Abstract
The intracellular signaling protein regulator of presynaptic morphology 1 (RPM-1) is a conserved regulator of synapse formation and axon termination in Caenorhabditis elegans. RPM-1 functions in a ubiquitin ligase complex with the F-box protein FSN-1 and functions through the microtubule binding protein RAE-1. Using a structure-function approach and positive selection for transgenic C. elegans, we explored the biochemical relationship between RPM-1, FSN-1, and RAE-1. This led to the identification of two new domains in RPM-1 that are sufficient for binding to FSN-1, called FSN-1 binding domain 2 (FBD2) and FBD3. Furthermore, we map the RAE-1 binding domain to a much smaller region of RPM-1. Point mutations in RPM-1 that reduce binding to RAE-1 did not affect FSN-1 binding, indicating that RPM-1 utilizes different biochemical mechanisms to bind these molecules. Analysis of RPM-1 protein complexes in the neurons of C. elegans elucidated two further discoveries: FSN-1 binds to RAE-1, and this interaction is not mediated by RPM-1, and RPM-1 binding to FSN-1 and RAE-1 reduces FSN-1·RAE-1 complex formation. These results indicate that RPM-1 uses different mechanisms to recruit FSN-1 and RAE-1 into independent signaling complexes in neurons.
Keywords: axon, Caenorhabditis elegans (C. elegans), neuron, structure-function, synapse, FSN-1, RAE-1, RPM-1
Introduction
The Pam/Highwire/RPM-1 (PHR)2 proteins are conserved intracellular signaling proteins that include human Pam/MYCBP2, Drosophila Highwire, and Caenorhabditis elegans regulator of presynaptic morphology 1 (RPM-1) (1, 2). The PHR proteins regulate various events in neuronal development, including synapse formation (3–6), axon guidance (6–9), and axon termination (7, 10–13). In C. elegans, defects in neuronal development caused by loss of RPM-1 result in mild defects in locomotion and severe defects in short-term learning (14). After completion of the developmental program, PHR proteins become important regulators of axon degeneration (15, 16).
One conserved mechanism by which PHR proteins regulate synapse and axon development is ubiquitination and negative regulation of the dual leucine zipper-bearing kinase (Dlk), also called DLK-1 in worms and Wallenda in flies (17–19). PHR proteins are RING family ubiquitin ligases that form complexes that include an F-box protein, which serves as a substrate recognition module. In C. elegans, RPM-1 functions in an Skp/Cullin/F-box (SCF) complex that includes the F-box protein FSN-1, the Skp protein SKR-1, and the Cullin CUL-1 (20). In mammals and flies, Phr1 and Highwire function in a non-canonical SCF complex that contains the F-box protein Fbxo45 and Skp1 but lacks a Cullin (21–23). Despite differences in the composition of PHR ubiquitin ligase complexes, the functional relationship between PHR proteins and FSN-1·Fbxo45 is evolutionarily conserved. Further, Fbxo45 is likely to be required for Phr1 to regulate axon degeneration in mice (16).
We previously analyzed the biochemical mechanism underlying the interaction between RPM-1 and FSN-1 (24). Using HEK 293 cells as a heterologous expression system, we found a single, conserved domain in RPM-1 that is sufficient for binding to FSN-1. We refer to this domain as FSN-1 binding domain 1 (FBD1). Point mutagenesis revealed several residues in FBD1 that are required for binding FSN-1. Unexpectedly, full-length RPM-1 containing these mutations did not show reduced binding to FSN-1 when expressed in the worm nervous system. One explanation for these findings is that further FSN-1 binding sites might be present in RPM-1. However, identification of these sites would likely require in vivo worm biochemistry, which is technically more difficult than biochemistry using 293 cells.
RPM-1 function is also mediated, in part, by binding to RAE-1, a microtubule binding protein (25, 26). Previous genetic results suggest that RAE-1 is not ubiquitinated and inhibited by RPM-1. Instead, RPM-1 positively regulates RAE-1 function (25). A single domain in RPM-1 is sufficient for binding to RAE-1 and is therefore called the RAE-1 binding domain (RBD). The RBD covers a relatively large region of RPM-1 and is very close to FBD1 in the primary RPM-1 protein sequence. At present, it remains uncertain whether residues in RPM-1 required for binding to RAE-1 impact binding to FSN-1 and vice versa.
Here we use transgenic C. elegans to expand our structure-function analysis of RPM-1 and FSN-1 in vivo. This approach revealed two new domains in RPM-1, FBD2 and FBD3, which are sufficient for binding to FSN-1. Because these interactions were not observed previously using 293 cells (24), our results show that binding of FSN-1 to portions of RPM-1 requires in vivo expression in worm neurons. Using a similar approach, we map the RBD to a significantly smaller region in RPM-1 and show that mutations in RPM-1 that reduce binding to RAE-1 do not affect binding to FSN-1. Transgenic overexpression experiments with fragments of RPM-1 and rescue experiments with RPM-1 deletion constructs indicate that the FBD2 and FBD3 are required for RPM-1 function in vivo.
During our structure-function analysis, we found that FSN-1 binds to RAE-1. This interaction is not dependent upon RPM-1. Rather, FSN-1 binding to RAE-1 is reduced in the presence of RPM-1, which forms separate complexes with both molecules. These results provide the first evidence that a PHR protein can form distinct signaling complexes in vivo.
Results
Multiple Domains in RPM-1 Are Sufficient for Binding to FSN-1
Our previous work showed that FBD1 of RPM-1 is sufficient for binding to FSN-1, and we identified point mutations in FBD1 that reduce binding in 293 cells (24). However, corresponding mutations in full-length RPM-1 did not affect binding to FSN-1 in worm neurons (24). This raised two points: there were likely to be other sites in RPM-1, besides FBD1, that were sufficient for binding to FSN-1, and identification of these sites was likely to require in vivo biochemistry from transgenic C. elegans.
Generally, heterologous expression systems, such as HEK 293 cells, are used for biochemistry with worm proteins because in vivo biochemistry from C. elegans can be technically challenging, particularly with proteins expressed in the nervous system. One reason for this is that extrachromosomal arrays with visible selection markers cannot be actively maintained while still growing sufficient quantities of worms for biochemistry, and generation of integrated strains is time-consuming and can reduce sensitivity. To deal with these issues, we used PHA-1 positive selection (see “Materials and Methods” for details), which allowed us to use a simple temperature shift to generate a pure population of transgenic animals. This facilitated significantly more sensitive biochemistry with proteins expressed in the nervous system. PHA-1 positive selection was necessary for all of the biochemistry described here.
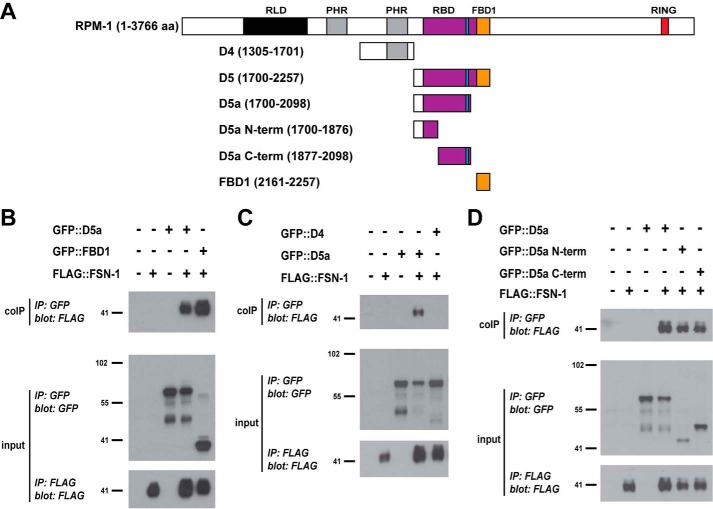
We began our biochemical mapping studies using domain 5 (D5) of RPM-1. FBD1 represents a small portion of D5, and D5 is sufficient for binding to FSN-1 in 293 cells (24). We generated worms carrying transgenic extrachromosomal arrays that used a pan-neuronal promoter (Prgef-1) to simultaneously express FLAG-tagged FSN-1 (FLAG::FSN-1) and GFP fusions with different RPM-1 constructs (Fig. 1A). These arrays also contained PHA-1 driven by its native promoter to allow for positive selection. To test for binding, whole worm lysates were generated from transgenic animals that were PHA-1 selected, and coIP was performed. As shown in Fig. 1B, FLAG::FSN-1 bound to GFP::FBD1. We also observed strong binding between FSN-1 and D5a (Fig. 1B), an interaction that was not observed in our previous experiments using 293 cells (24). RPM-1 D4 did not bind FSN-1 and acted as a negative control (Fig. 1C). These results demonstrate that D5a is sufficient for binding to FSN-1 in the neurons of C. elegans.
FIGURE 1.
Identification of two domains in RPM-1 that are sufficient for binding to FSN-1. A, schematic of RPM-1 and RPM-1 fragments tested. Annotated protein domains include RCC-1 like domain (RLD, black), PHR family-specific domain (PHR, gray), RBD (purple), FBD1 (orange), and RING-H2 ubiquitin ligase domain (RING, red). Highlighted in blue is a motif in the RBD that is necessary for binding to RAE-1. aa, amino acids. B–D, coIP (top panels), anti-GFP IP (input, center panels), and anti-FLAG IP (input, bottom panels) performed from lysates of transgenic C. elegans containing the indicated constructs. B, FLAG::FSN-1 coprecipitating with GFP::D5a and GFP::FBD1. C, FLAG::FSN-1 coprecipitating with GFP::D5a but not with GFP::D4. D, FLAG::FSN-1 coprecipitating with GFP::D5a N-term and GFP::D5a C-term. Shown are representatives of experiments that were performed at least three times using multiple transgenic lines for each genotype.
For further mapping, we generated two pieces of RPM-1 D5a: an N-terminal fragment (D5a N-term) and a C-terminal fragment (D5a C-term) (Fig. 1A). FSN-1 coprecipitated with both D5a N-term and D5a C-term (Fig. 1D). These results show that we have identified two new regions in RPM-1 that are sufficient for binding to FSN-1, which we refer to as FBD2 (D5a C-term) and FBD3 (D5a N-term).
Further Mapping of the RAE-1 Binding Domain in RPM-1
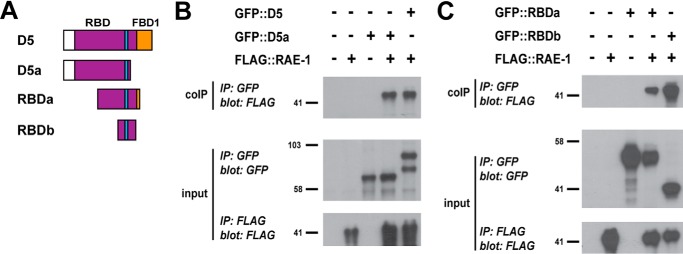
RPM-1 D5 contains a large region of sequence, called the RBD, that is sufficient for binding to RAE-1 (Figs. 1A and 2A) (25). The RBD overlaps with both FBD2 and FBD3. To more accurately distinguish the regions of RPM-1 that mediate binding to RAE-1 and FSN-1, we further mapped the RBD of RPM-1.
FIGURE 2.
Identification of RBDb, a small region of RPM-1 that is sufficient for binding to RAE-1. A, schematic of RPM-1 fragments tested. B and C, CoIP (top panels), anti-GFP IP (input, center panels), and anti-FLAG IP (input, bottom panels) performed from lysates of transgenic C. elegans containing the indicated constructs. B, FLAG::RAE-1 coprecipitating with GFP:: D5 and GFP ::D5a. C, FLAG::RAE-1 coprecipitating with GFP::RBDa and GFP::RBDb. Shown are representatives of experiments that were performed at least three times using multiple transgenic lines for each genotype.
To do so, we performed coIP from transgenic C. elegans with extrachromosomal arrays that pan-neuronally express FLAG:: RAE-1 and different RPM-1 constructs tagged with GFP. PHA-1 selection ensured that only animals containing transgenic arrays survived. Consistent with previous findings using 293 cells (25), FLAG::RAE-1 coprecipitated with GFP::D5 from whole worm lysates (Fig. 2B). Further, D5a was sufficient for binding to RAE-1 (Fig. 2B), as were two smaller RPM-1 fragments, termed RBDa (amino acids 1913–2178) and RBDb (amino acids 2034–2151) (Fig. 2C). These results indicate that a much smaller region in RPM-1, RBDb, is sufficient for binding to RAE-1. These results expand upon prior work on two levels: by further mapping the RBD and by showing that the RBD is sufficient to mediate binding to RAE-1 in the neurons of C. elegans when expressed using a physiologically relevant neuronal promoter.
FSN-1 and RAE-1 Bind to RPM-1 via Different Biochemical Mechanisms
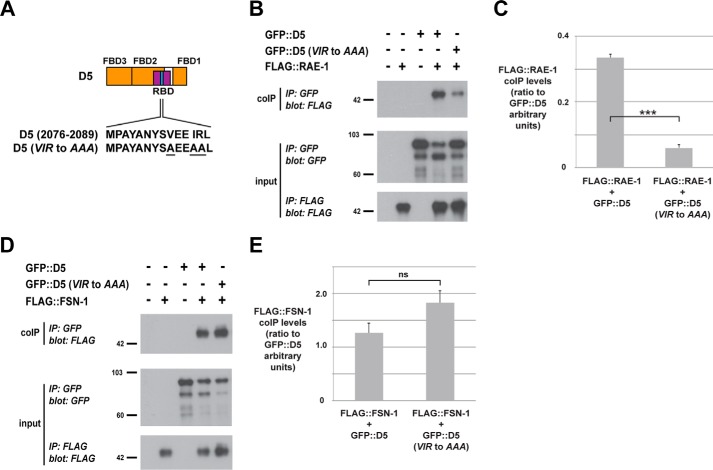
Our biochemical mapping showed that the RBD and all three FBDs are in close proximity in the primary sequence of RPM-1. Therefore, we wanted to test whether RAE-1 binding to RPM-1 might affect FSN-1 binding. A previous study in flies showed that Fsn and Rae1 are in protein complexes with the RPM-1 ortholog Highwire and that Fsn is dispensable for Rae1 binding to Highwire (26). However, it remained unclear whether RAE-1 affects binding of FSN-1 to RPM-1. We tested this by assessing how point mutations that reduce RAE-1 binding to RPM-1 affect FSN-1 binding (Fig. 3A) (25). As shown in Fig. 3B, FLAG::RAE-1 coprecipitated with GFP::D5 from lysates of transgenic worms expressing these constructs in the nervous system. The D5 (VIR to AAA) point mutant showed significantly reduced binding to RAE-1 (Fig. 3, B and C). In contrast, FSN-1 binding to D5 was not significantly altered in the D5 (VIR to AAA) point mutant (Fig. 3, D and E). These results show that FSN-1 and RAE-1 rely upon different biochemical mechanisms for binding to RPM-1.
FIGURE 3.
The VIR motif in RPM-1 D5 is necessary for binding to RAE-1 but not FSN-1. A, schematic of RPM-1 D5 with the expanded sequence showing the VIR motif that was mutated to AAA. B, coIP of FLAG::RAE-1 with D5 and D5 (VIR to AAA) from lysates of transgenic C. elegans. C, quantitation shows reduced FLAG::RAE-1 coprecipitation with D5 (VIR to AAA). D, coIP of FLAG::FSN-1 with D5 and D5 (VIR to AAA) from lysates of transgenic C. elegans. E, quantitation shows that FLAG::FSN-1 coprecipitation is similar for D5 and D5 (VIR to AAA). Quantitation was performed with data acquired from two or more transgenic lines for each genotype and represents data from two or more independent experiments. Histograms represent the ratio of FLAG::RAE-1 or FLAG::FSN-1 coprecipitated relative to the amount of GFP::D5 protein (all bands used in quantitation) that was immunoprecipitated. Images represent experiments that were performed at least three times using multiple transgenic lines for each genotype. Error bars represent the mean ± S.E., and significance was calculated using an unpaired t test. ***, p < 0.001; ns, not significant.
Overexpression of RPM-1 Fragments Inhibits Axon Termination and Synapse Formation
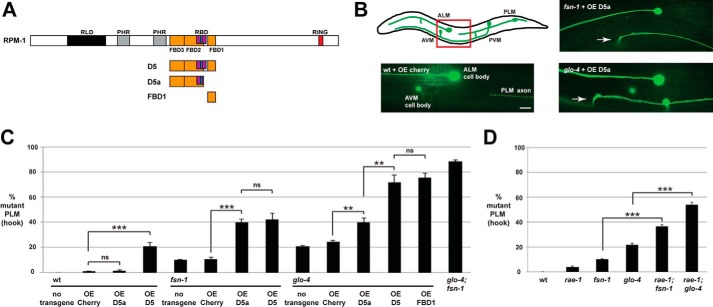
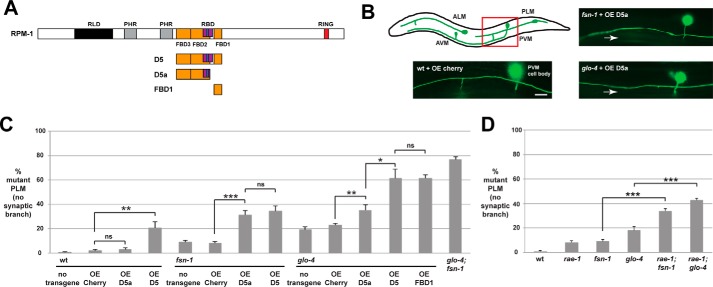
RPM-1 functions through several downstream signaling pathways to regulate axon termination and synapse formation in the mechanosensory neurons. FSN-1, RAE-1, and GLO-4 each mediate a portion of RPM-1 signaling and act in parallel genetic pathways (11, 25). Our biochemical results indicated that the D5 and D5a fragments of RPM-1 bound to both FSN-1 and RAE-1; therefore, we wanted to test whether transgenic overexpression of these RPM-1 fragments would affect axon termination and synapse formation.
Axon termination of the posterior lateral microtubule (PLM) mechanosensory neurons can be visualized using the transgene muIs32 (Pmec-7GFP). In C. elegans, there are two PLM mechanosensory neurons, each of which extends an axon that terminates before the cell body of the respective anterior lateral microtubule (ALM) mechanosensory neuron (Fig. 4B). Worms engineered with transgenic extrachromosomal arrays that used a pan-neuronal promoter (Prgef-1) to overexpress mCherry, a negative control protein, did not alter PLM axon termination (Fig. 4, B and C). Overexpression of D5a in wild-type animals did not alter axon termination, but overexpression in fsn-1 or glo-4 loss-of-function (lf) mutants resulted in an increased frequency of a severe axon termination defect in which the axon hooks toward the ventral side of the animal (Fig. 4, B and C). We refer to this severe axon termination defect as a “hook.” This is a similar phenotype to what occurs in glo-4; fsn-1 double mutants (Fig. 4C) and rpm-1 (lf) mutants (Fig. 8B) (11, 27). However, the enhanced frequency of defects caused by overexpression of D5a in fsn-1 or glo-4 single mutants was not as strong as that observed in glo-4; fsn-1 double mutants (Fig. 4C). D5 overexpression impaired axon termination in wild-type animals and enhanced defects in fsn-1 mutants to a comparable level as D5a (Fig. 4C). However, D5 overexpression in glo-4 mutants gave much stronger enhancer effects that were similar to glo-4; fsn-1 double mutants (Fig. 4C). Consistent with previous work (24), overexpression of FBD1 (also called RPM-1/FSN-1 complex inhibitory peptide) in glo-4 mutants gave a strong enhancer effect (Fig. 4C).
FIGURE 4.
Transgenic overexpression of RPM-1 D5a inhibits axon termination in the PLM mechanosensory neurons. A, schematic of RPM-1 and RPM-1 fragments tested. B, schematic (adapted from Ref. 28) showing the mechanosensory neurons of C. elegans (anterior left, dorsal top). The red box highlights the region of the animal shown in the images, which was visualized using muIs32 (Pmec-7GFP) for the indicated genotypes. The PLM axon terminates before the ALM cell body when the control protein mCherry is transgenically overexpressed in wild-type animals (wt + OE Cherry). Transgenic overexpression of RPM-1 D5a in an fsn-1 mutant (fsn-1 + OE D5a) or a glo-4 mutant (glo-4 + OE D5a) results in severe axon termination defects, referred to as hook defects (arrows). Scale bar = 10 μm. C, quantitation of axon termination defects (hook) for the indicated genotypes. Note that transgenic overexpression of RPM-1 D5 or D5a on an fsn-1 or glo-4 (lf) mutant background enhances the frequency of axon termination defects compared with overexpression of mCherry. D, quantitation of axon termination defects for the indicated genotypes. ***, p < 0.001; **, p < 0.005; ns, not significant.
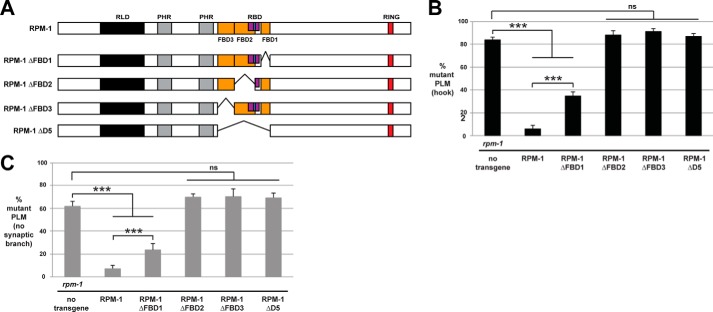
FIGURE 8.
RPM-1 deletion mutants lacking FBD1, FBD2, or FBD3 are functionally impaired. A, schematic of RPM-1 and RPM-1 deletion mutants tested. B and C, quantitation of PLM axon termination defects (B) or PLM synaptic branch defects (C) for the indicated genotypes. Transgenic expression of RPM-1 ΔFBD1 only partially rescues axon termination and synaptic branch defects in rpm-1 mutants. Transgenic expression of RPM-1 ΔFBD2, ΔFBD3, or ΔD5 fails to rescue rpm-1 (lf). ***, p < 0.001; ns, not significant.
RPM-1 also regulates synapse formation in the PLM mechanosensory neurons (10, 11). Therefore, we tested how transgenic overexpression of D5 and D5a affected PLM synapse formation. In wild-type animals, each PLM neuron extends a single synaptic branch that innervates interneurons of the ventral nerve cord, which can be visualized using muIs32 (Fig. 5B). Previous work showed that rpm-1 mutants have impaired synapse formation, which results in retraction of the synaptic branch (10, 11). Therefore, the synaptic branch is an accurate proxy for evaluating synapse formation in PLM neurons.
FIGURE 5.
Transgenic overexpression of RPM-1 D5a causes synaptic branch defects in PLM mechanosensory neurons. A, schematic of RPM-1 and RPM-1 fragments tested. B, schematic (adapted from adapted from Ref. 28) showing the mechanosensory neurons of C. elegans (anterior left, dorsal top). The red box highlights the region of the animal shown in the images, which were visualized using muIs32 (Pmec-7GFP) for the indicated genotypes. The PLM axons extends a synaptic branch when the control protein mCherry is transgenically overexpressed in wild-type animals (wt + OE Cherry). Transgenic overexpression of RPM-1 D5a in an fsn-1 mutant (fsn-1 + OE D5a) or a glo-4 mutant (glo-4 + OE D5a) results in loss of the synaptic branch (arrow), a phenotype that arises from impaired synapse formation. Scale bar = 10 μm. C, quantitation of synaptic branch defects (no synaptic branch) for the indicated genotypes. Note that transgenic overexpression of RPM-1 D5 or D5a on an fsn-1 or glo-4 (lf) mutant background enhances the frequency of synaptic branch defects compared with overexpression of mCherry. D, quantitation of synaptic branch defects for the indicated genotypes. ***, p < 0.001; **, p < 0.005; *, p < 0.02; ns, not significant.
We transgenically overexpressed D5 or D5a on various genetic backgrounds and analyzed the PLM synaptic branch. Overexpression of D5a or mCherry in wild-type animals had no phenotypic outcome, whereas overexpression of D5 resulted in low-frequency synaptic branch defects (Fig. 5C). Overexpression of D5 or D5a in either fsn-1 or glo-4 mutants resulted in an enhanced frequency of synaptic branch defects (Fig. 5, B and C). Similar to effects on axon termination, D5 and D5a enhanced fsn-1 (lf) at similar levels, but D5 gave much stronger enhancement of synaptic branch defects in glo-4 mutants (Fig. 5C).
Our observation that overexpression of D5 or D5a enhances both fsn-1 and glo-4 (lf) is consistent with two conclusions. First, D5 and D5a enhance fsn-1 (lf) to similar levels, which suggests that both impair the endogenous RPM-1·RAE-1 complex, as rae-1 (lf) moderately enhances fsn-1 and glo-4 (lf) (Figs. 4D and 5D) (25). Second, previous work showed that fsn-1 and glo-4 act in parallel pathways and strongly enhance one another (Figs. 4C and 5C) (11). Thus, D5a moderately enhances glo-4 (lf) because it partially impairs the function of the endogenous RPM-1·FSN-1 complex. In contrast, D5 strongly enhances glo-4 because it severely impairs endogenous RPM-1·FSN-1 function. These results show that overexpression of D5 or D5a enhance mutations that impair signaling downstream of RPM-1 and are consistent with our biochemical observations that D5 and D5a bind to both FSN-1 and RAE-1 in the neurons of C. elegans.
Defects Caused by Transgenic Overexpression of D5a Are Suppressed by dlk-1
Our biochemical results indicate that RPM-1 D5a binds to both FSN-1 and RAE-1 (Figs. 1B and 2B). Transgenic overexpression of D5a gives effects that are consistent with inhibition of endogenous RPM-1/RAE-1 function and partial inhibition of endogenous RPM-1·FSN-1 function (Figs. 4 and 5). To further support or refute this model, we tested how dlk-1 (lf) affects defects caused by D5a overexpression.
Previous work showed that axon termination and synapse formation defects caused by mutations in rpm-1 and fsn-1 are suppressed by loss of function in dlk-1 MAP3K (11, 17, 28). In contrast, dlk-1 does not suppress defects caused by rae-1 or glo-4 (lf) (Fig. 6) (11, 25). Therefore, dlk-1 (lf) provided a means to further test whether D5a overexpression impairs endogenous RPM-1·FSN-1 function.
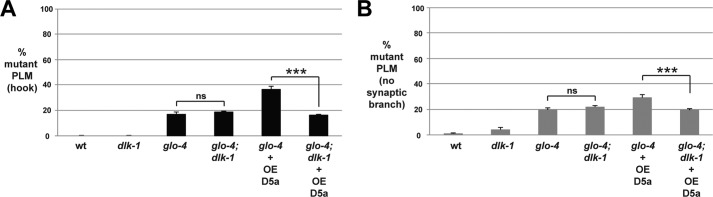
FIGURE 6.
Defects caused by transgenic overexpression of RPM-1 D5a are suppressed by dlk-1. A and B, quantitation of PLM axon termination defects (A) or PLM synaptic branch defects (B) for the indicated genotypes. The enhanced frequency of axon termination and synaptic branch defects caused by transgenic overexpression of D5a in glo-4 mutants is suppressed when D5a is overexpressed in glo-4; dlk-1 double mutants. ***, p < 0.001; ns, not significant.
Transgenic overexpression of D5a in glo-4 mutants resulted in an enhanced frequency of axon termination defects (Fig. 6A) and synaptic branch defects (Fig. 6B). Enhancer effects caused by D5a overexpression were completely suppressed on a glo-4; dlk-1 double mutant background (Fig. 6). These results are consistent with D5a impairing the function of the endogenous RPM-1·FSN-1 complex.
Overexpression of FBD2 or FBD3 Does Not Impair PLM Development
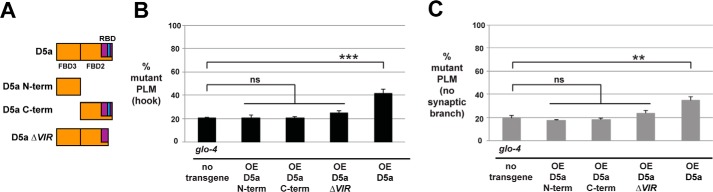
RPM-1 D5a is composed of two subdomains: the D5a C-term subdomain that contains FBD2 and a portion of RBD, including the VIR motif, which is required for binding to RAE-1 (Fig. 3), and the D5a N-term subdomain, which encompasses FBD3 (Fig. 7A). D5a C-term and D5a N-term are sufficient for binding to FSN-1 (Fig. 1). Therefore, we wanted to test whether enhancer effects caused by transgenic overexpression of D5a could be partitioned to the D5a N-term or D5a C-term fragments. As shown in Fig. 7B, overexpression of D5a N-term or D5a C-term in glo-4 mutants did not enhance the frequency of axon termination defects compared with non-transgenic glo-4 animals. Similarly, synaptic branch defects were not enhanced when D5a N-term or D5a C-term were overexpressed in glo-4 animals (Fig. 7C). These results show that only transgenic overexpression of full-length D5a enhances glo-4 (lf).
FIGURE 7.
Transgenic overexpression of D5a fragments does not enhance glo-4. A, schematic of RPM-1 fragments tested. B and C, quantitation of PLM axon termination defects (B) or PLM synaptic branch defects (C) for the indicated genotypes. Transgenic overexpression of D5a N-term, D5a C-term, and D5a ΔVIR in glo-4 mutants does not enhance the frequency of axon termination or synaptic branch defects. ***, p < 0.001; **, p < 0.005; ns, not significant.
Next we tested whether D5a that lacks the VIR motif and has reduced binding to RAE-1 can enhance glo-4 (lf). As shown in Fig. 7, B and C, transgenic overexpression of D5a lacking the VIR motif (D5a ΔVIR) in glo-4 animals did not enhance the frequency of axon termination or synaptic branch defects. These results provide further evidence that D5a inhibits a combination of endogenous RPM-1·FSN-1 and RPM-1·RAE-1 signaling in vivo and show that overexpression of FBD2 or FBD3 alone is insufficient to impair RPM-1·FSN-1 function.
Deletion of FBD2 or FBD3 Impairs RPM-1 Function
To complete our structure-function analysis of RPM-1 and FSN-1, we generated a series of deletion mutants of RPM-1 (Fig. 8A). These deletion mutants were tested for in vivo functional efficacy with transgenic rescue experiments using an rpm-1-null mutant (3). The native rpm-1 promoter was used to express RPM-1 constructs. As shown in Fig. 8B, severe axon termination defects (hook defects) occur with very high frequency in rpm-1 mutants and are rescued by transgenic expression of RPM-1. Consistent with previous work, RPM-1 lacking FBD1 (ΔFBD1) only partially rescued rpm-1 (lf) (Fig. 8B) (24). RPM-1 deletion mutants lacking D5 (ΔD5), FBD2 (ΔFBD2), or FBD3 (ΔFBD3) failed to rescue (Fig. 8B). Similar rescue outcomes occurred for synaptic branch defects (Fig. 8C). These results indicate that D5 is a critical functional region in RPM-1, and all three FBD domains are required for full RPM-1 function.
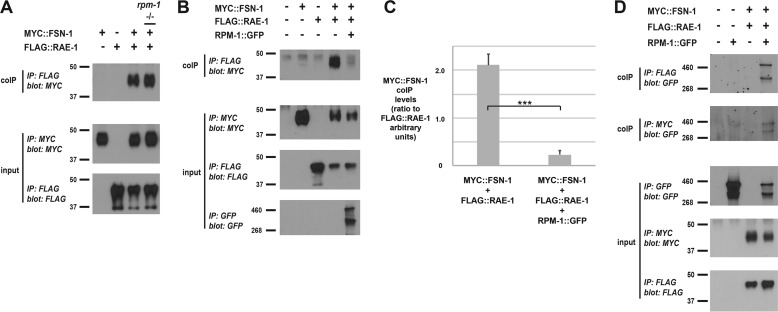
RPM-1 Reduces Binding of FSN-1 to RAE-1
Our finding that RPM-1 uses different biochemical mechanisms to bind FSN-1 and RAE-1 prompted us to test whether RPM-1 can form a tripartite complex with FSN-1 and RAE-1. FLAG:: RAE-1 and MYC ::FSN-1 were transgenically expressed using a pan-neuronal promoter and tested for binding by coIP. We observed strong binding of FSN-1 to RAE-1 (Fig. 9A). We thought this interaction might be mediated by endogenous RPM-1, but, unexpectedly, FSN-1 still bound to RAE-1 in an rpm-1 mutant background (Fig. 9A). A previous study showed that the rpm-1 allele we used, ju44, lacks RPM-1 protein expression (29).
FIGURE 9.
Coexpression of RPM-1 reduces binding of FSN-1 to RAE-1. Shown are coIPs (top panels) and IP inputs (center and bottom panels) from lysates of transgenic C. elegans containing the indicated constructs. A, MYC::FSN-1 coprecipitates with FLAG::RAE-1, and this interaction is not affected in an rpm-1 (lf) mutant. B, coprecipitation of MYC::FSN-1 with FLAG::RAE-1 is reduced by coexpression of RPM-1::GFP. C, quantitation of the reduction in MYC::FSN-1 binding to FLAG::RAE-1 with RPM-1::GFP coexpression. D, RPM-1::GFP coprecipitates with FLAG::RAE-1 or MYC::FSN-1 from transgenic worms simultaneously coexpressing all three constructs from a single transgenic array. Quantitation was performed with data acquired from four or more replicates from a single transgenic line. Histograms represent the ratio of MYC::FSN-1 coprecipitated relative to the amount of FLAG::RAE-1 that was immunoprecipitated. Images represent experiments that were performed at least three times using multiple transgenic lines for each genotype. Error bars represent the mean ± S.E., and significance was calculated using an unpaired t test. ***, p < 0.001.
Next we tested different biochemical interactions in transgenic worms that simultaneously coexpress MYC::FSN-1, FLAG:: RAE-1, and RPM-1 ::GFP. In these animals, FSN-1 and RAE-1 were expressed with a pan-neuronal promoter, and RPM-1 was expressed with its native promoter. Binding that occurs when only FSN-1 and RAE-1 are expressed together was strongly reduced by coexpression of RPM-1 (Fig. 9, B and C). Under these conditions, RPM-1::GFP still forms complexes with both FSN-1 and RAE-1 (Fig. 9D). These in vivo biochemical results indicate that RPM-1 reduces FSN-1·RAE-1 binding by forming two separate protein complexes with both molecules.
Discussion
C. elegans RPM-1 is a conserved regulator of axon termination and synapse formation (1, 2). Previous work showed that RPM-1 functions as an intracellular signaling hub that positively and negatively regulates numerous downstream signaling pathways (2, 11, 17, 25, 28, 30–32). At present, relatively little is known about the biochemistry that underpins formation of RPM-1 signaling complexes. Using positive selection of transgenic worms and in vivo biochemistry from the worm nervous system, we have generated new insights into the signaling complexes formed between RPM-1, FSN-1, and RAE-1.
Identification of FBD2 and FBD3
Our prior study identified a conserved region in RPM-1 that is sufficient for binding to FSN-1, called FBD1 (Fig. 10) (24). Mutation of several conserved residues in FBD1 reduced FSN-1 binding in 293 cells but did not reduce binding of FSN-1 to full-length RPM-1 in vivo in the neurons of C. elegans. These observations suggest that RPM-1 contains other sites that are sufficient for binding to FSN-1.
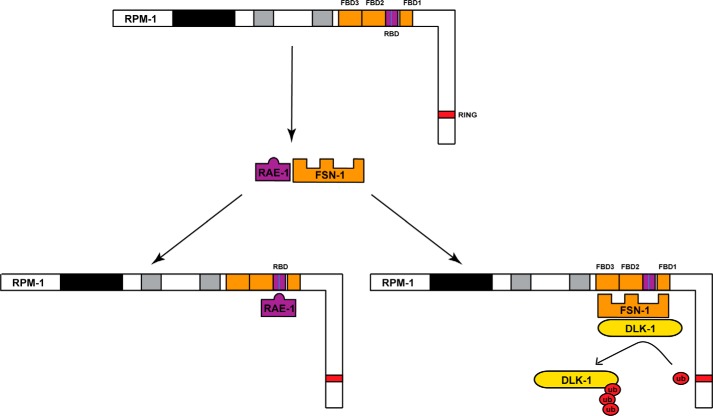
FIGURE 10.
Summary of structure-function analysis of the RPM-1/FSN-1 and RPM-1/RAE-1 signaling complexes. Schematic showing the binding of FSN-1 to RAE-1 and RPM-1 forming separate complexes with FSN-1 and RAE-1. Shown in RPM-1 are the new RBD (purple) and the two new FSN-1 binding domains we identified, FBD2 and FBD3 (orange). Highlighted in blue is the VIR motif in RPM-1 that is necessary for binding to RAE-1 but not FSN-1. The model shown incorporates DLK-1, a known ubiquitination target of the RPM-1·FSN-1 complex.
By using transgenic C. elegans, neuronal promoters for expression, and PHA-1 positive selection to increase the sensitivity of in vivo biochemistry, we found that the D5a fragment of RPM-1 binds to both FSN-1 and RAE-1 (Figs. 1 and 2). Biochemical mapping showed that D5a contains two domains that are sufficient for binding to FSN-1, which we have termed FBD2 and FBD3. We previously tested D5a for binding to FSN-1 in 293 cells but did not detect an interaction (24). This suggests that expression in neurons is required for FSN-1 to bind FBD2 and FBD3.
There are two likely explanations for why in vivo neuronal expression is required to detect these interactions. RPM-1, Highwire, and Pam/MYCBP2 are in SCF complexes that include Skp1 (20–22). It is plausible that mammalian Skp1 cannot facilitate binding of RPM-1 to FSN-1 in HEK 293 cells. In this case, FBD1 binding to FSN-1 would be detected in 293 cells if it is a direct interaction, whereas binding of FSN-1 to FBD2 and FBD3 would only be observed in C. elegans neurons because the native Skp protein, most likely Skr1, must be present. Alternatively, posttranslational modifications that only occur in the neurons of C. elegans might be needed for FSN-1 or Skp to bind FBD2 and FBD3. Future biochemical studies with different components of the RPM-1 and mammalian Phr1 ubiquitin ligase complexes will be needed to distinguish between these possibilities.
FSN-1 and RAE-1 Use Different Biochemical Mechanisms to Bind RPM-1
The RPM-1 sequence encoding FBD2 and FBD3 overlaps with the previously annotated RBD (25). This prompted us to further map the RBD to a smaller fragment, RBDb, that we show is sufficient for binding to RAE-1 (Fig. 2). Because of this finding, we have modified the annotation of protein domains in RPM-1 to reflect the boundaries of this much smaller RBD (Fig. 10).
It has been shown previously that Drosophila Fsn is not required for the interaction between Highwire and DRae1 (26). However, this study did not test whether DRae1 affects binding of DFsn to Highwire. We could not test whether RAE-1 is required for the interaction between RPM-1 and FSN-1 in vivo in worms because rae-1 (lf) animals are sterile, which prevented us from obtaining sufficient material for biochemistry. As an alternative, we show that mutation of the VIR motif in RPM-1 D5, which is necessary for binding to RAE-1, does not affect binding to FSN-1 (Fig. 3). This result supports the conclusion that RAE-1 and FSN-1 rely upon different biochemical mechanisms to bind RPM-1.
RPM-1 D5a Inhibits Both RPM-1/FSN-1 and RPM-1/RAE-1 Signaling in Vivo
To evaluate the functional relevance of the FSN-1 and RAE-1 binding sites we identified in RPM-1, we transgenically overexpressed RPM-1 D5a and assessed the effects on axon termination and synapse formation of the PLM mechanosensory neurons. A series of observations collectively support the conclusion that D5a impairs both RPM-1/FSN-1 and RPM-1/RAE-1 signaling in vivo.
First, as discussed earlier, RPM-1 D5a is sufficient to bind both FSN-1 and RAE-1. This suggests that transgenically overexpressed D5a would bind to endogenous FSN-1 and RAE-1 in neurons and potentially impair the formation or function of both endogenous RPM-1·FSN-1 and RPM-1·RAE-1 complexes.
Second, D5a overexpression enhances the frequency of axon termination and synapse formation defects in both fsn-1 and glo-4 (lf) single mutants (Figs. 4 and 5). If D5a overexpression solely impaired RPM-1/FSN-1 signaling, we would expect enhancement of glo-4 (lf) but not enhancement of fsn-1 (lf), similar to what occurs with overexpression of FBD1 (24). Likewise, if D5a overexpression completely impaired RPM-1/FSN-1 signaling, then we would expect much stronger enhancer effects with overexpression of D5a in glo-4 mutants, similar to what occurs with overexpression of FBD1 or D5 in glo-4 single mutants (Figs. 4 and 5). Further, D5a lacking the VIR motif, which is necessary for binding to RAE-1, fails to enhance glo-4 (lf) (Fig. 7). These genetic, transgenic, and biochemical results suggest that overexpression of D5a affects axon termination and synapse formation by impairing RPM-1·RAE-1 signaling.
Our results with transgenic overexpression of D5a C-term (FBD2) and D5a N-term (FBD3) suggest that D5a is unlikely to function solely by inhibiting RPM-1·RAE-1 signaling. If this were the case, we would expect D5a C-term (which contains the VIR motif that is required for RAE-1 binding) to give similar enhancer effects to full-length D5a. However, this did not occur (Fig. 7). Further, our observation that D5a enhancement of glo-4 (lf) is suppressed by dlk-1 suggests that D5a impairs FSN-1 function.
Rescue experiments with RPM-1 deletion mutants also support the conclusion that D5a overexpression impairs both RPM-1·RAE-1 and RPM-1·FSN-1 function. RPM-1 ΔFBD1 gave a strong but partial rescue of rpm-1 (lf) defects (Fig. 8). This is consistent with prior work suggesting that FBD1 only affects FSN-1 binding and function (24). In contrast, deletion of the entire D5 region, or FBD2 completely impairs rescue. Although this could reflect major structural problems caused by these deletions, it is also consistent with these deletions affecting key domains and residues that are required for both FSN-1 and RAE-1 binding to RPM-1. RPM-1 lacking FBD3 also completely failed to rescue rpm-1 (lf). Although FSN-1 is the only protein known to bind to this region of RPM-1, this result suggests that FBD3 could have further functional roles beyond simply binding to FSN-1. This combination of biochemical and genetic results demonstrates that overexpression of D5a impacts axon termination and synapse formation by inhibiting a combination of endogenous RPM-1·FSN-1 and RPM-1·RAE-1 signaling in neurons.
The recent discoveries that Drosophila Highwire and mouse Phr1 are required for in vivo axon degeneration has greatly increased interest in developing inhibitors of PHR ubiquitin ligase activity (15, 16). Our results showing that RPM-1 contains three functional FSN-1 binding domains (FBD1, FBD2, and FBD3) now provides several avenues for designing potential inhibitors of the mammalian Phr1·Fbxo45 ubiquitin ligase complex.
RPM-1 Forms Different Signaling Complexes with FSN-1 and RAE-1
Because RAE-1 and FSN-1 rely upon different mechanisms to bind RPM-1, we tested whether RPM-1, FSN-1, and RAE-1 form a single tripartite complex. Although we initially favored this hypothesis, the following observations indicate that this is not the case. RAE-1 and FSN-1 bind to one another in the neurons of C. elegans, and this interaction is not dependent upon RPM-1 (Fig. 9A). When RPM-1, FSN-1, and RAE-1 are simultaneously expressed from a single transgenic array, binding of FSN-1 to RAE-1 is strongly reduced (Fig. 9, B and C). Under these conditions, RPM-1 still binds to both FSN-1 and RAE-1 (Fig. 9D). These findings indicate that RPM-1 forms complexes with FSN-1 and RAE-1 that are likely to be mutually exclusive.
One interesting possibility that emerges from our observations is that RPM-1 forms different complexes to engage in positive versus inhibitory signaling. In this case, it is plausible that the positive regulatory complex would include RAE-1, GLO-4, and ANC-1 (11, 25, 31), whereas the negative regulatory complex would include FSN-1 and PPM-2 (28). Although this is an intriguing model, it is important to emphasize that we are only beginning to understand the biochemical nature of RPM-1 signaling complexes, and other more complex signaling models are also possible.
Our observation that RPM-1 reduces binding of FSN-1 to RAE-1 supports our prior genetic finding that RAE-1 acts downstream of RPM-1 (25). In contrast to findings in worms, DRae1 functions upstream of Highwire in flies (26). Further biochemistry with Highwire, DRae1, and DFsn1 in flies could prove helpful in understanding why these differences have been observed.
Why and how RAE-1 and FSN-1 bind to one another remains unclear. We noted that the RAE-1 binding motif in RPM-1 is not present in FSN-1 (data not shown) (25). This suggests that RAE-1 is unlikely to bind to FSN-1 and RPM-1 using the same mechanism. It is possible that RAE-1 binds to FSN-1 to keep FSN-1 inactive. RPM-1 would then form independent complexes with RAE-1 and FSN-1 to differentiate signaling during specific times in development or in particular subcellular compartments.
The emerging role of RPM-1 orthologs in axon degeneration has made developing inhibitors of the RPM-1·FSN-1 complex an important goal. Our findings now bring us one step closer to understanding how to best achieve this. Our results also suggest that facilitating the interaction between FSN-1 and RAE-1 could be an alternative way to inhibit the RPM-1·FSN-1 complex.
Materials and Methods
Genetics and Axon Morphology Analysis
Genetic analysis was performed using the N2 isolate of C. elegans and standard procedures (33). The alleles used in this study included fsn-1(gk429), rae-1(tm2784), glo-4(ok623), rpm-1(ju44), and dlk-1(ju476). To analyze axon termination and synaptic branch defects in the PLM mechanosensory neurons, the transgene muIs32 (Pmec-7GFP) was used. In keeping with prior studies, animals were cultivated at 23 °C, and 16–20 h after L4, young adult animals were anesthetized using either 1% (v/v) 1-phenoxy-2-propanol or 5 mm levamisole in M9 buffer and mounted on glass slides with 2% agarose. Animals were visualized using ×40 magnification, an oil immersion lens, and an epifluorescence microscope (Leica CRF 5000). Images were acquired using a charge coupled device (CCD) camera (Leica DFC345 FX). For quantitation, averages are shown for data collected from a minimum of six independent counts of 20–30 PLM neurons. For transgenes, data were pooled from five or more transgenic lines. Error bars represent the mean ± S.E., and significance was determined using an unpaired Student's t test.
Cloning
rpm-1 cDNAs were amplified by PCR using iProof DNA polymerase (Bio-Rad). Amplified cDNAs were inserted into pCR8 Topo GY (Invitrogen) and sequenced to ensure that they were error-free. The following pCR8 Topo clones were created using the indicated plasmids as templates: D5a N-term (pBG-GY586), D5a C-term (pBG-GY587), and D5a ΔVIR (pBG-GY595) were cloned from pCR8 RPM-1 D5a (pBG-GY363), and RBDa (pBG-GY599) and RBDb (pBG-GY600) were cloned from pCR8 RPM-1 D5 (pBG-GY175). For point mutagenesis, pCR8 RPM-1 D5 (pBG-GY175) was used as template. Primers encoding the desired changes and a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) were used to generate pCR8 RPM-1 D5 VIR (2084,87,88) AAA (pBG-GY626). This plasmid was sequenced to confirm point mutations. Clones in pCR8 Topo GY were recombined using LR clonase (Invitrogen) into one of the following destination plasmids: Prgef-1GFP GY (pBG-GY498), Prgef-1FLAG GY (pBG-GY134), Prgef-1MYC GY (pBG-GY525), or Prgef-1 GY (pBG-GY152). To clone rpm-1 deletion mutants, an rpm-1 fragment with unique flanking enzyme sites (HpaI-BssHII) was subcloned from pBG-205 (Prpm-1::rpm-1 genomic) and modified to remove different RPM-1 domains of interest. These deletion subclones were then cut with HpaI and BssHII and ligated back to pBG-205. Sequencing confirmed that deletions and reading frames were correct. This approach was used to generate Prpm-1::rpm-1 ΔD5 (Δaa 1700–2257, pBG-281), Prpm-1::rpm-1 ΔFBD2 (Δaa 1877–2098, pBG-286), and Prpm-1::rpm-1 ΔFBD3 (Δaa 1700–1876, pBG-282).
Transgenics
Transgenic animals were generated by standard microinjection procedures (34). Transgenic animals were constructed by injecting a mixture of plasmid DNA or PCR product of interest, a plasmid encoding Pmyo-2cherry (1 ng/μl), and pBluescript at a total DNA concentration of 100 ng/μl. For functional experiments (Figs. 4–7), PCR products were amplified by a long PCR kit (Roche) and injected at 5 ng/μl. The following plasmids were used as templates for long PCR: Prgef-1FLAG::mCherry (pBG-GY371), Prgef-1FLAG::FBD1 (pBG-GY440), Prgef-1FLAG::RPM-1 D5 (pBG-GY680), Prgef-1FLAG::RPM-1 D5a (pBG-GY557), Prgef-1RPM-1 D5a N-term (pBG-GY589), Prgef-1RPM-1 D5a C-term (pBG-GY590), and Prgef-1FLAG::RPM-1 D5a ΔVIR (pBG-GY598). For functional analysis of RPM-1 deletion mutants (Fig. 8), the following plasmids were injected at 20 ng/μl: Prpm-1::rpm-1 (pBG-205), Prpm-1::rpm-1 ΔFBD1 (pBG-210), Prpm-1::rpm-1 ΔFBD2 (pBG-286), Prpm-1::rpm-1 ΔFBD3 (pBG-282), and Prpm-1::rpm-1 ΔD5 (pBG-281).
For biochemical experiments (Figs. 1–3 and 9), transgenic animals were generated by injecting pha-1 (e2123) animals and adding pBX (genomic pha-1) at 20 ng/μl to the injection mixture as described previously (35). pha-1 (e2123) is a temperature-sensitive, lethal allele at 23 °C. Thus, the lethality of pha-1 allowed us to perform rapid positive selection and enrich a pure population of animals carrying transgenic extrachromosomal arrays that express PHA-1. PHA-1 selection resulted in a significant improvement in the sensitivity and quality of biochemical results compared with previous studies (11, 25, 28), which allowed us to use lower amounts of material and test more transgenic lines for expression levels. Transgenic worms for biochemistry were generated by injecting the following plasmids at 25–50 ng/μl: Prgef-1GFP::FBD1 (pBG-GY536), Prgef-1GFP::RPM-1 D4 (pBG-GY572), Prgef-1GFP::RPM-1 D5 (pBG-GY583), Prgef-1GFP::RPM-1 D5a (pBG-GY534), Prgef-1GFP::RPM-1 D5a N-term (pBG-GY592), Prgef-1GFP::RPM-1 D5a C-term (pBG-GY593), Prgef-1GFP::RPM-1 D5 VIR (2084,87,88) AAA (pBG-GY630), Prgef-1GFP::RBDa (pBG-GY601), Prgef-1GFP::RBDb (pBG-GY602), Prpm-1RPM-1::GFP (pBG-219), Prgef-1FLAG::FSN-1 (pBG-GY422), Prgef-1MYC::FSN-1 (pBG-GY736), and Prgef-1FLAG::RAE-1 (pBG-32). For the biochemical experiment shown in Fig. 9A, arrays were generated as described above and crossed into rpm-1 (ju44) mutants.
Biochemistry
To generate material for biochemistry, transgenic animals were placed on 10-cm nematode growth media agar plates seeded with Escherichia coli (OP-50) and grown for 2–3 days. Animals were fed as needed with E. coli HB101. At high density, mixed-stage animals were either harvested or transferred to M9 liquid culture containing HB101 and cholesterol and grown for 1–2 days to generate larger amounts of material. To ensure that a pure population of transgenic worms was obtained, animals were continuously grown at 23 °C to select for PHA-1 transgenic arrays (see above). Animals were harvested by centrifugation, washed three or more times in M9 buffer, and frozen at −80 °C. Frozen animal pellets were ground using a mortar and pestle under liquid N2. Ground whole, transgenic animals were lysed in 0.1% Nonidet P-40 lysis buffer (50 mm Tris (pH 7.5), 0.1% Nonidet P-40, 150 mm NaCl, 10% glycerol, 2 mm DTT, and EDTA-free protease inhibitor pellets). Total protein concentration was determined using the BCA protein assay kit (Pierce). For IP, 0.75–5 mg of total protein from transgenic worm lysates was used, and expression of transgenic components was normalized when needed. Lysates were incubated with one of the following antibodies for 30 min: 1.5 μl (1.6 μg) of M2 (anti-FLAG, mouse monoclonal, Sigma), 3 μl (0.6 μg) of 3E6 (anti-GFP, mouse monoclonal, MP Biomedicals), or 1.5 μl (4 μg) of 9E10 (anti-MYC, mouse monoclonal, Sigma). They were then precipitated for 4 h at 4 °C with 10 μl of protein G-agarose (Roche).
Precipitates were boiled in SDS Laemmli sample buffer (Bio-Rad) and run on a 4–12% BisTris gel with MOPS SDS running buffer or a 3–8% Tris acetate gel with Tris acetate SDS running buffer (Invitrogen). In all figures, when coIP was performed, 60% of the sample was run on the gel (shown as coIP, top panels) whereas 30% of the sample was run for input (shown as input, center and bottom panels). Gels were then transferred to PVDF membranes for 1 h at 90 V for proteins under 100 kDa or 16 h at 30 V for proteins over 100 kDa and immunoblotted. The following primary antibodies were diluted in 5% nonfat milk in tris-buffered saline plus tween (TBST) and applied to blots overnight at 4 °C: rabbit polyclonal anti-FLAG (Cell Signaling Technology), mouse monoclonal 9B11 anti-MYC (Sigma), or a mixture of mouse monoclonal anti-GFP antibodies (Roche).
Blots were visualized with HRP-conjugated anti-mouse or anti-rabbit secondary antibodies, Supersignal FemtoWest ECL (Pierce), and X-ray film. When proteins of interest migrated close to the antibody heavy chain, light chain-reactive secondary antibodies were used (Millipore). Quantitation of bands in immunoblots (Figs. 3 and 9) was performed using ImageJ software from National Institutes of Health (http://rsb.info.nih.gov/ij/). Quantitation was done from four or more replicates for a given genotype.
Author Contributions
S. T. B. performed and designed experiments, analyzed the data, and helped write the manuscript. B. G. designed experiments and wrote the manuscript.
Acknowledgments
We thank the C. elegans knockout consortium for generating alleles and the C. elegans Genetics Center for providing strains (National Institutes of Health Office of Research Infrastructure Programs, P40 OD010440).
This work was supported by National Institutes of Health Grant 2R01 NS072129-06 (to B. G.) and National Science Foundation Grant IOS-1121095 (to B. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PHR
- Pam/Highwire/RPM-1
- RPM
- regulator of presynaptic morphology
- FBD
- FSN-1 binding domain
- RBD
- RAE-1 binding domain
- IP
- immunoprecipitation
- N-term
- N-terminal
- C-term
- C-terminal
- PLM
- posterior lateral microtubule
- lf
- loss of function
- SCF
- Skp/Cullin/F-box.
References
- 1. Po M. D., Hwang C., and Zhen M. (2010) PHRs: bridging axon guidance, outgrowth and synapse development. Curr. Opin. Neurobiol. 20, 100–107 [DOI] [PubMed] [Google Scholar]
- 2. Grill B., Murphey R. K., and Borgen M. A. (2016) The PHR proteins: intracellular signaling hubs in neuronal development and axon degeneration. Neural Dev. 11, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhen M., Huang X., Bamber B., and Jin Y. (2000) Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron 26, 331–343 [DOI] [PubMed] [Google Scholar]
- 4. Wan H. I., DiAntonio A., Fetter R. D., Bergstrom K., Strauss R., and Goodman C. S. (2000) Highwire regulates synaptic growth in Drosophila. Neuron 26, 313–329 [DOI] [PubMed] [Google Scholar]
- 5. Burgess R. W., Peterson K. A., Johnson M. J., Roix J. J., Welsh I. C., and O'Brien T. P. (2004) Evidence for a conserved function in synapse formation reveals Phr1 as a candidate gene for respiratory failure in newborn mice. Mol. Cell. Biol. 24, 1096–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bloom A. J., Miller B. R., Sanes J. R., and DiAntonio A. (2007) The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev. 21, 2593–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lewcock J. W., Genoud N., Lettieri K., and Pfaff S. L. (2007) The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron 56, 604–620 [DOI] [PubMed] [Google Scholar]
- 8. Shin J. E., and DiAntonio A. (2011) Highwire regulates guidance of sister axons in the Drosophila mushroom body. J. Neurosci. 31, 17689–17700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li H., Kulkarni G., and Wadsworth W. G. (2008) RPM-1, a Caenorhabditis elegans protein that functions in presynaptic differentiation, negatively regulates axon outgrowth by controlling SAX-3/robo and UNC-5/UNC5 activity. J. Neurosci. 28, 3595–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schaefer A. M., Hadwiger G. D., and Nonet M. L. (2000) rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron 26, 345–356 [DOI] [PubMed] [Google Scholar]
- 11. Grill B., Bienvenut W. V., Brown H. M., Ackley B. D., Quadroni M., and Jin Y. (2007) C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron 55, 587–601 [DOI] [PubMed] [Google Scholar]
- 12. Kim J. H., Wang X., Coolon R., and Ye B. (2013) Dscam expression levels determine presynaptic arbor sizes in Drosophila sensory neurons. Neuron 78, 827–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Opperman K. J., and Grill B. (2014) RPM-1 is localized to distinct subcellular compartments and regulates axon length in GABAergic motor neurons. Neural Dev. 9, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giles A. C., Opperman K. J., Rankin C. H., and Grill B. (2015) Developmental function of the PHR protein RPM-1 is required for learning in Caenorhabditis elegans. G3 (Bethesda) 5, 2745–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiong X., Hao Y., Sun K., Li J., Li X., Mishra B., Soppina P., Wu C., Hume R. I., and Collins C. A. (2012) The Highwire ubiquitin ligase promotes axonal degeneration by tuning levels of Nmnat protein. PLoS Biol. 10, e1001440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Babetto E., Beirowski B., Russler E. V., Milbrandt J., and DiAntonio A. (2013) The Phr1 ubiquitin ligase promotes injury-induced axon self-destruction. Cell Rep. 3, 1422–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakata K., Abrams B., Grill B., Goncharov A., Huang X., Chisholm A. D., and Jin Y. (2005) Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell 120, 407–420 [DOI] [PubMed] [Google Scholar]
- 18. Collins C. A., Wairkar Y. P., Johnson S. L., and DiAntonio A. (2006) Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron 51, 57–69 [DOI] [PubMed] [Google Scholar]
- 19. Huntwork-Rodriguez S., Wang B., Watkins T., Ghosh A. S., Pozniak C. D., Bustos D., Newton K., Kirkpatrick D. S., and Lewcock J. W. (2013) JNK-mediated phosphorylation of DLK suppresses its ubiquitination to promote neuronal apoptosis. J. Cell Biol. 202, 747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liao E. H., Hung W., Abrams B., and Zhen M. (2004) An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature 430, 345–350 [DOI] [PubMed] [Google Scholar]
- 21. Saiga T., Fukuda T., Matsumoto M., Tada H., Okano H. J., Okano H., and Nakayama K. I. (2009) Fbxo45 forms a novel ubiquitin ligase complex and is required for neuronal development. Mol. Cell. Biol. 29, 3529–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brace E. J., Wu C., Valakh V., and DiAntonio A. (2014) SkpA restrains synaptic terminal growth during development and promotes axonal degeneration following injury. J. Neurosci. 34, 8398–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu C., Daniels R. W., and DiAntonio A. (2007) DFsn collaborates with Highwire to down-regulate the Wallenda/DLK kinase and restrain synaptic terminal growth. Neural Dev. 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sharma J., Baker S. T., Turgeon S. M., Gurney A. M., Opperman K. J., and Grill B. (2014) Identification of a peptide inhibitor of the RPM-1.FSN-1 ubiquitin ligase complex. J. Biol. Chem. 289, 34654–34666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grill B., Chen L., Tulgren E. D., Baker S. T., Bienvenut W., Anderson M., Quadroni M., Jin Y., and Garner C. C. (2012) RAE-1, a novel PHR binding protein, is required for axon termination and synapse formation in Caenorhabditis elegans. J. Neurosci. 32, 2628–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tian X., Li J., Valakh V., DiAntonio A., and Wu C. (2011) Drosophila Rae1 controls the abundance of the ubiquitin ligase Highwire in post-mitotic neurons. Nat. Neurosci. 14, 1267–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tulgren E. D., Baker S. T., Rapp L., Gurney A. M., and Grill B. (2011) PPM-1, a PP2Cα/β phosphatase, regulates axon termination and synapse formation in Caenorhabditis elegans. Genetics 189, 1297–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baker S. T., Opperman K. J., Tulgren E. D., Turgeon S. M., Bienvenut W., and Grill B. (2014) RPM-1 uses both ubiquitin ligase and phosphatase-based mechanisms to regulate DLK-1 during neuronal development. PLoS Genet. 10, e1004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abrams B., Grill B., Huang X., and Jin Y. (2008) Cellular and molecular determinants targeting the Caenorhabditis elegans PHR protein RPM-1 to perisynaptic regions. Dev. Dyn. 237, 630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yan D., Wu Z., Chisholm A. D., and Jin Y. (2009) The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell 138, 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tulgren E. D., Turgeon S. M., Opperman K. J., and Grill B. (2014) The Nesprin family member ANC-1 regulates synapse formation and axon termination by functioning in a pathway with RPM-1 and β-catenin. PLoS Genet. 10, e1004481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baker S. T., Turgeon S. M., Tulgren E. D., Wigant J., Rahimi O., Opperman K. J., and Grill B. (2015) Neuronal development in Caenorhabditis elegans is regulated by inhibition of an MLK MAP kinase pathway. Genetics 199, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mello C. C., Kramer J. M., Stinchcomb D., and Ambros V. (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Granato M., Schnabel H., and Schnabel R. (1994) pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 22, 1762–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]