Abstract
The pathogenesis and progression of many tumors, including hematologic malignancies is highly dependent on enhanced lipogenesis. De novo fatty-acid synthesis permits accelerated proliferation of tumor cells by providing membrane components but these may also alter physicochemical properties of lipid bilayers, which can impact signaling or even increase drug resistance in cancer cells. Cancer type-specific lipid profiles would permit us to monitor and interpret actual effects of lipid changes, potential fingerprints of individual tumors to be explored as diagnostic markers. We have used the shotgun MS approach to identify lipid patterns in different types of acute myeloid leukemia (AML) patients that either show no karyotype change or belong to t(8;21) or inv16 types. Differences in lipidomes of t(8;21) and inv(16) patients, as compared to AML patients without karyotype change, presented mostly as substantial modulation of ceramide/sphingolipid synthesis. Furthermore, between the t(8;21) and all other patients we observed significant changes in physicochemical membrane properties. These were related to a marked alteration in lipid saturation levels. The discovered differences in lipid profiles of various AML types improve our understanding of the pathobiochemical pathways involved and may serve in the development of diagnostic tools.
Introduction
Acute Myeloid Leukemia (AML) is a clonal bone marrow disorder resulting from diverse phenotypic and genetic alterations in the differentiation of hematopoietic stem cells and causing excessive proliferation and accumulation of abnormal immature leukemic neoplastic cells, called blasts[1–4]. The major causes of AML are deregulation in one or more of the numerous components of signaling networks that control cell growth, either by gain-of-function mutation or overexpression[5, 6]. Most efforts towards molecular characterization of AML have focused on genome, transcriptome or proteome changes [7–12] while evidence is growing that neoplastic pathogenesis and progression also involve, and might even be accelerated by, changes in cellular lipidomes[13–19]. In fact, one of the hallmarks of many tumors including hematologic malignancies is enhanced lipogenesis, arising from increased activities of fatty acid biosynthetic enzymes (Acc1, Fasn and Scd1)[20–23], necessary for accelerated growth. Furthermore, synthesis of mono-unsaturated fatty acids may have added benefit, since oleate can protect against saturated fatty acid toxicity and cellular stress[18, 22, 24, 25]. Thus, Acc1, Fasn and Scd1 have been identified as plausible targets for cancer therapy[14, 18, 20–23].
One of the lipid classes associated with cancer pathogenesis are the sphingolipids with their metabolites such as ceramide, hexosylceramide and sphingosine-1-phosphate, all of which act as effector molecules[15]. In particular, ceramides are collectively involved in the regulation of cancer-cell growth, differentiation, senescence and apoptosis[26–28]. Conversion of ceramide to glucosylceramide contributes to multidrug resistance of several human cancers, including leukemia, breast cancer, melanoma and neuroblastoma[17, 29–31]. Of equal importance, sphingolipids are structural lipids that tune membrane fluidity and subdomain structure of the lipid bilayer, especially lipid rafts[32]. These nanodomains act as regulatory platforms for a number of growth factor receptors e.g. EGFR, c-kit, VEGFR etc. controlling the initiation of signaling cascades in cell growth and differentiation[33, 34]. Therefore, deregulation of lipid metabolism threatens to perturb lateral membrane organization. Mechanisms that govern homeostasis within the enormous heterogeneity of distinct lipid species are largely unknown. Notably, metabolic transitions affecting lipid metabolism can prompt lipidome-wide ‘concentration waves’, in turn triggering compensatory metabolic responses[35, 36]. There is a growing interest in lipidome changes during pathogenesis, also in the hope of establishing unique cellular fingerprints useful as diagnostic markers.
The purpose of this study was to explore the lipidomic profiles of AML patients. We focused on three subsets of patients with specific clinical and biological AML characteristics: translocation t(8;21), inversion inv16 and patients with normal karyotype (AML-nk). Chromosomal abnormalities are detected in approximately 60% of newly diagnosed AML patients, among which t(8;21) and inv16 are the most common. They affect the expression of the Core Binding Factor (CBF)—a heterodimeric transcription factor controlling key programs in cell survival[37–39], at least in part by regulating sphingolipid metabolism [40, 41]. Both t(8;21) and inv16 are usually associated with good prognosis and survival. The ‘normal karyotype’ AML group is very heterogeneous as it emanates from diverse factors related to aberrant signal transduction pathways leading to uncontrolled growth and inhibition of apoptosis[2, 6, 42]. Moreover, these changes are often associated with increased activity of multidrug resistance proteins and the renewal of leukemia stem cells[30, 31].
Here, applying shotgun mass spectrometry we analyzed the cellular lipidomes of various AML types and identified salient differences. Major discordancies concerned the sphingolipids and their metabolites. We also observed pronounced shifts in membrane fluidity among the studied AML types. The recognized lipid patterns may guide the identification of druggable metabolic pathways for personalized anti-neoplastic therapies.
Materials and methods
Sample collection
All AML samples investigated in this study were bone marrow aspirates (anticoagulant lithium heparin) collected at the time of diagnosis. The blast fraction was enriched by density gradient centrifugation (density 1.077, BIOCOLL separating solution, Biochrom, Berlin Germany). Enriched samples were aliquoted (1–2 x106 cells/ ml), frozen viably in medium containing DMSO and fetal calf serum (FCS) and subsequently stored until analysis in liquid nitrogen. Cytogenetic and molecular analysis followed established methods as described [43, 44]. Patients were enrolled in prospective treatment protocols of the Study Alliance Leukemia (SAL). All patients gave written informed consent making available their material and data for scientific purposes. The studies were approved by the ethical board of the Medical Faculty TU Dresden (EK 98032010). Full description of patients can be found in Table 1.
Table 1. Description of patients.
| ID | karyotype | sex | age | FLT3-ITD mutation | FLT3-ITD ratio | NPM1 mutation | KIT mutation | first complete remission achieved | relapse after complete remission | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | normal | f | 81 | N | NA | Y | NA | N | N | |
| 2 | normal | m | 59 | Y | 0.92 | Y | NA | Y | Y | |
| 3 | normal | f | 62 | Y | 0.91 | Y | NA | N | N | |
| 4 | normal | f | 72 | Y | 5.9 | Y | NA | N | N | |
| 5 | normal | f | 72 | N | NA | Y | NA | Y | Y | |
| 6 | normal | m | 63 | Y | NA | Y | NA | Y | N | |
| 7 | inv16 | f | 53 | N | NA | N | N | N | N | |
| 8 | inv16 | f | 30 | N | NA | N | N | Y | Y | |
| 9 | inv16 | m | 28 | N | NA | N | Exon 8 | Y | Y | |
| 10 | inv16 | m | 36 | N | NA | N | Exon 8 | Y | N | |
| 11 | inv16 | m | 41 | N | NA | N | N | Y | Y | |
| 12 | t(8;21) | m | 37 | N | NA | N | N | Y | N | |
| 13 | t(8;21) | f | 26 | N | NA | N | D816V/H | Y | N | |
| 14 | t(8;21) | m | 41 | N | NA | N | N | Y | N | |
| 15 | t(8;21) | m | 53 | N | NA | N | D816V | Y | N | |
| 16 | t(8;21) | f | 34 | N | NA | N | N | Y | Y | |
F—female; m—male; NA-no information; FLT3-ITD—FMS-like tyrosine kinase 3 internal tandem duplication; NPM1—Nucleophosmin 1; N—no; Y—yes
Sample preparation
Synthetic lipid standards: cholesterol (Chol-d6) (deuterated), sterol ester (CE 20:0), triacylglycerol (TAG 17:0–17:0–17:0), diacylglycerols (DAG 17:0–17:0), phosphatidic acid (PA 17:0–17:0), phosphatidylcholine (PC 17:0–17:0), phosphatidylethanolamine (PE 17:0–17:0), phosphatidylglycerol (PG 17:0–17:0), phosphatidylserines (PS 17:0–17:0), phosphatidylinositol (PI 16:0–16:0), lysoglycerophospholipids (LPA 17:0, LPC 12:0, LPE 17:1, LPS 17:1, LPI 17:1), sphingomyelin (SM 18:1;2–12:0) and ceramides (Cer 18:1;2–17:0, GlcCer 18:1;2–12:0, LacCer 18:1;2–12:0) were purchased from Avanti Polar Lipids. Total protein concentration of leukemic cells was estimated using the microBCA Protein Assay Kit (Thermo Scientific). Aliquots of cells equivalent to 20–25 μg of protein that correspond to ~250 000 cells, were transferred into 2 ml tubes (Eppendorf AG), pelleted (1000g, 5’) washed twice with phosphate-buffered saline and spiked with 10 μl internal lipid standards mixture dissolved in methanol. All samples were subjected to two-step lipid extraction with 750 μl of 10:1 chloroform:methanol (C/M) (v/v) followed by a 2:1 C/M mixture at 4°C. Lipid extracts were collected from organic phases and evaporated under vacuum. Dry lipid extracts were immediately re-dissolved in 100 μl of chloroform/methanol 1:2 (v/v) and injected into the mass spectrometer Q-Exactive (Thermo Fisher Scientific) equipped with an electrospray ion source—ESI TriVersa Nanomate (Advion Biosciences). Three independent runs were done for each lipid extract.
Shotgun lipidomics analysis and data processing
Lipid extracts (7.5 μl) from the first extraction step were diluted in 13 mM ammonium acetate in propanol (10 μl) to achieve the final solvent composition of 7.5 mM ammonium acetate in chloroform/methanol/propanol 1:2:4 (v/v). Lipid extracts from the second extraction step were dissolved in 10 μl of 0.01% methylamine as ion modifier and subjected to negative mode MS analysis. Lipid extracts from the 10:1 organic phase were analyzed with polarity switching in both positive and negative ion mode, the 2:1 extracts in negative mode only. Positive ion mode analysis was performed using FTMS with target resolution of 280,000 at m/z 200 and MS/MS with 17,500 at m/z 200 to monitor CE, DAG and TAG species. Negative FTMS mode was chosen to monitor lyso-phospholipids (LPG, LPS, LPI, LPS, LPC, LPE) and combined with tandem MS/MS for selected phospholipids (PC, PC O-, PE, PE O-, PS, PA, PI, PG), sphingolipids (SM, Cer, Hex-Cer, diHex-Cer), globosides (Gb3 and Gb4) and ganglioside GM3. The same, targeted resolution settings were used for both polarities. Cholesterol was measured separately after chemical acetylation[45]. Briefly, 20 μl of 10:1 lipid extract was dried down and 75 μl of acetyl chloride in 1:2 C/M (v/v) added. After an hour incubation samples were dried down, re-dissolved in 50 μl C/M 1:2 and analyzed in positive ion mode with similar settings as described above. The molarity of lipid species was determined based on spiked-in, internal quantitative standards.
Samples were infused into the Q-Exactive instrument TriVersa NanoMate ESI source. Data acquisition was performed in both positive and negative mode with polarity switching where MS was used for quantification and MS/MS for fatty acid and lipid precursor identification. Data were analyzed and de-convoluted by LipidXplorer software. Data visualization and normalization (pmol, mol%) calculations were performed in Microsoft Excel, GraphPad Prism 6.0 and SIMCA 14.0.
GP measurements
For fluidity analysis, dried lipids were rehydrated in 150 mM NaCl, 25 mM HEPES pH 7.25. The resulting liposomes were subsequently subjected to 10 freezing/thawing cycles (liquid nitrogen/37°C) and extruded through 100 nm polycarbonate filters. Finally, vesicles were stained (15 min) with 2 μM C-laurdan. Fluorescence emission spectra were recorded (Ex. 385 nm, Em. 400–550 nm) and analyzed as described in Kaiser et al.[46].
Results
Lipidome analysis reveals significant differences among the AML subtypes
Total leukocyte fractions containing 250 000 cells, corresponding to approximately 20–25 μg of total protein content, were subjected to two-step lipid extraction, MS lipidomics and GP measurements. Shotgun lipidomics MS and MS/MS designated acquisition routine and Lipotype proprietary lipidomics software enabled profiling of more than 400 unique lipid species of 25 analyzed lipid classes (for details see S1 Table).
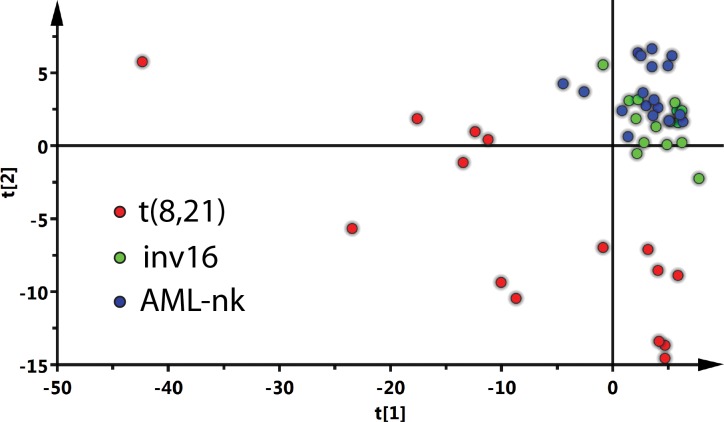
Lipidomics data were first normalized to internal standards and transformed to mol%. Multivariate statistical data analysis was applied to systematize and better understand the recorded lipidome variations. Dimension reduction and data visualization followed Principal Component Analysis (PCA) with Pareto scaling (SIMCA software). This statistical tool emphasizes variation and highlights robust patterns in a dataset. Subsequently, a two-dimensional score plot was generated (significance of the principal components based on the explained sum of squares was 0.253 and 0.094 for the first and second principal component respectively) (Fig 1), revealing t(8;21) samples clearly separated from inv16 and AML-nk patient samples—the latter clustered together. This suggested substantial differences between the lipidomes of t(8;21) and the other AML types.
Fig 1. Score plots of multivariate analysis by PCA.
The t(8;21) samples are separated from the inv16 and AML-nk samples. Each patient sample was measured in triplicates and each point on the plot represents an individual measurement. The calculated sum of squares was 0.253 and 0.094 for the first and second component, respectively. The analysis was performed using SIMCA 14.0 software.
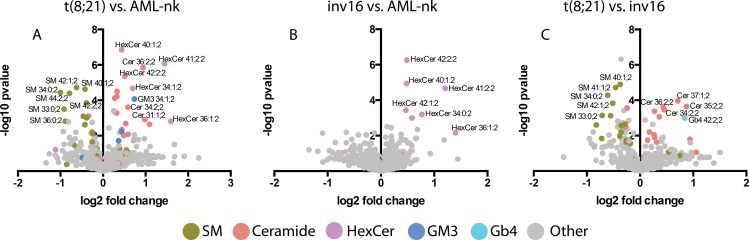
Next, lipid species differences between the AML types were analyzed. Briefly, lipid fold change followed by multiple t-tests analysis identified unique, species with the most pronounced deviations (Fig 2). The major difference between t(8;21) and AML-nk involved ceramide backbone-containing lipids: SM, Cer, Hex-Cer and GM3 ganglioside (Fig 2A). Decrease of SM in t(8;21) was correlated with an increase in the levels of ceramides (up to 3-fold), Hex-Cer (up to 5-fold), DiHex-Cer (up to 3-fold) and GM3 (up to 15-fold) suggesting a shift towards glycosphingolipid synthesis (S1 Table). Unfortunately GM3 derivatives are currently not accessible to quantitative shotgun MS. Similarly, the comparison of groups t(8;21) and inv16 (Fig 2B) mostly registered expression profile changes affecting sphingolipid pathways. We observed a decrease in SM levels and increased ceramide synthesis (Fig 2C), suggesting that this type of leukemic cells might be more prone to apoptosis[26–28]. Despite the fact that inv16 and AML-nk patients represent a mixed population in the PCA plot (Fig 1), direct comparison by orthogonal partial least-square displacement analysis brings to light considerable differences (p<0.001) between their lipidomes. The most prominent change was the increased abundance of various Hex-Cer species in the inv16 samples (Fig 2C). These lipids are metabolic precursors of other glycolipids, but were also ascribed an important role in multidrug resistance[17, 29, 31]. Inv16 is associated with a specific accumulation of one particular lipid class, rather than a shift in glycosphingolipid metabolism, as in the case of t(8;21) cells.
Fig 2. Comparison of lipid profiles of various AML types.
Each point on the volcano graph represents a single lipid specie. The analysis was performed using GraphPad PRISM 6.0.
Measurement of generalized polarization index reveals dramatic changes in membrane fluidity in t(8;21) cells
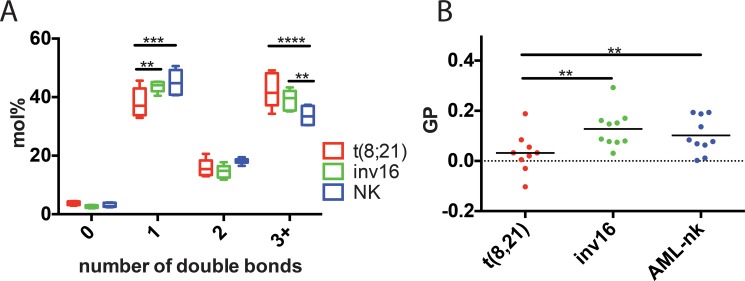
Activation of the lipogenic pathway at early stages of cancer pathogenesis is critical for proliferation of various types of tumors including hematologic malignancies[21, 23]. The majority of newly synthesized fatty acids in cancer cells are converted predominantly to membrane phospholipids. Specifically, the proportion of monounsaturated and polyunsaturated fatty acids in the major phospholipids was reported to be significantly higher in cancer compared to noncancerous tissues[47]. Here we compared each type of leukemia for differences in lipid saturation levels. A marked difference affected t(8;21) leukemia, a significant shift towards polyunsaturated at the cost of monounsaturated fatty acids (Fig 3A). This observation was specific to membrane lipids (phospholipids + sphingolipids), which comprise roughly 80% of the total measured lipids, but not to storage lipids (TAG, DAG, SE) (S1 Fig).
Fig 3. Lipid features from extracted AML samples.
(A) Saturation index of all lipids measured with shotgun MS. The analysis revealed significant changes of the mono- and polyunsaturated fatty acids in t(8;21) samples. (B) GP index of liposomes prepared from lipid extracts of various AML samples. GP is a measure of membrane order (higher GP equals more ordered membranes). t(8;21) samples exhibit lower GP values, indicating higher membrane fluidity.
Fatty acid saturation and length plus cholesterol content are the three major determinants of membrane fluidity. Having observed a significant shift of fatty acid saturation but not of cholesterol abundance or fatty acid length (Figs 3 and S1) we expected a substantial change in membrane fluidity of t(8;21) compared to the other AML types. Membrane fluidity (General Polarization Index, GP) was measured on small unilamellar vesicles prepared from the extracted lipids, using the fluorescent probe c-laurdan, as described [48]. Levels of unsaturation and GP values were closely correlated, confirming the greater fluidity of t(8;21) membranes. These findings underscore the importance of precisely measuring lipid and desaturation levels in cancer cells to resolve aberrantly regulated metabolic pathways and, ultimately, therapeutic targets.
Discussion
Similar to embryonic cells, cancer cells are highly dependent on de novo lipogenesis for their proliferation, and lipogenesis is activated at a relatively early stage in various types of tumors[18, 21, 49]. Moreover, there seems to be a correlation between fatty acid synthase expression, which is greatly elevated in metastatic tumors, increased membrane fluidity and decreased patient survival[50–53]. Proliferating cancer cells predominantly convert the majority of newly synthesized fatty acids to phospholipids for membrane incorporation [18]. Changes in lipid metabolism disequilibrating saturation levels of membrane fatty acids influence membrane fluidity and structure and composition of membrane domains (rafts) where many critical signaling cascades originate. In consequence, aberrant signal transduction may enhance the proliferation and survival of hematopoietic progenitor cells[2, 6]. An additional layer of complexity arises as a consequence of metabolic interconversion of one lipid to another—e.g. of PI(4,5)P2 to DAG or DAG/PC to PA—that impact signaling[54, 55]. Similarly, many pathways of sphingolipid metabolism constitute an interconnected network wherein individual levels and the balance of bioactive molecules can modulate cancer initiation, progression or treatment response. For example, ceramide hydrolysis produces sphingosine, the phosphorylation of which yields sphingosine-1-phosphate that regulates cell growth and suppresses programmed cell death[56–58]. Thus the difference in lipid metabolism between tumors and normal tissues renders lipogenic pathways attractive but so far only moderately exploited targets for the development of anti-cancer therapies [23, 59].
To evaluate the suitability of lipidomics for discriminating various types of cancer cells we analyzed the lipid profiles of AML cells of patients with (t(8;21), inv16) and without karyotype disorders (AML-nk). Importantly, in both t(8;21) and inv16 AML the karyotype changes result in deregulation of CBF expression, a factor that controls the expression of genes involved in sphingolipid metabolism[40, 41]. CBF overexpression has been shown to reduce intracellular long-chain ceramides in NIH3T3 fibroblasts and to elevate extracellular sphingosine-1-phosphate levels[40]. On the other hand, CBFβ-silenced cells exhibited substantial increases in ceramide species and decreases in lactosylceramides that correlated with enhanced cell death[41]. Here for the first time we demonstrate quantitative lipidome changes of cells with disorders in CBF expression. Indeed the major differences discovered between the various AML types affect the sphingolipids and ceramides. Interestingly, although both t(8;21) and inv16 share similarities (deregulation of CBF expression), significant differences in sphingolipid metabolism remain. Translocation t(8;21) unlike inv16 and AML-nk, is accompanied by substantial decrease of sphingomyelin (Fig 2), probably, as shown by others, due to decreased expression of SGMS1 and SGMS2[7], which detours ceramides to glycosphingolipid synthesis. In inv16 samples sphingolipid metabolism appears more at equilibrium and only hexosylceramides are specifically accumulated (Fig 2C), reported as a multidrug resistance phenotype in many cancer cells[17, 29–31]. The observed pattern is particularly revealing, as in t(8;21) and inv16 patients sensitive to the drugs ABT737 (Bcl-2 inhibitor) and BV6 (antagonist of IAP, ‘inhibitor of apoptosis proteins’), genes responsible for ceramide synthesis were upregulated, in contrast to patients resistant to these drugs[7]. Enzyme expression changes of sphingolipid metabolism in several cancers are consistent with the emerging tumor suppressor functions of ceramide. Our data provide evidence that ceramide levels significantly differing between patients, even those bearing similar genetic perturbations, might directly correlate with these patients’ susceptibility to chemotherapy.
Among all studied groups, the t(8;21) patients presented the highest membrane fluidity, predominantly due to increased unsaturation of membrane lipid fatty acid moieties. Elevated membrane fluidity has been associated with enhanced cell growth and proliferation in many cancers[60–62] as well as shown to modulate certain receptor tyrosine kinase proteins, resulting in aberrant signaling[63–66]. On the other hand the shift towards increased levels of saturated and monounsaturated phospholipids may protect cancer cells from oxidative damage by reducing lipid peroxidation[14, 18]. In line with this, it has been suggested of membrane active drugs that their ability to alter membrane fluidity correlates with pharmacological activity[67, 68].
The observed changes in membrane fluidity in t(8,21) samples might, to a lesser extent, be the consequence of lower sphingomyelin concentration. Yet, this appears to be compensated by the increase in glycosphingolipid (GSL) content (S1 Fig), making a dramatic effect on membrane fluidity unlikely. Interestingly however, the changes in t(8,21) samples in the ratio of SM and GSL might critically alter the physicochemical properties of rafts with important sequels for signal transduction pathways. Glycosphingolipids were among the first discovered tumor-associated antigens and are now appreciated as function modulators of several growth factor receptors that govern cell growth and differentiation [69, 70]. Although the current approach does not allow assaying of higher glycolipids (with more than three sugar residues), the observable ones (Hex-Cer, DiHex-Cer and GM3) indicated a significant shift in sphingomyelin-glycosphingolipid metabolism. Therefore, one can clearly expect significant, purely lipid-induced responses in signal transduction pathways not brought about by changes in expression levels or gain-of-function mutations in the components of the signaling cascades. The challenge for the future will be to unravel how, through modulation of the growth signaling cascades, individual differences in sphingolipid metabolism stimulate cancer development and progression. As shown here, quantification of cellular lipids, down to single species level allowed us to distinguish AML-type-specific lipid signatures. Such procedures pave the way towards new diagnostics and prognostics and should be integrated into a systemic approach towards the mechanistic understanding of cancer and the optimization of therapeutical targets.
Conclusion
Mass spectrometry-driven quantitative analysis of cellular lipids, especially the so-called shotgun lipidomics technique is increasingly being explored for its potential as a clinical diagnostic tool. A major benefit of this technology is that hundreds of lipid species can be directly identified and accurately quantified in a relatively short analysis time[71–73]. This work demonstrates that lipidomic profiling of acute myeloid leukemia cells supports the stratification of hematological malignancies. The observed imbalances of lipid synthesis pathways might directly contribute to disease progression.
Supporting information
(A) Lipid features (fatty acid saturation and length) measured with shotgun MS for “membrane” (PC, PC-O, PE, PE-O, PS, PI, PG, PA, SM, Cer, HexCer, DiHexCer, GM3, Gb3, Gb4) and “storage” lipids (SE, TAG, DAG). (B) Lipid profile of sphingolipids measured in various AML samples.
(PDF)
Mol% values of individual lipid species determined with shotgun MS for particular patient samples. Each sample was measured in three independent runs.
(XLSX)
Acknowledgments
We thank Christian Klose, Cornelia Schroeder and Patrycja Dubielecka for critical discussions and reading of the manuscript. We acknowledge the support of this research by the German Federal Ministry of Education and Research grant to the German Center for Diabetes Research (DZD e.V.) (M.G.).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The results of this research have been achieved using the German Federal Ministry of Education and Research grant to the German Center for Diabetes Research (DZD e.V.) (M.G.).
References
- 1.Pullarkat V, Aldoss I. Prognostic and therapeutic implications of early treatment response assessment in acute myeloid leukemia. Crit Rev Oncol Hematol. 2015;95(1):38–45. 10.1016/j.critrevonc.2015.01.005 [DOI] [PubMed] [Google Scholar]
- 2.Shipley JL, Butera JN. Acute myelogenous leukemia. Exp Hematol. 2009;37(6):649–58. 10.1016/j.exphem.2009.04.002 [DOI] [PubMed] [Google Scholar]
- 3.Hou HA, Lin CC, Chou WC, Liu CY, Chen CY, Tang JL, et al. Integration of cytogenetic and molecular alterations in risk stratification of 318 patients with de novo non-M3 acute myeloid leukemia. Leukemia. 2014;28(1):50–8. 10.1038/leu.2013.236 [DOI] [PubMed] [Google Scholar]
- 4.Martens JH, Stunnenberg HG. The molecular signature of oncofusion proteins in acute myeloid leukemia. FEBS Lett. 2010;584(12):2662–9. 10.1016/j.febslet.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 5.Scholl C, Gilliland DG, Frohling S. Deregulation of signaling pathways in acute myeloid leukemia. Semin Oncol. 2008;35(4):336–45. 10.1053/j.seminoncol.2008.04.004 [DOI] [PubMed] [Google Scholar]
- 6.Hatzimichael E, Georgiou G, Benetatos L, Briasoulis E. Gene mutations and molecularly targeted therapies in acute myeloid leukemia. Am J Blood Res. 2013;3(1):29–51. PubMed Central PMCID: PMCPMC3555190. [PMC free article] [PubMed] [Google Scholar]
- 7.Luck SC, Russ AC, Botzenhardt U, Paschka P, Schlenk RF, Dohner H, et al. Deregulated apoptosis signaling in core-binding factor leukemia differentiates clinically relevant, molecular marker-independent subgroups. Leukemia. 2011;25(11):1728–38. 10.1038/leu.2011.154 [DOI] [PubMed] [Google Scholar]
- 8.Mardis ER. Sequencing the AML genome, transcriptome, and epigenome. Semin Hematol. 2014;51(4):250–8. PubMed Central PMCID: PMCPMC4316686. 10.1053/j.seminhematol.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gosse G, Celton M, Lamontagne V, Forest A, Wilhelm BT. Whole genome and transcriptome analysis of a novel AML cell line with a normal karyotype. Leuk Res. 2015;39(7):709–18. 10.1016/j.leukres.2015.03.017 [DOI] [PubMed] [Google Scholar]
- 10.Luczak M, Kazmierczak M, Handschuh L, Lewandowski K, Komarnicki M, Figlerowicz M. Comparative proteome analysis of acute myeloid leukemia with and without maturation. J Proteomics. 2012;75(18):5734–48. 10.1016/j.jprot.2012.07.030 [DOI] [PubMed] [Google Scholar]
- 11.Kornblau SM, Tibes R, Qiu YH, Chen W, Kantarjian HM, Andreeff M, et al. Functional proteomic profiling of AML predicts response and survival. Blood. 2009;113(1):154–64. PubMed Central PMCID: PMCPMC2951831. 10.1182/blood-2007-10-119438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci U S A. 2009;106(31):12950–5. PubMed Central PMCID: PMCPMC2716381. 10.1073/pnas.0903091106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Mao J, Ai J, Deng Y, Roth MR, Pound C, et al. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One. 2012;7(11):e48889 PubMed Central PMCID: PMCPMC3495963. 10.1371/journal.pone.0048889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilvo M, Denkert C, Lehtinen L, Muller B, Brockmoller S, Seppanen-Laakso T, et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71(9):3236–45. 10.1158/0008-5472.CAN-10-3894 [DOI] [PubMed] [Google Scholar]
- 15.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4(8):604–16. 10.1038/nrc1411 [DOI] [PubMed] [Google Scholar]
- 16.Zhang F, Du G. Dysregulated lipid metabolism in cancer. World J Biol Chem. 2012;3(8):167–74. PubMed Central PMCID: PMCPMC3430731. 10.4331/wjbc.v3.i8.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouaze V, Liu YY, Prickett CS, Yu JY, Giuliano AE, Cabot MC. Glucosylceramide synthase blockade down-regulates P-glycoprotein and resensitizes multidrug-resistant breast cancer cells to anticancer drugs. Cancer Res. 2005;65(9):3861–7. 10.1158/0008-5472.CAN-04-2329 [DOI] [PubMed] [Google Scholar]
- 18.Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010;70(20):8117–26. 10.1158/0008-5472.CAN-09-3871 [DOI] [PubMed] [Google Scholar]
- 19.Marien E, Meister M, Muley T, Fieuws S, Bordel S, Derua R, et al. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int J Cancer. 2015;137(7):1539–48. PubMed Central PMCID: PMCPMC4503522. 10.1002/ijc.29517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corominas-Faja B, Cuyas E, Gumuzio J, Bosch-Barrera J, Leis O, Martin AG, et al. Chemical inhibition of acetyl-CoA carboxylase suppresses self-renewal growth of cancer stem cells. Oncotarget. 2014;5(18):8306–16. PubMed Central PMCID: PMCPMC4226684. 10.18632/oncotarget.2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763–77. 10.1038/nrc2222 [DOI] [PubMed] [Google Scholar]
- 22.Mason P, Liang B, Li L, Fremgen T, Murphy E, Quinn A, et al. SCD1 inhibition causes cancer cell death by depleting mono-unsaturated fatty acids. PLoS One. 2012;7(3):e33823 PubMed Central PMCID: PMCPMC3310881. 10.1371/journal.pone.0033823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Southam AD, Khanim FL, Hayden RE, Constantinou JK, Koczula KM, Michell RH, et al. Drug Redeployment to Kill Leukemia and Lymphoma Cells by Disrupting SCD1-Mediated Synthesis of Monounsaturated Fatty Acids. Cancer Res. 2015;75(12):2530–40. 10.1158/0008-5472.CAN-15-0202 [DOI] [PubMed] [Google Scholar]
- 24.Listenberger LL, Han X, Lewis SE, Cases S, Farese RV Jr., Ory DS, et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci U S A. 2003;100(6):3077–82. PubMed Central PMCID: PMCPMC152249. 10.1073/pnas.0630588100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon B, Lee HK, Querfurth HW. Oleate prevents palmitate-induced mitochondrial dysfunction, insulin resistance and inflammatory signaling in neuronal cells. Biochim Biophys Acta. 2014;1843(7):1402–13. 10.1016/j.bbamcr.2014.04.004 [DOI] [PubMed] [Google Scholar]
- 26.Heinrich M, Neumeyer J, Jakob M, Hallas C, Tchikov V, Winoto-Morbach S, et al. Cathepsin D links TNF-induced acid sphingomyelinase to Bid-mediated caspase-9 and -3 activation. Cell Death Differ. 2004;11(5):550–63. 10.1038/sj.cdd.4401382 [DOI] [PubMed] [Google Scholar]
- 27.Bose R, Verheij M, Haimovitz-Friedman A, Scotto K, Fuks Z, Kolesnick R. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell. 1995;82(3):405–14. [DOI] [PubMed] [Google Scholar]
- 28.Riboni L, Campanella R, Bassi R, Villani R, Gaini SM, Martinelli-Boneschi F, et al. Ceramide levels are inversely associated with malignant progression of human glial tumors. Glia. 2002;39(2):105–13. 10.1002/glia.10087 [DOI] [PubMed] [Google Scholar]
- 29.Liu YY, Han TY, Giuliano AE, Cabot MC. Ceramide glycosylation potentiates cellular multidrug resistance. FASEB J. 2001;15(3):719–30. 10.1096/fj.00-0223com [DOI] [PubMed] [Google Scholar]
- 30.Xie P, Shen YF, Shi YP, Ge SM, Gu ZH, Wang J, et al. Overexpression of glucosylceramide synthase in associated with multidrug resistance of leukemia cells. Leuk Res. 2008;32(3):475–80. 10.1016/j.leukres.2007.07.006 [DOI] [PubMed] [Google Scholar]
- 31.Lavie Y, Cao H, Bursten SL, Giuliano AE, Cabot MC. Accumulation of glucosylceramides in multidrug-resistant cancer cells. J Biol Chem. 1996;271(32):19530–6. [DOI] [PubMed] [Google Scholar]
- 32.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327(5961):46 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- 33.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1(1):31–9. 10.1038/35036052 [DOI] [PubMed] [Google Scholar]
- 34.Ratajczak MZ, Adamiak M. Membrane lipid rafts, master regulators of hematopoietic stem cell retention in bone marrow and their trafficking. Leukemia. 2015;29(7):1452–7. 10.1038/leu.2015.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarasov K, Stefanko A, Casanovas A, Surma MA, Berzina Z, Hannibal-Bach HK, et al. High-content screening of yeast mutant libraries by shotgun lipidomics. Mol Biosyst. 2014;10(6):1364–76. 10.1039/c3mb70599d [DOI] [PubMed] [Google Scholar]
- 36.Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106(7):2136–41. PubMed Central PMCID: PMCPMC2650121. 10.1073/pnas.0811700106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Bruijn MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23(24):4238–48. 10.1038/sj.onc.1207763 [DOI] [PubMed] [Google Scholar]
- 38.Kilbey A, Terry A, Cameron ER, Neil JC. Oncogene-induced senescence: an essential role for Runx. Cell Cycle. 2008;7(15):2333–40. PubMed Central PMCID: PMCPMC2562501. 10.4161/cc.6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.San Martin IA, Varela N, Gaete M, Villegas K, Osorio M, Tapia JC, et al. Impaired cell cycle regulation of the osteoblast-related heterodimeric transcription factor Runx2-Cbfbeta in osteosarcoma cells. J Cell Physiol. 2009;221(3):560–71. PubMed Central PMCID: PMCPMC3066433. 10.1002/jcp.21894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kilbey A, Terry A, Jenkins A, Borland G, Zhang Q, Wakelam MJ, et al. Runx regulation of sphingolipid metabolism and survival signaling. Cancer Res. 2010;70(14):5860–9. PubMed Central PMCID: PMCPMC2906707. 10.1158/0008-5472.CAN-10-0726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greer AH, Yong T, Fennell K, Moustafa YW, Fowler M, Galiano F, et al. Knockdown of core binding factorbeta alters sphingolipid metabolism. J Cell Physiol. 2013;228(12):2350–64. 10.1002/jcp.24406 [DOI] [PubMed] [Google Scholar]
- 42.Park S, Chapuis N, Tamburini J, Bardet V, Cornillet-Lefebvre P, Willems L, et al. Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia. Haematologica. 2010;95(5):819–28. PubMed Central PMCID: PMCPMC2864389. 10.3324/haematol.2009.013797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiede C, Steudel C, Mohr B, Schaich M, Schakel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99(12):4326–35. [DOI] [PubMed] [Google Scholar]
- 44.Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood. 2006;107(10):4011–20. 10.1182/blood-2005-08-3167 [DOI] [PubMed] [Google Scholar]
- 45.Liebisch G, Binder M, Schifferer R, Langmann T, Schulz B, Schmitz G. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Biochim Biophys Acta. 2006;1761(1):121–8. 10.1016/j.bbalip.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 46.Kaiser HJ, Lingwood D, Levental I, Sampaio JL, Kalvodova L, Rajendran L, et al. Order of lipid phases in model and plasma membranes. Proc Natl Acad Sci U S A. 2009;106(39):16645–50. 10.1073/pnas.0908987106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakai K, Okuyama H, Yura J, Takeyama H, Shinagawa N, Tsuruga N, et al. Composition and turnover of phospholipids and neutral lipids in human breast cancer and reference tissues. Carcinogenesis. 1992;13(4):579–84. [DOI] [PubMed] [Google Scholar]
- 48.Kaiser HJ, Lingwood D, Levental I, Sampaio JL, Kalvodova L, Rajendran L, et al. Order of lipid phases in model and plasma membranes. Proc Natl Acad Sci U S A. 2009;106(39):16645–50. PubMed Central PMCID: PMCPMC2757813. 10.1073/pnas.0908987106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medes G, Thomas A, Weinhouse S. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 1953;13(1):27–9. [PubMed] [Google Scholar]
- 50.Epstein JI, Carmichael M, Partin AW. OA-519 (fatty acid synthase) as an independent predictor of pathologic state in adenocarcinoma of the prostate. Urology. 1995;45(1):81–6. [DOI] [PubMed] [Google Scholar]
- 51.Alo PL, Visca P, Marci A, Mangoni A, Botti C, Di Tondo U. Expression of fatty acid synthase (FAS) as a predictor of recurrence in stage I breast carcinoma patients. Cancer. 1996;77(3):474–82. [DOI] [PubMed] [Google Scholar]
- 52.Alo PL, Visca P, Trombetta G, Mangoni A, Lenti L, Monaco S, et al. Fatty acid synthase (FAS) predictive strength in poorly differentiated early breast carcinomas. Tumori. 1999;85(1):35–40. [DOI] [PubMed] [Google Scholar]
- 53.Campanella R. Membrane lipids modifications in human gliomas of different degree of malignancy. J Neurosurg Sci. 1992;36(1):11–25. [PubMed] [Google Scholar]
- 54.Ariotti N, Liang H, Xu Y, Zhang Y, Yonekubo Y, Inder K, et al. Epidermal growth factor receptor activation remodels the plasma membrane lipid environment to induce nanocluster formation. Mol Cell Biol. 2010;30(15):3795–804. Epub 2010/06/03. PubMed Central PMCID: PMC2916403. 10.1128/MCB.01615-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–7. Epub 2006/10/13. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Desai NN, Olivera A, Seki T, Brooker G, Spiegel S. Sphingosine-1-phosphate, a novel lipid, involved in cellular proliferation. J Cell Biol. 1991;114(1):155–67. PubMed Central PMCID: PMCPMC2289065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olivera A, Spiegel S. Sphingosine-1-phosphate as second messenger in cell proliferation induced by PDGF and FCS mitogens. Nature. 1993;365(6446):557–60. 10.1038/365557a0 [DOI] [PubMed] [Google Scholar]
- 58.Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind S, et al. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature. 1996;381(6585):800–3. 10.1038/381800a0 [DOI] [PubMed] [Google Scholar]
- 59.Abramson HN. The lipogenesis pathway as a cancer target. J Med Chem. 2011;54(16):5615–38. 10.1021/jm2005805 [DOI] [PubMed] [Google Scholar]
- 60.Sok M, Sentjurc M, Schara M, Stare J, Rott T. Cell membrane fluidity and prognosis of lung cancer. Ann Thorac Surg. 2002;73(5):1567–71. [DOI] [PubMed] [Google Scholar]
- 61.Funaki NO, Tanaka J, Kohmoto M, Sugiyama T, Ohshio G, Nonaka A, et al. Membrane fluidity correlates with liver cancer cell proliferation and infiltration potential. Oncol Rep. 2001;8(3):527–32. [DOI] [PubMed] [Google Scholar]
- 62.Zhao W, Prijic S, Urban BC, Tisza MJ, Zuo Y, Li L, et al. Candidate anti-metastasis drugs suppress the metastatic capacity of breast cancer cells by reducing membrane fluidity. Cancer Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Casalou C, Costa A, Carvalho T, Gomes AL, Zhu Z, Wu Y, et al. Cholesterol regulates VEGFR-1 (FLT-1) expression and signaling in acute leukemia cells. Mol Cancer Res. 2011;9(2):215–24. 10.1158/1541-7786.MCR-10-0155 [DOI] [PubMed] [Google Scholar]
- 64.Labrecque L, Royal I, Surprenant DS, Patterson C, Gingras D, Beliveau R. Regulation of vascular endothelial growth factor receptor-2 activity by caveolin-1 and plasma membrane cholesterol. Mol Biol Cell. 2003;14(1):334–47. PubMed Central PMCID: PMCPMC140249. 10.1091/mbc.E02-07-0379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams AB, Li L, Nguyen B, Brown P, Levis M, Small D. Fluvastatin inhibits FLT3 glycosylation in human and murine cells and prolongs survival of mice with FLT3/ITD leukemia. Blood. 2012;120(15):3069–79. PubMed Central PMCID: PMCPMC3471516. 10.1182/blood-2012-01-403493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drube S, Heink S, Walter S, Lohn T, Grusser M, Gerbaulet A, et al. The receptor tyrosine kinase c-Kit controls IL-33 receptor signaling in mast cells. Blood. 2010;115(19):3899–906. 10.1182/blood-2009-10-247411 [DOI] [PubMed] [Google Scholar]
- 67.Jendrossek V, Handrick R. Membrane targeted anticancer drugs: potent inducers of apoptosis and putative radiosensitisers. Curr Med Chem Anticancer Agents. 2003;3(5):343–53. [DOI] [PubMed] [Google Scholar]
- 68.Horan KL, Lutzke BS, Cazers AR, McCall JM, Epps DE. Kinetic evaluation of lipophilic inhibitors of lipid peroxidation in DLPC liposomes. Free Radic Biol Med. 1994;17(6):587–96. [DOI] [PubMed] [Google Scholar]
- 69.Miljan EA, Bremer EG. Regulation of growth factor receptors by gangliosides. Science. 2002;2002(160):10. [DOI] [PubMed] [Google Scholar]
- 70.Coskun Ü, Grzybek M, Drechsel D, Simons K. Regulation of human EGF receptor by lipids. Proc Natl Acad Sci U S A. 2011;108(22):9044–8. PubMed Central PMCID: PMCPMC3107302. 10.1073/pnas.1105666108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11(8):593–8. 10.1038/nrm2934 [DOI] [PubMed] [Google Scholar]
- 72.Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, et al. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12(1):R8 PubMed Central PMCID: PMCPMC3091306. 10.1186/gb-2011-12-1-r8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stahlman M, Ejsing CS, Tarasov K, Perman J, Boren J, Ekroos K. High-throughput shotgun lipidomics by quadrupole time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(26):2664–72. 10.1016/j.jchromb.2009.02.037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Lipid features (fatty acid saturation and length) measured with shotgun MS for “membrane” (PC, PC-O, PE, PE-O, PS, PI, PG, PA, SM, Cer, HexCer, DiHexCer, GM3, Gb3, Gb4) and “storage” lipids (SE, TAG, DAG). (B) Lipid profile of sphingolipids measured in various AML samples.
(PDF)
Mol% values of individual lipid species determined with shotgun MS for particular patient samples. Each sample was measured in three independent runs.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.