Abstract
Protein-protein interactions are either through direct contacts between two binding partners or mediated by structural waters. Both direct contacts and water-mediated interactions are crucial to the formation of a protein-protein complex. During the recent CAPRI rounds, a novel parallel searching strategy for predicting water-mediated interactions is introduced into our protein-protein docking method, MDockPP. Briefly, a FFT-based docking algorithm is employed in generating putative binding modes, and an iteratively derived statistical potential-based scoring function, ITScorePP, in conjunction with biological information is used to assess and rank the binding modes. Up to 10 binding modes are selected as the initial protein-protein complex structures for MD simulations in explicit solvent. Water molecules near the interface are clustered based on the snapshots extracted from independent equilibrated trajectories. Then, protein-ligand docking is employed for a parallel search for water molecules near the protein-protein interface. The water molecules generated by ligand docking and the clustered water molecules generated by MD simulations are merged, referred to as the predicted structural water molecules. Here, we report the performance of this protocol for CAPRI rounds 28–29 and 31–35 containing 20 valid docking targets and 11 scoring targets. In the docking experiments, we predicted correct binding modes for nine targets, including one high-accuracy, two medium-accuracy, and six acceptable predictions. Regarding the two targets for the prediction of water-mediated interactions, we achieved models ranked as “excellent” in accordance with the CAPRI evaluation criteria; one of these two targets is considered as a difficult target for structural water prediction.
Keywords: Protein-protein interactions, Molecular docking, Water-mediated interactions, Molecular dynamics simulations, Scoring function
INTRODUCTION
Protein-protein interactions play a key role in cellular functions. Experimental methods for determination of protein complex structures, such as X-ray, NMR, and cryo-EM, are costly and time-consuming. Computational methods, such as molecular docking, provide an alternative way for predicting structural details of protein complexes at the atomic level.1–5 In promotion of the development of computational methods for predicting protein complexes, the Critical Assessment of PRediction of Interactions (CAPRI) was initiated in 2001 and more than one hundred targets have been successfully tested to date.6–11
The interactions between proteins are either through direct contacts between binding partners or mediated by water molecules. Both direct contacts and water-mediated interactions are crucial to the formation of a protein-protein complex. Almost all the current protein-protein docking programs focus on direct-contact interactions among proteins but ignore water-mediated interactions. Since water molecules interact with flexible protein side-chains at the binding interface through non-covalent interactions, the degrees of freedom of the search space for these structural water molecules are enormous, making the prediction of water-mediated interactions a great challenge. A recent CAPRI round posted two targets (T104 and T105) for prediction of both direct and water-mediated interactions in protein-protein complexes, directing us to address this important issue. In addition, two other underemphasized problems, protein-peptide structure prediction (T60–64) and protein-DNA structure prediction (T95), were also raised in recent CAPRI rounds.
To tackle the task of predicting water-mediated interactions in protein-protein complexes, we introduced a novel strategy into MDockPP12–13, a hierarchical protein-protein structure prediction protocol developed by our group during the past CAPRI experiments. In its original form, MDockPP starts with an exhaustive sampling of the conformational space of a protein-protein complex using a Fast-Fourier Transform (FFT)-based method14–17. Then, it scores and ranks the generated putative binding modes by a statistical potential-based protein-protein scoring function, ITScorePP, which was iteratively derived to circumvent the challenging reference state problem18. If available, biological information is also used in the selection of the final models. In this study, for the prediction of water-mediated interactions, a new protocol integrating protein-protein docking, protein-ligand docking and molecular dynamics (MD) simulations was developed. Specifically, up to 10 predicted models constructed by MDockPP were used as the initial protein-protein structures for explicit solvent MD simulations. For each model, the positions of water molecules were extracted from independently equilibrated trajectories, and water molecules near the protein-protein interface were clustered. Meanwhile, a parallel search for water-mediating protein-protein interactions was carried out through protein-ligand docking. The water molecules generated by the ligand docking and the clustered water molecules generated by the MD simulations were merged to represent the predicted structural water molecules. Finally, the resulting protein-protein-water complex was further relaxed by MD simulations. The predictive power of this protocol for water-mediated protein-protein complexes was demonstrated in the recent CAPRI rounds. We achieved “excellent” water models in terms of the CAPRI criteria with the two targets (T104 and T105) for which the prediction of water-mediated interactions is required. Particularly, only four groups achieved a total of 15 “excellent” water-mediated models for T104, among which our group achieved 7 “excellent” models out of 10 predictions for this target.
MATERIALS AND METHODS
Homology modeling
For targets with only sequence information available, the program MODELLER 9.1319 was employed to build monomeric structures based on the resolved structures of homologous proteins (also known as templates). Sequence alignments were prepared by either the program fasta3620 or the MODELLER alignment program align2d. A total of 100 models were generated for each modeling unless otherwise specified. The model refinement level (the md_level option) was set to “slow”. The model with the lowest value of the DOPE-HR score was selected for docking.
In the cases in which the homologs of the receptor and the ligand formed a complex in the template structure, the complex template was also used to build the target complex structure with the program MODELLER. The parameters were identical to those used in building monomer structures. Crystal water molecules contained in the template (if available) that were located in the protein-protein interface were retained in the target complex structure. The generated complex structure was used as one of the putative binding modes, which will be described in the next docking and scoring step.
Docking and scoring protocol
A hierarchical protocol, MDockPP, was developed during the previous CAPRI experiments (rounds from 2007 to 2013) for the protein-protein complex structure prediction.12–13 We used a similar strategy as described in MDockPP for CAPRI rounds 28–29 and 31–35.
In the docking experiment, first, an FFT-based docking algorithm, either ZDOCK 3.016 or a modified 3D-DOCK17, was used to generate putative binding modes. The interval of the Euler angles was set to 6° and 3000 putative binding modes were generated for each docking. Second, the created binding modes were optimized and scored by an atomic-level, statistical potential-based scoring function for protein-protein interactions, ITScorePP, which was developed by our group using an efficient iterative method based on crystal structures of dimeric protein complexes18. Available biological information would be used as a filter in this step. Afterwards, the putative binding modes were ranked and then clustered based on the backbone root-mean-square deviation (b-RMSD) of the complexes. For any two binding modes with a b-RMSD less than a cutoff value (Rclu), only the one with the better score was kept. The value of Rclu was set to 5 Å unless otherwise specified. After clustering, up to 100 binding modes were kept for manual inspection, and ten models were selected and submitted to CAPRI.
In the scoring experiment, the same protocol was used except for the generation of the putative binding modes. The putative binding modes were collected from the groups participating in the docking experiment and redistributed by CAPRI. As with docking, ten best binding modes were selected and submitted to CAPRI.
Predicting water-mediated interactions
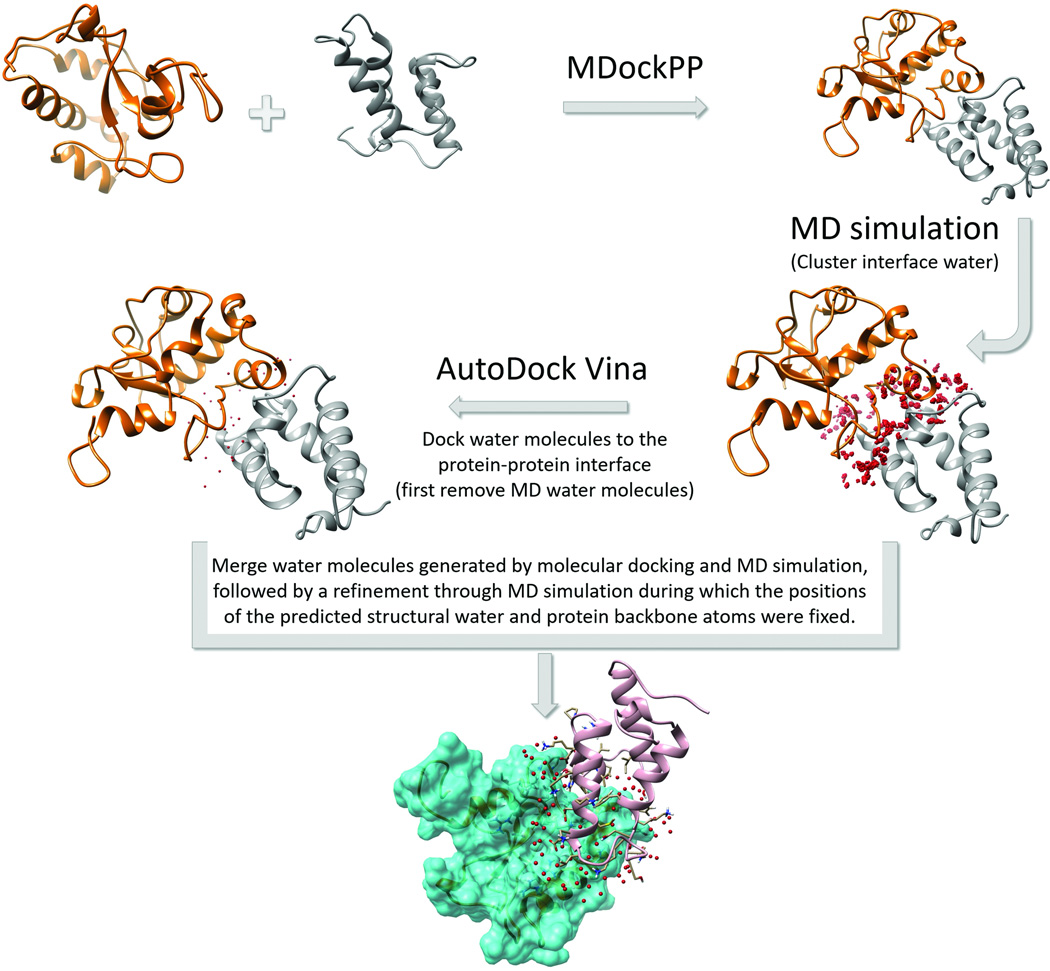
In this work, we introduced a novel strategy for the prediction of water-mediated interactions by combining protein-protein docking, protein-ligand docking and molecular dynamics (MD) simulations. The schematic diagram depicting the workflow of the strategy is shown in Fig. 1. Specifically, first, for a protein and its binding partner, the protein-protein complex structure was predicted using MDockPP. Then, the following strategy was used to predict water-mediated interactions in protein-protein complexes:
-
MD simulation. The selected models were used as the initial protein-protein structures for MD simulations in explicit solvent through the MD engine Gromacs 5.0.221 with GROMOS 53a6 force field22. The complexes were solvated by simple point charge (SPC) water molecules23 in a cubic box extending at least 1.5 nm in all directions from the solute. A twin-range van der Waal (vdW) scheme was used to account for the non-electrostatic non-bonded interactions between atoms. The vdW cutoff and neighbor list cutoff were set to 1.4 nm and 0.9 nm, respectively. The neighbor list was updated every 5 steps. The electrostatic energies were calculated through the particle mesh Evald (PME) method24, with a coulomb cutoff of 1.4 nm. The bond length was fixed by LINCS algorithm25. The periodic boundary condition was imposed in xyz directions. The temperature of the system was coupled by using the velocity rescaling method26. The integration time step was set to 2 fs.
For each model, the initial system was relaxed by 50000 steps minimization using the steepest descent algorithm. Simulated annealing was applied to the minimized system in order to identify potential structural water molecules that are critical to the structural integrity of the protein complex. The simulation time for the simulated annealing was set to 600ps and was divided into three intervals. In the first 200 ps interval, the temperature was maintained at 300 K; in the second 200 ps interval, the temperature was linearly decreases to 100 K; in the last 200 ps interval, the simulation continued at the temperature of 100 K. The positions of the backbone atoms were fixed during annealing, by using a non-equilibrium MD feature of Gromacs called “freeze groups”21. The side-chains were free to adjust their conformations. Five independent trajectories were produced. Then, the water molecules near the protein-protein interface were clustered using the DBScan (Density-based spatial clustering of applications with noise) algorithm27 based on the snapshots from the 5 independent equilibrated trajectories. The water molecule closest to the geometric center of a cluster, referred to as the clustered water molecule, was kept for each cluster. The protein-protein complex structure from the snapshots that contained a large number of contacts and few clashes with the clustered water molecules was kept as an initial protein-protein-water complex.
Protein-ligand docking. In parallel to the aforementioned MD simulations, protein-ligand docking was employed to search for water molecules near the protein-protein interface for the same protein-protein-water complex. Specifically, after the removal of the water molecules from the complex, a water probe was docked into the protein-protein interface using our modified version of the docking program AutoDock Vina28, with an emphasis on hydrogen bonding interactions. The docking box was set to a size large enough to cover the whole protein-protein interface. When this Vina docking was performed, the exhaustiveness value was set to 30, and up to 500 protein-ligand binding modes were produced as output.
The water molecules generated by the ligand docking and the clustered water molecules generated by MD simulations were merged, referred to as the predicted structural water molecules.
The resulting protein-protein-water complex was further refined by an MD simulation, during which the positions of the predicted structural water molecules and of the protein backbone atoms were fixed as described previously in the first step.
FIGURE 1.
A flowchart of our strategy for predicting water-mediated interactions in protein-protein complexes.
Protein-peptide structure prediction
Predicting the binding mode of a peptide on a protein is a challenging problem, because of the flexibility of the peptide and because of the lack of a reliable scoring function for protein-peptide complexes. The standard strategy of MDockPP is not suitable for the protein-peptide structure prediction. Consequently, the template-based method and the protein-ligand docking method were employed. For Targets 60–64 and Target 67, the homology modeling method was used to generate 1000 complex structures. For other protein-peptide targets (65 and 66) without templates, first, a protein-peptide binding site prediction tool, ACCLUSTER29, was used to downsize the possible binding regions on the protein surface. Second, our modified AutoDock Vina was employed to dock the flexible peptide (3~4 amino acids in length) into the predicted binding sites. The exhaustiveness value was set to 30, and up to 500 protein-ligand binding modes were produced as output. The generated binding modes were evaluated by the scoring function ITScore30, which is an atomic, statistical potential-based scoring function for protein-ligand interactions. To remove redundancy, for any two modes with b-RMSD smaller than 3 Å, only the one with the lower ITScore was kept. Finally, top 10 modes after clustering were manually selected for CAPRI submission.
RESULTS AND DISCUSSION
Overall performance
Here, we report our performance in CAPRI Rounds 28–29 and 31–35, which were held between 2013 and 2015. Results for Round 30 (the first CASP/CAPRI joint experiment) were excluded, because they have been presented in a recent paper11. Targets 102 and 106 were also excluded, because Target 102 was recycled as Target 107 and because Target 106 was canceled. In total, 20 targets were posted for the docking experiments and 11 targets were posted for the scoring experiments. Among these targets, 12 are the targets for predicting only protein-protein interactions, with T104 and T105 highlighting water-mediated interactions and T95 involving protein-protein-DNA interactions. The remaining 8 targets are protein-peptide complexes. A summary of our results is listed in Table 1.
Table 1.
Performance of our docking and scoring methods in CAPRI rounds 28–29 and 31–35
| Predicting | Scoring | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Targeta | Complex | Typeb | Bio. Info.c | fnat (%) | Lrmsd (Å) | Irmsd (Å) | Accuracyd | fnat (%) | Lrmsd (Å) | Irmsd (Å) | Accuracye |
| 59 | Edc3/Rps28b | H/H | ? | 0.00 | 19.37 | 7.77 | 0 | 0.10 | 10.64 | 4.04 | 0 |
| 60 | Impα/peptide | U/H | Y | 0.33 | 5.06 | 1.80 | 5 | - | - | - | - |
| 61 | Impα/peptide | U/H | Y | 0.24 | 3.96 | 1.49 | 5 | - | - | - | - |
| 62 | Impα/peptide | U/H | Y | 0.30 | 4.76 | 1.63 | 5 | - | - | - | - |
| 63 | Impα/peptide | U/H | Y | 0.29 | 4.99 | 2.17 | 0 | - | - | - | - |
| 64 | Impα/peptide | U/H | Y | 0.22 | 4.92 | 1.73 | 6 | - | - | - | - |
| 65 | RNase HI/SSB-Ct | U/H | - | 0.00 | 37.87 | 13.75 | 0 | - | - | - | - |
| 66 | PriA/SSB-Ct | U/H | - | 0.00 | 23.97 | 4.42 | 0 | - | - | - | - |
| 67 | Nedd4/peptide | U/H | Y | 0.83 | 2.30 | 1.32 | 10 | - | - | - | - |
| 95 | NCP/PRC1 | U/U | Y | 0.51 | 5.03 | 2.74 | 5 | - | - | - | - |
| 96 | GFP/αRep | H/H | - | 0.07 | 18.67 | 8.02 | 0 | 0.00 | 18.83 | 8.04 | 0 |
| 97 | GFP/αRep | H/H | - | 0.45 | 2.87 | 1.33 | 2**/3 | 0.45 | 2.87 | 1.33 | 2**/4 |
| 98 | UCH-L5/RPN13 | H/H | Y | 0.00 | 18.32 | 7.34 | 0 | 0.00 | 20.77 | 9.10 | 0 |
| 99 | UCH-L5~UbPrg/RPN13 | H/H | Y | 0.02 | 21.37 | 6.93 | 0 | 0.01 | 20.47 | 9.29 | 0 |
| 100 | UCH-L5~UbPrg/INO80G | H/H | Y | 0.02 | 15.18 | 12.40 | 0 | 0.02 | 15.23 | 12.42 | 0 |
| 101 | UCH-L5/INO80G | H/H | Y | 0.00 | 41.28 | 22.34 | 0 | 0.00 | 40.91 | 20.03 | 0 |
| 103 | UBE2Z/FAT10 | H/H | - | 0.25 | 50.27 | 13.57 | 0 | 0.23 | 52.06 | 13.83 | 0 |
| 104 | PyoAP41/ImAP41 | H/H | Y | 0.63 | 2.72 | 0.97 | 4***/6** | 0.67 | 2.77 | 0.98 | 3***/7** |
| 104w | PyoAP41/ImAP41/water | H/H | - | 0.58 | - | - | 7+++/3++ | 0.54 | - | - | 2+++/8++ |
| 105 | PyoS2/ImS2 | H/H | Y | 0.76 | 1.82 | 1.38 | 10** | 0.76 | 1.34 | 0.94 | 3***/7** |
| 105w | PyoS2/ImS2/water | H/H | - | 0.77 | - | - | 10+++ | 0.88 | - | - | 2++++/7+++/1++ |
| 107 | HxuA/Hemopexin-Nt | U/U | - | 0.00 | 38.15 | 19.05 | 0 | 0.00 | 38.16 | 19.04 | 0 |
Target 102 was re-held in Target 107, and T106 was cancelled. Therefore, T102 and T106 are not listed in this table. The results for the prediction of water-mediated interactions are marked by “w”.
The symbol “U” stands for the unbound experimental structure and “H” for the homology-modeled structure.
“Y” means that valid biological information was available for the binding site, “-” means that no or little useful biological information was available, and the question mark “?” means that the available experimental information about the binding site is not consistent with the crystal structure.
The accuracy is categorized by three parameters following the CAPRI criteria6,7: The percentage of the native residue-residue contacts (fnat), the ligand RMSD (Lrmsd), and the interface RMSD (Irmsd). “***” stands for high-accuracy, “**” for medium-accuracy, “*” for acceptable accuracy, and “0” for no correct prediction, respectively. For the water prediction targets 104w and 105w, the accuracy is categorized by fwmc(nat) and the fraction of water-mediated contacts53. “++++” stands for outstanding-accuracy, “+++” for excellent-accuracy, “++” for good-accuracy, “+” for fair-accuracy, and “0” for no correct prediction, respectively.
There were no scoring experiments for Targets 60–67 and 95.
For the docking experiments, we predicted at least one acceptable binding mode for nine targets, among which there is one high-accuracy prediction (Target 104), two medium-accuracy predictions (Targets 97 and 105), and six acceptable predictions (Targets 60, 61, 62, 64, 67, and 95). For the prediction of water-mediated interactions (Targets 104 and 105), we achieved excellent models for both targets. For the scoring experiments, we identified high-accuracy models for Targets 104 and 105, and medium-accuracy models for Target 97. The details of the results for each target are presented as follows, except for T59 and T103 whose crystal structures have not been released yet.
Targets 60–64 (Impα/peptides)
Targets 60–64 are the complexes formed by the import receptor importin-α (Imp α) bound with five peptides (PDB IDs: 3ZIN, 3ZIO, 3ZIP, 3ZIQ, and 3ZIR)31. The unbound structures were provided for the protein receptors by CAPRI, but only the sequence information was given for the peptides. While the peptide in Target 60 is the nuclear localization signal (NLS) from mouse RNA helicase II/Guα, the peptides in other targets are peptide-library derived peptides. Two NLS-binding sites (major and minor) on importin-α were observed. The major site is on the armadillo (ARM) repeats 2–4 and the minor site is on the ARM repeats 6–8. These NLS peptides bind to both major and minor NLS-binding sites, but the minor site is the primary binding site.
Due to the high flexibility of the peptide, the standard strategy of MDockPP is not effective for predicting protein-peptide complex structures. Therefore, we used templated-based methods for Targets 60–64. Depending on the sequence similarity, either 1EJL or 1EJY32 was selected as the template for the query peptide. Peptides in our predicted binding modes bind at the minor site. Unfortunately, we did not predict any acceptable binding modes with the full-length peptides for all the five targets. Nevertheless, when the peptides were shortened to the same number of residues as those on the major site, we recovered at least one acceptable binding mode for four out of five targets, except Target 64. For Target 64, the best binding mode in our submission was actually close to the criteria for acceptable accuracy, with fnat = 28.6%, Lrmsd = 4.99 Å, and Irmsd = 2.17 Å.
Targets 65 and 66 (RNase HI/SSB-Ct and PriA/SSB-Ct)
Targets 65 and 66 share the same peptide sequence, but distinguish in protein receptors (PDB IDs: 4Z0U33 and 4NL834). The peptide is the intrinsically disordered C terminus of the single-stranded DNA-binding protein (SSB-Ct). The proteins in Targets 65 and 66 are ribonuclease HI (RNase HI) and PriA DNA helicase, respectively. We did not find any templates for these two targets. Consequently, a flexible docking strategy based on the predicted binding sites was used, as described in the MATERIALS AND METHODS. Since AutoDock Vina is not capable of docking a peptide with many rotatable bonds, the four key amino acids (DIPF) resolved in the crystal structures were used for docking.
For Target 65, we selected only the top predicted binding site for docking. However, this binding site is in the protein-protein homodimer interface. It is worth mentioning that although only one chain of the protein receptor was provided by CAPRI, the protein receptors actually form a homodimer in the crystal structure. In retrospect, our third predicted binding site is the true binding site for the peptide; moreover, our second predicted binding site is also in the protein-protein interface.
The receptor in Target 66 is a large protein with more than 700 amino acids, making the binding site prediction very difficult. We selected the top 3 predicted binding sites for docking. Unfortunately, the true binding site of the peptide turned out to be our fourth predicted binding site.
Target 67 (Nedd4/peptide)
Target 67 is the complex formed by a WW domain of Nedd4 with a peptide fragment from arrestin-related domain-containing protein-3 (ARRDC3) (PDB ID: 4N7H)35. The peptide contains a PPXY motif, which is crucial to the structural formation of the complex. The template-based method was used for this target. Two templates, 2KPZ36 and 2KQ037, were used. A total of 2000 putative binding modes were generated for the optimization and evaluation. We did not succeed in predicting any acceptable binding modes for the full-length peptide (13 amino acids). However, when the peptide was truncated to contain only the PPXY motif, all our submitted binding modes became acceptable with fnat = 83.3%, Lrmsd = 2.30 Å, and Irmsd = 1.32 Å.
Target 95 (NCP/ PRC1)
Target 95 is a complex of the Polycomb repressive complex 1 (PRC1) ubiquitylation module bound to the nucleosome core particle (NCP) (PDB ID: 4R8P)38. PRC1 is the complex of E3 ubiquitin ligase and E2 enzyme UbcH5c. E3 is a heterodimer formed by Bmi1 and Ring1b. NCP is the fundamental unit of the eukaryotic genome consisting of about 146 base pairs (bp) of DNA wrapped around a histone octamer (2 copies each of H2A, H2B, H3, and H4). The unbound structures for both PRC1 and NCP were provided by CAPRI, with PDB IDs of 3RPG39 and 3LZ040, respectively. This target is difficult due to the complexity and the huge size of the system and the involvement of protein-DNA interactions. In the docking experiment, a total of only 13 correct binding modes were submitted from three groups (including our group). No scoring experiment was organized for this target.
To predict this complex structure, we parameterized the protein-DNA interactions in our docking protocol. First, in the sampling stage, ZDOCK2.315 with the DNA parameters provided by Fanelli and Ferrari41 was employed to generate decoys. Then, in the scoring stage, these decoys were ranked based on ITScorePP. Specifically, to simultaneously evaluate the protein-protein interactions and protein-DNA interactions of each decoy, the DNA atoms were assigned to the atom types of ITScorePP based on their chemical connections.
Meanwhile, we noticed that the NCP binding proteins, LANA (PDB ID: 1ZLA)42, RCC1 (PDB ID: 3MVD)43, and Sir3 (PDB ID: 3TU4)44 bind to an acidic patch in the H2A/H2B dimer interface through positive residues on a loop. In our predictions, the two positive residues (K97 and R98) on the Ring1b of E3 bind to the acidic patch, which consists of four residues on H2A (E61, E64, E92, and D90) and three residues on H2B (V48, E105, and H109). We achieved five acceptable predictions for this target.
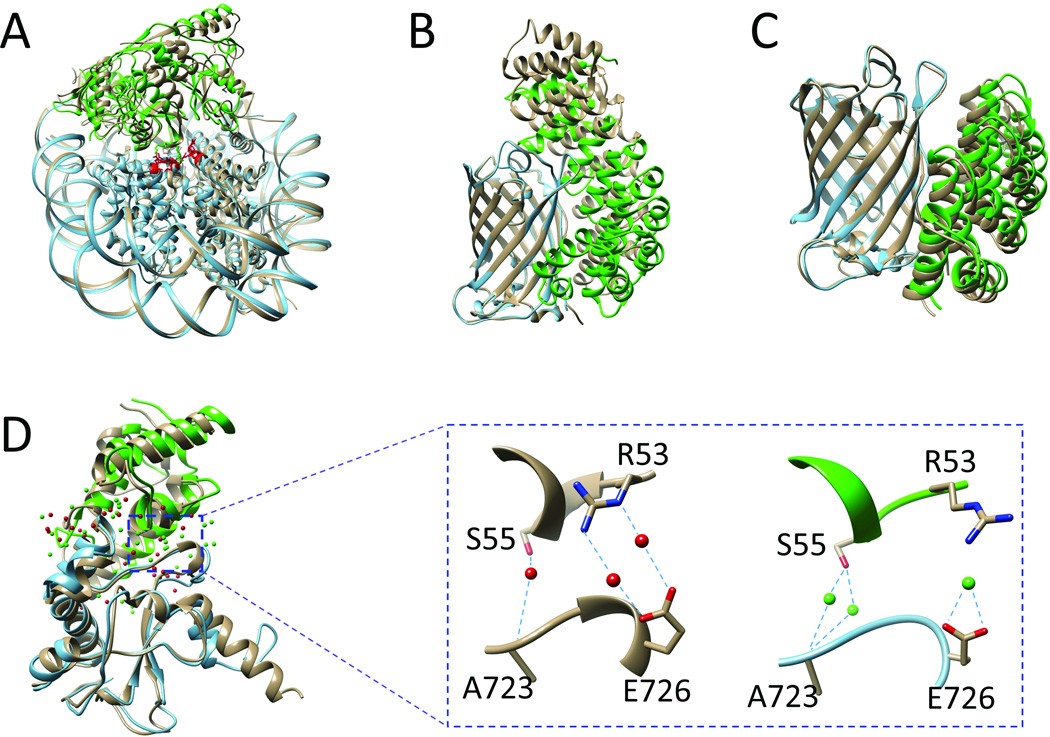
A comparison between the crystal structure and our predictions is depicted in Fig. 2A. The crystal structure is colored tan. One of our predicted binding modes (acceptable-accuracy of fnat = 50.8%, Lrmsd = 5.03 Å, and Irmsd = 2.74 Å) is matched to the crystal structure by superimposing the NCPs. The PRC1 in our predicted binding mode is colored green. Residues in the acidic patch on NCP are plotted in stick representation and highlighted in red, and the corresponding two positive residues on PRC1 are also shown in stick representation.
FIGURE 2.
Our predictions for four targets of protein-protein complexes in comparison with bound, crystal structures (represented by ribbon and colored tan). For each target, one binding partner (colored cyan) in the predicted structure was aligned to the corresponding part in the crystal structure. The other binding partner in the predicted structure is colored green. A. Target 95 is the NCP/ PRC1. Residues in the acidic patch on NCP are represented by stick model and colored red. B. Target 96 is the complex formed by GFP with an αRep containing 8 repeats of αhelices pairs. C. Target 97 is the complex formed by GFP with an αRep containing 5 repeats of αhelices pairs. D. T104 is the PyoAP41/ImAP41 complex. Crystal water molecules are colored red and predicted water molecules are colored green. The zoom-in panel shows several conserved water molecules in the crystal structure as well as our prediction results. Hydrogen bonds are represented by cyan dashed lines.
Targets 96 and 97 (GFP/αReps)
Targets 96 and 97 are complexes formed by green fluorescent protein (GFP) with two αRep members (PDB ID: 4XL5 and 4XVP)45. αRep is a family of artificial proteins that was designed based on a natural family of helical repeat. The αReps in T96 and T97 contain 8 and 5 repeats, respectively. Each repeat consists of about 30 residues and forms a pair of α helices. GFP was built based the template 1JBZ46, and the two αReps were built based on the template 3LTJ47. Because the template 3LTJ contains only 6 repeats, we generated a new template with 8 repeats for T96 using two 3LTJs. Specifically, two repeats at the N-terminal of a 3LTJ was superimposed on the two repeats at the C-terminal of the other 3LTJ, and only two repeats were kept for the overlapped region. Because αReps are artificial proteins, there is no biological information for these two targets.
In the docking experiment, our predicted binding modes for T96 locate in the correct binding site, as shown in Fig. 2B. However, there exists a shift and rotation between our predicted modes and the crystal structure. We did not predict any acceptable-accuracy binding modes for this target. The failure could be attributed to the bad quality of the modeled monomeric structure of αRep, for which the 8 repeats was constructed based on a template with only 6 repeats. Comparison of our predicted αRep monomeric structure with the corresponding crystal structure shows a different bending curvature formed by the repeats.
For T97, we predicted two binding modes with medium-accuracy and three binding modes with acceptable-accuracy. Fig. 2C shows a predicted binding mode with medium-accuracy of fnat = 45.1%, Lrmsd = 2.87 Å, and Irmsd = 1.33 Å.
Similarly, for the scoring experiment, no acceptable binding mode was achieved for Target 96, and two medium-accuracy and four acceptable-accuracy binding modes were achieved for T97.
Targets 98–101 (UCH-L5/RPN13, UCH-L5~UbPrg/RPN13, UCH-L5~UbPrg/INO80G, and UCH-L5/INO80G)
Targets 98–101 are four complexes related to the activation (activated by the DEUBAD domain in PRN13) and inhibition (inhibited by the DEUBAD domain in INO80G) of a deubiquitinating enzyme UCH-L5 (PDB ID: 4UEM, 4UEL, 4UF6, and 4UF5)48. Targets 98 and 99 are complexes of UCH-L5/RPN13 without and with the binding of ubiquitin-propargyl (UbPrg), respectively. Targets 100 and 101 are complexes of UCH-L5/INO80G with and without the binding of UbPrg, respectively. UCH-L5 was built based on the template 3IHR. Biological information indicated that both RPN13 and INO80G bind to a long fragment at the C-terminal of the UCH-L5. However, the structure of the fragment was missing in the template 3IHR49. PRN13 and INO80G were built based on the template 2KQZ50. According to the released crystal structures of the four complexes, there exists large conformational change for both PRN13 and INO80G upon the binding with the long fragment at the C-terminal of the UCH-L5. Consequently, no group achieved any correct binding modes in both docking and scoring experiments for these difficult targets.
Targets 104 and 105 (PyoAP41/ImAP41 and PyoS2/ImS2)
Targets 104 and 105 are complexes of Pyocin DNases AP41 and S2 binding with their immunity (Im) proteins ImAP41 and ImS2, respectively (PDB ID: 4UHP and 4QKO)51. In addition to direct contacts between proteins, water-mediated interactions are essential for high-affinity DNase-Im complexes formation. There are several homologous protein complexes in PDB for the two targets. In our prediction, 3U43 (structure of colicin E2 DNase-Im2 complex)52 were selected as the template for the two targets. Specifically, both PyoAP41 and PyoS2 were constructed based on chain B (colicin E2 DNase) of the template 3U43. ImAP41 and ImS2 were built based on chain A (Im2) of the same template 3U43. For each target, modeled monomeric structures were used as inputs for MDockPP to generate putative binding modes (docking-based models). In addition, for each target, a complex structure was directly built based the template, namely the templated-based model. Then, the templated-based model and docking-based models were merged and evaluated by the scoring function. Up to 10 binding modes were selected as initial structures for the prediction of both direct and water-mediated interactions.
The evaluation for the two targets was composed of two parts, the evaluation for the binding mode and the evaluation for the water-mediated interactions. The binding mode evaluation used the same criteria (fnat, Lrmsd, and Irmsd) as other targets. For the assessment of water-mediated interactions, the criterion was described in a previous CAPRI paper53. Briefly, “water-mediated contacts are defined whenever residues from both the ligand and the receptor proteins have one or more heavy atoms within a 3.5 Å distance of the same water molecule”. The quantity fwmc(nat) is defined as the fraction of water-mediated contacts in the target that is recalled by the predicted model.
Because the homologous complex structures were available and the binding sites of the two targets were clear, Targets 104 and 105 are considered as easy targets for binding mode prediction. For the binding mode prediction, all of our submitted models were correct. For Target 104, we predicted four high-accuracy and six medium-accuracy binding modes in the docking experiment. Similar results were achieved in the scoring experiments. Fig. 2D shows one of our best models. For Target 105, we predicted ten medium-accuracy binding modes in the docking experiment. Better performance was achieved in the scoring experiment than the docking experiment, three high-accuracy and seven medium-accuracy binding modes were recovered.
As compared to the binding mode prediction, predicting water-mediated interactions is much more challenging, as reflected by the overall performance of the CAPRI community on these targets. Using the docking experiment of Target 104 as an example, 11 groups predicted 53 models with high-accuracy for binding mode prediction. In contrast, among all the submitted models, none was classified into the “outstanding” category (fwmc(nat) ≥ 0.8) of the water prediction and only 15 models from 4 groups (nearly half of these models were submitted by our group) were categorized as “excellent” (0.5 ≤ fwmc(nat) < 0.8).
By using a novel strategy for predicting water-mediated interactions (as described in the MATERIALS AND METHODS), we achieved good performance for both Targets 104 and 105. 7 (2) of our 10 submitted models in the docking (scoring) experiment of Target 104 were graded into the “excellent” category, and the remaining models graded into the “good” category (0.3 ≤ fwmc(nat) < 0.5). Regarding T105, we were the only group that submitted 10 “excellent” models in the docking experiment. In the scoring experiment, 2 models were ranked into the “outstanding” category, 7 models into the “excellent” category, and the remaining model into the “good” category.
Next, we analyzed whether the MD simulation method and the protein-ligand docking method complement each other for the prediction of interfacial water molecules. The results for each method before their combination were analyzed as follows. For Target 104, the complex formed by chains E and F in PDB entry 4UHP50 was used as the reference to characterize the water-mediated interactions, because E:F contains more interfacial water molecules than the complexes formed by other chains in the same PDB entry. Using one of our best submitted models as an example, the MD method generated more interface water molecules (38) than protein-ligand docking (27), but the protein-ligand docking method achieved higher fwmc(nat) (0.46) than MD (fwmc(nat) = 0.35). Interestingly, the merge of the water molecules by combining the two methods did not improve fwmc(nat) (=0.46), which indicates that water-mediated interactions correctly predicted by the MD were also predicted by protein-ligand docking. Therefore, protein-ligand docking is a useful tool for the prediction of water molecules at a protein-protein interface. The lower value of fwmc(nat) for the MD method may result from the fact that the interfacial water molecules generated by the MD were clustered based on multiple snapshots from independent trajectories while the protein-protein complex was selected from only one snapshot (see MATERIALS AND METHODS). The lower fwmc(nat) values do not mean the MD method is not needed, because the possible atomic clashes and other inaccuracies due to the clustering of water molecules from multiple snapshots can be improved by the later refinement step (see below). In contrast, for the protein-ligand docking method, only the submitted protein-protein complex was used for docking to search for the interfacial water molecules. Therefore, these two methods provide a balance between a single selected protein-protein conformation and multiple protein-protein conformations and are complementary for interfacial water prediction. Similar results were obtained for Target 105.
In the MD refinement step after the combination of the two above methods, the predicted interfacial water molecules and the protein backbone atoms were frozen whereas the protein sidechain atoms were movable. The fwmc(nat) was improved to 0.58. However, the MD refinement did not always improve the results. Regarding our 10 submitted models, the MD refinement improved the fwmc(nat) values for 3 models, made no changes for 3 other models, and reduced the fwmc(nat) values for the remaining 4 models. Nonetheless, clashes between the clustered water molecules and the protein atoms resulted from the MD method were removed in the refinement step. Similar results were found for Target 105.
In addition to water-mediated interactions, direct interactions at the protein-protein interface were also analyzed to find whether the MD simulation would help improve the accuracy of predicted interface structures. Because the backbone atoms were fixed during the MD simulations, fnat was used as the criterion. Unfortunately, the MD simulation did not improve the direct interactions for all the cases except one submitted model. For example, for the model with the highest fwmc(nat), the fnat of the initially docked protein-protein complex was 0.62, but this value was reduced to 0.58 after the MD simulation. Similar results were also found for Target 105.
Several conserved water molecules forming hydrogen bonds with the residues from both the receptor and the ligand were observed in the target complex structures. It is interesting to see if we correctly predicted these conserved water molecules and the corresponding hydrogen bonds. Here, we use one of our best models in the T104 docking experiment as an example. In the crystal structure, the side chain of S55 in ImAP41 and the carbonyl of A723 in PyoAP41 are bridged by a conserved water molecule through hydrogen bonds, as shown in the middle panel of Fig. 2D. In addition, two water molecules form hydrogen bonds with the side chain of R53 in ImAP41 and the side chain of E726 in PyoAP41. In our predicted model, as shown in the right panel of Fig. 2D, we successfully predicted the water-mediated interactions for both ImAP41-S55/PyoAP41-A723 and ImAP41-R53/PyoAP41-E726 in accordance with the CAPRI criterion (i.e., only considering the distance but ignoring hydrogen bond formation). We also analyzed the hydrogen bonds formed in our predicted models. Two water molecules were predicted to form four hydrogen bonds between ImAP41-S55 and PyoAP41-A723. Only one water molecule was predicted to form two hydrogen bonds with the carboxylic acid of PyoAP41-E726. The distance between the water molecule and the guanidinium group of ImAP41-R53 was about 3.1 Å. However, they did not form any hydrogen bonds. There is no doubt that predicting hydrogen bond formation is much more difficult than predicting contacts using only a distance cutoff. Similar performance was achieved for Target 105. The results indicate that our method is feasible for predicting water-mediated interactions.
Target 107 (Haemopexin-Nt/HxuA)
Target 107 is a protein-protein complex formed by HxuA and the N-terminal domain of haemopexin (Haemopexin-Nt) (PDB ID: 4RT6)54. The unbound structures were provided by CAPRI for the two proteins. No reliable binding site information was found for this target. We did not predict any correct binding modes in both docking and scoring experiments. In the released crystal structure of the complex, Haemopexin-Nt binds to the C-terminal of HxuA, and a long loop, called M loop (residues 711–728), on HxuA plays a key role in the complex formation. The structure of the M loop is missing in the unbound structure, making Target 107 one of the most difficult targets. No group made any correct prediction for this target in both docking and scoring experiments.
CONCLUSION AND DISCUSSIONS
In this article, we report the performance of our latest hierarchical approach for protein-protein docking (MDockPP) employed in CAPRI rounds 28–29 and 31–35. In its current form, MDockPP has been augmented to include a novel strategy for predicting water-mediated interactions. In the docking experiments, we made correct predictions for 9 targets out of 20 targets, including one high-accuracy, two medium-accuracy, and six acceptable predictions. In addition, we achieved excellent-accuracy models for the two targets focusing on water-mediated interactions according to CAPRI and one of these targets is considered as a difficult target for structural water prediction. In the scoring experiments, we made correct predictions for 3 out of 11 targets, including two high-accuracy and one medium-accuracy predictions.
One of the most prominent and long-standing challenges in protein-protein docking is the treatment of protein flexibility. It is reflected by the poor performance of the CAPRI community on the targets with large conformational changes induced by the protein-protein complex formation. There are at least six targets (98–101, 103, and 107) attributable to the protein flexibility problem. No group made correct predictions in either docking or scoring experiments for these challenging targets. In our protocol, proteins were treated as rigid bodies when the binding modes were generated and minor protein flexibility was considered only implicitly. This approximation is definitely inadequate for the targets characterized by large conformational changes.
Another challenge is the prediction of water-mediated interactions. During the CAPRI experiments, we developed a novel strategy for predicting water-mediated interactions by incorporating protein-protein docking, protein-ligand docking and MD simulations. Good performance was achieved for the two water-prediction targets, T104 and T105. Nonetheless, false positive of the water-mediated interactions was found in our predicted models. Using Target 104 as an example, in our best predicted model with “excellent” accuracy of fwmc(nat) = 0.58 for water predictions, the fraction of non-native water-mediated contacts [fwmc(nonnat)] was as high as 0.72, which could be the reason that some of our predicted models were not able to pass the high-accuracy criteria. As in the docking experiment for Target 105, although all of our submitted models were graded into the “excellent” category for the water prediction, they were only medium-accuracy models as evaluated by the criteria for protein-protein binding mode prediction. Our future study will stress on reducing fwmc(nonnat) and balancing the accuracy in the prediction of direct interactions and water-mediated interactions.
Acknowledgments
This work was supported by NSF CAREER Award DBI-0953839, NIH R01GM109980, and the American Heart Association (Midwest Affiliate) [13GRNT16990076] (XZ). The computations were performed on the high performance computing infrastructure supported by NSF CNS-1429294 (PI: Chi-Ren Shyu) and the HPC resources supported by the University of Missouri Bioinformatics Consortium (UMBC).
REFERENCES
- 1.Wodak SJ, Janin J. Computer analysis of protein–protein interaction. J Mol Biol. 1978;124:323–342. doi: 10.1016/0022-2836(78)90302-9. [DOI] [PubMed] [Google Scholar]
- 2.Smith GR, Sternberg MJ. Prediction of protein–protein interactions by docking methods. Curr Opin Struct Biol. 2002;12:28–35. doi: 10.1016/s0959-440x(02)00285-3. [DOI] [PubMed] [Google Scholar]
- 3.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 4.Gray JJ. High-resolution protein–protein docking. Curr Opin Struct Biol. 2006;16:183–193. doi: 10.1016/j.sbi.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Bonvin AM. Flexible protein–protein docking. Curr Opin Struct Biol. 2006;16:194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Janin J, Henrick K, Moult J, Ten Eyck L, Sternberg MJE, Vajda S, Vasker I, Wodak SJ. CAPRI: a critical assessment of predicted interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 7.Mendez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3–5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 8.Lensink MF, Wodak SJ, Mendez R. Docking and scoring protein complexes: CAPRI 3rd edition. Proteins. 2007;69:704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 9.Lensink MF, Wodak SJ. Blind predictions of protein interfaces by docking calculations in CAPRI. Proteins. 2010;78:3085–3095. doi: 10.1002/prot.22850. [DOI] [PubMed] [Google Scholar]
- 10.Lensink MF, Wodak SJ. Docking, scoring, and affinity prediction in CAPRI. Proteins. 2013;81:2082–2095. doi: 10.1002/prot.24428. [DOI] [PubMed] [Google Scholar]
- 11.Lensink MF, Velankar S, Kryshtafovych A, Huang S-Y, Schneidman-Duhovny D, Sali A, Segura J, Fernandez-Fuentes N, Viswanath S, Elber R, Grudinin S, Popov P, Neveu E, Lee H, Baek M, Park S, Heo L, Rie Lee G, Seok C, Qin S, Zhou H-X, Ritchie DW, Maigret B, Devignes M-D, Ghoorah A, Torchala M, Chaleil RAG, Bates PA, Ben-Zeev E, Eisenstein M, Negi SS, Weng Z, Vreven T, Pierce BG, Borrman TM, Yu J, Ochsenbein F, Guerois R, Vangone A, Rodrigues JPGLM, van Zundert G, Nellen M, Xue L, Karaca E, Melquiond ASJ, Visscher K, Kastritis PL, Bonvin AMJJ, Xu X, Qiu L, Yan C, Li J, Ma Z, Cheng J, Zou X, Shen Y, Peterson LX, Kim H-R, Roy A, Han X, Esquivel-Rodriguez J, Kihara D, Yu X, Bruce NJ, Fuller JC, Wade RC, Anishchenko I, Kundrotas PJ, Vakser IA, Imai K, Yamada K, Oda T, Nakamura T, Tomii K, Pallara C, Romero-Durana M, Jiménez-García B, Moal IH, Férnandez-Recio J, Joung JY, Kim JY, Joo K, Lee J, Kozakov D, Vajda S, Mottarella S, Hall DR, Beglov D, Mamonov A, Xia B, Bohnuud T, Del Carpio CA, Ichiishi E, Marze N, Kuroda D, Roy Burman SS, Gray JJ, Chermak E, Cavallo L, Oliva R, Tovchigrechko A, Wodak SJ. Prediction of homoprotein and heteroprotein complexes by protein docking and template-based modeling: A CASP-CAPRI experiment. Proteins. 2016 doi: 10.1002/prot.25007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S-Y, Zou X. MDockPP: A hierarchical approach for protein-protein docking and its application to CAPRI rounds 15–19. Proteins. 2010;78:3096–3103. doi: 10.1002/prot.22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang SY, Yan C, Grinter SZ, Chang S, Jiang L, Zou X. Inclusion of the orientational entropic effect and low-resolution experimental information for protein–protein docking in Critical Assessment of PRedicted Interactions (CAPRI) Proteins. 2013;81:2183–2191. doi: 10.1002/prot.24435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 16.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PloS one. 2011;6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabb HA, Jackson RM, Sternberg MJ. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J Mol Biol. 1997;272:106–120. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 18.Huang S-Y, Zou X. An iterative knowledge-based scoring function for protein–protein recognition. Proteins. 2008;72:557–579. doi: 10.1002/prot.21949. [DOI] [PubMed] [Google Scholar]
- 19.Marti-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Šali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 20.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 22.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 23.Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J Phys Chem-Us. 1987;91(24):6269–6271. [Google Scholar]
- 24.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 25.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. [Google Scholar]
- 26.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 27.Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. InKdd. 1996;96:226–231. [Google Scholar]
- 28.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan C, Zou X. Predicting peptide binding sites on protein surfaces by clustering chemical interactions. J Comput Chem. 2015;36:49–61. doi: 10.1002/jcc.23771. [DOI] [PubMed] [Google Scholar]
- 30.Huang SY, Zou X. Scoring and lessons learned with the CSAR benchmark using an improved iterative knowledge-based scoring function. J Chem Inf Model. 2011;51:2097–2106. doi: 10.1021/ci2000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang CW, Couñago RM, Williams SJ, Bodén M, Kobe B. Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-α. Traffic. 2013;14:1144–1154. doi: 10.1111/tra.12098. [DOI] [PubMed] [Google Scholar]
- 32.Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J Mol Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 33.Petzold C, Marceau AH, Miller KH, Marqusee S, Keck JL. Interaction with single-stranded DNA-binding protein stimulates Escherichia coli Ribonuclease HI enzymatic activity. J Biol Chem. 2015;290:14626–14636. doi: 10.1074/jbc.M115.655134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharyya B, George NP, Thurmes TM, Zhou R, Jani N, Wessel SR, Sandler SJ, Ha T, Keck JL. Structural mechanisms of PriA-mediated DNA replication restart. Proc Natl Acad Sci USA. 2014;111:1373–1378. doi: 10.1073/pnas.1318001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi S, O'Hayre M, Gutkind JS, Hurley JH. Structural and biochemical basis for ubiquitin ligase recruitment by arrestin-related domain-containing protein-3 (ARRDC3) J Biol Chem. 2014;289(8):4743–4752. doi: 10.1074/jbc.M113.527473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iglesias-Bexiga M, Luque I, Macias M. Human NEDD4 3RD WW Domain Complex with Human T-cell Leukemia virus GAP-Pro Poliprotein Derived Peptide. In preparation [Google Scholar]
- 37.Iglesias-Bexiga M. Human NEDD4 3RD WW Domain Complex with Human T-cell Leukemia virus GAP-Pro Poliprotein Derived Peptide. In preparation [Google Scholar]
- 38.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasudevan D, Chua EY, Davey CA. Crystal structures of nucleosome core particles containing the ‘601’strong positioning sequence. J Mol Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 41.Fanelli F, Ferrari S. Prediction of MEF2A–DNA interface by rigid body docking: a tool for fast estimation of protein mutational effects on DNA binding. J Struct Biol. 2006;153:278–283. doi: 10.1016/j.jsb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 43.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chevrel A, Urvoas A, De La Sierra-gallay IL, Aumont-Nicaise M, Moutel S, Desmadril M, Perez F, Gautreau A, van Tilbeurgh H, Minard P, Valerio-Lepiniec M. Specific GFP-binding artificial proteins (αRep): a new tool for in vitro to live cell applications. Biosci Rep. 2015;35:e00223. doi: 10.1042/BSR20150080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanson GT, McAnaney TB, Park ES, Rendell ME, Yarbrough DK, Chu S, Xi L, Boxer SG, Montrose MH, Remington SJ. Green fluorescent protein variants as ratiometric dual emission pH sensors. 1. Structural characterization and preliminary application. Biochemistry. 2002;41:15477–15488. doi: 10.1021/bi026609p. [DOI] [PubMed] [Google Scholar]
- 47.Urvoas A, Guellouz A, Valerio-Lepiniec M, Graille M, Durand D, Desravines DC, van Tilbeurgh H, Desmadril M, Minard P. Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins (αRep) based on thermostable HEAT-like repeats. J Mol Biol. 2010;404:307–327. doi: 10.1016/j.jmb.2010.09.048. [DOI] [PubMed] [Google Scholar]
- 48.Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H, Sixma TK. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell. 2015;57:887–900. doi: 10.1016/j.molcel.2014.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgie SE, Bingman CA, Soni AB, Phillips GN. Structural characterization of human Uch37. Proteins. 2012;80:649–654. doi: 10.1002/prot.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, Lee BH, Finley D, Walters KJ. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol Cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joshi A, Grinter R, Josts I, Chen S, Wojdyla JA, Lowe ED, Kaminska R, Sharp C, McCaughey L, Roszak AW, Cogdell RJ. Structures of the Ultra-High-Affinity Protein–Protein Complexes of Pyocins S2 and AP41 and Their Cognate Immunity Proteins from Pseudomonas aeruginosa. J Mol Biol. 2015;427:2852–2866. doi: 10.1016/j.jmb.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wojdyla JA, Fleishman SJ, Baker D, Kleanthous C. Structure of the ultra-high-affinity colicin E2 DNase–Im2 complex. J Mol Biol. 2012;417:79–94. doi: 10.1016/j.jmb.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 53.Lensink MF, Moal IH, Bates PA, Kastritis PL, Melquiond AS, Karaca E, Schmitz C, Dijk M, Bonvin AM, Eisenstein M, Jiménez-García B, Grosdidier S, Solernou A, Pérez-Cano L, Pallara C, Fernández-Recio J, Xu J, Muthu P, Praneeth Kilambi K, Gray JJ, Grudinin S, Derevyanko G, Mitchell JC, Wieting J, Kanamori E, Tsuchiya Y, Murakami Y, Sarmiento J, Standley DM, Shirota M, Kinoshita K, Nakamura H, Chavent M, Ritchie DW, Park H, Ko J, Lee H, Seok C, Shen Y, Kozakov D, Vajda S, Kundrotas PJ, Vakser IA, Pierce BG, Hwang H, Vreven T, Weng Z, Buch I, Farkash E, Wolfson HJ, Zacharias M, Qin S, Zhou HX, Huang SY, Zou X, Wojdyla JA, Kleanthous C, Wodak SJ. Blind prediction of interfacial water positions in CAPRI. Proteins. 2014;82:620–632. doi: 10.1002/prot.24439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zambolin S, Clantin B, Chami M, Hoos S, Haouz A, Villeret V, Delepelaire P. Structural basis for haem piracy from host haemopexin by Haemophilus influenzae. Nat commun. 2016;18:7. doi: 10.1038/ncomms11590. [DOI] [PMC free article] [PubMed] [Google Scholar]