Abstract
The emergence and spread of antibiotic resistance among Acinetobacter spp. have been investigated extensively. Most studies focused on the multiple antibiotic resistance genes located on plasmids or genomic resistance islands. On the other hand, the mechanisms controlling intrinsic resistance are still not well understood. In this study, we identified the novel subclass of aminoglycoside nucleotidyltransferase ANT(3")-II in Acinetobacter spp., which comprised numerous variants distributed among three main clades. All members of this subclass can inactivate streptomycin and spectinomycin. The three ant(3")-II genes, encoding for the three ANT(3")-II clades, are widely distributed in the genus Acinetobacter and always located in the same conserved genomic region. According to their prevalence, these genes are intrinsic in Acinetobacter baumannii, Acinetobacter pittii, and Acinetobacter gyllenbergii. We also demonstrated that the ant(3")-II genes are located in a homologous recombination hotspot and were recurrently transferred among Acinetobacter species. In conclusion, our findings demonstrated a novel mechanism of natural resistance in Acinetobacter spp., identified a novel subclass of aminoglycoside nucleotidyltransferase and provided new insight into the evolutionary history of intrinsic resistance genes.
Author summary
The level of interest in intrinsic resistance genes has increased recently, and one of reasons is that their mobilization could lead to emergence of resistant pathogens. Insertion sequences (ISs) or plasmids can capture intrinsic resistance genes and disseminate them in bacterial populations. In this study, we identified a novel subclass of aminoglycoside nucleotidyltransferases which are intrinsic in A. baumannii and other Acinetobacter species. The genes encoding the aminoglycoside nucleotidyltransferase were frequently horizontally transferred between different Acinetobacter species by homologous recombination. This work reports a novel mechanism of natural resistance in Acinetobacter and an overlooked pathway for the dissemination of resistance among species in this genus.
Introduction
The genus Acinetobacter includes a broad group of biochemically and physiologically versatile bacteria that are ubiquitous in many environments [1]. Acinetobacter baumannii is the most clinically significant Acinetobacter species, and is among the most concerning causes of hospital-acquired bacterial infections [2]. Several other species of the genus may also cause human infections, such as A. nosocomialis and A. pittii, and less frequently A. ursingii, A. haemolyticus, A. lwoffii, A. parvus, and A. junii [3]. In the last few decades, the rate of hospital-acquired infections caused by Acinetobacter spp. have increased markedly worldwide [4, 5]. Of more concern is the rapidly increasing rate of multidrug resistance among Acinetobacter isolates, which has reduced the number of effective therapeutic options [1]. A study in US hospitals demonstrated that the rate of multidrug-resistant (MDR) Acinetobacter isolates increased from 7 to 30% between 1993 and 2004 [6]. In Chinese hospitals, more than half of A. baumannii isolates in 2014 were resistant to at least 10 frequently used antibiotics, and the rate of resistance to imipenem and meropenem has increased markedly (from 31–39% in 2005 to 62.4–66.7% in 2014) [7]. Additionally, the frequency of detection of multi-resistant non-baumannii isolates such as A. pittii, A. junii, and A. johnsonii has increased considerably, which demonstrated the exceptional adaptation of Acinetobacter spp. to antibiotic pressure.
The emergence and spread of antibiotic resistance among Acinetobacter spp. have been investigated extensively. Many antibiotic resistance genes (ARGs) are located on plasmids or genomic resistance islands in Acinetobacter isolates [8, 9]. In addition to ARGs located on mobile elements, several chromosomally encoded efflux systems and β-lactamases have been reported to contribute to the resistance of A. baumannii. The corresponding genes confer resistance when their expression changes due to mobilization or mutation of their associated regulatory elements. For instance, the adeABC operon, which encodes an efflux pump, is not expressed in environmental A. baumannii isolates; and the MDR phenotype is caused by mutations in adeRS located upstream of adeABC, leading to over-expression of the pump [10]. By providing a strong promoter, insertion of ISAba1 upstream of blaOXA-51 in A. baumannii also increased the expression of the β-lactamase gene, which conferred resistance to carbapenems [11, 12]. Interestingly, ISAba1 was also responsible for blaOXA-51 mobilization, as indicated by detection of a plasmid-borne ISAba1-blaOXA-51-like gene in baumannii and non-baumannii species of Acinetobacter [13, 14]. Another example is the emergence of the mobile blaNDM-1 carbapenemase gene, which was initially captured by the genome of an Acinetobacter species where it was integrated in the transposon Tn125, enabling its efficient spread in various pathogens [15]. Finally, recent works have shown that the aphA6 and aac(6')-Ih genes, which encode aminoglycoside-modifying enzymes, are intrinsic in A. guillouiae and A. gyllenbergii, respectively, but were acquired by A. baumannii by means of IS mobile elements [16, 17]. When captured by mobile elements, chromosomally-encoded conserved genes can thus act as a source of acquired resistance. Despite the important role of intrinsic genes in resistance, a substantial gap remains in our understanding of the intrinsic mechanisms responsible for the antibiotic resistance of Acinetobacter spp. Good candidates for intrinsic resistance are transferases, which are present in bacterial chromosomes at a high frequency, and which have a diverse range of antibiotic substrates. For example, nucleotidyltransferases, phosphotransferases, and acetyltransferases can modify aminoglycosides, macrolides, chloramphenicols, and lincosamides [18].
In the present study, we scanned the publicly available bacterial genomes of Acinetobacter spp. to detect putative antibiotic transferases. Putative aminoglycoside nucleotidyltransferase (ant) genes were found in conserved chromosomal regions in various Acinetobacter spp. Functional analyses indicated that the putative encoded transferases from different species can increase streptomycin and spectinomycin resistance of Escherichia coli. According to the distant phylogenetic relationship between these proteins and the reported ANT(3")-I nucleotidyltransferases, we propose that the Acinetobacter ANT(3") should refer to a new subclass of aminoglycoside nucleotidyltransferases, designated ANT(3")-II. The genes encoding for ANT(3")-II are intrinsic in at least Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter gyllenbergii. Moreover, we also demonstrated that these intrinsic genes recurrently transferred among Acinetobacter species by homologous recombination.
Results
Putative aminoglycoside nucleotidyltransferases are widely distributed in the genus Acinetobacter
Comparison of the amino acid sequences of known aminoglycoside, macrolide, chloramphenicol, and lincosamide transferases with complete and draft genomes of Acinetobacter spp. from GenBank resulted in identification of 90 protein sequences with more than 30% amino acid identity with known resistance genes and less than 80% amino acid identity with each other. Among them, five were encoded by genes located in conserved chromosomal regions and had not hitherto been reported as antibiotic resistance determinants. These putative proteins shared the highest amino acid sequence identity (39–44%) with aminoglycoside nucleotidyltransferases/adenylytransferases (ANT) of the class ANT(3")-I (sometimes designated Aad), which catalyze adenylylation at the 3'' [ANT(3'')] position [19] (S1 Table). They are present in A. baumannii, A. pittii, A. junii, A. parvus, A. gyllenbergii, and A. ursingii, as well as 24 Acinetobacter isolates that have not been identified to the species level.
We generated neighbor-joining, maximum-parsimony, and maximum-likelihood phylogenetic trees to evaluate the relationships between the putative ANTs and all known reported ANT(3")-I enzymes (Fig 1, S1 Fig). We used ANT(9) sequences from Staphylococcus aureus and Enterococcus faecalis as outgroups, which catalyze adenylylation at the 9[ANT(9)] position [19]. The three phylogenetic methods yielded trees with similar topologies, in which all protein sequences of the genus Acinetobacter clustered apart from known ANT(3")-I enzymes, as a highly supported monophyletic clade (with 100% bootstrap confidence), designated ANT(3")-II. These proteins share 64–72% amino acid identity with each other and 35–44% amino acid identity with known ANT(3")-Ia enzymes. ANT(3")-II formed three highly supported monophyletic clades, which were designated Cluster IIa, IIb, and IIc (Fig 1). Cluster IIa included putative ANTs of A. baumannii, A. pittii, and A. junii. All IIa members share at least 96% amino acid identities with each other. Cluster IIb consisted of only four sequences, all of which were from unknown Acinetobacter species, and shared more than 96% amino acid identities with each other. Finally, Cluster IIc included all remaining ANT(3")-II homolog sequences detected in Acinetobacter genomes, and shared 68–100% amino acid identities with each other. These three groups shared at most 72% amino acid identity with each other and their phylogenetic relationships was not consistent depending on the reconstruction method used (S1 Fig), supporting the clustering in three independent groups.
Fig 1. Phylogenetic neighbor-joining tree of aminoglycoside nucleotidyltransferases (ANT(3")).
Numbers above each node are the percentages of tree configurations that occurred during 1000 bootstrap replicates. Only values greater than 70% are shown. The abbreviation of bacteria denoted as follow: E. coli, Escherichia coli; P. multocida, Pasteurella multocida; P. aeruginosa, Pseudomonas aeruginosa; C. glutamicum, Corynebacterium glutamicum; V. fluvialis, Vibrio fluvialis; P. rettgeri, Providencia rettgeri; S. enterica, Salmonella enterica; A. media, Aeromonas media; Y. pestis, Yersinia pestis; K. pneumoniae, Klebsiella pneumoniae; E. faecalis, Enterococcus faecalis; S. aureus, Staphylococcus aureus. Red dots represent the ANT(3")-II from Acinetobacter species expressed in E. coli.
Putative aminoglycoside nucleotidyltransferases provide streptomycin and spectinomycin resistance when expressed in E. coli and in their natural host
The putative ant(3")-II genes from A. baumannii, A. junii, and A. pittii (IIa); Acinetobacter sp. NIPH 758 (IIb); and A. parvus, A. gyllenbergii, and A. ursingii (IIc) were functionally expressed in E. coli (Fig 1). All of the above candidate genes increased the resistance of E. coli to streptomycin and spectinomycin by 32–128-fold, compared to the control (E. coli harboring the null vector pUC18 only) (Table 1). However, the expressed ANTs did not result in increased resistance to other aminoglycosides, suggesting a substrate specificity for the transferase, consistent with other enzymes of the ANT(3") class [19].
Table 1. Functional analysis of putative ANT(3")-II from Acinetobacter expressed in E. coli.
| Original strains | Protein ID | Expression host | MIC (μg/ml) | |
|---|---|---|---|---|
| streptomycin | spectinomycin | |||
| E. coli DH5α | 4 | 32 | ||
| A. baumannii ATCC 19606 | EEX02086 | E. coli DH5α | 512 | 2048 |
| A. baumannii AYE | CAJ77832 | E. coli DH5α | 512 | 1024 |
| A. baumannii ATCC 17978 | ABO10616 | E. coli DH5α | 512 | 1024 |
| A. junii CIP 64.5 | ENV65326 | E. coli DH5α | 512 | 2048 |
| A. pittii CIP 70.29 | ENW14756 | E. coli DH5α | 512 | 2048 |
| A. pittii PHEA-2 | ADY83642 | E. coli DH5α | 256 | 2048 |
| A. parvus CIP 108168 | ENU37733 | E. coli DH5α | 256 | 2048 |
| A. ursingii ANC 3649 | ENV80847 | E. coli DH5α | 128 | 1024 |
| A. gyllenbergii NIPH 230 | ESK39014 | E. coli DH5α | 256 | 2048 |
| Gen. sp. 13BJ/14TU NIPH1859 | ENX32354 | E. coli DH5α | 256 | 2048 |
| Taxon 20 NIPH 758 | ENU91137 | E. coli DH5α | 256 | 2048 |
To assess the contribution of ANT(3")-IIa to intrinsic resistance in A. baumannii, the ant(3")-IIa gene was inactivated in A. baumannii ATCC 19606 and the susceptibility of the mutant strain to aminoglycosides was evaluated (Table 2). Inactivation of ant(3")-IIa decreased streptomycin and spectinomycin resistance by 16-fold and 2-fold, respectively, which can be fully restored by a complementation plasmid expressing ant(3")-IIa (Table 2), demonstrating that ANT(3")-IIa contributes to intrinsic resistance in A. baumannii.
Table 2. Resistance levels of ant(3")-IIa deficient mutant and complementation assay of the mutant.
| Strains | MIC (μg/ml) | |||||
|---|---|---|---|---|---|---|
| Km | Sm | Spe | Gm | Ak | Tb | |
| A. baumannii ATCC 19606 (wild-type) | 16 | 1024 | 5120 | 32 | 32 | 8 |
| A. baumannii ATCC 19606 (△ant) | 16 | 64 | 2560 | 32 | 32 | 8 |
| △ant/pKFAb::ant(3")-IIa | - | >1024 | >5120 | - | - | - |
Km, kanamycin; Sm, streptomycin; Spe, Spectinomycin; Gm, gentamycin; Ak, amikacin; Tb, tobramycin. △ant, ant(3")-IIa deficient mutant of wild-type A. baumannii ATCC 19606; △ant/pKFAb::ant(3")-IIa, complementation plasmid pKFAb::ant(3")-IIa expressed in △ant.
ANT(3")-IIa is a valid streptomycin nucleotidyltransferase
To characterize the products of enzymatic inactivation, ANT(3")-IIa from A. baumannii ATCC 19606 was over-expressed in E. coli BL21 (DE3) and purified. The molecular weight of ANT(3")-IIa was 32 kDa (S2 Fig). Enzyme activity was assayed using liquid chromatography-mass spectrometry (LC-MS). With streptomycin [m/z 582.4 (M+H)+] as a substrate, a reaction product with an (M+H)+ ion at m/z 911.3 was detected in the reaction system (S3 Fig), indicative of addition of a single adenyl group to the antibiotic, which was confirmed by the product ion at m/z 911.3. This result suggests that the ANT(3")-IIa enzyme of A. baumannii can adenylate streptomycin, and validates its functional classification as a streptomycin nucleotidyltransferase.
ant(3")-IIa and ant(3")-IIc are intrinsic in A. baumannii, A. pittii, and A. gyllenbergii
The ant(3")-IIa gene in A. baumannii isolates was located at the same chromosomal locus, within a more than 20-kbp-long region highly conserved among most strains (S4 Fig), and with no evidence of mobile element in the vicinity of the gene. Moreover, the gene was present in 1027 of 1406 A. baumannii genomes. The conserved location and high prevalence of the gene among A. baumannii isolates (73%) suggest it to be intrinsic in this species. Similarly, ant(3")-IIa in A. pittii, and ant(3")-IIc in A. gyllenbergii are located in conserved chromosomal regions and are also present in most sequenced isolates, again indicating that they are intrinsic in the corresponding species. Although the ant(3")-II genes are also located in the same chromosomal region in all other Acinetobacter genome sequences we investigated, determination of whether these genes are intrinsic is hampered by the impossibility to cluster the sequenced isolates into well defined species.
The intrinsic ant(3")-IIa gene of A. baumannii was horizontally transferred to A. junii
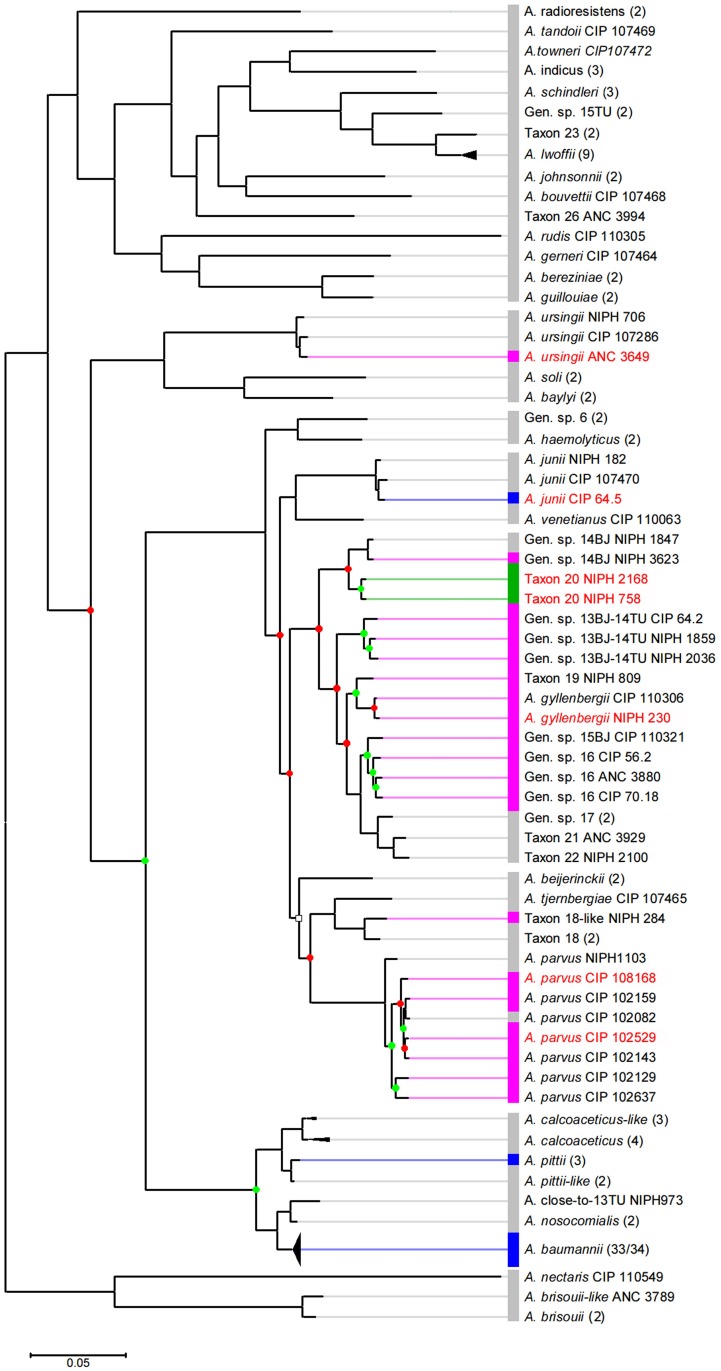
The unexpected inclusion of A. junii ANT(3")-IIa sequences among A. baumannii sequences in the phylogenetic tree (Fig 1) prompted further investigation of the phylogenetic relationships among ANT-carrying Acinetobacter strains. The position of strains carrying ant(3")-II genes was mapped on the Acinetobacter phylogenetic tree based on the whole core-genome proteins [3]. First, the observed pattern revealed that almost all strains carrying an ant(3")-II gene (with the exception of A. ursignii ANC3649) cluster in a single clade of the phylogeny (Fig 2). Second, numerous strains lacking an ant(3")-II gene are also scattered in this clade. Third, even when looking at branches including ant(3")-II-carrying strains only, the ANT(3")-II distribution of Fig 1 is globally inconsistent with the strains' phylogeny. Nearly half of the nodes linking ant(3")-II-carrying strains do not match the phylogenetic relationships observed for the ANT(3")-II sequences (red nodes in Fig 2). Such discrepancies between accessory and core gene phylogenies are generally indicative of horizontal gene transfer (HGT), usually mediated by mobile genetic elements. However, no mobile element or mobile element remnant could have been detected in the vicinity of the ant(3")-II genes in any of the genomes investigated.
Fig 2. Phylogeny of the Acinetobacter genus based on the alignment of the core-genome proteins.
Modified from Touchon et al. with permission [3]. The white squared node have a 67% bootstrap value; all other nodes show >95% bootstrap values. Green and red nodes are consistent and inconsistent with the ANT(3")-II phylogenetic tree of Fig 1, respectively. The ant(3")-II gene present in each taxon is indicated by a colored box: blue, ant(3")-IIa; green, ant(3")-IIb; pink, ant(3")-IIc; grey, no gene. Taxa written in red are those discussed in the text. Numbers in parenthesis indicate the number of genomes concatenated under the same taxon; they are all consistent in term of ant(3")-II variant, except in A. baumannii where the strain TYTH-1 lacks a 13.8 kbp region including ant(3")-IIa.
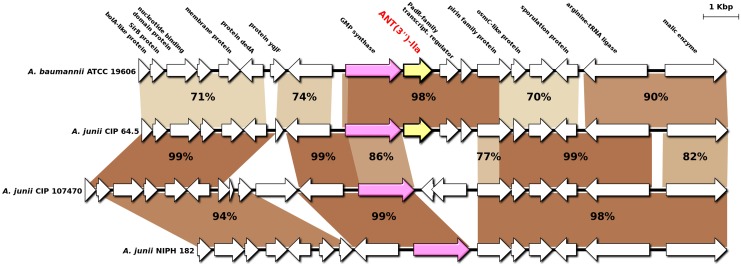
To explore whether HGT caused the observed inconsistencies even so, we first focused on the unexpected position of A. junii ANT(3")-IIa among A. baumannii ANT(3")-IIa proteins, considering that both species are distantly related in the core-genome tree (Fig 2). We aligned the 10-kbp downstream and upstream regions of ant(3")-IIa from the A. baumannii ATCC19606 genome, the A. junii CIP 64.5 genome, and two other representative A. junii genomes devoid of the gene (Fig 3). Consistent with the phylogenetic analysis, the 4.3-kbp region including ant(3")-IIa in A. junii CIP 64.5 was 98% identical to those of A. baumannii, while the flanking regions were only 70–77% identical. More interestingly, an inner portion of the 4.3-kbp region, which included ant(3")-IIa and the following two genes, was replaced by an unrelated 1.8-kbp DNA sequence in A. junii CIP 107470 and by a small, unrelated 300-bp sequence in A. junii NIPH 182 (Fig 3). Such pattern is typical of gene acquisition through homologous recombination (HR). To validate the occurrence of a HR event between A. baumannii and A. junii at the ant(3")-IIa locus, we performed an HR analysis on the 14-kbp region surrounding ant(3")-IIa by recombination detection program (RDP) analysis. Seven independent HR detection algorithms were selected, all of which provided highly significant support for a 4.3-kbp recombination event in A. junii CIP 64.5 with A. baumannii ATCC 19606 as the donor (S2 Table). All methods located the maximal recombined region between positions 103,874 and 108,115 bp in the A. junii CIP 64.5 sequence, which included ant(3")-IIa. These results confirm that ant(3")-IIa in A. junii CIP 64.5 originated from A. baumannii and was integrated in the genome through homologous recombination.
Fig 3. Genomic context and homologous recombination (HR) analysis of ant(3")-IIa in A. baumannii and A. junii.
Deep brown indicates high nucleotide similarity, and light brown indicates low nucleotide similarity. Arrows indicate open reading frames and the direction of transcription. Yellow arrow, ant(3")-IIa; Pink arrow, GMP synthase gene. Boldfaced text above the arrows indicates the protein product of each gene. The displayed sequences of A. baumannii ATCC19606, A. junii CIP 64.5, CIP 107470 and NIPH 182 are subregions of Genbank accessions, JMRY01000011, APPX01000040, APPS01000002 and APPW01000012, respectively.
The ant(3")-II genes are in a recombination hotspot in the genus Acinetobacter
Another clear-cut inconsistency between the ANT(3")-II and Acinetobacter phylogenies is the clustering of the A. gyllenbergii NIPH 230 ANT(3")-IIc with A. parvus-related ANT(3")-IIc. Analysis of the genomic context of ant(3")-IIc in A. gyllenbergii NIPH 230 and A. parvus CIP 102637 showed that the sequence of the gene and its surrounding region (1.5 kbp) is 100% identical between the two strains, while the remaining flanking regions are only 74–84% identical (S5 Fig). Other discrepancies include the presence of ant(3")-IIc in A. ursingii ANC 3649, the presence of the strains carrying ant(3")-IIb in an inner branch of the ant(3")-IIc clade, and the position of A. parvus CIP 108168 and CIP 102529 away from the other A. parvus strains in the ANT(3")-II phylogeny (Figs 1 and 2). All these discrepancies can be attributed to HGT mediated by HR, although the lack of representative genomes and sequence incompleteness prevented us from fully resolving the HR events leading to the observed patterns (S6–S8 Figs).
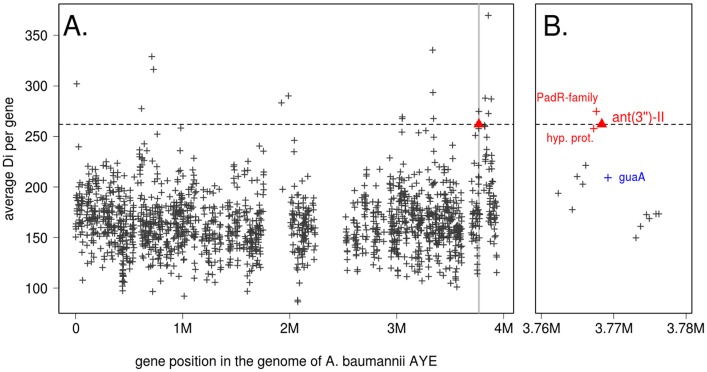
The high number of observed HR events involving ant(3")-II genes across Acinetobacter species prompted us to investigate the rate of recombination (precisely, the intensity of DNA transfer) at this locus compared to the global genome recombination rate. We first ran the ordered Painting algorithm [20] on all the A. baumannii genomes present in Fig 1 (see Methods). In A. baumanni, ant(3")-IIa ranked as the 126th most recombined gene out of the 2282 genes present in all investigated genomes. Although such rank is indicative of a high recombination rate, it cannot be considered as a recombination hotspot according to the criterion proposed in [20]. When the analysis was performed on all Acinetobacter genomes carrying an ant(3")-II gene, the locus ranked 17th most recombined gene out of the 1694 genes present in all genomes, and fell in the range of the predicted recombination hotspots (Fig 4A). Interestingly, the two genes directly adjacent to ant(3")-IIa in A. baumannii AYE (encoding a PadR-family transcriptional regulator and a small conserved hypothetical protein) ranked 12th and 21th most recombined genes, respectively (Fig 4B). On the contrary, guaA (encoding a GMP synthase) ranked 118th despite being also adjacent to ant(3")-II genes (Fig 4B). These results indicate that the ant(3")-II genes are part of a small recombination hotspot of ca. 2 kbp involved in the frequent horizontal transfer of the resistance gene across Acinetobacter species.
Fig 4. Intensity of homologous recombination (HR) along Acinetobacter genomes.
The X axis indicates the position in the reference genome A. baumanni AYE, and the Y axis the gene-average value of the orderedPainting statistics Di, measuring the intensity of recombination at nucleotide i. The dotted line represents the top percentile value. The ant(3")-II locus is marked by a red triangle. (A) Recombination intensity along the whole genome. (B) HR intensity in the 20 kbp surrounding ant(3")-II shaded in grey in (A). Genes considered as in a recombination hotspot are marked in red, and guaA is marked in blue.
Discussion
Intrinsic resistance genes are defined as conserved chromosomal genes, conferring phenotypic resistance in a given species [21]. Traditional intrinsic resistance genes encode either multidrug resistance efflux pumps that pump out intracellular antibiotics or outer membrane proteins that reduce the permeability of the outer membrane, decreasing antibiotic uptake [22]. Recently, a large number of other types of intrinsic resistance genes have been identified in E. coli, Pseudomonas aeruginosa, and Staphylococcus aureus by screening libraries of mutants obtained through precise gene deletions or transposon-tagged insertions that belong to a variety of cellular functional categories [23–26]. In the present study, bioinformatics screening of complete genome sequences and extensive molecular biology and biochemistry experiments were used to identify a new subclass of aminoglycoside nucleotidyltransferases, ANT(3")-II, in Acinetobacter spp. ANT(3")-II enzymes are phylogenetically distant from all known ANT(3")-I and the genes encoding for ANT(3")-II are always located at the same chromosomal locus in Acinetobacter spp., while the genes of ANT(3")-I are part of mobile genetic elements widely distributed in gram-negative bacteria. Furthermore, our findings revealed that ANT(3")-II is intrinsic in A. baumannii, A. pittii and A. gyllenbergii, bringing to light a novel natural mechanism of resistance in these clinically important pathogens.
Understanding the origins of antibiotic resistance would enable tracing their evolution, and predicting the emergence of clinical resistance. It has been proposed that certain antibiotic resistance genes in human bacterial pathogens originated from antibiotic-producing microorganisms, which use these genes to prevent suicide during drug production [27, 28]. Recent studies have shown that intrinsic genes, particularly in the genus Acinetobacter, can become mobile. The intrinsic genes aph(3’)-VI and aac(6’)-Ih, which encode aminoglycoside-modifying enzymes that originated from A. guillouiae and A. gyllenbergii, respectively, were captured by an IS element and disseminated in other Acinetobacter species [16, 17]. Similarly, A. radioresistens might be the origin of the blaOXA-23 carbapenem resistance determinant in A. baumannii [29]. The findings of our study demonstrated that, contrary to the above examples, the ant(3")-II genes were not mobilized through mobile elements but are located in a recombination hotspot, leading to frequent allele replacement or ant(3")-II gene acquisition in various Acinetobacter species. The impact of HR on the diversification of surface molecules and on the transfer of two resistance genes (an activated cephalosporin resistance gene and the blaNDM-1 gene) have been reported previously in A. baumannii [30–32]. However, the role of this molecular process in antibiotic resistance dissemination between Acinetobacter species is shown here for the first time. Moreover, the results suggest that this dissemination can occur even between very distantly related species of the genus, as exemplified by the ant(3")-IIc acquisition in A. ursingii. This phenomenon parallels the extensive horizontal exchanges of an intrinsic macrolide resistance gene among environmental and pathogenic species of the Bacillus cereus group we reported previously [33]. In the B. cereus group, acquisition of the intrinsic resistance gene was also mediated by HR. The fact that this process occurs in both Gram-positive and Gram-negative bacteria, and in both environmental and clinical isolates, suggests that transfer of resistance via HR may be more common than previously appreciated.
However, the high divergence of ant(3")-II sequences and their distribution mostly among non-clinical Acinetobacter spp. indicate that the first acquisition of this gene in a subgroup of Acinetobacter and its high rate of horizontal transfer likely predates the development of antibiotic for therapeutic use. It has become clear that clinical antibiotic resistance genes originated from intrinsic genes of environmental bacterial species, for which their primary function in the natural environment have been questioned [18, 28]. These general considerations would also apply for the ant(3")-II genes. On the one hand, ant(3")-II may function as a streptomycin/spectinomycin resistance gene in nature, since these two antibiotics are naturally produced by soil-dwelling bacteria [34, 35]. The high rate of horizontal transfer would thus be an ecological response to the patchy distribution of antibiotic producers in the soil. Similarly, the sequence diversification of ANT(3")-II may have been an evolutionary response to the production of new antibiotics variants by the producer, following the arms race hypothesis [36]. Exchange of ant(3")-II variants or even creation of mosaic variants would thus help the Acinetobacter spp. to adapt to the antibiotic compounds produced locally. On the other hand, the ant(3")-II genes primary function in nature may be unrelated to antibiotics resistance. More and more examples are now reported in the literature, suggesting that it could be a general trend [37–39]. Under this hypothesis, the underlying reasons of the diversification and high rate of horizontal transfer of ant(3")-II genes remains the same, except that the adaptive pressure is not known. A last possibility could be that the selection pressure actually acts on the flanking region rather than on the ant(3")-II locus itself. Indeed, the recombination hotspot region expands over 1.5 kbp downstream of ant(3")-II genes and include two other genes. The closest one particularly (ABAYE3738 in A. baumannii AYE, encoding a PadR-family transcriptional regulator), show even higher recombination intensity than ant(3")-II. The high rate of ant(3")-II genes horizontal transfers could thus be just a by-product of its physical position before this gene.
HR facilitates only permanent integration of a gene from an extra chromosomal DNA fragment into the chromosome, but the mechanism that could mediates cellular uptake of DNA fragments in Acinetobacer spp. remains unclear. Natural transformation has been described in some strains of A. baylyi [40], A. baumannii [41, 3], and A. calcoaceticus [42], suggesting that DNA uptake could occur via transformation. Genomic analysis of the genus Acinetobacter indicated the presence of most of the 13 key T4P (type IV pili) and competence-associated components in all genomes [3]. These studies suggest that members of the genus Acinetobacter have the potential to develop natural competence. Alternatively, general transduction has been suggested between A. baumannii strains, facilitated by one of the prophages present in the chromosome [32]. The extent to which these two processes provide the raw DNA material for HR and contribute to the genome plasticity observed in this genus is of great interest and requires further investigation.
Intrinsic resistance genes could provide an attractive therapeutic target to encourage the development of novel drugs that rejuvenate the activity of existing antibiotics [43]. We have shown that the deletion of ant(3")-II significantly decreases the resistance level of A. baumannii to streptomycin. Although no inhibitor of aminoglycoside-modifying enzymes is currently available, there is extensive effort to find how to overcome the action of these enzymes in pathogens [44]. Therefore, the demonstration that A. baumannii become more susceptible to streptomycin when ant(3")-IIa is silenced blazes a trail for future treatments against this extremely multi-resistant pathogen.
This work identified a novel subclass of intrinsic aminoglycoside nucleotidyltransferases in environmental and clinically important Acinetobacter species. The corresponding genes ant(3”)-II not only conferred phenotypic resistance in a given species but were also frequently horizontally transferred between different Acinetobacter species by homologous recombination. Overall, these results provide insight of Acinetobacer’s natural resistance and enhance our understanding of the evolutionary history of intrinsic resistance determinants in bacteria.
Materials and methods
Bacterial strains and culture conditions
Escherichia coli DH5α was used as a host for the expression of putative transferase resistance genes in vitro. The genomic DNA of A. baumannii ATCC 19606 was extracted to use as template for polymerase chain reaction (PCR). All strains were grown on Luria-Bertani (LB) broth at 37°C. The MIC for each strain was determined using broth microdilution assays, as recommended by the Clinical and Laboratory Standards Institute (CLSI, http://www.clsi.org/; CLSI 2010).
Construction of recombinant plasmids carrying putative transferase resistance genes
Eleven open reading frames, representative of the putative ant(3")-II subclass were chosen from various Acinetobacter to determine their antibiotic resistance phenotypes. One DNA sequence, corresponding to the putative ant(3")-IIa from A. baumannii ATCC 19606, was amplified using the primers antF/R listed in S3 Table. Other genes from A. baumannii ATCC 17978, A. baumannii AYE, A. junii CIP 64.5, A. pittii CIP 70.29, A. pittii PHEA-2, A. parvus CIP 108168, A. ursingii ANC 3649, A. gyllenbergii NIPH 230, A. sp. NIPH 1859, and A. sp. NIPH 758 were synthesized by Sangon Biotech (Shanghai, China). The PCR product and synthesized DNA were cloned into multiple cloning sites of the pUC18 vector, resulting in recombinant plasmids carrying the target genes. E. coli DH5α was transformed with these recombinant plasmids, and clones were selected by blue-white screening on LB agar plates supplemented with 100 μg/ml ampicillin. The transformants containing the inserted fragments were confirmed by enzyme digestion and sequencing. Resistance of transformants to aminoglycosides (streptomycin, spectinomycin, kanamycin, gentamycin, amikacin, and tobramycin) were determined using the microdilution method, with induction by 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG), as recommended by the CLSI (http://www.clsi.org/; CLSI 2010).
Construction of resistance gene-deficient mutants
The plasmid pEX18Gm was used as a suicide vector to knock out ant(3'')-IIa in A. baumannii. The upstream and downstream regions (~500 bp) of the gene were amplified using genomic DNA of A. baumannii ATCC 19606 as a template, and combined by overlap PCR. The desirable fusion PCR product was ligated to pEX18Gm, resulting in a recombinant plasmid. The plasmid was transformed into E. coli S17-1λpir, to create a donor strain. A. baumannii ATCC 19606 was used as a recipient strain. Using a conjugation experiment, a merodiploid (containing the plasmid integrated with one HR event) was obtained by plating cells on LB plates containing 100 μg/ml ampicillin and 20 μg/ml gentamycin. The merodiploid was inoculated sequentially into LB liquid medium free of antibiotic for propagation at 37°C. After several rounds of propagation, serial dilutions were spread on LB agar plates with 20% sucrose. After overnight incubation at 37°C, colonies were selected on gentamycin-containing LB agar plates and LB agar without antibiotics. The gentamycin-sensitive clones, which have lost pEX18Gm through a second HR event, were verified as correct ant(3'')-IIa-deficient mutants by PCR and sequencing. The primers used for construction of gene-deficient mutants are listed in S3 Table.
Construction of complementation plasmid
A 1337-bp DNA fragment containing the A. baumannii origin of replication was synthesized by Sangon Biotech (Shanghai, China) and then cloned into plasmid pKF18k-2 (Takara) using PvuII restriction site. Inserts were selected in kanamycin 50 μg/ml and verified with PCR and enzyme digestion. The resulting plasmid was designated pKFAb, which can replicate in E. coli and A. baumannii. To construct the complementation plasmid the ant(3")-II gene was amplified from the A. baumannii ATCC 19606 chromosome using antF/R primers (S3 Table) and cloned into plasmid pKFAb with EcoRI and HindIII restriction sites by transformation of E. coli Top10. The resulting recombinant plasmid pKFAb::ant was verified by enzyme digestion and sequencing. At last the construct pKFAb::ant was introduced into competent A. baumannii (Δant) by electroporation, and transformants were selected on LB plates supplemented with streptomycin 150 μg/ml.
ANT(3")-IIa enzyme overexpression and purification
ant(3")-IIa from A. baumannii ATCC 19606 (EEX02086) was inserted into pET28a (+) expression plasmid. The resulting recombinant plasmid was transformed into E. coli BL21 (DE3). A fresh overnight culture was used to inoculate 500 ml LB media in a 1/100 inoculum size. Culture was incubated at 37°C with shaking at 200 rpm up to OD600 of 0.6–0.8, followed by continued cultivation for an additional 16 h by addition of 0.5 mM IPTG at 25°C. Subsequently, cells were harvested by centrifugation, washed with 50 mM Tris-HCl buffer (pH 7.5). Then cells were resuspended in 30 ml of 50 mM Tris-HCl buffer (pH 7.5) and disrupted by using a ultrasonic cell disruptor (SCIENTZ, Ningbo). The cell lysate was clarified by centrifugation at 12,000×g for 20 min, and the supernatant (soluble protein fraction) was used for protein purification. Recombinant protein was purified with the His-Bind protein purification kit (Novagen, Madison, WI, USA) according to the manufacturer's instructions. To remove any residual proteins, imidazole and salts from the collected fractions, these fractions were pooled and re-separated with Superdex 200 gel chromatography with Tris-HCl buffer using a fast protein liquid chromatography system (Äkta FPLC, Amersham Biosciences, USA). The purified protein was concentrated by ultrafiltration using a Millipore Ultra-15 (10 kDa) centrifugal filter (Millipore, USA) and stored at −70°C.
Detection of reaction product by LC-MS
The ANT(3")-IIa enzymatic reaction consisted of 50 mM HEPES (pH7.5), 1 mM dithiothreitol, 15 mM MgCl2, 1 mg streptomycin, 3 mM ATP, 0.5 mg of purified ANT(3")-IIa protein, and 2 units of inorganic pyrophosphatase in a total volume of 1 ml [45]. The reaction was performed at 37°C for 24 h and stopped by the addition of cold acetonitrile. The supernanant fraction was then applied onto a Millipore Ultra15 (10 kDa) centrifugal filter tube. After centrifugation at 12,000×g for 20 min, the protein was removed. The streptomycin and reaction product was eluted with deionized water. Prior to analyses, samples were filtered through a 0.22 μm filter and then subjected to LC-MS analysis on an Agilent 1260/6460 Triple quadrupole LC/MS system. The analytic column was ZORBAX SB-Aq (Agilent, 2.1×100 mm, 3.5 μm). A mixture of 2% methanol and 0.1% formic acid (v/v) in water under an isocratic elution program was used as the mobile phase. The flow rate was set at 0.1 ml min-1, and the column temperature was set at 30°C. The fragmentor voltage was 260 V. A capillary voltage of 4.0 kV and atomizing gas pressure of 35 psi were used. The flow rate of drying gas was 12 ml min-1, and the temperature of the solvent removal was 350°C. N2 was used as the collision gas and a collision voltage of 45 V was used for product ion scan. The LC-MS data were acquired and analyzed using a MassHunter Workstation Software (Version B.04.00, Agilent, Santa Clara, CA, USA).
Bioinformatic methods and phylogenetic analysis
To discover new antibiotic transferases, two databases were constructed. One database consisted of a collection of amino acid sequences of known antibiotic transferases, including aminoglycoside, macrolide, chloramphenicol, and lincosamides transferases obtained from the Antibiotic Resistance Genes Database (ARDB; http://ardb.cbcb.umd.edu/index.html), and completed with more recently described transferase resistance genes. The other was the complete collection of assembled and draft Acinetobacter genome sequences downloaded from the NCBI FTP Site (ftp://ftp.ncbi.nlm.nih.gov/) in summer 2014. The antibiotic transferase sequences were searched against the genomes using TBLASTN. And the proteins sharing more than 30% amino acid identity and 50% coverage with known antibiotic transferases were selected for expression analyses.
Putative ANT(3")-II amino-acid sequences from Acinetobacter species were extracted from the respective genomes and the full set of ANT(3'')-I (AadA) protein sequences described in [19] were downloaded from Genbank. The protein dataset was completed with the recently described AadA25 protein [46], four additional AadA sequences located in Acinetobacter species, and two ANT(9)-I sequences to serve as outgroups. The accession numbers for all these proteins are listed in S4 Table. The sequence dataset was aligned using the CLUSTALW software with standard parameters [47]. Phylogenetic trees were constructed using NJ, MP and ML methods implemented in MEGA 6 software [48]. All phylogenetic trees were constructed using the partial deletion parameter set to 80% and 1000 bootstrap replicates. Other parameters were set to default for NJ and MP analyses, while the WAG+F model, with gamma-distributed rates among sites, was used for ML analysis.
The construction of the Acinetobacter core-proteins phylogenetic tree was described previously [3]. Briefly, orthologous core proteins shared by the analyzed Acinetobacter genomes were retrieved from bi-directional best hits. The 950 resulting families of proteins were concatenated and aligned to construct an approximated maximum-likelihood using FastTree with the WAG matrix of protein evolution, a gamma correction for variable evolutionary rates, and 100 bootstraps.
Detection of homologous recombination events
HR detection at the ant(3")-IIa locus was performed using the RDP4 package, which includes several HR detection methods [49], on a 14 kbp conserved region surrounding the gene in A. baumannii and A. junii genomes. The region was extracted from the four sequences presented in Fig 1 and aligned using CLUSTALW, with a gap penalty of 20. The alignment was then carefully corrected visually, as alignment errors might critically alter HR detections, as explained in the RDP4 manual. The alignment was examined using the RDP [50], GENECONV [51], BootScan [52, 53], Maximum Chi Square (MAXCHI) [54], Chimaera [55], SisterScan [56], and PhylPro [57] methods. We retained only events unambiguously detected by the seven methods, using the following RDP4 parameters: a corrected P-value < 0.000001, with phylogenetic evidence, and with overlapping signals disentangled. HR investigations were made by running automated RDP analysis of the alignment of four sequences, and significant recombination events were detected.
Inference of landscape and hot regions of homologous recombination
First, complete and draft genome sequences of the 31 A. baumannii strains carrying ant(3")-IIa (Fig 1) were downloaded from the Genbank database. The genome sequence of A. baumannii AYE was selected as reference, and each of the remaining genomes were aligned on this reference using progressiveMauve v2.4.0 [58]. Resulting pairwise alignments were then combined, and all positions in AYE identical in all strains as well as positions with missing data (gaps in one or more sequence) were discarded. The SNPs and their position in the remaining core genomic regions were provided as input to orderedPainting [20]. Next, the same analysis was performed on the whole Acinetobacter genus, again with A. baumannii AYE as reference. Genome sequence of all Acinetobacter from Fig 1 carrying ant(3")-II were downloaded from Genbank, except that only three A. baumannii (AYE, ATCC19606, and NIPH601) and three A. pittii (PHEA-2, ABBL015, and NBRC110504) were retained. The output of orderedPainting is a recombination intensity at each polymorphic site measured as a statistic Di. We therefore averaged the intensity value for each gene annotated in AYE. The top one percentile of gene-based recombination intensity was selected as indicative of recombination hotspot, as proposed in Yahara et al.'s report [20].
Supporting information
Numbers above each node are the percentages of tree configurations that occurred during 1000 bootstrap replicates. Only values greater than 70% are provided.
(TIF)
lane 1, molecular weight markers (10 kDa–200 kDa); lane 2, E. coli BL21 (DE3) harboring null vector pET28a; lane 3, extract of E. coli BL21 (DE3) harboring pET28a-ant; 4, Purified ANT(3")-IIa protein.
(TIF)
(TIF)
Aqua green indicates high nucleotide similarity (>98%). Arrows indicate open reading frames and the direction of transcription. Yellow arrow, ant(3")-IIa; Red arrow, GMP synthase gene. Boldfaced text above the arrows indicates the protein product of each gene. The displayed sequences of A. baumannii ATCC 19606, 6200, AB0057, AB307-0294, ANC 4097, 17978, AYE, MDR-TJ, NIPH 1669, NIPH 290, NIPH 329, NIPH 527, NIPH 67, TCDC-AB0715, MDR-ZJ06, NCGM 237, 1656–2 and NIPH 201 are subregions of Genbank accessions, JMRY01000011, NZ_CP010397, NC_011586, NC_011595, APRF01000002, CP000521, NC_010410, NC_017847, APOQ01000016, APRD01000022, APQY01000009, APQW01000014, APRA01000009, CP002522, NC_017171, NZ_AP013357, NC_017162 and APQV01000013, respectively.
(TIF)
Deep orange indicates high nucleotide similarity, and light orange indicates low nucleotide similarity. Arrows indicate open reading frames and the direction of transcription. Boldfaced text above the arrows indicates the protein product of each gene. Yellow arrow, ant(3")-IIc; pink arrow, GMP synthase gene; genes without label, hypothetical proteins. The displayed sequences of A. gyllenbergii CIP 110306, A. gyllenbergii NIPH 230, A. parvus CIP 102637, and A. parvus CIP 102129 are subregions of Genbank accessions ATGG01000028, AYEQ01000163, APPG01000010, and APPA01000001, respectively.
(TIF)
Blue indicates high nucleotide similarity, and light green indicates low nucleotide similarity. Arrows indicate open reading frames and the direction of transcription. Yellow arrow, ant(3")-II genes; Pink arrow, GMP synthase gene; genes without label, hypothetical proteins. The putative HR event encompassed ant(3")-IIb and the downstream inserted region in Taxon 20 NIPH 2168 and NIPH 758, leading to a low similarity region inside an otherwise more highly similar region, when compared to Gen. sp. 14BJ NIPH 3623. Sequences of A. sp. neg1 and B-65365 are essentially identical to sequences of Taxon 20 NIPH 2168 and NIPH 758 and are not represented here. The displayed sequences of Taxon 20 NIPH 2168, NIPH 758, and Gen. sp. 14BJ NIPH 3623 are subregions of Genbank accessions APRW01000008, APPC01000022, and APSA01000001, respectively.
(TIF)
Blue indicates high nucleotide similarity, and light green indicates low nucleotide similarity. Arrows indicate open reading frames and the direction of transcription. Yellow arrow, ant(3")-IIc; Pink arrow, GMP synthetase gene; genes without label, hypothetical proteins. The putative HR region encompasses the ant(3")-IIc gene and the GMP synthase-encoding gene in A. ursignii ANC 3649. The displayed sequences of A. ursingii CIP 107286, ANC 3649, Gen. sp. 15BJ CIP 110321, and Gen. sp. 16 ANC 3880 are subregions of Genbank accessions APQA01000027, APQC01000003, AQFL01000027, and APSD01000005, respectively.
(TIF)
The phylogenetic relationship between strains as reported in Fig 2 is shown on the left side. Deep Orange indicates high nucleotide similarity, and light orange indicates low nucleotide similarity. Arrows indicate open reading frames and the direction of transcription. Yellow arrow, ant(3")-IIc; pink arrow, GMP synthase gene; striped arrow, restriction-modification gene; genes without label, hypothetical proteins. A. parvus CIP 102637 is repeated at the bottom of the figure to show its similarity with A. parvus CIP 102143 at the ant(3")-IIc locus. The nucleotide pairwise similarity at the ant(3")-IIc locus and the directly adjacent genes (including the GMP synthase and the PadR-family transcriptional regulator) is consistently lower than at the flanking regions, indicating multiple events of allele exchange through homologous recombination. ant(3")-IIc have also been precisely replaced by an unrelated gene in A. parvus CIP 102082. The displayed sequences of A. sp. A. parvus CIP 102637, CIP 108168, CIP 102159, CIP 102082, CIP 102529, and CIP 102143 are subregions of Genbank accessions APPG01000010, APOM01000001, KB851223, KB849372, KB849355, and APSE01000019, respectively.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We want to thank Dr. Marie Touchon from Institut Pasteur who kindly provided us the core-proteins-based phylogenetic tree of Acinetobacter. We are also grateful to Dr. Gerard D Wright of McMaster University for the critical reading of the manuscript and valuable suggestions (prior to submission), and to the three anonymous reviewers which helped us to improve the quality of our study.
Data availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the National Key Basic Research Development Plan of China (973 Program) (grant 2015CB554202) and the National Natural Science Foundation of China (grant 31500061). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Towner KJ. Acinetobacter: an old friend, but a new enemy. J Hosp Infect. 2009;73: 355–363. 10.1016/j.jhin.2009.03.032 [DOI] [PubMed] [Google Scholar]
- 2.Nordqvist H, Nilsson LE, Claesson C. Mutant prevention concentration of colistin alone and in combination with rifampicin for multidrug-resistant Acinetobacter baumannii. Eur J Clin Microbiol Infect Dis. 2016;35: 1845–1850. 10.1007/s10096-016-2736-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Touchon M, Cury J, Yoon EJ, Krizova L, Cerqueira GC, Murphy C, et al. The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol Evol. 2014;6: 2866–2882. 10.1093/gbe/evu225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergogne-Bérézin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9: 148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almasaudi SB. Acinetobacter spp. as nosocomial pathogens: Epidemiology and resistance features. Saudi J Biol Sci. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockhart SR, Abramson MA, Beekmann SE, Gallagher G, Riedel S, Diekema DJ, et al. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J Clin Microbiol. 2007;45: 3352–3359. 10.1128/JCM.01284-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu FP, Guo Y, Zhu DM, Wang F, Jiang XF, Xu YC, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin Microbiol Infect. 2016;22: Suppl 1:S9–S14. [DOI] [PubMed] [Google Scholar]
- 8.Nigro SJ, Hall RM. Loss and gain of aminoglycoside resistance in global clone 2 Acinetobacter baumannii in Australia via modification of genomic resistance islands and acquisition of plasmids. J Antimicrob Chemother. 2016;71: 2432–2440. 10.1093/jac/dkw176 [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Yang P, Wang X, Zong Z. Characterization of Acinetobacter johnsonii isolate XBB1 carrying nine plasmids and encoding NDM-1, OXA-58 and PER-1 by genome sequencing. J Antimicrob Chemother. 2016;71: 71–75. 10.1093/jac/dkv324 [DOI] [PubMed] [Google Scholar]
- 10.Coyne S, Courvalin P, Perichon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents. Chemother. 2011;55: 947–953. 10.1128/AAC.01388-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez P, Mattar S. Imipenem-resistant Acinetobacter baumannii carrying the ISAba1-blaOXA-23,51 and ISAba1-blaADC-7 genes in Monteria, Colombia. Braz J Microbiol. 2012;43: 1274–1280. 10.1590/S1517-83822012000400006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mu X, Nakano R, Nakano A, Ubagai T, Kikuchi-Ueda T, Tansho-Nagakawa S, et al. Loop-mediated isothermal amplification: Rapid and sensitive detection of the antibiotic resistance gene ISAba1-blaOXA-51-like in Acinetobacter baumannii. J Microbiol Methods. 2016;121: 36–40. 10.1016/j.mimet.2015.12.011 [DOI] [PubMed] [Google Scholar]
- 13.Chen TL, Lee YT, Kuo SC, Hsueh PR, Chang FY, Siu LK, et al. Emergence and Distribution of Plasmids Bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob Agents Chemother. 2010;54: 4575–4581. 10.1128/AAC.00764-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YT, Kuo SC, Chiang MC, Yang SP, Chen CP, Chen TL, et al. Emergence of carbapenem-resistant non-baumannii species of Acinetobacter harboring a blaOXA-51-like gene that is intrinsic to A. baumannii. Antimicrob Agents Chemother. 2012;56: 1124–1127. 10.1128/AAC.00622-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnin RA, Poirel L, Nordmann P. New Delhi metallo-β-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol. 2014;9: 33–41. 10.2217/fmb.13.69 [DOI] [PubMed] [Google Scholar]
- 16.Yoon EJ, Goussard S, Touchon M, Krizova L, Cerqueira G, Murphy C, et al. Origin in Acinetobacter guillouiae and dissemination of the aminoglycoside -modifying enzyme APH(3')-VI. MBio. 2014;5: e01972–14. 10.1128/mBio.01972-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon EJ, Goussard S, Nemec A, Lambert T, Courvalin P, Grillot-Courvalin C. Origin in Acinetobacter gyllenbergii and dissemination of aminoglycoside -modifying enzyme AAC(6')-Ih. J Antimicrob Chemother. 2015;71: 601–606. 10.1093/jac/dkv390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morar M, Wright GD. The genomic enzymology of antibiotic resistance. Annu Rev Genet. 2010;44: 25–51. 10.1146/annurev-genet-102209-163517 [DOI] [PubMed] [Google Scholar]
- 19.Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13: 151–171. 10.1016/j.drup.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yahara K, Didelot X, Ansari MA, Sheppard SK, Falush D. Efficient inference of recombination hot regions in bacterial genomes. Mol Biol Evol. 2014;31: 1593–1605. 10.1093/molbev/msu082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fajardo A, Martínez-Martín N, Mercadillo M, Galán JC, Ghysels B, Matthijs S, et al. The neglected intrinsic resistome of bacterial pathogens. PLoS One. 2008;3: e1619 10.1371/journal.pone.0001619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivares J, Bernardini A, Garcia-Leon G, Corona F, B Sanchez M, Martinez JL. The intrinsic resistome of bacterial pathogens. Front Microbiol. 2013;4: 103 10.3389/fmicb.2013.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breidenstein EB, Khaira BK, Wiegand I, Overhage J, Hancock RE. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob Agents Chemother. 2008;52: 4486–4491. 10.1128/AAC.00222-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock RE, Martínez JL. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob Agents Chemother. 2010;54: 4159–4167. 10.1128/AAC.00257-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, et al. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob Agents Chemother. 2010;54: 1393–1403. 10.1128/AAC.00906-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blake KL, O'Neill AJ. Transposon library screening for identification of genetic loci participating in intrinsic susceptibility and acquired resistance to antistaphylococcal agents. J Antimicrob Chemother. 2013;68: 12–16. 10.1093/jac/dks373 [DOI] [PubMed] [Google Scholar]
- 27.Cundliffe E. How antibiotic-producing organisms avoid suicide. Annu Rev Microbiol. 1989;43: 207–233. 10.1146/annurev.mi.43.100189.001231 [DOI] [PubMed] [Google Scholar]
- 28.Wright GD. The origins of antibiotic resistance. Handb Exp Pharmacol. 2012;211: 13–30. [DOI] [PubMed] [Google Scholar]
- 29.Poirel L, Figueiredo S, Cattoir V, Carattoli A, Nordmann P. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob Agents Chemother. 2008;52: 1252–1256. 10.1128/AAC.01304-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snitkin ES, Zelazny AM, Montero CI, Stock F, Mijares L; NISC Comparative Sequence Program, et al. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc Natl Acad Sci USA. 2011;108: 13758–13763. 10.1073/pnas.1104404108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamidian M, Hall RM. Resistance to third-generation cephalosporins in Acinetobacter baumannii due to horizontal transfer of a chromosomal segment containing ISAba1-ampC. J Antimicrob Chemother. 2014;69: 2865–1866. 10.1093/jac/dku202 [DOI] [PubMed] [Google Scholar]
- 32.Krahn T, Wibberg D, Maus I, Winkler A, Bontron S, Sczyrba A, et al. Intraspecies transfer of the chromosomally encoded Acinetobacter baumannii blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 2016;60: 3032–3040. 10.1128/AAC.00124-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Sui Z, Leclercq SO, Zhang G, Zhao M, Chen W, et al. Functional characterization and phylogenetic analysis of acquired and intrinsic macrolide phosphotransferases in the Bacillus cereus group. Environ Microbiol. 2015;17: 1560–1573. 10.1111/1462-2920.12578 [DOI] [PubMed] [Google Scholar]
- 34.Distler J, Ebert A, Mansouri K, Pissowotzki K, Stockmann M, Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987;15: 8041–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim KR, Kim TJ, Suh JW. The gene cluster for spectinomycin biosynthesis and the aminoglycoside-resistance function of spcM in Streptomyces spectabilis. Curr Microbiol. 2008;57: 371–374. 10.1007/s00284-008-9204-y [DOI] [PubMed] [Google Scholar]
- 36.Van Valen L. A new evolutionary law. Evolutionary Theory. 1973;1: 1–30. [Google Scholar]
- 37.Magnet S, Courvalin P, Lambert T. Activation of the cryptic aac(6')-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J Bacteriol. 1999;181: 6650–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vetting MW, Magnet S, Nieves E, Roderick SL, Blanchard JS. A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chem Biol. 2004;11: 565–573. 10.1016/j.chembiol.2004.03.017 [DOI] [PubMed] [Google Scholar]
- 39.Martínez JL, Coque TM, Baquero F. What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol. 2015;13: 116–123. 10.1038/nrmicro3399 [DOI] [PubMed] [Google Scholar]
- 40.Gerischer U, Ornston LN. Dependence of linkage of alleles on their physical distance in natural transformation of Acinetobacter sp. strain ADP1. Arch Microbiol. 2001;176: 465–469. 10.1007/s00203-001-0353-7 [DOI] [PubMed] [Google Scholar]
- 41.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS Jr. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. MBio. 2013;4: pii: e00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen KM, Bones AM, Van Elsas JD. Induced Natural Transformation of Acinetobacter calcoaceticus in Soil Microcosms. Appl Environ Microbiol. 1997;63: 3972–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cox G, Wright GD. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol. 2013;303: 287–292. 10.1016/j.ijmm.2013.02.009 [DOI] [PubMed] [Google Scholar]
- 44.Labby KJ, Garneau-Tsodikova S. Strategies to overcome the action of aminoglycoside -modifying enzymes for treating resistant bacterial infections. Future Med Chem. 2013;5: 1285–309. 10.4155/fmc.13.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim C, Hesek D, Zajicek J, Vakulenko SB, Mobashery S. Characterization of the bifunctional aminoglycoside-modifying enzyme ANT(3")-Ii/AAC(6')-IId from Serratia marcescens. Biochemistry. 2006;45: 8368–8377. 10.1021/bi060723g [DOI] [PubMed] [Google Scholar]
- 46.Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, et al. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J Antimicrob Chemother. 2012;67: 84–90. 10.1093/jac/dkr406 [DOI] [PubMed] [Google Scholar]
- 47.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23: 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution. 2015;1: vev003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16: 562–563. [DOI] [PubMed] [Google Scholar]
- 51.Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265: 218–225. 10.1006/viro.1999.0056 [DOI] [PubMed] [Google Scholar]
- 52.Salminen MO, Carr JK, Burke DS, McCutchan FE. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by BOOTSCANning. AIDS Res Hum Retroviruses. 1995; 11: 1423–1425. 10.1089/aid.1995.11.1423 [DOI] [PubMed] [Google Scholar]
- 53.Martin DP, Posada D, Crandall KA, Williamson C. A modified BOOTSCAN algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses. 2005;21: 98–102. 10.1089/aid.2005.21.98 [DOI] [PubMed] [Google Scholar]
- 54.Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992; 34: 126–129. [DOI] [PubMed] [Google Scholar]
- 55.Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA. 2001;98: 13757–13762. 10.1073/pnas.241370698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibbs MJ, Armstrong JS, Gibbs AJ. Sisterscanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 2000;16: 573–582. [DOI] [PubMed] [Google Scholar]
- 57.Weiller GF. Phylogenetic profiles: a graphical method for detecting genetic recombinations in homologous sequences. Mol Biol Evol. 1998;15: 326–335. [DOI] [PubMed] [Google Scholar]
- 58.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5: e11147 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Numbers above each node are the percentages of tree configurations that occurred during 1000 bootstrap replicates. Only values greater than 70% are provided.
(TIF)
lane 1, molecular weight markers (10 kDa–200 kDa); lane 2, E. coli BL21 (DE3) harboring null vector pET28a; lane 3, extract of E. coli BL21 (DE3) harboring pET28a-ant; 4, Purified ANT(3")-IIa protein.
(TIF)
(TIF)
Aqua green indicates high nucleotide similarity (>98%). Arrows indicate open reading frames and the direction of transcription. Yellow arrow, ant(3")-IIa; Red arrow, GMP synthase gene. Boldfaced text above the arrows indicates the protein product of each gene. The displayed sequences of A. baumannii ATCC 19606, 6200, AB0057, AB307-0294, ANC 4097, 17978, AYE, MDR-TJ, NIPH 1669, NIPH 290, NIPH 329, NIPH 527, NIPH 67, TCDC-AB0715, MDR-ZJ06, NCGM 237, 1656–2 and NIPH 201 are subregions of Genbank accessions, JMRY01000011, NZ_CP010397, NC_011586, NC_011595, APRF01000002, CP000521, NC_010410, NC_017847, APOQ01000016, APRD01000022, APQY01000009, APQW01000014, APRA01000009, CP002522, NC_017171, NZ_AP013357, NC_017162 and APQV01000013, respectively.
(TIF)
Deep orange indicates high nucleotide similarity, and light orange indicates low nucleotide similarity. Arrows indicate open reading frames and the direction of transcription. Boldfaced text above the arrows indicates the protein product of each gene. Yellow arrow, ant(3")-IIc; pink arrow, GMP synthase gene; genes without label, hypothetical proteins. The displayed sequences of A. gyllenbergii CIP 110306, A. gyllenbergii NIPH 230, A. parvus CIP 102637, and A. parvus CIP 102129 are subregions of Genbank accessions ATGG01000028, AYEQ01000163, APPG01000010, and APPA01000001, respectively.
(TIF)
Blue indicates high nucleotide similarity, and light green indicates low nucleotide similarity. Arrows indicate open reading frames and the direction of transcription. Yellow arrow, ant(3")-II genes; Pink arrow, GMP synthase gene; genes without label, hypothetical proteins. The putative HR event encompassed ant(3")-IIb and the downstream inserted region in Taxon 20 NIPH 2168 and NIPH 758, leading to a low similarity region inside an otherwise more highly similar region, when compared to Gen. sp. 14BJ NIPH 3623. Sequences of A. sp. neg1 and B-65365 are essentially identical to sequences of Taxon 20 NIPH 2168 and NIPH 758 and are not represented here. The displayed sequences of Taxon 20 NIPH 2168, NIPH 758, and Gen. sp. 14BJ NIPH 3623 are subregions of Genbank accessions APRW01000008, APPC01000022, and APSA01000001, respectively.
(TIF)
Blue indicates high nucleotide similarity, and light green indicates low nucleotide similarity. Arrows indicate open reading frames and the direction of transcription. Yellow arrow, ant(3")-IIc; Pink arrow, GMP synthetase gene; genes without label, hypothetical proteins. The putative HR region encompasses the ant(3")-IIc gene and the GMP synthase-encoding gene in A. ursignii ANC 3649. The displayed sequences of A. ursingii CIP 107286, ANC 3649, Gen. sp. 15BJ CIP 110321, and Gen. sp. 16 ANC 3880 are subregions of Genbank accessions APQA01000027, APQC01000003, AQFL01000027, and APSD01000005, respectively.
(TIF)
The phylogenetic relationship between strains as reported in Fig 2 is shown on the left side. Deep Orange indicates high nucleotide similarity, and light orange indicates low nucleotide similarity. Arrows indicate open reading frames and the direction of transcription. Yellow arrow, ant(3")-IIc; pink arrow, GMP synthase gene; striped arrow, restriction-modification gene; genes without label, hypothetical proteins. A. parvus CIP 102637 is repeated at the bottom of the figure to show its similarity with A. parvus CIP 102143 at the ant(3")-IIc locus. The nucleotide pairwise similarity at the ant(3")-IIc locus and the directly adjacent genes (including the GMP synthase and the PadR-family transcriptional regulator) is consistently lower than at the flanking regions, indicating multiple events of allele exchange through homologous recombination. ant(3")-IIc have also been precisely replaced by an unrelated gene in A. parvus CIP 102082. The displayed sequences of A. sp. A. parvus CIP 102637, CIP 108168, CIP 102159, CIP 102082, CIP 102529, and CIP 102143 are subregions of Genbank accessions APPG01000010, APOM01000001, KB851223, KB849372, KB849355, and APSE01000019, respectively.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.