of HDACs or LSD1 could be for reprogramming cell phenotype during development as our studies show that, in parallel with a decrease in expression of rod photoreceptor-specific genes, genes specific for other retina cell types are up-regulated. This reprogramming could be a useful tool to fight a number of degenerative diseases of the CNS.
Experimental Procedures
Mice
C57Bl/6j mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All animal experiments were approved by the Animal Care and Use Committee of the Pennsylvania State University School of Medicine (Protocol number 2009-061) and were conducted in accordance with National Institutes of Health guidelines.
Antibodies, HDAC Inhibitors, and Reagents
Anti-H3K27ac (catalog number 07-360; lot numbers 2197510, 2239804, and 2455678) was from Millipore (Temecula, CA). Anti-H3K9ac (ab4441; lot number GR205018-2), anti-H4K12ac (ab46983; lot numbers GR41233-16 and GR41233-35), and anti-H3K4me2 (ab1220; lot number GR32351-1) were from Abcam (Cambridge, MA). Anti-H3K4me3 (CS200604; lot number DAM1462601) was from Upstate (Charlottesville, VA). Antibodies against histone modifications were used before and have passed validation (Antibody Validation Database and Fig. 14) (74). Anti-PCNA (16D10; lot number 090428) was from Chromotec (Hauppauge, NY). Anti-rhodopsin monoclonal antibody RET-P1 was described previously (75). Alexa Fluor 488 mouse anti-BrdU was from BD Biosciences (558599; lot number 84784). Secondary antibodies were Alexa Fluor-conjugated anti-mouse IgG (594-A11005, lot number 1024061 and 488-A11001, lot number 1484573) or anti-rabbit IgG (594-A11012, lot number 1084427 and 488-A11008, lot number 913909) from BD Biosciences. HDACis are listed in Table 1 (23–34). Entinostat, MC-1568, and sodium butyrate were purchased from Sigma. Apicidin and CAS 193551-00-7 were from Santa Cruz Biotechnology (Dallas, TX). Romidepsin was from Apexbio (Houston, TX).
FIGURE 14.
Western blots of nuclear samples isolated from mouse retina during development and probed with antibodies against H3K4me2, H3K9ac, and H4K12ac. Coomassie Blue staining of core histones was used as a loading control.
Retina Isolation and Explant Culture
Retinas from pups at PN1 were isolated by removing the sclera and most of the retinal pigmented epithelium and cultured in 1 ml of UltraCultureTM serum-free medium (Cambrex Bio Science, Rockland, ME) supplemented with gentamycin (10 μg/ml) as described previously (76, 77). Retinas were treated with different concentrations of the specific HDACi for 96 h unless otherwise mentioned. Medium was changed every other day by replacing 0.5 ml with fresh medium and adding the proper concentration of the inhibitor. For the “washout” experiments, retinas were incubated in the presence of the inhibitor for 96 h, washed with medium for five times, and incubated another 96 h with fresh medium with 1% DMSO.
Immunofluorescence Staining
Retina explants were fixed in 4% paraformaldehyde overnight at 4 °C; washed three times in PBS; incubated in 5% sucrose, PBS for 30 min; and then dehydrated in 20% sucrose, PBS overnight at 4 °C. Retinas were embedded in a 2:1 mixture of 20% sucrose and OCT (Optimal Cutting Temperature Compound) (Sakura Finetek, Torrance, CA) and stored at −80 °C. Blocks with tissue samples were sectioned to 8 μm on a Cryostat Microtome HM550 (Thermo Fisher Scientific) and stored at −20 °C. Antigen retrieval was performed by incubating the slides in 10 mm sodium citrate, pH 6, for 30 min at 80 °C. Double labeling immunohistochemistry was performed as described previously (64, 65) using fluorescent dye-conjugated secondary antibodies diluted 1:500 (BD Biosciences). Primary antibodies were diluted as follow: anti-H3K9ac, ant-H3K27ac, and anti-H4K12ac, 1:500; anti-rhodopsin, 1:100; anti-BrdU, 1:20; and anti-PCNA, 1:1000. Slides were counterstained with Hoechst 33258 (1 μg/ml) and visualized using an Olympus Fluoview FV1000 confocal microscope (Olympus, Center Valley, PA). The acquisition parameters were maintained constant for each set of experiments. Fluorescence intensity was assessed and quantitated using the ImageJ (1.48v/Java1.6.0–20) software package.
Western Blotting
PN1 retina explants were cultured in the presence of 54 nm romidepsin, 1 μm CAS 193551-00-7, 5 μm entinostat, 36 μm MC-1568, 16 nm apicidin, or 5 μm sodium butyrate. Proteins were extracted with lysis buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1% SDS, 1 mm PMSF, 1× Halt proteinase inhibitor mixture (Thermo Fisher Scientific)). Samples were sonicated twice, and protein concentration was measure using Bio-Rad DCTM protein assay according to the manufacturer's instructions. Samples were diluted to 1 mg/ml, resolved on a Criterion TGX precast gel Any kD (Bio-Rad), and transferred to nitrocellulose membranes followed by immunoblotting with antibodies anti-H3K9ac, anti-H3K27ac, and anti-H4K12ac diluted 1:5000. Secondary goat anti-rabbit HRP (Jackson ImmunoResearch Laboratory, West Grove, PA; 111-035-144; lot number 87644) was diluted 1:5000. An ECL Western blot detection system (Thermo Fisher Scientific) was used for visualization according to the manufacturer's instructions. The membrane was exposed to HyperfilmTM film (GE Healthcare) for 20 s for H4K12ac, 5 s for H3K9ac, and 30 s for H3K27ac. For quantitative analysis of relative protein levels in samples, the Coomassie-stained gels and autoradiographs after ECL detection were scanned and digitized, and the intensity of protein bands was quantitated using the ImageJ (ImageJ 1.48v/Java1.6.0–20) software package.
Isolation of nuclei from mouse retinas was carried out according to Popova et al. (67). For Western blots presented in Fig. 14, isolated nuclei from retina samples were dissolved in SDS-containing loading buffer, and electrophoresis was carried out in 15% polyacrylamide-SDS gels. Proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore) and detected with primary and secondary HRP-conjugated antibodies.
BrdU Staining
Cell proliferation assays were performed as described previously (67). PN1 retina explants were treated with 0.5 or 1.0 μm CAS 193551-00-7; 8, 16, or 32 nm apicidin; or 1% DMSO for 24 h and then labeled for another 24 h with 10 μm BrdU. BrdU incorporation was detected using the Alexa Fluor 488-labeled mouse anti-BrdU antibody diluted 1:20 (BD Biosciences). Sections were counterstained with Hoechst 33258 (1 μg/ml) and analyzed by confocal microscopy.
TUNEL Assay
PN1 retina explants were treated with 0.5 or 1.0 μm CAS 193551-00-7 or 1% DMSO for 48 h. The TUNEL assay was carried out using the In Situ Cell Death Detection kit (fluorescein) from Roche Applied Science according to the manufacturer's instructions. Sections were counterstained with Hoechst 33258 (1 μg/ml) and analyzed by confocal microscopy.
cDNA Synthesis and RT-PCR
Total RNA was isolated using QIAshredder and an RNeasy mini kit (Qiagen, Germantown, MD) according to the manufacturer's instructions. RNA concentration and purity were determined using a GeneSpectIII (Hitachi, Tokyo, Japan) spectrophotometer. cDNA was synthesized using the SuperscriptIII First-strand Synthesis System (Invitrogen) according to the manufacturer's instructions. Quantitative real time PCRs were made in triplicates using the 2× iQ-SYBR Green PCR Supermix (Bio-Rad) on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Lists of primers used are in Tables 2 and 3. The relative expression level for each gene was calculated by the 2−ΔΔCt method (78, 79) where ΔCt = Ct(target gene) −Ct(Gapdh) and ΔΔCt =ΔCt(target sample) − ΔCt(reference sample), and the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as a reference gene. During retina development in PN1–PN4 explant cultures, the Gapdh expression level is stable (76). Genes were considered up- or down-regulated if the p value was <0.05.
TABLE 2.
PCR primers used in this work
| Gene | Sequence | Fragment size |
|---|---|---|
| bp | ||
| cdna_Apaf1-Fa | GTA CAC CCC CTG AAA AGC AA | 108 |
| cdna_Apaf1-Ra | CAG GGT GGG TCA CCA TCT AT | |
| cdna_Bcl2-Fa | AGC CCG TGT TTG TAA TGG AG | 476 |
| cdna_Bcl2-Ra | CAC AGC CTT GAT TTT GCT GA | |
| cdna_Caspase3-Fa | AGG GGT CAT TTA TGG GAC AAA | 422 |
| cdna_Caspase3-Ra | TAC ACG GGA TCT GTT TCT TTG | |
| cdna_Caspase9-Fa | CAG GCC CGT GGA CAT TGG TT | 438 |
| cdna_Caspase9-Ra | CAG CCG CTC CCG TTG AAG ATA | |
| cdna Ccnd1 | Qiagen catalog number QT00154595 | 108 |
| cdna Crx | Qiagen catalog number QT00115402 | 127 |
| cdna_Dll1-F | TCG ACT CCT TCA GCC TGC | 233 |
| cdna_Dll1-R | GCT ACT GTG AAG GTC CTG AGA C | |
| cdna_Foxn4-F | GTG AGA TCT ACA GCT TCA TGA AGG | 275 |
| cdna_Foxn4-R | TGA GAT GAG CTT GTC CAA CTC C | |
| cdna GAPDH | Qiagen catalog number QT01658692 | 144 |
| cdna_Gnat2-F | ACC ATG CCT CCT GAG TTG | 187 |
| cdna_Gnat2-R | TGA CTC TGG ATC GAA GCA C | |
| cdna_Hes1-F | TTC CAA GCT AGA GAA GGC AGA C | 191 |
| cdna_Hes1-R | GCA CCT CGG TGT TAA CGC | |
| cdna_Hes5-F | AAG CTG CTG CTG GAG CAG | 229 |
| cdna_Hes5-R | GCA GCT TCA TCT GCG TGT C | |
| cdna Isl1 | Qiagen catalog number QT01048691 | 79 |
| cdna_NeuroD1-F | CCT GTG ACC TTT CCC ATG C | 219 |
| cdna_NeuroD1-R | AGA AGT GCT AAG GCA ACG C | |
| cdna_Notch1-F | GCT CCG AGG AGA TCA ACG AG | 268 |
| cdna_Notch1-R | TTG ACA TCA CCC TCA CAC CG | |
| cdna_Nrl-F | GTG CCT CCT TCA CCC ACC TTC AGT GA | 361 |
| cdna_Nrl-R | GCG TGC GGC GCC TCT GCT TCA GCC G | |
| cdna_Otx2-F | TCG CCA CCT CTA CTT TGA TAG | 85 |
| cdna_Otx2-R | AGC CGC ATT GGA CGT TAG | |
| cdna_Pax6-F | TTT AAC CAA GGG CGG TGA GCA G | 475 |
| cdna_Pax6-R | TCT CGG ATT TCC CAA GCA AAG ATG | |
| cdna_Pde6b-F | CTG ACG AGT ATG AGG CCA AAG | 204 |
| cdna_Pde6b- R | TAG GCA GAG TCC GTA TGC AGT | |
| cdna_Pde6c-F | CCT TAT GTG GTC AGC CAA TAA AG | 210 |
| cdna_Pde6c-R | CCA TCT GGA GTC TTT GGT CC | |
| cdna_Rcvrn-F | GCA GCT TCG ATG CCA ACA G | 66 |
| cdna_Rcvrn-R | TCA TGT GCA GAG CAA TCA CGT A | |
| cdna_Rgr-F | TTG TGT GGA TGT CAT CTG C | 184 |
| cdna_Rgr-R | GAA GTG TGT GTG ATG AAC AGG | |
| cdna_Rho-F | CTT CTC CAA CGT CAC AGG CGT | 346 |
| cdna_Rho-R | GGA CCA CAG GGC GAT TTC AC | |
| cdna_Sag-F | GCC ATG AGT GTC CTC ACC | 255 |
| cdna_Sag-R | GGC ATG CTG CAC TTT CC | |
| cdna_Kcnv2-F | CTT CTC TGC AGC TGT CTA CTC G | 226 |
| cdna_Kcnv2-R | GTA ATC GGA GAA CTT GTT GTA GAG G | |
| cdna Opn1w | Qiagen catalog number QT00163723 | 1221 |
| cdna_Opn1sw-F | CAG CAT CCG CTT CAA CTC CAA | 229 |
| cdna_Opn1sw-R | GCA GAT GAG GGA AAG AGG AAT GA | |
| cdna PKCα | Qiagen catalog number QT00146384 | 101 |
| cdna Rom1 | Qiagen catalog number QT00172165 | 132 |
| cdna_Samd11-F | CTT TCT GGC TGT GGC GAG | 173 |
| cdna_Samd11-R | GCC ATG TAG AAG ACA CGG C | |
| cdna Tfap2b | Qiagen catalog number QT00135478 | 83 |
| cdna_Vsx2-F1 | ACG GAG CTC CCA GAA GAC | 176 |
| cdna_Vsx2-R1 | CCA TCC TTG GCA GAC TTG | |
| chip_Ccnd1-F | TTA GAA TAA AGC GGT TCC ACC | 284 |
| chip_Ccnd1-R | TTC GGA GCT ACA GTG GAA TC | |
| chip_Crx-F | GCT CAG GTT GGC CTC AGA C | 179 |
| chip_Crx-R | CCA CAC TAG TGC GAG ACC TGA G | |
| chip_Foxn4-F | GGG AGA GAC TAC TAC CTT TGA CTT CTC | 248 |
| chip_Foxn4-R | CCC TAC GTG CTC TAT AGA GAA TAA TAG C | |
| chip_Gapdh-F | CCT CAA CTT TTC CGC AGC | 331 |
| chip_Gapdh-R | CCT CCT CCC TCT CTT TGG | |
| chip_Gnat2-F | TTT GAA CTG AGA GTG CAT TTC TAA C | 185 |
| chip_Gnat2-R | GAC TCC TAT TCT GCT ACT GCT CTG | |
| chip_Hes1-F | GCC TGG CCA CAA AAG AAA TA | 238 |
| chip_Hes1-R | CCC AAA CTT TCT TTC CCA CA | |
| chip_Notch1-F | CTA TAG GCA TCA GGA GGA TTG AG | 204 |
| chip_Notch1-R | CCG TGG AAC GTC TAG ACT CG | |
| chip_Nrl-F | TGC CTT GGA GAG CCT AGC | 160 |
| chip_Nrl-R | AAG CAG GAG TCC TAT CCA CTT C | |
| chip_Rho-F | CCA CTT CAG ACT CTA GGC CTC | 114 |
| chip_Rho-R | GTG TTG GCT CAG TGA GAC AAG | |
| chip_Tfap2b-F | CAT CAT TGT AGA CTT ACT TCT GGA GC | 73 |
| chip_Tfap2b-R | TTC TTT AAG GGC TTG GCT ACC | |
| chip_Vsx2-F1 | CAG AGC CCA CCA CAC TTG | 190 |
| chip_Vsx2-R1 | GGT GTC TAA TGC TCT GGA GC | |
| chip_Rho_F1 | GGA ATT CCC AGA GGA CTC TG | 287 |
| chip_Rho_R1 | CTC TTC GTA GAC AGA GAC C | |
| chip_Rho_F2 | TGA CCT CTT CAT GGT CTT CG | 134 |
| chip_Rho_R2 | CAG TCT CTC TGC TCA TAC CTC C | |
| chip_Rho_F3 | CCA GGA GTG AGC TCT AGC TT | 117 |
| chip_Rho_R3 | GGC AGT GAG ATG TAC AAG TTT AC | |
| chip_Vsx2_F2 | AAC AGT CCC TAT GCA CCT GTA TC | 139 |
| chip_Vsx2_R2 | CAC ACA GTA CTT GGA GTT TGG G | |
| chip_Vsx2_F3 | CCA TAC AGA TGT TGT TCC TGC | 206 |
| chip_Vsx2_R3 | CCT TAC AAC AGG AGA GAA CCC |
a Ref. 46.
TABLE 3.
PCR primers for HDACs used in this work
| Gene | Sequence | Fragment size |
|---|---|---|
| bp | ||
| cdna_HDAC1-F | CATCAGCCCTTCTAACATGACC | 253 |
| cdna_HDAC1-R | TTCTCCCTCCTCATCTGAGTCC | |
| cdna_HDAC2-F | AAGGAGGTCGTAGGAATGTTG | 260 |
| cdna_HDAC2-R | CAATGTCCTCAAACAGGGAAG | |
| cdna_HDAC3-F | GTGGAGATTTAGAGCAGCATGG | 253 |
| cdna_HDAC3-R | GGCTCATTACCCATAACGTTCC | |
| cdna_HDAC4-F | AGCACAGAGGTGAAGATGAAGC | 267 |
| cdna_HDAC4-R | AAGCCTTGAGCGTAATTTCAGG | |
| cdna_HDAC5-F | GGGATTCTGCTTCTTCAACTCC | 252 |
| cdna_HDAC5-R | CACGCCACATTTACGTTGTACC | |
| cdna_HDAC6-F | AGGAGGCAAGTTGATTCTGTC | 250 |
| cdna_HDAC6-R | CTCTTCTAGCACGGCTTCTTCC | |
| cdna_HDAC7-F | CCTTGCCTTCAAAGTAGCTTCC | 253 |
| cdna_HDAC7-R | GCGATGAAGGGAAATGTAGAGC | |
| cdna_HDAC9-F | TTCTCTGGATGCACTTGAAAGG | 250 |
| cdna_HDAC9-R | CTGGTCAGAATGTCAGGAATGG | |
| cdna_HDAC10-F | GGATGTACTCATTCAGCGATGC | 254 |
| cdna_HDAC10-R | ACACCATATCAGGCTGGAAACC | |
| cdna_HDAC11-F | ATCAAGAGTCAGGGAGGACAGC | 273 |
| cdna_HDAC11-R | AGAATCCCAGCTTCCTTCTTCC | |
| cdna_b-actin-Fa | GTGGGGCGCCCCAGGCACCA | 452 |
| cdna_b-actin-Ra | CTCCTTATTGTCACGCACGGATTT C |
a Ref. 82.
Chromatin Immunoprecipitation
PN1 retina explants were treated for 96 h with either 1.0 μm CAS 193551-00-7, 16 nm apicidin, or DMSO. ChIPs were done in triplicates for the HDAC1i and in duplicate for the HDAC3i. ChIP was done as previously described (37) and subjected to quantitative PCR. Absolute quantification was done by the standard curve method using primers listed in Table 2 (80, 81). For quantitative real time PCR, we used 2× iQ-SYBR Green PCR Supermix. Samples in triplicate were run on the iQ5 Multicolor Real Time PCR Detection System (Bio-Rad). Each run included suitable standard curve samples using a wide range of concentrations for genomic DNA prepared similarly to inputs.
Statistical Analysis
Graphs were generated using GraphPad Prism 5 software. Results are presented as means ± S.E. Unpaired, one-tailed Student's t test was used to evaluate statistical significance between groups. If variances between samples were significantly different, Welch's correction was applied. A p value <0.05 was considered significant. To compare more than two groups, a one-way analysis of variance with Bonferroni correction was used.
Author Contributions
R. C. F. and E. Y. P. designed, performed, analyzed the experiments shown in Figs. 3–11 and wrote the paper. S. S. Z. designed, performed, and analyzed the experiment shown in Fig. 1. R. C. F., E. Y. P., and J. J. designed, performed, and analyzed the experiment shown in Fig. 2. M. R. S. B. conceive the study of HDAC3 and wrote the paper. C. J. B. and S. S. Z. conceived and coordinated the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
The authors declare that they have no conflicts of interest with the contents of this article.
- H3
- histone 3
- H4
- histone 4
- CAS 193551-00-7
- 4-(dimethylamino)-N-[6-(hydroxyamino)-6-oxohexyl]benzamide
- CoREST
- corepressor for element-1-silencing transcription factor
- DHS
- DNase-hypersensitive sites
- E
- embryonic day
- H3K9ac
- acetylation of lysine 9 of histone 3
- H4K12ac
- acetylation of lysine 12 of histone 4
- H3K27ac
- acetylation of lysine 27 of histone 3
- H3K4me2
- histone H3 dimethyllysine 4
- H3K9
- histone H3 lysine 9
- H3K27
- histone H3 lysine 27
- H3K27me3
- histone H3 trimethyllysine 27
- H4K12
- histone H4 lysine 12
- HDAC
- histone deacetylase
- HDACi
- HDAC inhibitor
- LSD1
- lysine demethylase 1
- NuRD
- nucleosome remodeling and histone deacetylation
- PCNA
- proliferating cell nuclear antigen
- PN
- postnatal day
- Pol
- polymerase
- rhHDAC
- recombinant human histone deacetylase
- TSS
- transcription start site
- TSA
- trichostatin A.
References
- 1. Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 [DOI] [PubMed] [Google Scholar]
- 2. Cosgrove M. S., and Wolberger C. (2005) How does the histone code work? Biochem. Cell Biol. 83, 468–476 [DOI] [PubMed] [Google Scholar]
- 3. Jenuwein T., and Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 4. Kuo M. H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., Edmondson D. G., Roth S. Y., and Allis C. D. (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272 [DOI] [PubMed] [Google Scholar]
- 5. Kuo M. H., and Allis C. D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays 20, 615–626 [DOI] [PubMed] [Google Scholar]
- 6. Davie J. R., and Candido E. P. (1978) Acetylated histone H4 is preferentially associated with template-active chromatin. Proc. Natl. Acad. Sci. U.S.A. 75, 3574–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pogo B. G., Pogo A. O., Allfrey V. G., and Mirsky A. E. (1968) Changing patterns of histone acetylation and RNA synthesis in regeneration of the liver. Proc. Natl. Acad. Sci. U.S.A. 59, 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12, 599–606 [DOI] [PubMed] [Google Scholar]
- 9. Wang Z., Zang C., Cui K., Schones D. E., Barski A., Peng W., and Zhao K. (2009) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haberland M., Montgomery R. L., and Olson E. N. (2009) The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morris M. J., and Monteggia L. M. (2013) Unique functional roles for class I and class II histone deacetylases in central nervous system development and function. Int. J. Dev. Neurosci. 31, 370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kelly R. D., and Cowley S. M. (2013) The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem. Soc. Trans. 41, 741–749 [DOI] [PubMed] [Google Scholar]
- 13. Sun G., Fu C., Shen C., and Shi Y. (2011) (2011). Histone deacetylases in neural stem cells and induced pluripotent stem cells. J. Biomed. Biotechnol. 2011, 835968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reichert N., Choukrallah M.-A., and Matthias P. (2012) Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell. Mol. Life Sci. 69, 2173–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cho Y., and Cavalli V. (2014) HDAC signaling in neuronal development and axon regeneration. Curr. Opin. Neurobiol. 27, 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hsieh J., Nakashima K., Kuwabara T., Mejia E., and Gage F. H. (2004) Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 101, 16659–16664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Montgomery R. L., Hsieh J., Barbosa A. C., Richardson J. A., and Olson E. N. (2009) Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci. U.S.A. 106, 7876–7881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitrousis N., Tropepe V., and Hermanson O. (2015) Post-translational modifications of histones in vertebrate neurogenesis. Front. Neurosci. 9, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Finnin M. S., Donigian J. R., Cohen A., Richon V. M., Rifkind R. A., Marks P. A., Breslow R., and Pavletich N. P. (1999) Structures of a histone deacetylase homolog bound to the TSA and SAHA inhibitors. Nature 401, 188–193 [DOI] [PubMed] [Google Scholar]
- 20. Riggs M. G., Whittaker R. G., Neumann J. R., and Ingram V. M. (1977) n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 268, 462–464 [DOI] [PubMed] [Google Scholar]
- 21. Candido E. P., Reeves R., and Davie J. R. (1978) Sodium butyrate inhibits histone deacetylation in cultured cells. Cell 14, 105–113 [DOI] [PubMed] [Google Scholar]
- 22. Khan N., Jeffers M., Kumar S., Hackett C., Boldog F., Khramtsov N., Qian X., Mills E., Berghs S. C., Carey N., Finn P. W., Collins L. S., Tumber A., Ritchie J. W., Jensen P. B., et al. (2008) Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem. J. 409, 581–589 [DOI] [PubMed] [Google Scholar]
- 23. Jung M., Brosch G., Kölle D., Scherf H., Gerhäuser C., and Loidl P. (1999) Amide analogues of trichostatin A as inhibitors of histone deacetylase and inducers of terminal cell differentiation. J. Med. Chem. 42, 4669–4679 [DOI] [PubMed] [Google Scholar]
- 24. Galletti M., Cantoni S., Zambelli F., Valente S., Palazzini M., Manes A., Pasquinelli G., Mai A., Galiè N., and Ventura C. (2014) Dissecting histone deacetylase role in pulmonary arterial smooth muscle cell proliferation and migration. Biochem. Pharmacol. 91, 181–190 [DOI] [PubMed] [Google Scholar]
- 25. Greene T. T., Tokuyama M., Knudsen G. M., Kunz M., Lin J., Greninger A. L., DeFilippis V. R., DeRisi J. L., Raulet D. H., and Coscoy L. (2016) A herpesviral induction of RAE-1 NKG2D ligand expression occurs through release of HDAC mediated repression. Elife 5, e14749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Furumai R., Matsuyama A., Kobashi N., Lee K. H., Nishiyama M., Nakajima H., Tanaka A., Komatsu Y., Nishino N., Yoshida M., and Horinouchi S. (2002) FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 62, 4916–4921 [PubMed] [Google Scholar]
- 27. Newbold A., Lindemann R. K., Cluse L. A., Whitecross K. F., Dear A. E., and Johnstone R. W. (2008) Characterisation of the novel apoptotic and therapeutic activities of the histone deacetylase inhibitor romidepsin. Mol. Cancer Ther. 7, 1066–1079 [DOI] [PubMed] [Google Scholar]
- 28. VanderMolen K. M., McCulloch W., Pearce C. J., and Oberlies N. H. (2011) Romidepsin (Istodax, NSC 630176, FR901228, FK228, depsipeptide): a natural product recently approved for cutaneous T-cell lymphoma. J. Antibiot. 64, 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mai A., Massa S., Pezzi R., Simeoni S., Rotili D., Nebbioso A., Scognamiglio A., Altucci L., Loidl P., and Brosch G. (2005) Class II (IIa)-selective histone deacetylase inhibitors. 1. Synthesis and biological evaluation of novel (aryloxopropenyl)pyrrolyl hydroxyamides. J. Med. Chem. 48, 3344–3353 [DOI] [PubMed] [Google Scholar]
- 30. Lamb T. D. (2013) Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 36, 52–119 [DOI] [PubMed] [Google Scholar]
- 31. Chen B., and Cepko C. L. (2007) Requirement of histone deacetylase activity for the expression of critical photoreceptor genes. BMC Dev. Biol. 7, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wallace D. M., Donovan M., and Cotter T. G. (2006) Histone deacetylase activity regulates apaf-1 and caspase 3 expression in the developing mouse retina. Invest. Ophthalmol. Vis. Sci. 47, 2765–2772 [DOI] [PubMed] [Google Scholar]
- 33. Colletti S. L., Myers R. W., Darkin-Rattray S. J., Gurnett A. M., Dulski P. M., Galuska S., Allocco J. J., Ayer M. B., Li C., Lim J., Crumley T. M., Cannova C., Schmatz D. M., Wyvratt M. J., Fisher M. H., et al. (2001) Broad spectrum antiprotozoal agents that inhibit histone deacetylase: structure-activity relationships of apicidin. Part 1. Bioorg. Med. Chem. Lett. 11, 107–111 [DOI] [PubMed] [Google Scholar]
- 34. Darkin-Rattray S. J., Gurnett A. M., Myers R. W., Dulski P. M., Crumley T. M., Allocco J. J., Cannova C., Meinke P. T., Colletti S. L., Bednarek M. A., Singh S. B., Goetz M. A., Dombrowski A. W., Polishook J. D., and Schmatz D. M. (1996) Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl. Acad. Sci. U.S.A. 93, 13143–13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shi P., Scott M. A., Ghosh B., Wan D., Wissner-Gross Z., Mazitschek R., Haggarty S. J., and Yanik M. F. (2011) Synapse microarray identification of small molecules that enhance synaptogenesis. Nat. Commun. 2, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saito A., Yamashita T., Mariko Y., Nosaka Y., Tsuchiya K., Ando T., Suzuki T., Tsuruo T., and Nakanishi O. (1999) A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc. Natl. Acad. Sci. U.S.A. 96, 4592–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tatamiya T., Saito A., Sugawara T., and Nakanishi O. (2004) Isozyme-selective activity of the HDAC inhibitor MS-275. Cancer Res. 64, 567–567 [Google Scholar]
- 38. Duong V., Bret C., Altucci L., Mai A., Duraffourd C., Loubersac J., Harmand P.-O., Bonnet S., Valente S., Maudelonde T., Cavailles V., and Boulle N. (2008) Specific activity of class II histone deacetylases in human breast cancer cells. Mol. Cancer Res. 6, 1908–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scognamiglio A., Nebbioso A., Manzo F., Valente S., Mai A., and Altucci L. (2008) HDAC-class II specific inhibition involves HDAC proteasome-dependent degradation mediated by RANBP2. Biochim. Biophys. Acta 1783, 2030–2038 [DOI] [PubMed] [Google Scholar]
- 40. Davie J. R. (2003) Inhibition of histone deacetylase activity by butyrate. J. Nutr. 133, 2485S–2493S [DOI] [PubMed] [Google Scholar]
- 41. Hague A., Manning A. M., Hanlon K. A., Huschtscha L. I., Hart D., and Paraskeva C. (1993) Sodium butyrate induces apoptosis in human colonic tumour cell lines in a p53-independent pathway: implications for the possible role of dietary fibre in the prevention of large-bowel cancer. Int. J. Cancer 55, 498–505 [DOI] [PubMed] [Google Scholar]
- 42. Kim D. S., Ross S. E., Trimarchi J. M., Aach J., Greenberg M. E., and Cepko C. L. (2008) Identification of molecular markers of bipolar cells in the murine retina. J. Comp. Neurol. 507, 1795–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hojo M., Ohtsuka T., Hashimoto N., Gradwohl G., Guillemot F., and Kageyama R. (2000) Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development 127, 2515–2522 [DOI] [PubMed] [Google Scholar]
- 44. Mu X., Fu X., Beremand P. D., Thomas T. L., and Klein W. H. (2008) Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc. Natl. Acad. Sci. U.S.A. 105, 6942–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Siegert S., Cabuy E., Scherf B. G., Kohler H., Panda S., Le Y. Z., Fehling H. J., Gaidatzis D., Stadler M. B., and Roska B. (2012) Transcriptional code and disease map for adult retinal cell types. Nat. Neurosci. 15, 487–495 [DOI] [PubMed] [Google Scholar]
- 46. Tiwari P., Sahay S., Pandey M., Qadri S. S., and Gupta K. P. (2015) Combinatorial chemopreventive effect of butyric acid, nicotinamide and calcium glucarate against the 7,12-dimethylbenz(a)anthracene induced mouse skin tumorigenesis attained by enhancing the induction of intrinsic apoptotic events. Chem. Biol. Interact. 226, 1–11 [DOI] [PubMed] [Google Scholar]
- 47. Popova E. Y., Xu X., DeWan A. T., Salzberg A. C., Berg A., Hoh J., Zhang S. S., and Barnstable C. J. (2012) Stage and gene specific signatures defined by histones H3K4me2 and H3K27me3 accompany mammalian retina maturation in vivo. PLoS One. 7, e46867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Corbo J. C., Lawrence K. A., Karlstetter M., Myers C. A., Abdelaziz M., Dirkes W., Weigelt K., Seifert M., Benes V., Fritsche L. G., Weber B. H., and Langmann T. (2010) CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 20, 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hao H., Kim D. S., Klocke B., Johnson K. R., Cui K., Gotoh N., Zang C., Gregorski J., Gieser L., Peng W., Fann Y., Seifert M., Zhao K., and Swaroop A. (2012) Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 8, e1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tummala P., Mali R. S., Guzman E., Zhang X., and Mitton K. P. (2010) Temporal ChIP-on-Chip of RNA-polymerase-II to detect novel gene activation events during photoreceptor maturation. Mol. Vis. 16, 252–271 [PMC free article] [PubMed] [Google Scholar]
- 51. Wilken M. S., Brzezinski J. A., La Torre A., Siebenthall K., Thurman R., Sabo P., Sandstrom R. S., Vierstra J., Canfield T. K., Hansen R. S., Bender M. A., Stamatoyannopoulos J., and Reh T. A. (2015) DNase I hypersensitivity analysis of the mouse brain and retina identifies region-specific regulatory elements. Epigenetics Chromatin 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Balasubramaniyan V., Boddeke E., Bakels R., Küst B., Kooistra S., Veneman A., and Copray S. (2006) Effects of histone deacetylation inhibition on neuronal differentiation of embryonic mouse neural stem cells. Neuroscience 143, 939–951 [DOI] [PubMed] [Google Scholar]
- 53. Kim H. J., Leeds P., and Chuang D. M. (2009) The HDAC inhibitor, sodium butyrate, stimulates neurogenesis in the ischemic brain. J. Neurochem. 110, 1226–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fan J., Alsarraf O., Chou C. J., Yates P. W., Goodwin N. C., Rice D. S., and Crosson C. E. (2016) Ischemic preconditioning, retinal neuroprotection and histone deacetylase activities. Exp. Eye Res. 146, 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anderson K. W., Chen J., Wang M., Mast N., Pikuleva I. A., and Turko I. V. (2015) Quantification of histone deacetylase isoforms in human frontal cortex, human retina, and mouse brain. PLoS One 10, e0126592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tiwari S., Dharmarajan S., Shivanna M., Otteson D. C., and Belecky-Adams T. L. (2014) Histone deacetylase expression patterns in developing murine optic nerve. BMC Dev. Biol. 14, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lagger G., O'Carroll D., Rembold M., Khier H., Tischler J., Weitzer G., Schuettengruber B., Hauser C., Brunmeir R., Jenuwein T., and Seiser C. (2002) Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 21, 2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cunliffe V. T. (2004) Histone deacetylase 1 is required to repress Notch target gene expression during zebrafish neurogenesis and to maintain the production of motoneurones in response to hedgehog signalling. Development 131, 2983–2995 [DOI] [PubMed] [Google Scholar]
- 59. Nambiar R. M., Ignatius M. S., and Henion P. D. (2007) Zebrafish colgate/hdac1 functions in the non-canonical Wnt pathway during axial extension and in Wnt-independent branchiomotor neuron migration. Mech. Dev. 124, 682–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yamaguchi M., Tonou-Fujimori N., Komori A., Maeda R., Nojima Y., Li H., Okamoto H., and Masai I. (2005) Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development 132, 3027–3043 [DOI] [PubMed] [Google Scholar]
- 61. Kranenburg O., van der Eb A. J., and Zantema A. (1996) Cyclin D1 is an essential mediator of apoptotic neuronal cell death. EMBO J. 15, 46–54 [PMC free article] [PubMed] [Google Scholar]
- 62. Spange S., Wagner T., Heinzel T., and Krämer O. H. (2009) Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 41, 185–198 [DOI] [PubMed] [Google Scholar]
- 63. Togi S., Kamitani S., Kawakami S., Ikeda O., Muromoto R., Nanbo A., and Matsuda T. (2009) HDAC3 influences phosphorylation of STAT3 at serine 727 by interacting with PP2A. Biochem. Biophys. Res. Commun. 379, 616–620 [DOI] [PubMed] [Google Scholar]
- 64. Zhang S. S., Wei J., Qin H., Zhang L., Xie B., Hui P., Deisseroth A., Barnstable C. J., and Fu X.-Y. (2004) STAT3-mediated signaling in the determination of rod photoreceptor cell fate in mouse retina. Invest. Ophthalmol. Vis. Sci. 45, 2407–2412 [DOI] [PubMed] [Google Scholar]
- 65. Zhang S. S., Liu M. G., Kano A., Zhang C., Fu X. Y., and Barnstable C. J. (2005) STAT3 activation in response to growth factors or cytokines participates in retina precursor proliferation. Exp. Eye Res. 81, 103–115 [DOI] [PubMed] [Google Scholar]
- 66. Ozawa Y., Nakao K., Shimazaki T., Takeda J., Akira S., Ishihara K., Hirano T., Oguchi Y., and Okano H. (2004) Downregulation of STAT3 activation is required for presumptive rod photoreceptor cells to differentiate in the postnatal retina. Mol. Cell. Neurosci. 26, 258–270 [DOI] [PubMed] [Google Scholar]
- 67. Popova E. Y., Pinzon-Guzman C., Salzberg A. C., Zhang S. S., and Barnstable C. J. (2016) LSD1-mediated demethylation of H3K4me2 is required for the transition from late progenitor to differentiated mouse rod photoreceptor. Mol. Neurobiol. 53, 4563–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sáez J. E., Gómez A. V., Barrios Á. P., Parada G. E., Galdames L., González M., and Andrés M. E. (2015) Decreased expression of CoREST1 and CoREST2 together with LSD1 and HDAC1/2 during neuronal differentiation. PLoS One 10, e0131760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Barrios ÁP, Gómez A. V., Sáez J. E., Ciossani G., Toffolo E., Battaglioli E., Mattevi A., and Andrés M. E. (2014) Differential properties of transcriptional complexes formed by the CoREST family. Mol. Cell. Biol. 34, 2760–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Whyte W. A., Bilodeau S., Orlando D. A., Hoke H. A., Frampton G. M., Foster C. T., Cowley S. M., and Young R. A. (2012) Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hakimi M. A., Bochar D. A., Chenoweth J., Lane W. S., Mandel G., and Shiekhattar R. (2002) A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. U.S.A. 99, 7420–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hakimi M. A., Dong Y., Lane W. S., Speicher D. W., and Shiekhattar R. (2003) A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 278, 7234–7239 [DOI] [PubMed] [Google Scholar]
- 73. Yokoyama A., Takezawa S., Schüle R., Kitagawa H., and Kato S. (2008) Transrepressive function of TLX requires the histone demethylase LSD1. Mol. Cell. Biol. 28, 3995–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Egelhofer T. A., Minoda A., Klugman S., Lee K., Kolasinska-Zwierz P., Alekseyenko A. A., Cheung M. S., Day D. S., Gadel S., Gorchakov A. A., Gu T., Kharchenko P. V., Kuan S., Latorre I., Linder-Basso D., et al. (2011) An assessment of histone-modification antibody quality. Nat. Struct. Mol. Biol. 18, 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Barnstable C. J. (1980) Monoclonal antibodies which recognize different cell types in the rat retina. Nature 286, 231–235 [DOI] [PubMed] [Google Scholar]
- 76. Liu M. G., Li H., Xu X., Barnstable C. J., and Zhang S. S. (2008) Comparison of gene expression during in vivo and in vitro postnatal retina development. J. Ocul. Biol. Dis. Infor. 1, 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang S. S., Fu X. Y., and Barnstable C. J. (2002) Tissue culture studies of retinal development. Methods 28, 439–447 [DOI] [PubMed] [Google Scholar]
- 78. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 79. Schmittgen T. D., and Livak K. J. (2008) Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 80. Muller P. Y., Janovjak H., Miserez A. R., and Dobbie Z. (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques 32, 1372–1374 [PubMed] [Google Scholar]
- 81. Larionov A., Krause A., and Miller W. (2005) A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pinzon-Guzman C., Zhang S. S., and Barnstable C. J. (2011) Specific protein kinase C isoforms are required for rod photoreceptor differentiation. J. Neurosci. 31, 18606–18617 [DOI] [PMC free article] [PubMed] [Google Scholar]