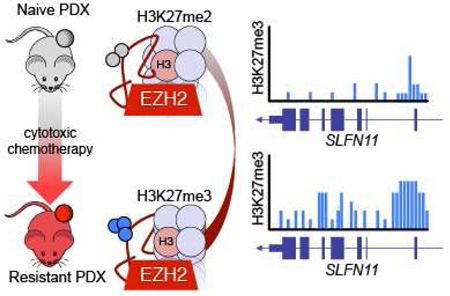
SUMMARY
Small cell lung cancer is initially highly responsive to cisplatin and etoposide but in almost every case becomes rapidly chemoresistant, leading to death within one year. We modeled acquired chemoresistance in vivo using a series of patient-derived xenografts to generate paired chemosensitive and chemoresistant cancers. Multiple chemoresistant models demonstrated suppression of SLFN11, a factor implicated in DNA damage repair deficiency. In vivo silencing of SLFN11 was associated with marked deposition of H3K27me3, a histone modification placed by EZH2, within the gene body of SLFN11, inducing local chromatin condensation and gene silencing. Inclusion of an EZH2 inhibitor with standard cytotoxic therapies prevented emergence of acquired resistance and augmented chemotherapeutic efficacy in both chemosensitive and chemoresistant models of small cell lung cancer.
Keywords: small cell lung cancer, EZH2, acquired resistance, SLFN11, TWIST1, patient-derived xenograft, EPZ011989, cisplatin, etoposide
Graphical abstract
eTOC blurb/In brief
By generating paired chemonaive and chemoresistant small cell lung cancer (SCLC) patient derived xenograft models, Gardner et al. find that EZH2 promotes chemoresistance by epigenetically silencing SLFN11. EZH2 inhibition prevents acquisition of chemoresistance and improves chemotherapeutic efficacy in SCLC.
INTRODUCTION
Small cell lung cancer (SCLC) affects an estimated 270,000 individuals per year worldwide and is metastatic at the time of diagnosis in approximately two thirds of cases (Shepherd et al., 2007; Torre et al., 2015). Metastatic SCLC is exceptionally lethal, associated with a median survival of 9 to 10 months from the time of diagnosis and a 5-year survival of less than 2% (Shepherd et al., 2007). Even when detected prior to metastasis, most patients with localized disease will suffer disease recurrence and death within the first 2 years. More effective treatment approaches to SCLC are desperately needed.
The standard first line treatment for metastatic SCLC consists of a platinum doublet, cisplatin or carboplatin, generally paired with the topoisomerase II inhibitor etoposide (Kalemkerian et al., 2013). Standard approaches to recurrent SCLC include treatment with a topoisomerase I inhibitor, topotecan or irinotecan (IRI). There are no approved therapies for SCLC after progression on a second line regimen. De novo SCLC is remarkably sensitive to first line platinum doublet chemotherapy, with objective response rates of over 50% in patients without substantial co-morbid conditions. These impressive responses are also disappointingly transient: median progression-free survival in current trials remains less than 5 months (Belani et al., 2016). The response rates to second line topoisomerase I inhibitor therapy are substantially lower, below 20% overall (Horita et al., 2015).
The molecular mechanisms responsible for the remarkable shift between de novo chemosensitive disease and rapidly emergent chemoresistant disease in SCLC have not been defined. Defining these mechanisms would both provide insights into the biology of SCLC and inform clinical strategies to prevent or delay therapeutic resistance. More broadly, characterizing mechanisms of acquired resistance in this cancer that undergoes a dramatic shift between chemosensitivity and chemoresistance could have implications for understanding acquired resistance to DNA damaging cytotoxic therapy in other malignancies.
Here, we sought to discover mechanisms of acquired resistance to first line cisplatin and etoposide therapy in SCLC by mimicking clinical practice as closely as possible through in vivo treatment of a set of chemosensitive SCLC patient-derived xenograft (PDX) models. We hypothesized that an approach to detect causal alterations against a high background of tobacco carcinogen-induced passenger mutations would be to conduct a pair-wise comparison of changes in individual tumor models prior to, and following, acquired chemotherapy resistance (George et al., 2015; Rudin et al., 2012).
RESULTS
Modeling acquired resistance in vivo
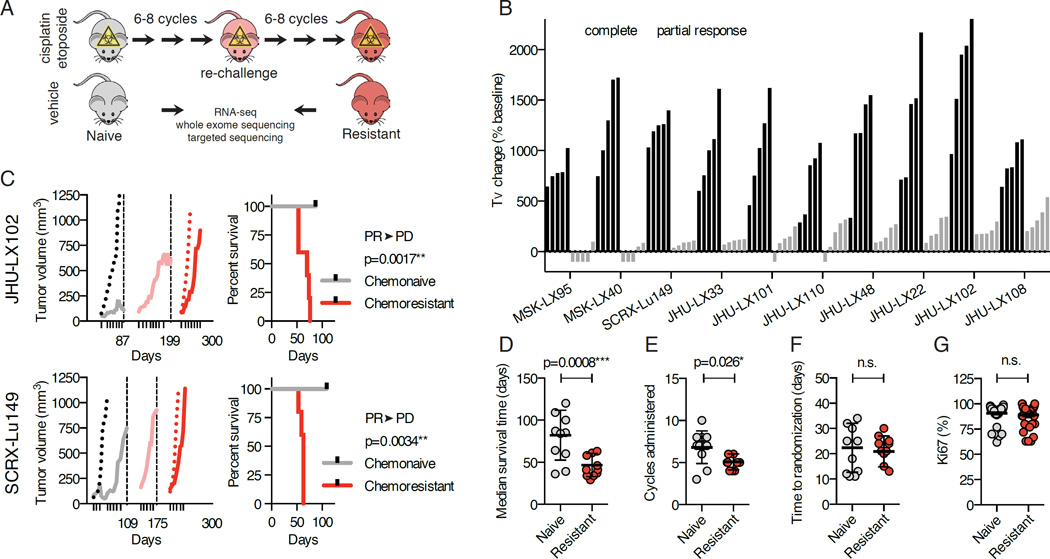
Patients with SCLC are typically treated with a regimen of up to 6 cycles of chemotherapy, each cycle consisting of cisplatin on day 1 and etoposide on days 1, 2, and 3, at near maximally tolerated doses. To study mechanisms that may govern acquired chemoresistance in vivo, we adopted an analogous approach of repeated chemotherapy cycles in tumor bearing animals in order to select populations of tumor cells that could effectively grow through chemotherapy (Figure 1A). We determined that we could safely administer 6–8 cycles of cisplatin and etoposide (C/E) on a weekly schedule in mice. We applied this approach to 10 independent PDX models of SCLC, the majority of which were derived from chemonaive patients (Table S1). We observed a spectrum of response; 2/10 models achieved complete responses, while 8/10 showed a broad index of partial responses, ranging from 65% to 95% tumor growth inhibition, without any dose-limiting toxicity as measured by animal weights (Figures 1B, S1A–S1C). To ensure that tumors progressing through multiple cycles of chemotherapy would develop intrinsic chemoresistance, these tumors were disaggregated, re-implanted into a second generation of mice, and selected again through multiple cycles of chemotherapy (Figure 1C). To assess the extent of acquired chemoresistance, progressing tumors re-implanted in a third generation of mice were again randomized to chemotherapy or vehicle control arms. Comparing the chemonaive to the chemoresistant state across all models, we observed a significant (p=0.0008) difference in the median survival time, defined as time to reach a volumetric endpoint of 1,000 mm3 (Figure 1D). Further, we observed a clear difference between the total cycles of C/E administered to naive versus resistant models (p=0.026, Figure 1E). This was not due to a change in the time to randomization of these models (Figure 1F) or increased mitotic index as measured by Ki67 staining (Figure 1G), but rather it reflects the ability of the chemoresistant derivative tumors to grow through the selective pressure (Figures 1C and S1).
Figure 1. Modeling acquired resistance C/E in SCLC.
(A) Model generation and workflow. Tumor-bearing animals were administered weekly cycles of cisplatin/etoposide (C/E) or vehicle through three sequential engraftments to generate chemoresistant models.
(B) Responses to C/E across 10 SCLC PDX models. Change in tumor volume (Tv change) compared to volumes at treatment initiation (baseline). Data shown as individual bars for vehicle (black) and C/E-treated (grey) at vehicle volumetric endpoint; n=5 per group.
(C) Tumor growth kinetics of representative models JHU-LX102 and SCRX-Lu149. Average tumor volumes during the initial C/E challenge, vehicle (black dotted line) and treated (grey solid line). X-axis ticks indicate day 1 of weekly C/E cycle. Dashed vertical lines and x-axis days indicate time on study treated tumors were collected and re-engrafted into secondary treated cohorts (pink solid line) and then tertiary (red; vehicle-dotted, C/E-solid line) cohorts. Survival analysis of chemonaive and chemoresistant cohorts performed using a log-rank (Mantle-Cox) test. Chemosensitivity conversion status indicated above p value; PR=partial response, PD=progressive disease.
(D) Median survival time for chemonaive and chemoresistant pairs. Median survival times were calculated from survival curves of treated (naive-grey; resistant-red) cohorts. P value is for paired t-test; n=10 per group (1 data point per model). Median ± SD.
(E) Cycles of C/E administered to naive and resistant cohorts. P value for paired t-test; n=10 per group. Mean ± SD.
(F) Time to randomization in naive and resistant cohorts. Paired t-test; n.s., nonsignificant; n=10 per group. Mean ± SD.
(G) Ki67 IHC positive cells in tumors from naive and resistant cohorts. Three independent tumor cores were evaluated per model; n=30 per group. Paired t-test; n.s., non-significant.
See also Figure S1 and Table S1.
Acquired chemoresistance is not associated with emergence of recurrent mutations
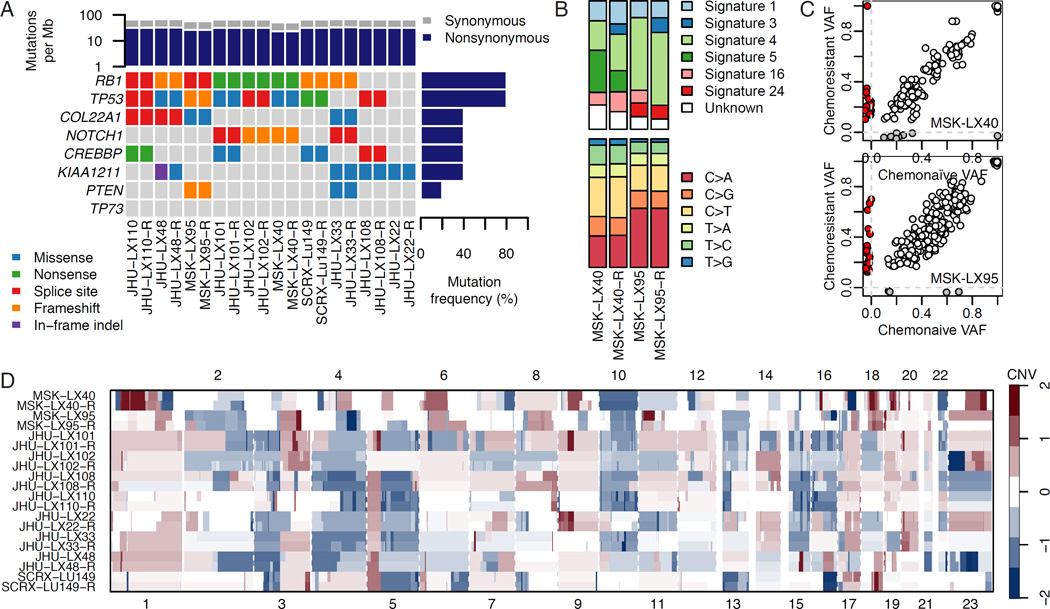
We hypothesized that the development of acquired resistance to chemotherapy in these models could be caused by changes in their genetic landscapes. To investigate this, we performed whole exome sequencing in the 10 paired models. We first confirmed by examining more than 1,000 common single nucleotide polymorphisms (SNPs) that the resistant models were derived from the parental models and did not arise from cross-contamination with other cell lines, PDX, or spontaneous murine malignancies (Figure S2A). In all cases, >90% of bases could be called based on a read depth of ≥15 (Figure S2B). We observed the expected pattern of genetic alterations consistent with SCLC, including frequent alterations in TP53 and RB1 (Figure 2A). Importantly, in each case the key genetic alterations identified in the chemonaive model were maintained through acquisition of chemoresistance. We next sought to determine to what extent the total mutational burden was shared between chemonaive and chemoresistant models. A focused analysis on MSK-LX40 and MSK-LX95, two models with matched normal DNA, revealed the expected mutational pattern enriched in C>A transversions, consistent with tobacco smoke induced mutational signature (Alexandrov et al., 2013) (Figure 2B). The mutational signatures of these tumors were relatively stable; however, both models acquired a minor component of mutational Signature 3, which has been associated with impaired double-strand break-repair by homologous recombination (Rosenthal et al., 2016). A majority of called mutations were shared, while only a minority of mutations was private to either the chemonaive or chemoresistant setting (Figure 2C). The number of private mutations was greater in the chemoresistant setting, suggesting that additional mutations were acquired during treatment. However, we were not able to identify recurrent acquired mutations across independent tumor models; no putative causal mutations of acquired resistance were found (Figure S2C), leading us to conclude that private mutations in the chemoresistant setting are passengers, not direct drivers of chemoresistance. Consistent with apparent mutational stability, we found copy number alterations to be concordant between chemonaive and chemoresistant models and failed to identify any significant focal copy number alterations or evidence of treatment induced genome doubling events that could be indicative of chromosomal instability (Figure 2D). Without clear evidence of a genetic basis for the chemoresistance phenotype, we focused subsequent analyses on potential epigenetic mechanisms of acquired chemoresistance.
Figure 2. Genomic characterization of paired chemonaive and chemoresistant SCLC PDX models.
(A) Mutational analysis of paired naive and resistant PDX models by whole exome sequencing. Shown are identifiable mutations by class among the most frequently mutated genes in primary SCLC (George et al., 2015).
(B) Analysis of mutational profiles and their associated COSMIC mutational signatures in two paired chemonaive and chemoresistant (-R) PDX models.
(C) Private versus shared mutations between paired chemonaive and chemoresistant PDX models. Colors indicate mutations variant allele frequencies (VAF) called as unique to the naive (grey) or resistant (red) tumor, with shared mutations in white.
(D) Copy number variation (CNV) in somatic chromosomes for paired chemonaive and chemoresistant models.
See also Figure S2.
SLFN11 suppression and TWIST1 induction characterize distinct subsets of chemoresistant disease
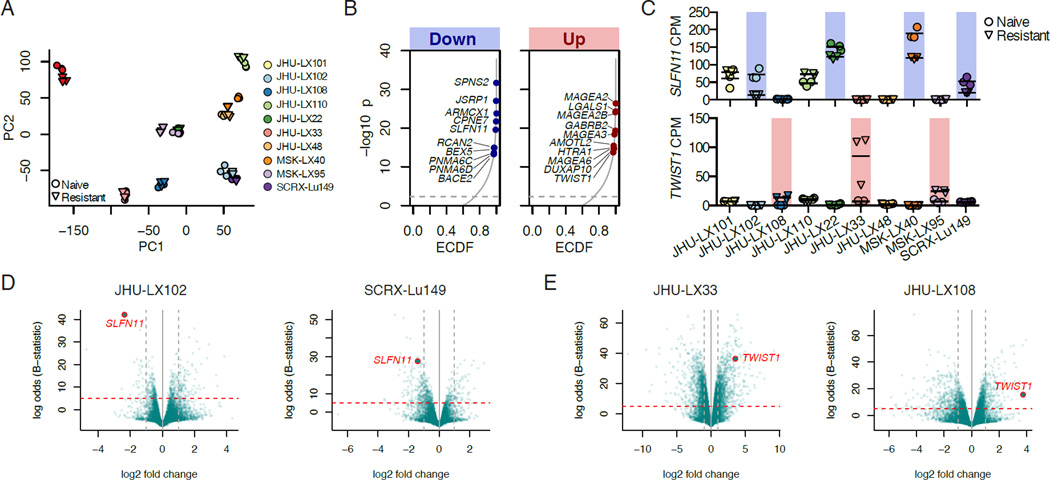
We hypothesized that there could be epigenetically driven recurrent changes in gene expression in the setting of acquired chemoresistance. To pursue this hypothesis, we performed RNA-sequencing (RNA-seq) on each of the 10 paired models. Principal component analysis suggested that gene expression patterns were remarkably consistent between chemonaive and chemoresistant settings and could easily discriminate between PDX models (Figures 3A and S3A). Due to the high degree of similarity between paired models, we hypothesized that changes in the expression of a minority of genes, as opposed to broad transcriptional changes, could drive chemoresistance. To identify recurrently differentially expressed genes, we generated a meta p value for each gene based on the degree of differential expression in each of the individual models (Figure 3B). Schlafen family member 11 (SLFN11), a gene that we and others have reported as being critical to sensitivity to DNA damaging agents (Barretina et al., 2012; Lok et al., 2016; Sousa et al., 2015; Stewart et al., 2014; Tang et al., 2015; Zoppoli et al., 2012), was among the most significantly downregulated genes. Cancer-testis antigens, a family of genes that are highly sensitive to epigenetic perturbation (De Smet et al., 1999), were significantly upregulated, as well as Twist family bHLH transcription factor 1 (TWIST1), a gene previously described to play an important role in acquired resistance to a variety of agents (Fischer et al., 2015; Zheng et al., 2015). In addition to its role in therapeutic resistance, TWIST1 is a mediator of epithelial-mesenchymal transition (EMT), metastasis, and stemness (Beck et al., 2015; Schmidt et al., 2015; Yang et al., 2004; Yang et al., 2012). The potential mechanisms identified are mutually exclusive and together are represented in 7 of 10 chemoresistance models (Figures 3C-3E).
Figure 3. Paired RNA-seq identifies conserved changes in SLFN11 and TWIST1.
(A) Principal Component (PC) Analysis of RNA-seq data from chemonaive (circles) and chemoresistant (downward triangles) replicate samples from 10 paired models.
(B) Empirical cumulative distribution function (ECDF) of combined p values of differentially down or upregulated genes. The top ten significantly downregulated (left) or upregulated (right) genes that occur in at least 3 of 10 models with a fold change >1.5 are indicated.
(C) Individual gene expression changes in SLFN11 (top panel) and TWIST1 (bottom panel) counts per million (CPM) by RNA-seq. Models with downregulated SLFN11 and upregulated TWIST1 are indicated by blue and red backlighting, respectively. Individual data points per tumor shown with a horizontal line for the mean.
(D) Volcano plots demonstrating downregulation of SLFN11 in the chemoresistant setting. Horizontal lines indicate a Beta (B)-statistic cutoff of 5; vertical lines indicate a fold change cutoff of 2.
(E) Volcano plots demonstrating upregulation of TWIST1 in the resistant setting. Horizontal lines indicate a Best (B)-statistic cutoff of 5; vertical lines indicate a fold change cutoff of 2.
See also Figure S3.
We extended these results by modeling acquired resistance to C/E in murine models of SCLC (mSCLC), including two cell lines (Rb1/Trp53 null, DKO and Rb1/Rbl2/Trp53 null, TKO) previously generated by others (Park et al., 2011) and one chemonaive allograft directly isolated from a tumor and passaged exclusively in vivo (Rb1/Rbl2/Trp53 null, TKO-A). Robust in vitro acquired resistance to etoposide was associated with a change in phenotype, converting from a suspension, spheroid culture to an exclusively adherent culture (Figure S3B). Using the same schedule of C/E to generate chemoresistant PDXs, we generated a chemoresistant allograft (TKO-AR). Again, the schedule of C/E was well-tolerated based on animals weights and provided significant tumor growth control in the naive, but not resistant allograft (p=0.0026, Figure S3C). Principal component and differential gene expression analysis suggested EMT-like changes in all three models tested, with the first principal component strongly separating parental and resistant versions of the models (Figures S3D–S3F).
Given the upregulation of TWIST1/Twist1 in both human and mouse models of SCLC upon acquired resistance, we assessed whether direct gain or loss could affect chemosensitivity. We infected the parental and resistant versions of the allograft ex vivo with lentiviruses expressing doxycycline-inducible murine Twist1 constructs and generated stable cell lines in culture. Notably, conditional gain of WT or K145E DNA-binding mutant Twist1 (Maia et al., 2012) did not robustly change the sensitivity to etoposide, in contrast to the shift we observed ex vivo between naive and resistant allograft lines (IC50 naive TKO-A ∼0.15 µM, versus IC50 resistant TKO-AR ∼3.0 µM; Figure S3G). Moreover, conditional suppression of Twist1/TWIST1 by shRNA could neither rescue chemosensitivity in mouse or TWIST1HIGH human SCLC cell lines nor influence other features of EMT observed in the resistant mSCLC cell lines, such as downregulation of E-cadherin (Figures S3G–S3I). Taken together, these results suggest that, while increases in TWIST1 may be associated with acquired resistance to chemotherapy in SCLC, this gene does not directly promote the acquired resistance observed in our models. However, we cannot rule out the possibility an event upstream of TWIST1 expression may play a more direct role in acquired chemoresistance.
SLFN11 expression is decreased in cell lines and clinical samples from previously treated patients
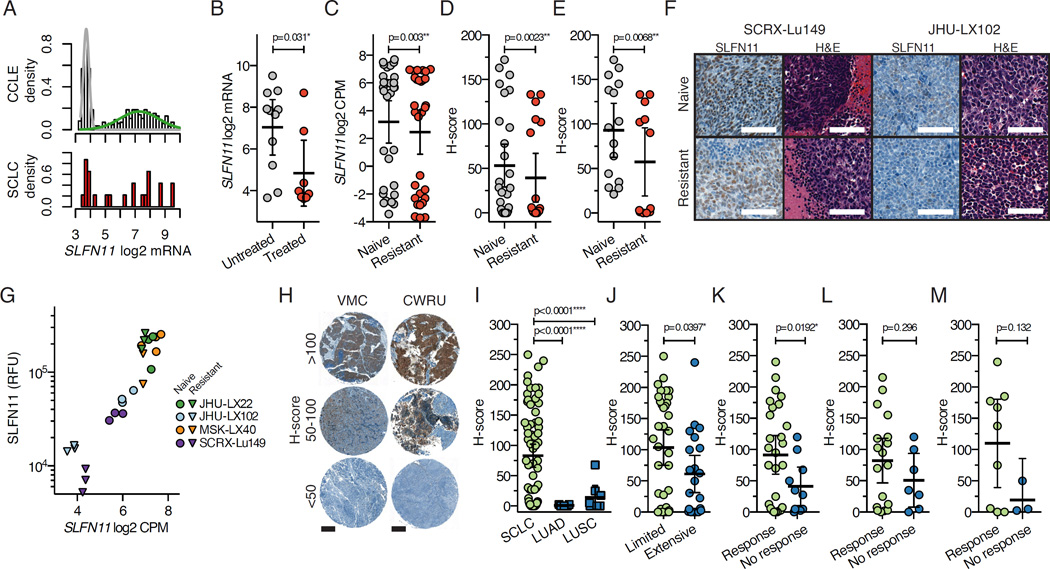
We then interrogated the role of SLFN11 in the context of acquired chemoresistance, as it had been previously implicated as a factor regulating DNA damage repair (Mu et al., 2016) and was shown to correlate with responses to DNA damaging agents in vitro (Barretina et al., 2012; Sousa et al., 2015) and in vivo (Tang et al., 2015). SLFN11 is bi-modally expressed when examined across cancer cell lines within the Cancer Cell Line Encyclopedia (CCLE), as well as within SCLC in both primary tumor (Lok et al., 2016) and cell lines (Figure 4A). Many SCLC cell lines have been established and annotated with regards to their primary source (Carney et al., 1985). Cell lines generated from treated patients have lower levels of SLFN11 expression relative to lines generated from untreated patients (p=0.031; Figure 4B), which also held true when comparing chemonaive to chemoresistant PDX models (p=0.003; Figure 4C). We observed similar results when examining SLFN11 protein expression by IHC using an H-score as the comparative metric, including either all models or only models with detectable SLFN11 at baseline (Figures 4D and 4E). In 2 models where chemoresistance was associated with a substantial decrease in SLFN11 (Figure 4F), we confirmed quantitative decreases in SLFN11 expression at both the transcript level by RNA-seq and protein level by quantitative Western blotting (Figures 4G and S4A–S4C). Interestingly, an endpoint analysis of vehicle and C/E treatment groups from chemonaive and chemoresistant cohorts in one model revealed a significant decrease in SLFN11 in progressing tumors (p=0.029; Figure S4D).
Figure 4. SLFN11 is downregulated at the transcript and protein level in SCLC following exposure to chemotherapy.
(A) Density histograms of SLFN11 gene expression across the Cancer Cell Line Encyclopedia (CCLE) dataset and SCLC cell lines within CCLE. Cell lines in CCLE are shown as density distributions of those that express little to no SLFN11 (grey) versus the broad distribution of SLFN11 levels in other lines (green). SCLC cell lines are shown in red (lower panel).
(B) Comparison of SLFN11 expression in SCLC cell lines derived from untreated or treated patients; p=0.031, Fisher’s exact test. Treatment history available in (Polley et al., 2016).
(C) SLFN11 gene expression in all chemonaive and chemoresistant PDX models. Three replicates per model, per condition are plotted; n=30 per group. Mean ± SD. P value for paired Student’s t-test.
(D) SLFN11 IHC scoring metric (H-score) compared between all chemonaive and chemoresistant PDX models. Three independent core samples per model, per condition; n=30 per group. Mean ± SD. P value from Mann-Whitney test.
(E) H-score comparison including only models with detectable SLFN11 by IHC (IHC positivity ≥1+ and one core with H-score >20; 5/10 PDX models included). P value from Mann-Whitney test.
(F) Representative SLFN11 IHC and H&E sections from two models (SCRX-Lu149 and JHU-LX102) in the chemonaive and chemoresistant cohorts. Scale bars 100 µm.
(G) Concordance between SLFN11 gene expression by RNA-seq and SLFN11 protein expression by quantitative Western blot. Relative fluorescence units (RFU).
(H) Representative IHC staining for SLFN11 in clinically-annotated SCLC TMAs. VMC=Vanderbilt Medical Center, CWRU=Case Western Reserve University. Scale bars (200 µm) for entire 1 mm cores.
(I) SLFN11 expression by IHC in SCLC, lung adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC). 60/215 untreated VMC and 12/22 treated CWRU cores were evaluable for comparison. 7 LUAD and 7 LUSC samples were evaluated. P values from unpaired two-tailed t-tests.
(J) Comparison of SLFN11 H-score by stage of patient from pooled analysis of VMC and CWRU TMAs. P values from unpaired t-tests
(K) Comparison of SLFN11 H-score by response of patients from both cohorts. Responses include complete or partial response, where no response includes progressive or stable disease. P values from unpaired t-tests
(L) Comparison of SLFN11 H-score by response of patients from the untreated cohort. P values from unpaired t-tests
(M) Comparison of SLFN11 H-score by response of patients from the previously treated cohort. P values from unpaired t-tests
See also Figure S4.
To assess whether SLFN11 expression was correlated with clinical response in patients with SCLC, SLFN11 immunohistochemistry was performed on clinically annotated tumor microarrays from untreated (Vanderbilt Medical Center; VMC) and previously treated (Case Western Reserve University; CWRU) SCLC patients, with H-scores for each intact core determined by a pathologist blinded to sample identity (Figure 4H). Immunostaining for SLFN11 was low to nearly absent in lung squamous cell carcinoma and adenocarcinoma in contrast to SCLC (Figure 4I). SLFN11 expression was modestly associated with stage of disease when viewed in aggregate, with SLFN11 greater in limited stage versus extensive stage patients (p=0.0397; Figure 4J). Consistent with a role in determining chemosensitivity, among all treated patients, SLFN11 expression was higher in tumors from patients who responded to therapy versus those who did not (p=0.0192; Figure 4K). When evaluating SLFN11 as a pretreatment predictor of response in untreated patients (Figure 4L) and as a post-treatment correlate of response (Figure 4M), we found a trend for greater SLFN11 expression in patients categorized as treatment responsive; however, statistical significance was not reached. Notably, among untreated patients, 83% (10/12) of patients with a SLFN11 H-score >75 responded to treatment (Figure 4L); using the same threshold among previously treated patients, all 6 patients with an H-score >75 responded to treatment (Figure 4M). However, applying a dichotomized H-score of 68.8 using Youden’s index (Youden, 1950) did not show a statistically significant difference in overall survival (p=0.884, Log-rank test; Figure S4E). Taken together, these data suggest that high SLFN11 expression in SCLC confers greater sensitivity to chemotherapy but does not confer an overall survival benefit in this clinical cohort.
EZH2 inhibition restores SLFN11 expression and chemosensitivity in vitro
A recent report suggested that SLFN11 expression is partly regulated by an epigenetic silencing mechanism reversed by broad DNA methylation inhibitors such as 5-Azacitidine (5-Aza) (Nogales et al., 2016). We chose to examine whether EZH2 (Enhancer of zeste homology 2) could have a role in silencing SLFN11 in SCLC for several reasons: 1) there is evidence for an EZH2 binding site upstream of the first exon of SLFN11 from cell line data reported by the ENCODE project (Gerstein et al., 2012; Wang et al., 2013), 2) we have shown EZH2 expression levels to be higher in SCLC than in any of the tumor types included in TCGA (Poirier et al., 2015), and 3) others have shown that PRC2 target gene repression correlates with shorter survival in primary SCLC (Sato et al., 2013). Overall, these lines of evidence prompted us to examine whether chemical inhibitors targeting DNA or histone methyltransferase enzymes could play a role in regulating SLFN11 expression.
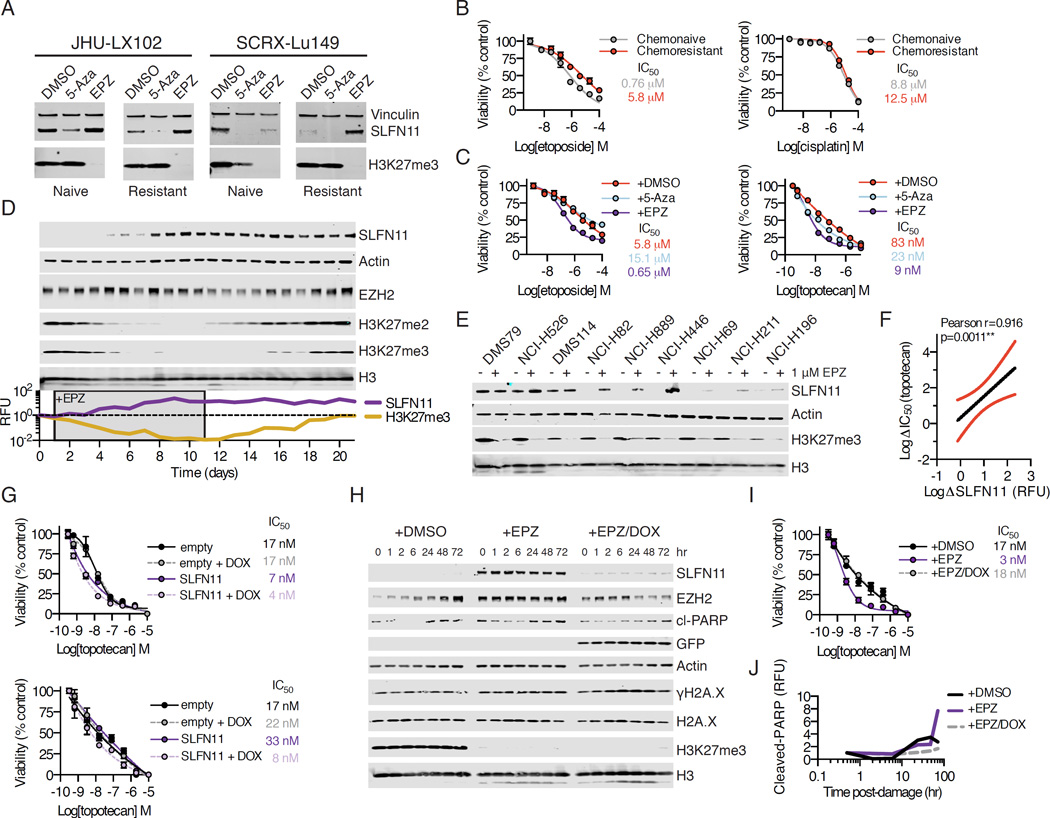
Using short term ex vivo culture, we exposed naive and resistant versions of select PDX models to either 5-Aza or the EZH2 inhibitor EPZ011989 (abbreviated EPZ). After 7 days of continuous daily exposure, we noted striking increases in SLFN11 expression in the EPZ-treated cells versus DMSO or 5-Aza-treated cells, suggesting that SLFN11 expression may be regulated at the level of histone methylation, rather than DNA methylation, in SCLC (Figure 5A). Moreover, treatment with EPZ, but not 5-Aza, effectively restored ex vivo etoposide sensitivity of the chemoresistant model to that of the chemonaive model, where ex vivo resistance was most pronounced for topoisomerase inhibitors and less for cisplatin (Figures 5B and 5C). We next interrogated the kinetics of SLFN11 re-expression in the SCLC cell line NCI-H82, which was derived from a previously treated patient and shows minimal expression of SLFN11 protein. We monitored the re-expression of SLFN11 over a 21 day period in suspension culture, during which time cells were treated daily with 1 µM EPZ for 10 days and then cultured in fresh media without compound for an additional 10 days. We noted striking re-expression of SLFN11 after ∼7–10 days of drug exposure, with concomitant suppression of H3K27me2/3, consistent with on-target inhibition of EZH2 (Figure 5D). Re-expression of SLFN11 was dose-dependent and stable under acute DNA damage (Figure S5A). This re-expression was sustained even as global H3K27me2/3 levels returned to baseline levels during a 10-day washout period (Figure 5D). In 7-day treatment assays, we noted a dramatic increase in SLFN11 protein expression in cell lines with little to no detectable SLFN11 (Figure 5E). This was in contrast to cell lines with high de novo expression of SLFN11, in which no further increase above baseline was observed. The extent to which SLFN11 protein levels were increased by EPZ was strongly correlated to an increase in topotecan sensitivity across all cell lines tested (Pearson r=0.916; Figures 5F and S5B). SLFN11 protein re-expression by EPZ was suppressible with shRNAs targeting SLFN11 (Figure S5C and S5D). SLFN11 expression could also be induced with GSK126, a chemically distinct EZH2 inhibitor, although EPZ showed greater capacity for SLFN11 re-expression when compared to GSK126, which had comparatively less robust H3K27me3 suppression in our experience (McCabe et al., 2012).
Figure 5. Chemical EZH2 inhibition rescues SLFN11 expression and sensitizes SCLC to DNA damage.
(A) Chemonaive and chemoresistant PDX models JHU-LX102 and SCRX-Lu149 cultured ex vivo and treated for 7 days with 1 µM of 5-azacitidine (5-Aza) or EPZ daily and then assayed by Western blot.
(B) Viability of chemonaive and chemoresistant JHU-LX102 upon treatment with etoposide and cisplatin for 72 hr. n=3/data point; mean viability ± SEM.
(C) Viability for etoposide (left) and topotecan (right) treated chemoresistant JHU-LX102 with or without co-treatment with 5-Aza or EPZ. Cells were treated as in (A) for 7 days before re-plating and exposing to either etoposide or topotecan for 72 hr as above. n=3/data point; mean viability ± SEM.
(D) NCI-H82 cells treated daily for 10 days with 1 µM EPZ then washed and released into fresh media for 10 days. Media was changed every other day. Samples collected every day for 21 days and assayed by Western blot. Relative fluorescence units (RFU) for SLFN11 and H3K27me3 shown.
(E) SCLC cell lines cultured with 1 µM EPZ for 7 days, refreshing compound every day and media every other day prior to collection of samples and Western blot.
(F) Correlation of SLFN11 expression upon chemical EZH2 inhibition to sensitivity to topotecan. Pearson correlation r=0.916. Red lines represent 95% confidence intervals.
(G) Conditional re-expression of SLFN11 and topotecan sensitivity in NCI-H82 (top) and NCI-H446 (bottom). Cells were transduced and expression was induced with doxycycline (DOX) at 1 µg/mL added every other day for 4 days before re-plating for 72 hr with topotecan. n=3/data point; mean viability ± SEM.
(H) NCI-H82 cells were transduced with DOX-inducible shRNA against SLFN11 and exposed to DMSO, 1 µM EPZ or EPZ/DOX for 7 days before exposure to 1 µM topotecan for 1 hr. Cells then released into fresh media and collected at time points for Western blot. cl=cleaved
(I) Effect of topotecan on NCI-H82 cells expressing shRNA against SLFN11 after 7 days of treatment with DMSO, 1 µM EPZ or 1 µM EPZ and EPZ/DOX. Following treatment, cells were exposed to the indicated amounts of topotecan for 72 hr. n=3/data point; mean viability ± SEM
(J) Cleaved-PARP relative fluorescence units (RFU) from (H). RFU normalized to undamaged controls following 7 days of treatment (0 hr for DMSO, EPZ, or EPZ/DOX). See also Figure S5.
A shift in global repressive histone methylation may have pleiotropic effects on gene expression that manifest in a cell line specific manner (Jadhav et al., 2016). To directly address whether re-expression of SLFN11 was sufficient to sensitize SCLC cell lines with low levels of SLFN11 to DNA damaging agents, such as topotecan, we used a doxycycline-inducible expression vector to express SLFN11 in NCI-H82 and NCI-H446. Both of these cell lines were derived from previously treated patients and are highly resistant to etoposide. Exogenous expression of SLFN11 was capable of sensitizing these cell lines to topotecan relative to non-induced or empty vector controls (Figure 5G), although this effect was less pronounced than that of the EPZ-mediated sensitization (Figure 5C). Consistent with this observation, shRNA-mediated suppression of SLFN11 in NCI-H526, a cell line with high baseline SLFN11 expression, modestly decreased sensitivity of this line to topotecan, although we note that complete suppression of SLFN11 was not possible even with a potent shRNA (Figure S5E). Remarkably, while EPZ-treatment shifted the sensitivity of NCI-H82 cells to topotecan by approximately one log, this was reversed by concurrent shRNA suppression of SLFN11 (Figures 5H and 5I). While we did not note changes in markers of acute DNA damage response (γH2A.X), we found that EPZ-treated cells showed increased markers of programmed cell death (cleaved-PARP) upon acute topotecan exposure that were reduced when suppressing SLFN11 by shRNA (Figure 5J). Taken together, these data strongly implicate SLFN11 as a determining factor in sensitivity to DNA damaging agents in SCLC.
EZH2 silences SLFN11 expression in SCLC
To further define the mechanistic relationship between EZH2 and SLFN11 expression, we sought to explore the effects of EZH2 inhibition on local chromatin structure in the vicinity of the SLFN11 locus. We examined the efficacy of EPZ in 4 PDX models at two dose schedules over a treatment period of 3 weeks (Table S2). This schedule slowed tumor growth modestly in 3 of 4 models tested (Figure S6A), and EPZ was well tolerated in animals based on weight measurements when administered at 250 mg/kg orally, twice a day (PO bid) (Figure S6B). This analysis included a chemorefractory model, JHU-LX44, that was omitted from the analysis of chemosensitive models (Figure S6C). Our ability to observe single agent efficacy in vivo may have been limited by the duration we were able to treat animals relative to the time required to remodel the epigenome, which is thought to require weeks to months (LaFave et al., 2015). We further assessed the contribution of time-dependence for efficacy in vivo by re-engrafting equivalent viable cell numbers from tumors that had previously been treated for 21 days with or without EPZ and then re-treating tumors with vehicle or a secondary round of EPZ as soon as tumors became palpable (Figure S6D). These results suggest that prolonged exposure of SCLC tumors to EZH2 inhibition may have a greater effect on decreasing proliferative capacity; however, continuous treatment led to weight loss that approached protocol limits.
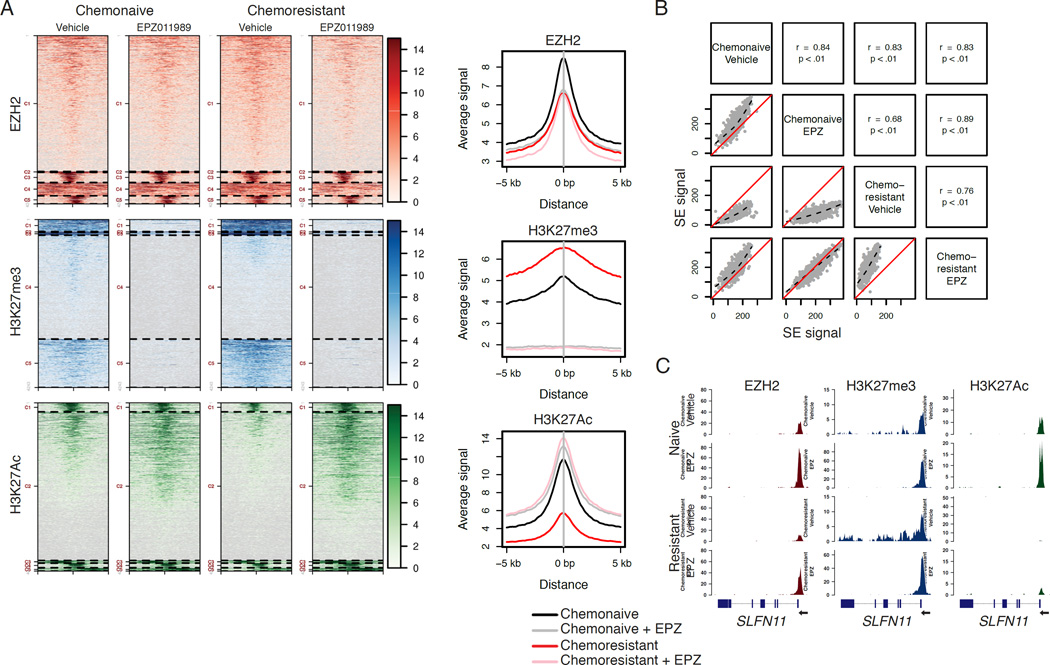
To demonstrate the ability of EPZ to modulate the SCLC chemoresistant epigenome, we performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) from flash frozen tissue collected from chemonaive or chemoresistant SCRX-Lu149 tumors treated for 21 days with vehicle or EPZ. ChIP-seq was performed on three targets: EZH2, H3K27me3, and H3K27Ac (Figure 6A). While total EZH2 levels were not significantly altered in any condition, we observed increased global H3K27me3 in the chemoresistant setting that could be abolished by EPZ, as well as decreased global H3K27Ac in the chemoresistant setting that could be rescued by EPZ. Global H3K27Ac levels were increased by EPZ in both the chemonaive and chemoresistant settings with concomitant loss of H3K27me3. The global reduction of H3K27Ac in the chemoresistant setting prompted us to explore differential intensities within genomic regions termed super-enhancers (Loven et al., 2013; Pott and Lieb, 2015), thought to be critical for SCLC proliferation (Christensen et al., 2014; Lin et al., 2012). We observed differential signal intensities in such regions, consistent with global loss of H3K27Ac in the chemoresistant setting, which could be restored with EPZ. However, selective loss or rescue of discrete super-enhancer regions was not observed (Figure 6B).
Figure 6. Epigenetic changes acquired in chemoresistance are reversible with chemical EZH2 inhibition in vivo.
(A) Cluster analysis of ChIP-seq results from four SCRX-Lu149 tumor groups: chemonaive or chemoresistant treated with either vehicle or 250 mg/kg EPZ PO bid for 21 days. Chromatin was pooled from three independent tumor samples per arm prior to IP.
(B) Scatter plots of super-enhancer signal intensity. Spearman’s rank correlation coefficient and p value are calculated for each comparison. Dotted black lines are locally-weighted polynomial regressions. Solid red lines indicate unity.
(C) SLFN11 ChIP-seq gene tracks for the indicated sample treatment arms. Y-axes limits are scaled to drosophila spike-in for each sample for peak height comparisons. The SLFN11 gene model is oriented right-to-left along the x-axis as indicated.
See also Figure S6 and Table S2.
Focusing specifically on the SLFN11 locus, ChIP-seq data demonstrate focally concentrated EZH2 and H3K27me3 in the immediate vicinity of the transcription start site (TSS) in vehicle-treated tumors, with spreading of H3K27me3 across the gene body in the context of acquired chemoresistance (Figure 6C). Coordinate with the increase in H3K27me3 across the gene body in the chemoresistant versions, there is almost complete loss of H3K27Ac, a mark associated with transcriptionally active chromatin, at the TSS of SLFN11, which we confirmed by ChIP-qPCR (p<0.0001; Figure S6E). Interestingly, EPZ treatment increased the density of EZH2 bound near the SLFN11 TSS and largely erased H3K27me3 throughout the gene body of SLFN11. Levels of H3K27me3 near the TSS remained largely unchanged. We further confirmed gene body erasure of H3K27me3 within exons 2–7 of SLFN11 by exon-by-exon ChIP-qPCR (Figure S6F). EPZ treatment also resulted in an increase in levels of H3K27Ac near the SLFN11 TSS and throughout the gene body. These data suggest that re-expression of this EZH2 target gene via chemical inhibition may also require cooperative histone modifications to promote gene expression. While we observed rescue of SLFN11 in PDX models that downregulated the gene upon acquiring resistance to C/E, expression was not rescued in models that expressed low to undetectable levels of SLFN11 at baseline (Figure S6G).
Pharmacologic EZH2 inhibition prevents emergence of chemoresistance and augments chemoresponse in vivo
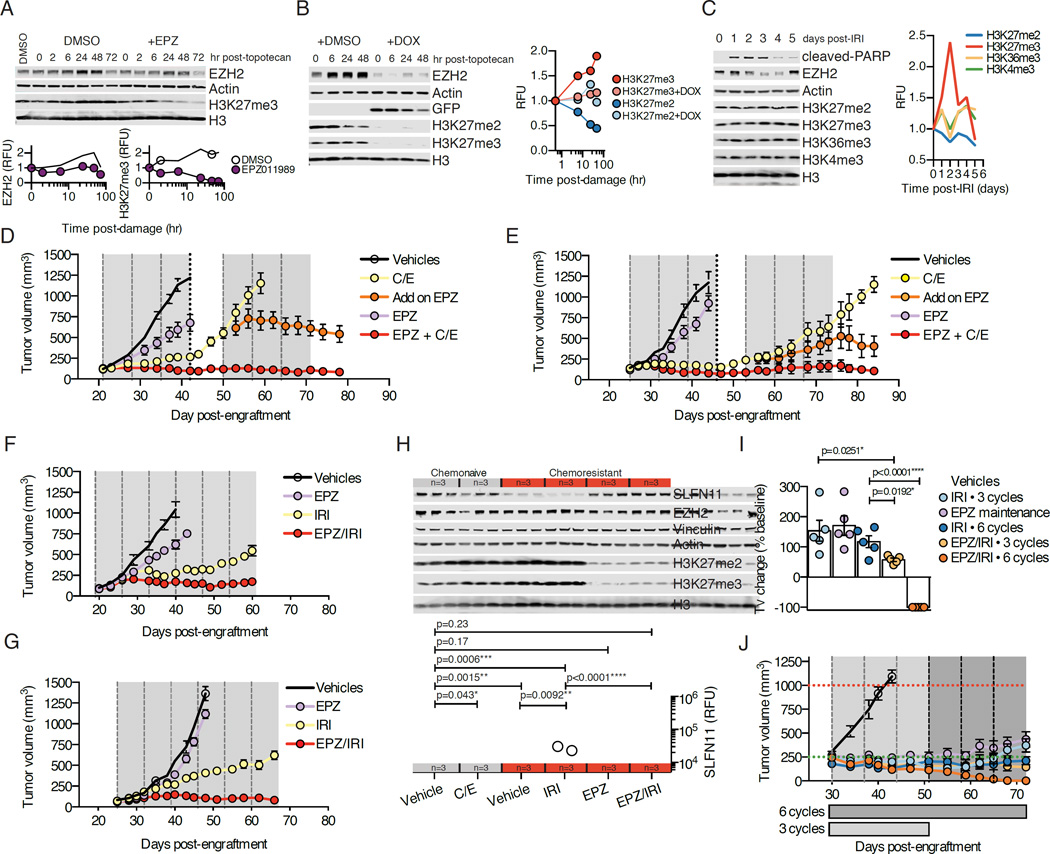
The downregulation of SLFN11 in acquired chemoresistance and its apparent regulation by EZH2 suggest a model in which EZH2 is induced by cytotoxic chemotherapy, resulting in deposition of repressive chromatin marks in the SLFN11 gene body, decreasing expression and promoting chemoresistance. To test this hypothesis and evaluate time-dependent changes in EZH2 activity, we performed DNA damage time course experiments in NCI-H446, following exposure to topotecan in the presence or absence of EPZ. Over the course of 48 hr following DNA damage, we noted a progressive increase in EZH2, as well as H3K27me3, that was suppressed by EPZ (Figure 7A). Further, we could demonstrate that the effect on H3K27me3 was a function of EZH2, as shRNA suppression of EZH2 during a similar DNA damage time course abolished any increase in H3K27me3 and the accompanying loss of H3K27me2 (Figure 7B). These data support a global methylation event where di-methylated H3K27 is converted to the tri-methylated state. The effect of DNA damage increasing global H3K27me3 levels was not cell line specific (Figure S7A), demonstrated dose-dependence (Figure S7B), and was more pronounced for topotecan when compared to either cisplatin or etoposide on an equimolar basis (Figure S7C). These shifts in EZH2 activity and resultant H3K27me3 were also observed in vivo, using a single dose-chase approach in tumor-bearing animals treated with IRI and quantitatively evaluating time-dependent changes in several H3 methyl marks, demonstrating specificity for the induction of H3K27me3, with contemporaneous decreases in H3K27me2 (Figure 7C).
Figure 7. EZH2 is an actionable target in combination with standard of care in SLFN11HIGH SCLC.
(A) NCI-H446 cells exposed to 1 µM topotecan for 1 hr ± 1 µM EPZ. Cells were then washed and collected at time points for Western blot. Quantified EZH2 and H3K27me3 fluorescence units (RFU) over time.
(B) NCI-H446 cells transduced with EZH2 shRNA treated ± 1 µg/mL doxycycline (DOX) for 48 hr before adding 1 µM topotecan, as in (A). On right, quantified H3K27me3 and H3K27me2 (RFU) over time.
(C) Size-matched chemonaive JHU-LX102 tumor-bearing mice (∼400mm3) treated IP with 100 mg/kg IRI and tumors collected at the indicated time points post-dose for Western blot.
(D) SLFN11HIGH chemonaive JHU-LX102 tumor volume responses. Dashed vertical lines indicate day 1 of a weekly C/E cycle. Dotted line indicates 5 of the 10 animals within C/E group now randomized to receive EPZ with 3 additional cycles of C/E. Grey areas indicate the dosing periods for EPZ (250 mg/kg PO bid). n=5 per arm (n=10 in C/E arm through 3 cycles, before adding on EPZ). Tumor volume mean ± SEM.
(E) Results for chemonaive SCRX-Lu149 treated as in (D). Tumor volume mean ± SEM
(F) Chemoresistant JHU-LX102 tumor volume responses. Dashed vertical lines indicate day 1 of a cycle. 6 weekly cycles of IRI ± EPZ or single agent EPZ. n=5 per arm. Tumor volume mean ± SEM.
(G) Results for mice engrafted with chemoresistant SCRX-Lu149 as in (F). n=5 per arm. Tumor volume mean ± SEM.
(H) Western blot analysis for SLFN11 suppression and rescue in chemonaive (grey bar; n=6) or chemoresistant (red bar; n=12) JHU-LX102 treated as labeled and collected at day 21. Tumors size matched (∼400 mm3) per cohort. SLFN11 (RFU) quantified below; n=3/arm. Three cycles of each indicated treatment arm were administered before collecting tumors. values from paired t-tests.
(I) Change in tumor volume (Tv) from volumes at treatment initiation (baseline) in chemonaive JHU-LX102 treated with 3 or 6 weekly cycles of 100 mg/kg IRI and/or EPZ. Comparison at 6 weeks post-treatment (day 72). n=5/arm. Dots indicate Tv change of individual tumors and bars indicate average ± SEM. P values from paired t-tests.
(J) Chemonaive JHU-LX102 response (n=5 per arm) to 3 or 6 weekly cycles of 100 mg/kg IRI and/or EPZ.. Horizontal dashed lines indicate starting (green) and ending (red) tumor volumes. Group duration along x-axis. Vertical dashed lines indicate a weekly cycle of IRI. Tumor volume mean ± SEM.
These data support the hypothesis that EZH2 activity is an important determinant of acquired chemotherapy resistance in SCLC. To examine the efficacy of EPZ with cytotoxic regimens in vivo, we chose two clinically translatable scenarios: 1) combining EPZ with standard of care in the first line setting for SLFN11 expressing (SLFN11HIGH) chemonaive models and, 2) combining EPZ with a standard second line agent in the chemoresistant setting, in which the silencing of SLFN11 has been established. Addition of EPZ to 6 cycles of C/E strongly enhanced disease control in the chemonaive setting in both JHU-LX102 and SCRX-Lu149 relative to either EPZ or C/E alone without increasing animal weight loss (Figures 7D, 7E, S7D and S7E). To examine the benefit of EZH2 inhibition in tumors actively progressing on C/E, after three cycles we randomized 5 of 10 animals in the C/E arm to receive EPZ in combination with three additional cycles of C/E. Remarkably, the addition of EPZ potently induced tumor regression relative to C/E alone during the remaining 3 cycles, supporting a role for EZH2 in promoting chemoresistance (Figures 7D and 7E).
We next assessed the efficacy of IRI, a topoisomerase I poison administered in the setting of relapsed SCLC, with or without EPZ, in chemoresistant JHU-LX102 and SCRX-Lu149. Importantly, we observed strong cross-resistance to IRI, but not ionizing radiation, in the chemoresistant setting, further supporting the mechanism of acquired resistance operant in these models had specificity to the selection agents used (cisplatin and/or etoposide) (Figures S7F and S7G). The addition of EPZ to IRI in the chemoresistant setting resulted in potent combinatorial activity that could control disease to a greater extent than either agent alone through 6 weekly cycles of treatment without increasing animal weight loss (Figures 7F, 7G, S7H and S7I).
To further assess active repression and de-repression of SLFN11 in vivo, we used chemonaive and chemoresistant versions of JHU-LX102, randomizing size-matched tumors (∼400 mm3) to 3 cycles of chemotherapy ± EPZ. Strikingly, we noted quantitative suppression of SLFN11 in tumors after as few as 3 cycles of C/E in the chemonaive setting, with further suppression after 3 cycles of IRI in the chemoresistant setting (Figure 7H). Critically, EZH2 inhibition could rescue SLFN11 to baseline levels in the chemoresistant setting, even in the presence of concurrent chemotherapy (chemonaive vehicle vs. chemoresistant EPZ/IRI).
In combining EPZ with IRI in the chemonaive setting, we could completely ablate hind flank tumors through 6 cycles of combination (complete response in 5/5 animals; Figures 7I and 7J). Moreover, the activity of the combination was greater, even when administered for a shorter duration of time (3 cycles), than the equivalent schedule of IRI administered for 6 cycles. However, we did not observe additional benefit when initiating adjuvant EPZ as a single agent at the point of maximal consolidative effect of single agent IRI (post cycle 3), as this was inferior to 3 additional cycles of IRI at the point of comparison. When comparing the responses of a SLFN11HIGH to a SLFN11LOW model, both chemonaive models displayed sensitivity to IRI, but only in the SLFN11HIGH model is the combined activity of EPZ and IRI strikingly different (Figures S7J and S7K). Moreover, while all SLFN11HIGH JHU-LX102 tumors in the combination arm were ablated after 6 cycles, tumors eventually recurred yet still maintained comparable sensitivity to the combination (Figure S7K). Recurrent tumors appeared similar to untreated tumors, not displaying differentiation away from their proliferative, neuroendocrine features (Figures S7L). Body weight and repeat blood chemistry monitoring during 2 weeks of treatment revealed no significant toxicity of the combination as compared to single agents (Figure S7M and Table S3). Further, gross necropsy also suggested no apparent major organ toxicities (Figure S7N). Taken together, combining a clinical stage EZH2 inhibitor with a topoisomerase I poison is well-tolerated and highly effective in SLFN11-expressing SCLC.
DISCUSSION
SCLC has been singled out by the US National Cancer Institute (NCI) as a designated “recalcitrant” cancer based on incidence rate, exceptionally high lethality, and the lack of substantial therapeutic progress made over several decades. By mimicking the clinical experience of repeated cycles of cisplatin and etoposide exposure in PDXs, we have generated a set of paired models representing initial chemosensitive and subsequent chemoresistant disease. We used these models to define two mutually exclusive mechanistic classes of acquired resistance, including an EMT shift associated with TWIST1 upregulation and epigenetically mediated suppression of SLFN11. Finally, we have identified a therapeutically tractable vulnerability in SCLC: dependence on EZH2’s activity for the development of acquired chemoresistance.
We found that consistent gene expression and epigenetic changes, not mutations, are associated with acquired chemoresistance across independent SCLC models and indeed are reflected in primary human tumor samples. In a set of 10 PDX models, TWIST1 induction was observed in three; this pathway was additionally reflected in multiple murine models of SCLC. While initial data point to TWIST1 as a biomarker rather than a driver of resistance per se, further exploration of therapeutic vulnerabilities associated with an EMT signature in this set of tumors is warranted. In 4 of the 10 PDX models, acquired resistance was associated with specific suppression of SLFN11. Most notably, our data point to SLFN11 suppression as a primary contributor to acquired chemotherapy resistance in SCLC – one that can be prevented and/or actively remodeled through targeting EZH2.
We have demonstrated here that SLFN11 suppression during selection for acquired resistance in SCLC is associated with a global increase in H3K27me3 with modest reductions in global H3K27Ac and that SLFN11 gene expression can be restored and/or maintained by pharmacological inhibition of EZH2, even in the presence of DNA damaging agents. While we have shown that SLFN11 is both necessary and sufficient for sensitivity to DNA damaging agents in SCLC, we recognize that EZH2 inhibition alters expression of many genes. We anticipate that factors beyond SLFN11 will be identified that contribute to the ability of EZH2 inhibition to restore chemosensitivity in SCLC. Defining the relative contributions of SLFN11 versus other EZH2-modified factors remains an area for future investigation.
Several potent and selective EZH2 inhibitors are now in different stages of clinical development, including phase II (Epizyme) and phase I (Constellation, GSK) trials in multiple solid tumor and hematological indications. Epizyme recently updated the phase I experience and ongoing phase II experience for their lead inhibitor tazemetostat (EPZ-6438) (Ribrag et al. and, Morschhauser et al., ASH Lymphoma 2016), which is a closely related structural homolog of the tool compound inhibitor EPZ011989 described in this paper (Fillmore et al., 2015). The safety profile of tazemetostat in 82 patients with non-Hodgkin lymphoma on the ongoing phase II study was favorable, with the most frequent treatment-related adverse events being grade 1 or 2 nausea and asthenia. Myelosuppression was observed infrequently with only 11% and 6% of patients experiencing grade ≥3 thrombocytopenia and grade ≥3 neutropenia, respectively, suggesting tazemetostat may be safely combined with cytotoxic chemotherapy in SCLC. Importantly, the work presented here suggests that targeted EZH2 inhibitors should be anticipated to have minimal single agent activity in SCLC, and that evaluation of relevant combination therapies should be considered early in clinical development of these agents in SCLC. Clinical trials testing the therapeutic strategies defined here are now being designed with these considerations in mind.
EXPERIMENTAL PROCEDURES
Patient-derived xenografts
All animal experiments were approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Animal Care and Use Committee. Primary tumors and whole blood samples collected for generation of patient-derived xenograft (PDX) models were obtained with informed consent from patients under protocols approved by MSKCC and JHMI Institutional Review Boards (IRB). Subcutaneous flank tumors were generated as previously described (Daniel et al., 2009). A list of PDX model details can be found in Table S1.
DNA/RNA sequence alignment
Raw reads were aligned to custom human/mouse hybrid references as previously described (Schneeberger et al., 2016). Briefly, hybrid indexes were generated using a FASTA file consisting of all human GRCh38 and mouse GRCm38.p3 reference contigs and in the case of RNA-seq, assisted by the human GENCODE gene set release 20 transcript model. Downstream analysis was performed with reads mapping to human reference contigs.
Statistics
Sample sizes per in vivo groups were at a minimum of 5 per condition, unless specified within the text for purposes of endpoint analysis. Power calculations and sample size estimates were not performed. Student’s t-tests, one-way ANOVA, Pearson’s correlation and log-rank tests were performed using GraphPad Prism version 6.00 for Mac. GraphPad Software, La Jolla California USA, www.graphpad.com. For in vitro / ex vivo dose-response curves, best fit IC50 values are were reported from normalized datasets using the equation, “log(inhibitor) versus response – variable slope” with an ordinary fitting method. At least three data points per dose were used, with experiments being performed at least twice. Error bars for SD or SEM are shown. Where indicated in Figures, degrees of p value significance are as follows: <0.05*, <0.005**, <0.0005***, and <0.0001****.
Supplementary Material
EZH2 drives acquired resistance to chemotherapy in small cell lung cancer (SCLC)
DNA damage induces EZH2 activity and H3K27me3 deposition genome-wide in SCLC
SLFN11 is an EZH2 target gene in SCLC
Combining an EZH2 inhibitor with standard of care controls SCLC in vivo
Significance.
Small cell lung cancer is among the most lethal human malignancies. Typical progression of this disease is characterized by a rapid shift between initial chemoresponsive and subsequent chemoresistant states. The mechanisms responsible for acquired therapeutic resistance in small cell lung cancer have not been defined. Using patient-derived tumor xenografts to closely model clinical acquired resistance, this work: (1) defines a mechanism of chemoresistance operant across multiple, independent cancers, (2) identifies an epigenetic regulator controlling the mechanism of acquired resistance, and (3) establishes a therapeutic strategy to both prevent and treat acquired resistance in vivo. These observations have immediate clinical implications, describing an approach that may lead to durable and effective treatment for patients with this disease.
Acknowledgments
We thank Ralph Garippa and members of the MSKCC RNAi core facility, past and present members of the Antitumor Assessment Core Facility for assistance with in vivo work, members of the Molecular Cytology and Pathology core facilities for assistance with TMA construction and automated staining. We acknowledge Madeleine Craske of Active Motif, Inc. for assistance with ChIP-qPCR experiments. We thank all members of the Rudin Lab for collaborative efforts.
FUNDING
This work was supported by a grant from the LUNGevity Foundation and the American Lung Association (JTP), a grant from Free to Breathe (JTP), R01 CA197936-01A1 (CMR), U54 OD020355 (EdS), the MSKCC Support Grant/Core Grant P30 CA008748, Conquer Cancer Foundation of ASCO (BHL), Radiological Society of North America (RR1634) (BHL), and NIH UL1TR00457 administered by the Clinical and Translational Science Center at Weill Cornell Medical Center and MSKCC (BHL). This work was supported in part by a VA Merit review I01CX001425-01 (PPM). Additional research funding provided by the Van Andel Research Institute through the Van Andel Research Institute – Stand Up To Cancer Epigenetics Dream Team (CMR). Stand Up To Cancer is a program of the Entertainment Industry Foundation, administered by AACR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number
All raw data resulting from RNA-seq, whole exome sequencing, and targeted sequencing is available from the database of Genotypes and Phenotypes (dbGaP) under accession number phs001249.v1.p1.
AUTHOR CONTRIBUTIONS
Conceptualization, EEG, JTP, CMR; Methodology, EEG, BHL, VES, EdS, NR, JTP; Formal analysis: EEG, AN, JTP; Investigation: EEG, BHL, VES, LAM, PKA, PD, IK, NR; Resources: TN, JS, JEC, SR, AD, PPM; Writing – original draft, EEG, JTP, CMR; Supervision and Funding Acquisition, JTP and CMR.
REFERENCES
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B, Lapouge G, Rorive S, Drogat B, Desaedelaere K, Delafaille S, Dubois C, Salmon I, Willekens K, Marine JC, Blanpain C. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell. 2015;16:67–79. doi: 10.1016/j.stem.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Belani CP, Dahlberg SE, Rudin CM, Fleisher M, Chen HX, Takebe N, Velasco MR, Jr, Tester WJ, Sturtz K, Hann CL, et al. Vismodegib or cixutumumab in combination with standard chemotherapy for patients with extensive-stage small cell lung cancer: A trial of the ECOG-ACRIN Cancer Research Group (E1508) Cancer. 2016 doi: 10.1002/cncr.30062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, Zweig MH, Minna JD. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer research. 1985;45:2913–2923. [PubMed] [Google Scholar]
- Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, Chipumuro E, Herter-Sprie GS, Akbay EA, Altabef A, et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell. 2014;26:909–922. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M, Peacock CD, Watkins DN. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, Zhang H, Marquez VE, Hammerman PS, Wong KK, Kim CF. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520:239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, Leenders F, Lu X, Fernandez-Cuesta L, Bosco G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita N, Yamamoto M, Sato T, Tsukahara T, Nagakura H, Tashiro K, Shibata Y, Watanabe H, Nagai K, Inoue M, et al. Topotecan for Relapsed Small-cell Lung Cancer: Systematic Review and Meta-Analysis of 1347 Patients. Sci Rep. 2015;5:15437. doi: 10.1038/srep15437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav U, Nalapareddy K, Saxena M, O’Neill NK, Pinello L, Yuan GC, Orkin SH, Shivdasani RA. Acquired Tissue-Specific Promoter Bivalency Is a Basis for PRC2 Necessity in Adult Cells. Cell. 2016;165:1389–1400. doi: 10.1016/j.cell.2016.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalemkerian GP, Akerley W, Bogner P, Borghaei H, Chow LQ, Downey RJ, Gandhi L, Ganti AK, Govindan R, Grecula JC, et al. Small cell lung cancer. J Natl Compr Canc Netw. 2013;11:78–98. doi: 10.6004/jnccn.2013.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFave LM, Beguelin W, Koche R, Teater M, Spitzer B, Chramiec A, Papalexi E, Keller MD, Hricik T, Konstantinoff K, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21:1344–1349. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok BH, Gardner EE, Schneeberger VE, Ni A, Desmeules P, Rekhtman N, de Stanchina E, Teicher BA, Riaz N, Powell SN, et al. PARP Inhibitor Activity Correlates with SLFN11 Expression and Demonstrates Synergy with Temozolomide in Small Cell Lung Cancer. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia AM, da Silva JH, Mencalha AL, Caffarena ER, Abdelhay E. Computational modeling of the bHLH domain of the transcription factor TWIST1 and R118C, S144R and K145E mutants. BMC Bioinformatics. 2012;13:184. doi: 10.1186/1471-2105-13-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Mu Y, Lou J, Srivastava M, Zhao B, Feng XH, Liu T, Chen J, Huang J. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 2016;17:94–109. doi: 10.15252/embr.201540964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales V, Reinhold WC, Varma S, Martinez-Cardus A, Moutinho C, Moran S, Heyn H, Sebio A, Barnadas A, Pommier Y, Esteller M. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget. 2016;7:3084–3097. doi: 10.18632/oncotarget.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, Bernard K, Conklin JF, Szczepny A, Yuan J, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med. 2011;17:1504–1508. doi: 10.1038/nm.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL, Rudin CM. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene. 2015;34:5869–5878. doi: 10.1038/onc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley E, Kunkel M, Evans D, Silvers T, Delosh R, Laudeman J, Ogle C, Reinhart R, Selby M, Connelly J, et al. Small Cell Lung Cancer Screen of Oncology Drugs, Investigational Agents, and Gene and microRNA Expression. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Kaneda A, Tsuji S, Isagawa T, Yamamoto S, Fujita T, Yamanaka R, Tanaka Y, Nukiwa T, Marquez VE, et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep. 2013;3:1911. doi: 10.1038/srep01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Panzilius E, Bartsch HS, Irmler M, Beckers J, Kari V, Linnemann JR, Dragoi D, Hirschi B, Kloos UJ, et al. Stem-cell-like properties and epithelial plasticity arise as stable traits after transient Twist1 activation. Cell Rep. 2015;10:131–139. doi: 10.1016/j.celrep.2014.12.032. [DOI] [PubMed] [Google Scholar]
- Schneeberger VE, Allaj V, Gardner EE, Poirier JT, Rudin CM. Quantitation of Murine Stroma and Selective Purification of the Human Tumor Component of Patient-Derived Xenografts for Genomic Analysis. PLoS One. 2016;11:e0160587. doi: 10.1371/journal.pone.0160587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd FA, Crowley J, Van Houtte P, Postmus PE, Carney D, Chansky K, Shaikh Z, Goldstraw P International Association for the Study of Lung Cancer International Staging, C., and Participating, I. The International Association for the Study of Lung Cancer lung cancer staging project: proposals regarding the clinical staging of small cell lung cancer in the forthcoming (seventh) edition of the tumor, node, metastasis classification for lung cancer. J Thorac Oncol. 2007;2:1067–1077. doi: 10.1097/JTO.0b013e31815bdc0d. [DOI] [PubMed] [Google Scholar]
- Sousa FG, Matuo R, Tang SW, Rajapakse VN, Luna A, Sander C, Varma S, Simon PH, Doroshow JH, Reinhold WC, Pommier Y. Alterations of DNA repair genes in the NCI-60 cell lines and their predictive value for anticancer drug activity. DNA Repair (Amst) 2015;28:107–115. doi: 10.1016/j.dnarep.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Goshorn R, Bradley C, Griffiths LM, Benavente C, Twarog NR, Miller GM, Caufield W, Freeman BB, 3rd, Bahrami A, et al. Targeting the DNA repair pathway in Ewing sarcoma. Cell Rep. 2014;9:829–841. doi: 10.1016/j.celrep.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SW, Bilke S, Cao L, Murai J, Sousa FG, Yamade M, Rajapakse V, Varma S, Helman LJ, Khan J, et al. SLFN11 Is a Transcriptional Target of EWS-FLI1 and a Determinant of Drug Response in Ewing Sarcoma. Clin Cancer Res. 2015;21:4184–4193. doi: 10.1158/1078-0432.CCR-14-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin XY, Greven MC, Kim BH, Moore J, Pierce BG, Dong X, Virgil D, et al. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013;41:D171–D176. doi: 10.1093/nar/gks1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yang WH, Lan HY, Huang CH, Tai SK, Tzeng CH, Kao SY, Wu KJ, Hung MC, Yang MH. RAC1 activation mediates Twist1-induced cancer cell migration. Nat Cell Biol. 2012;14:366–374. doi: 10.1038/ncb2455. [DOI] [PubMed] [Google Scholar]
- Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, Doroshow JH, Pommier Y. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A. 2012;109:15030–15035. doi: 10.1073/pnas.1205943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.