Abstract
Motivation
Model simulation exchange has been standardized with the Simulation Experiment Description Markup Language (SED-ML), but specialized software is needed to generate simulations in this format. Text-based languages allow researchers to create and modify experimental protocols quickly and easily, and export them to a common machine-readable format.
Results
phraSED-ML language allows modelers to use simple text commands to encode various elements of SED-ML (models, tasks, simulations, and results) in a format easy to read and modify. The library can translate this script to SED-ML for use in other softwares.
Availability
phraSED-ML language specification, libphrasedml library, and source code are available under BSD license from http://phrasedml.sourceforge.net/.
Keywords: SED-ML, SBML, simulation, reproducibility
1. Introduction
Reproducibility is a cornerstone of scientific research. In systems biology, the ability to losslessly encode and share computational models of biological systems is enabled with the development of Systems Biology Markup Language (SBML)1 and CellML.2 More recently, the Simulation Experiment Description Markup Language (SED-ML) was released.3 This new markup language expanded the field of computer-exchangeable modeling from the models to the experiments performed on those models. The first version of SED-ML (Level 1 version 1) included the ability to encode uniform time course experiments only. Version 2 expanded SED-ML’s capabilities to include ‘repeated tasks’, which allow more complicated experiments such as parameter scans. SED-ML defines five basic classes: the Model references a particular XML-based model (usually in SBML or CellML), or a modified version of the same; the Simulation defines a particular algorithm; the Task relates an algorithm to a model; and an Output defines how the results of a Task are to be conveyed to the user, either as a plot or a report. The RepeatedTask introduced in Version 2 of the specification is a more specialized version of a Task that defines how to repeat another Task over a specified range. Elements inside referenced models are referred to the use of XML Path Language (XPath) strings,4 and algorithms and their parameters are referenced by their Kinetic Simulation Algorithm Ontology (KiSAO) ids.5
There are several programs that support SED-ML by importing and performing simulation described in a SED-ML file. Our own Tellurium (tellurium.analogma-chine.org) translates SED-ML to Python code that utilizes Roadrunner’s Python bindings.6 Other simulators include the SBW Simulation Tool7 and the Systems Biology Simulation Core Library.8 Apart from generic XML editors, some others let you edit or create SED-ML. SED-ML Script9 defines a script-based language that is closely tied to SED-ML itself: each script function adds a particular SED-ML element to a document, with the arguments of that function setting all of the particular attributes and child elements of that root element. However, no particular attempt is made to hide the complexity or to error-check: all arguments to the script functions are passed as-is to the SED-ML creator, including XPath strings. While simpler than editing the XML directly, this approach leaves a burden on the user to comprehend the details of raw SED-ML.
On the other end of the complexity spectrum, Frank Bergmann created a SED-ML creator ‘wizard’ (http://sysbioapps.dyndns.org/SED-ML/Web/Tools/) which creates a ‘standard’ SED-ML file from an uploaded SBML file with few basic options. Another GUI-based SED-ML creator is SED-ED,10 which lets the user build up a SED-ML file element by element, with error-checking along the way, while providing a graphical overview of the relationships between the elements. It also provides automated methods for creating XPath strings and labeled fields for the necessary SED-ML attributes.
Another tool that allows the creation of SED-ML files is ‘Copasi2SEDML’, which allows conversion of a COPASI file11 with a time course simulation to the equivalent SED-ML. Similarly, CellDesigner12 allows both import and export of SED-ML.
Here, we provide phraSED-ML as a balance between these options, occupying a niche similar to that inhabited by Antimony,13 which is a text-based language to define the models themselves. We distribute libraries for the language to integrate it with any other simulation software that can understand SED-ML, providing the users with a new option for SED-ML creation and exploration. Python bindings provide a unified scripting environment that can both execute and export their simulation experiment.
2. Features
2.1. Simple abstractions
The phraSED-ML language is designed to be easily readable and writable without the need to reference the documentation at each turn. The format of each line is declarative, and because the subject of SED-ML is the act of simulation, all keywords are verbs. Model objects are declared with the keyword ‘model’, simulations with the keyword ‘simulate’, tasks with the keyword ‘run’, repeated tasks with the keyword ‘repeat’, and output with the keyword ‘plot’ and ‘report’. The DataGenerator object is abstracted away entirely — objects are created as needed according to user directives parsed in the requested outputs. The following illustrates a typical phraSED-ML code.
mod1 = model “sbml_model.xml” sim1 = simulate uniform(0,10,100) task1 = run sim1 on mod1 plot time vs S1
XPath expressions are avoided on the user end by allowing model elements to be referenced by ID alone (e.g. ‘S1’), which the libphrasedml library translates to an appropriate XPath string. Element attribute values are similarly translated behind the scenes, made possible by the use of libsbml library to parse the referenced model to determine which element attribute must be used. This restricts the scope of phraSED-ML somewhat from the broader abilities of SED-ML, which can perform arbitrary XML transformations. However, this restriction was not deemed too onerous, particularly in light of the simplification it offered.
2.2. Allowed complexity
Some of the more advanced features of SED-ML may still be accessed with phraSED-ML. KiSAO terms are not necessary, but if the user wishes to use backward differentiation formula for solving ordinary differential equation (ODE), for example, they may use the ‘bdf’ keyword:
sim1.algorithm = bdf
which will be translated to SED-ML as KiSAO id 288. The KiSAO id may also be used directly:
sim1.algorithm = kisao.288
which is useful for algorithms for which there are no built-in keywords. Similarly, algorithm parameters may be defined either by keyword or KiSAO id. Here, the relative tolerance (KiSAO id 209) is set:
sim1.algorithm.relative_tolerance = 0.001 sim1.algorithm.209 = 0.001
SED-ML ‘repeated tasks’ also have phraSED-ML equivalents:
task2 = repeat task1 for S1 in [1, 10, 15] task3 = repeat task1 for S2 in uniform(0,10,100)
which set up tasks looping S1 through a vector of values, and S2 through a ‘uniformRange’ of evenly spaced values.
3. Implementation and Distribution
The cross-platform library is written in Bison (https://www.gnu.org/software/bison) and C++, with a simple C API, along with Python bindings. It uses lib-SEDML (https://github.com/fbergmann/libSEDML) to parse and create SED-ML files, and libSBML14 to parse the SBML files to which the SED-ML documents refer. It also uses the check library (http://libcheck.github.io/check/) for unit tests. A standalone command-line tool (phrasedml-convert) is provided for easy conversion between phraSED-ML and SED-ML. Source code, executables, libraries, and documentation are available at http://phrasedml.sf.net.
4. Integration into Tellurium
Additionally, Python bindings for libphrasedml have been incorporated into Tellurium, a Python-based environment for systems and synthetic biology modeling. Tellurium users can define a simulation experiment in phraSED-ML, and have it executed directly via libRoadRunner.6 Tellurium provides wrapper functions for handling of phraSED-ML code through the experiment module. The experiment module takes lists of models in Antimony and simulation setups in phraSED-ML string, which can be simulated through the execute function. Conversion between phraSED-ML and SED-ML code is straightforward as the library provides a function that accepts both phraSED-ML and SED-ML and translates to the other. Because Tellurium can translate SED-ML to Python scripts, users can define simulation setups in phraSED-ML and execute them directly.
5. Examples
Several examples of application involving phraSED-ML are demonstrated below. We use the Mitogen-Activated Protein (MAP) kinase cascade model15 as the model of interest for most of the examples.
5.1. Simple time course simulation
The simplest example illustrating the use of phraSED-ML is a simple time course simulation. The code below performs a time course simulation on the model from 0 to 4000 with 1000 time points, and plots time versus MAP kinase, phosphorylated MAP kinase, and double phosphorylated MAP kinase as the output.
The output of the phraSED-ML string is shown in Fig. 1. The Python translation which is used internally by Tellurium to execute the setup is also provided in the Supplementary Material as a reference.
Fig. 1.
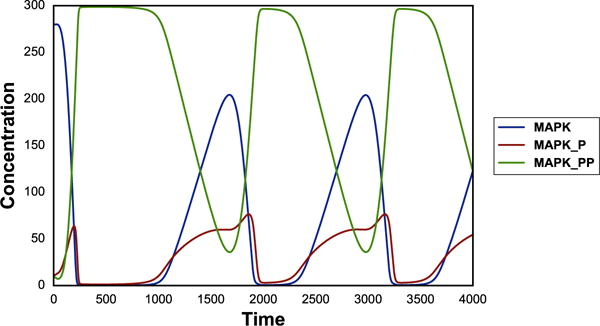
Output of a simple time course simulation. Blue line represents MAP kinase, red line represents phosphorylated MAP kinase, and green line represents double phosphorylated MAP kinase.
5.2. Phase portrait
The following example shows how it is possible to specify a simple phase portrait. In this case we run a simulation of the Lorenz attractor16 which under certain parameter values exhibits chaotic behavior. In Fig. 2, we plot the variable z versus x.
Fig. 2.
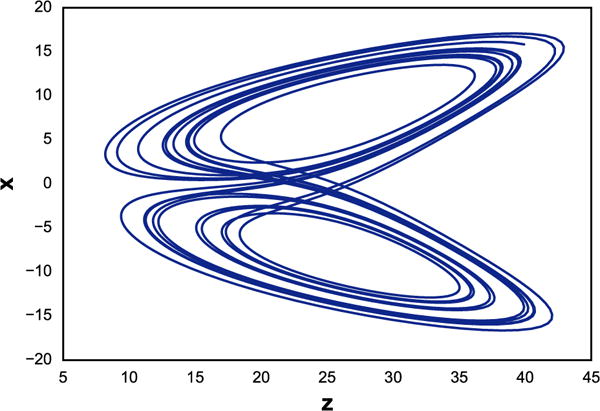
Phase plot of Lorenz attractor resulted from running the phraSED-ML code in Listing 2.
5.3. 1-dimensional parameter scan
Running parameter scan is simple and intuitive using phraSED-ML by using ‘repeat’. The example below runs a 1-Dimensional parameter scan on parameter ‘J1_KK2’ with values 1, 10, and 100, while resetting the model back to initial condition every time. The output is shown in Fig. 3.
Fig. 3.
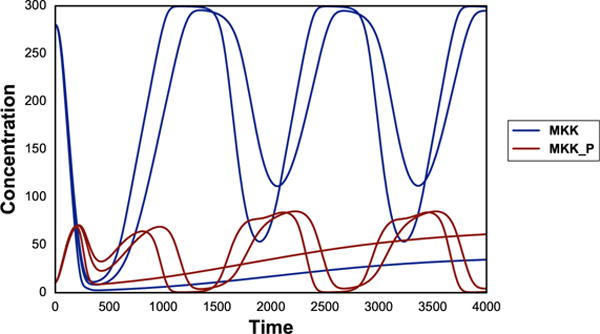
Typical output of phraSED-ML string running 1-dimensional parameter scan. The blue lines represent MAP kinase kinase and red lines represent phosphorylated MAP kinase kinase.
5.4. Multi-dimensional parameter scan
Expanding from 1-Dimensional parameter scan, parameter scan on two different parameters can be achieved through repeats of ‘repeat’. The code above illustrates 2-Dimensional parameter scan where parameter ‘J1_KK1’ is varied as before while parameter ‘J4_KK5’ is changed from 0 to 100 in 10 uniform steps. The output is plotted in Fig. 4.
Fig. 4.
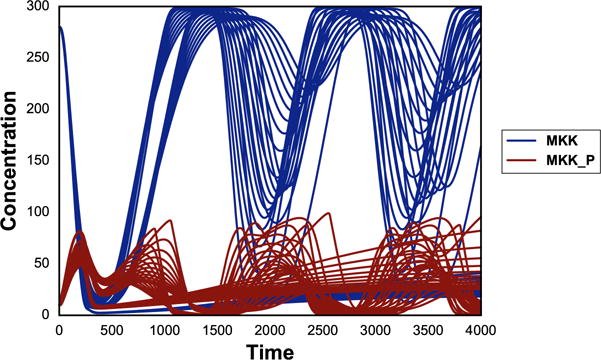
Typical output of phraSED-ML string running 2-dimensional parameter scan. The blue lines represent MAP kinase kinase and red lines represent phosphorylated MAP kinase kinase.
5.5. Repeated stochastic simulations
Another application of phraSED-ML shows how to run a repeated stochastic simulations on models. The following phraSED-ML string demonstrates the differences between stochastic simulations with and without a given seed. Typical output of the simulation setup is shown in Fig. 5.
Fig. 5.
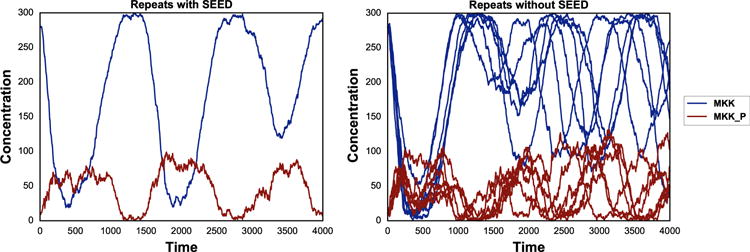
The same plot as Fig. 3 but using the stochastic Gillespie algorithm. The blue lines represent MAP kinase kinase and red lines represent phosphorylated MAP kinase kinase. The left panel shows stochastic simulations with a single seed. The right panel shows stochastic simulations with varying seeds.
6. Future Developments
SED-ML continues to be developed by the community (http://sed-ml.org/) and changes to phraSED-ML will follow this development. Of particular interest are planned changes to SED-ML that will allow a richer set of symbols to be specified when generating output, i.e. eigenvalues or sensitivity coefficients. These new symbols will be added to phraSED-ML once the new SED-ML specification has been approved.
Listing 1.

Simple time course, see Fig. 1.
Listing 2.

Lorenz attractor phase plot, see Fig. 2.
Listing 3.

1-Dimensional parameter scan, see Fig. 3.
Listing 4.

Multi-dimensional parameter scan, see Fig. 4.
Listing 5.

Repeated stochastic simulations, see Fig. 5.
Acknowledgments
We would like to thank the SED-ML community in general and Frank Bergmann, Ion Moraru, David Nickerson, and other SED-ML editors in particular for helpful feedback.
Funding
Research reported in this publication was supported by NIGMS of the National Institutes of Health under award number R01GM081070.
Biographies

Kiri Choi received the B.Sc. in physics from University of Washington, Seattle, in 2013. Currently, he is in Biological Physics, Structure and Design program working towards the Ph.D. in bioengineering at University of Washington. His research interests include systems and synthetic biology modeling, complex systems, algorithm development, and machine learning. He is currently working on a Python-based modeling platform for systems and synthetic biology called Tellurium.

Lucian P. Smith began work in computational systems biology with Dr. Sauro writing tools for biologists, and then later working for Dr. Hucka at Caltech supporting libraries and specifications for the tool writers themselves. Over the past decade, he has been the developer and maintainer of the Antimony language, writing several model translators between Antimony, SBML, CellML, and JSim, and updating the Antimony library over time to incorporate the SBML Level 3 ‘Flux Balance Constraints’ and ‘Distributions’ packages. He regularly participates in the SBML and COMBINE communities, having been a community-elected SBML Editor from 2010 to 2013, and having continued to participate in the SBML family of standards development since then. He has also been directly involved with writing SBML Level 3 packages, being first author on the ‘Hierarchical Model Composition’ package, taking the role of specification developer for the ‘Distributions’ and ‘Spatial’ packages when the original authors moved on, and contributing in other ways to other package specifications. He has chaired several sessions at the biannual COMBINE and HARMONY 5-day workshops on standard interoperability and standardization, and negotiating the package contents. In addition, he has created several hundred tests for the recent releases of the SBML Test Suite. He served as primary developer of the phraSED-ML project library.

J. Kyle Medley obtained his B.Sc. in Mechanical Engineering from the University of Missouri-Kansas City and is currently pursuing a Ph.D. in Bioengineering from the University of Washington, Seattle. His research interests include modeling dynamical biophysical processes such as receptor tyrosine kinase signaling pathways. He currently develops libRoadRunner, a software package for simulating dynamical models encoded in SBML.

Herbert M. Sauro is an Associate Professor in the Department of Bioengineering at the University of Washington, Seattle. He earned his Ph.D. in Computational Biochemistry from Oxford Brookes University, M.Sc. in Biological Computation from University of York, and B.Sc. in Biochemistry and Microbiology from University of Kent. His interests include understanding cellular control systems, development of standards such as SBML and SBOL and software provision for systems and synthetic biology. He has published three textbooks on enzyme kinetics, modeling and linear algebra.
Footnotes
Conflict of Interest
None declared.
References
- 1.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (sbml): A medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd CM, Halstead MD, Nielsen PF. Cellml: Its future, present and past. Prog Biophys Mol Biol. 2004;85(2):433–450. doi: 10.1016/j.pbiomolbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Waltemath D, Adams R, Bergmann FT, Hucka M, Kolpakov F, Miller AK, Moraru II, Nickerson D, Sahle S, Snoep JL, et al. Reproducible computational biology experiments with sed-ml-the simulation experiment description markup language. BMC Syst Biol. 2011;5(1):1. doi: 10.1186/1752-0509-5-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark J, DeRose S, et al. XML path language (XPath) version 1.0. :1999. [Google Scholar]
- 5.Courtot M, Juty N, Knüpfer C, Waltemath D, Zhukova A, Dräger A, Dumontier M, Finney A, Golebiewski M, Hastings J, et al. Controlled vocabularies and semantics in systems biology. Mol Syst Biol. 2011;7(1):543. doi: 10.1038/msb.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somogyi ET, Bouteiller JM, Glazier JA, König M, Medley JK, Swat MH, Sauro HM. libroadrunner: A high performance sbml simulation and analysis library. Bioinformatics. 2015:btv363. doi: 10.1093/bioinformatics/btv363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergmann FT, Sauro HM. Sbw-a modular framework for systems biology. Proc 38th Conf Winter Simulation. 2006:1637–1645. [Google Scholar]
- 8.Keller R, Dörr A, Tabira A, Funahashi A, Ziller MJ, Adams R, Rodriguez N, Nov ere NL, Hiroi N, Planatscher H, et al. The systems biology simulation core algorithm. BMC Syst Biol. 2013;7(1):55. doi: 10.1186/1752-0509-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergmann FT. Sed-ml script language. Nature Precedings. 2011 [Google Scholar]
- 10.Adams RR. Sed-ed, a workflow editor for computational biology experiments written in sed-ml. Bioinformatics. 2012;28(8):1180–1181. doi: 10.1093/bioinformatics/bts101. [DOI] [PubMed] [Google Scholar]
- 11.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. Copasia complex pathway simulator. Bioinformatics. 2006;22(24):3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 12.Funahashi A, Morohashi M, Kitano H, Tanimura N. Celldesigner: A process diagram editor for gene-regulatory and biochemical networks. Biosilico. 2003;1(5):159–162. [Google Scholar]
- 13.Smith LP, Bergmann FT, Chandran D, Sauro HM. Antimony: A modular model definition language. Bioinformatics. 2009;25(18):2452–2454. doi: 10.1093/bioinformatics/btp401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bornstein BJ, Keating SM, Jouraku A, Hucka M. Libsbml: An api library for sbml. Bioinformatics. 2008;24(6):880–881. doi: 10.1093/bioinformatics/btn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kholodenko BN. Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades. Eur J Biochem. 2000;267(6):1583–1588. doi: 10.1046/j.1432-1327.2000.01197.x. [DOI] [PubMed] [Google Scholar]
- 16.Lorenz EN. Deterministic nonperiodic flow. J Atmos Sci. 1963;20(2):130–141. [Google Scholar]


