Abstract
Structural characterization of proteins is essential for understanding life processes at the molecular level. However, only a fraction of known proteins have experimentally determined structures. This fraction is even smaller for protein-protein complexes. Thus, structural modeling of protein-protein interactions (docking) primarily has to rely on modeled structures of the individual proteins, which typically are less accurate than the experimentally determined ones. Such “double” modeling is the Grand Challenge of structural reconstruction of the interactome. Yet it remains so far largely untested in a systematic way. We present a comprehensive validation of template-based and free docking on a set of 165 complexes, where each protein model has six levels of structural accuracy, from 1 to 6 Å Cα RMSD. Many template-based docking predictions fall into acceptable quality category, according to the CAPRI criteria, even for highly inaccurate proteins (5 – 6 Å RMSD), although the number of such models (and, consequently, the docking success rate) drops significantly for models with RMSD > 4 Å. The results show that the existing docking methodologies can be successfully applied to protein models with a broad range of structural accuracy, and the template-based docking is much less sensitive to inaccuracies of protein models than the free docking.
Keywords: protein recognition, protein docking, protein modeling, structure prediction, interactome
Introduction
Protein-protein interactions (PPI) drive many cellular processes. Structural characterization of PPI is important for better understanding of these processes and for our ability to manipulate them. Experimental techniques for structure determination of PPI have limited capabilities. The X-ray crystallography, the major source of today's knowledge of atomic-level structures of PPI, accounts only for 26% of known PPI in E. coli and 6.7% in human.1 Thus, the structure of most known protein interactions has to be determined by computational PPI modeling (protein docking).2
Current protein docking methods generally belong to two major categories: (a) free docking, where relative positions of the two proteins are systematically sampled and, generally, no information other than the structure of the two proteins, is assumed to be known a priori; and (b) template-based docking, where the prediction is made according to the sequence or the structure similarity of the target proteins to the ones in co-crystallized complexes.3–6 Although the co-crystallized protein-protein structures are still few, our earlier study1 showed that valid templates for the PPI modeling by structure alignment can be found for almost all known PPI that involve proteins for which the structure is known or can be built by homology (templates are available for the homology modeling of a significant part of the individual proteins7). A serious obstacle to the docking of protein structures is the conformational changes upon complex formation.8 Whereas the ultra-low resolution docking may be applicable to cases with large inaccuracies,9 the problem is explicitly addressed by docking methods that allow structure flexibility.10 The community-wide experiment on Critical Assessment of Predicted Interactions, CAPRI10,11 offers an objective comparative evaluation of the existing docking approaches.
Most proteins in the interactome are themselves models of limited accuracy.3 An important question asked by protein modelers,12 and biological researchers in general, is: what kind of structural information can be obtained from the docking of protein models and what is the reliability of such predictions? Protein models were shown to have significant utility in studies of protein-ligand interactions and characterization of functional sites.13–15 Protein-protein docking of models by information driven approach was validated on a set of CAPRI targets.16 High-resolution free docking was recently tested on a diverse set of protein models to reveal that meaningful predictions can be obtained even for models with significant distortions, although at significantly lower success rates.17 The systematic benchmarking on arrays of protein models at different accuracy levels was performed by the ultra-low resolution approach.18 The results showed that such docking determines the gross structural features of the complex for a significant portion of protein models, including highly inaccurate ones. However, because of limited availability of the templates for modeling of individual proteins, the study was based on “simulated models” of the proteins, which reflected the general structural accuracy of the homology models, but were not necessarily structurally similar to them. The study also was restricted to the ultra-low resolution free docking,9 effectively predicting the binding sites only. In this paper, we address the problem of models' utility in protein docking using our recent benchmark sets of actual protein models.19,20 The quality of the free and the template-based docking predictions built from these models was thoroughly assessed to reveal the tolerance limits of docking to structural inaccuracies of the protein models. The predictive power of currently available rigid-body and flexible docking approaches is similar.10 Thus in this study we used basic rigid-body approaches, developed in our group, that would clearly reveal the general similarities and differences in the free and the template-based docking performance depending on the modeling accuracy of the interacting proteins.
The results show that the existing docking methodologies can be successfully applied to protein models with a broad range of structural accuracy; the template-based docking is much less sensitive to the inaccuracies of protein models than the free docking; and docking can be successfully applied to the entire proteomes where most proteins are models of different accuracy.
Methods
Benchmark sets of protein models
The sensitivity of docking protocols to the inherent inaccuracies of protein models was tested on our specialized, carefully curated Models Docking Benchmark Set 2.20 The set contains 165 binary protein-protein complexes from the bound DOCKGROUND part21 with each monomer represented by six models with the increasing levels of inaccuracy (model-to-native Cα RMSD within 1 ± 0.2 Å, 2 ± 0.2 Å, … 6 ± 0.2 Å intervals). All monomer structures are bona fide models, generated by I-TASSER22,23, thus adequately reflecting the reality of modeling in the real case scenario.
Docking protocols
The free docking was performed by the FFT (Fast Fourier Transform) program GRAMM24,25 at low resolution, with 3.5 Å grid step and 10° angular interval. Top 100,000 matches were scored by the Miyazawa-Jernigan statistical potential26 and clustered. The clustering procedure utilized a simple greedy approach where the models are ranked according to their energy, and all matches within 10 Å ligand RMSD from the lowest energy one are assigned to the cluster. The procedure is repeated to exclude the clustered models (except for the lowest energy one).
The template-based docking was performed by the full structure alignment protocol,27 using template library28 of 4,950 co-crystallized binary complexes from DOCKGROUND.21 The target proteins were structurally aligned to the template monomers by TM-align.29 The resulting models (target/template TM-score > 0.4 only) were scored by the average of the two TM-scores.30
Metrics for docking accuracy
The accuracy of the predicted model-model complex combines the accuracy of the docking with the accuracy of the monomers modeling. Thus the docking assessment in this case is more complicated than in traditional docking of the X-ray structures.
To quantify the difference between docking modes, we calculated the fraction of shared residue contacts. Residues were considered in contact if the distance between their closest atoms was < 12 Å. Each configuration i of the protein-protein complex is characterized by a set Si of Ni pairwise contacts
| (1) |
where (a, b) is a pair of residues a of the receptor (the larger protein in the complex) and b of the ligand (the smaller protein in the complex) interacting across the interface. The similarity between configurations i and j, FSCij (fraction of shared contacts), can be calculated as the Jaccard index of the two sets Si and Sj
| (2) |
As opposed to ligand RMSD (RMSD between ligands in two docking modes with receptors superimposed), the FSCij between similar docking modes does not have substantial variation from complex to complex (see Supporting Information, Figure S1). The fraction of the native contacts (fnat), in CAPRI definition,31 cannot be directly used for pairwise comparison of model-model docking predictions because of the required reference set of the native interface residues/contacts, which varies in different docking models. In this regard, FSCij (Eq. 2) can be considered a modification of fnat, such that the number of shared contacts is normalized by the number of contacts in either of the two models, making the score symmetric (FSCij = FSCji).
Assessing docking predictions by CAPRI criteria
The docking predictions were assigned to four accuracy categories (high, medium, acceptable, incorrect) according to the CAPRI criteria31 (Table S1). A docked model-model complex was compared to a reference complex built by superimposition of two protein models with the same model-to-native RMSD onto the corresponding monomers from the native X-ray structure.20 Such “ideal” model-model complexes provide an estimate of the highest level of accuracy that can be achieved in the rigid-body docking of protein models. The co-crystallized X-ray structures were also used as the reference structures.
Assessing template-based docking predictions
In addition to the CAPRI criteria and the TM-score, we assessed the quality of the template-based docking using FSC-score, defined similarly to FSCij (Eq. 2)
| (3) |
where Stempl and Smodel are contact sets in the template and in the model built from that template, respectively. However, as opposed to the FSCij the FSC-score needs an additional rule for finding contacts shared by the two complexes with different monomers. We considered the template contacts (atempl, btempl) and the model contacts (amodel, bmodel) shared if in the alignments used to build the model, residues atempl and btempl are aligned to the residues amodel and bmodel, correspondingly. Equation 3 then can be rewritten in a simpler form
| (4) |
where Ntempl = |Stempl| and Nmodel = |Smodel| are the total number of contacts in the template and the model, respectively, and Nshared = |Stempl ∩ Smodel| is the number of contacts shared by the template and the model built on that template. Almost all models with FSC-score ≤ 0.05 are incorrect (L-RMSD > 10 Å), as shown in Figure S2, upper left panel, and thus were excluded from further consideration. Such simple filtering not only eliminated > 50% of bad predictions, but also ensures that any template-based docking prediction has a certain amount of contacts. Unlike the CAPRI criteria, the FSC-score can be used for the assessment of the docking models in the real-case modeling scenario when the reference native structure is not available.
Results and Discussions
Detection of near-native solutions
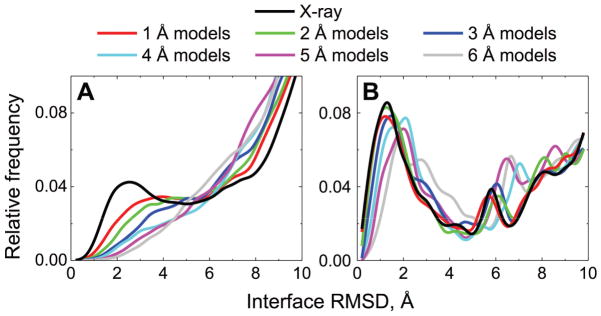
Protein interactions are driven by a funnel-like energy landscape, with the native structure of the complex inside the funnel.32 Thus the success of docking depends directly on the ability to detect the funnel. Since the energy landscape is a function of atomic coordinates, the landscapes of inherently inaccurate protein models differs from the landscapes of the corresponding X-ray structures. Thus the question is: whether the funnels can still be detected in the case of models. We addressed this question by analyzing spatial distributions of the top 1000 free and all template-based docking predictions for each model accuracy level for Benchmark 2 (Figure 1).
Figure 1. Distribution of near-native and false-positive matches according to the accuracy of protein models.
The top 1000 free docking (A) and all template-based docking (B) predictions, for each of the 165 complexes from the Models Docking Benchmark 2, at each of the six accuracy levels, were compared to the corresponding “ideal” complexes (see Methods) in terms of I-RMSD. In the docking of the X-ray structures, comparisons were made to the corresponding native X-ray structures. Near-native matches were defined as those with I-RMSD < 4 Å.
The bimodal distribution of interface RMSD between docking predictions and corresponding reference complex indicates detection of the funnels by the free32 and the template-based33 docking. As expected, the native peak at interface RMSD < 4 Å (in line with the CAPRI criteria, see Table S1) is clearly observed if the bound X-ray structures are docked by both the free and the template-based methods (black lines in Figure 1). With the decrease of models' accuracy, the peak in the free docking results diffuses and is no longer detectable for models with distortions ≥4 Å RMSD (Figure 1A). The near-native cluster of the free docking solutions, corresponding to this peak, decays exponentially (Figure S3) due to large structural distortions at the interface regions in the dataset (RMSD between interface Cα atoms of the model and the native structures for about half of the models is larger than RMSD calculated over all Cα atoms, see inset in Figure S3).
The template-based docking yields I-RMSD distributions with the distinct peak, corresponding to the near-native solutions, at all levels of monomer accuracy (Figure 1B). Unlike the free docking, which is based on the protein surface complementarity (and as a consequence, is sensitive to the local structural distortions), the template-based algorithm accounts for the entire protein fold. Thus the observed bimodality reflects the link between protein structure and function, which implies similar binding modes of structurally related proteins.
Because of the high sensitivity to the local structural inaccuracies, the success rate of the free docking decreases much more rapidly with the increasing level of model inaccuracy, compared to the template-based docking (dark gray and dashed bars in Figure 2). Interestingly, for some complexes, the free docking yielded good predictions for the models, but not for the X-ray structures (dashed bars in Figure 2A), due to the degree of noise inherent to the free docking. The template-based docking has almost no such cases (the dashed parts are hardly distinguishable in Figure 2B), which is related to the high degree of template conservation.
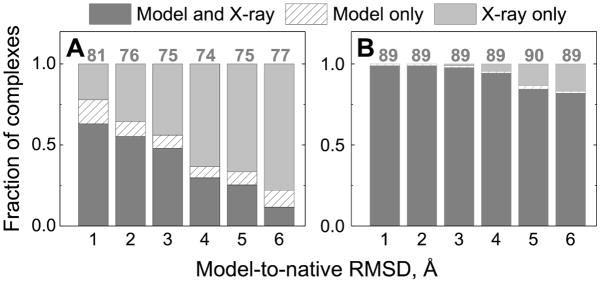
Figure 2. Docking success rates for protein models compared to the success rates for X-ray structures.
Successfully predicted complexes (those for which at least one acceptable or better quality prediction is among the top 10 docking poses), in the free docking (left hand panel) and the template-based docking (right hand panel) are in dark gray. Complexes with successful predictions by the X-ray docking only are in light gray. Complexes with successful predictions by the models docking only are in dashed bars. The quality of the models docking was accessed relative to the “ideal” complexes (see Methods). The data are normalized by the total numbers of complexes in all three categories shown on top of the bars.
In the above analysis, all docking predictions of models were compared to the corresponding “ideal” complexes (see Methods). If the native bound conformations were used instead, docking success rates decreased slightly along with the quality of models assessed by the CAPRI criteria, but the observed trends remained the same (Figure S4).
Stability of the solutions space
In addition to the analysis of top predictions, built into the traditional “success rates” metrics (Figure 2), analysis of a much broader range of predictions adds to the assessment of the docking quality. Docking with consistent hits near the correct prediction is more reliable than the one where the hits are widely dispersed.
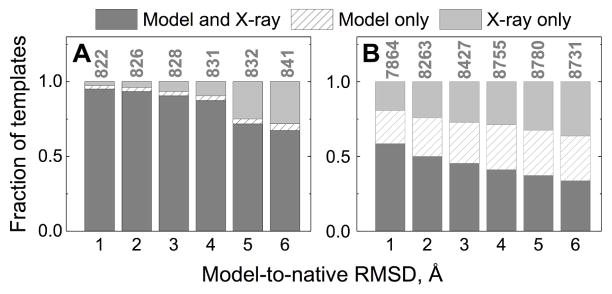
In the template-based docking, the number of predictions is limited by the number of detected templates, which varies from zero to several hundred. Most good (acceptable or better quality) model and X-ray docking predictions were built on the same templates (Figure 3A). Despite large local distortions (inset in Figure S3), global folds of the native structures are preserved in the majority of the models in the benchmark set (inset in Figure S6). Most templates yielding good models in X-ray template-based docking have target/template TM-scores > 0.6 (Figure S5). Distortions in models, albeit reducing target/template structural similarity (distributions for good models in Figure S5 shift to the left as the distortions in monomers increase) are, in most cases, not sufficient to bring the model under the detection threshold (TM-score 0.4). Thus, if a template yields a good X-ray docking prediction (typically with high TM-score), there is a high probability that the same template would be selected in the model docking, yielding a good docking prediction as well, although often in a lower quality category. The significant drop in the template-based docking performance is correlated with the loss of the native folds at large inaccuracy levels (Figure S6).
Figure 3. Conservation of templates in template-based docking of models.
Dark gray bars show templates common for the docking predictions of the X-ray structures and docking predictions of the corresponding models. Light gray bars show templates for the docking of the X-ray structures predictions only. Dashed bars show templates for the docking of the models predictions only. Data for good (acceptable or higher quality) predictions (A), and incorrect predictions (B) is normalized by the total number of templates shown on top of the bars.
Templates for incorrect docking predictions usually share less structural similarity to the target, resulting in TM-scores closer to the detection threshold, already seen for the X-ray-template pairs (Figure S5). Typically the alignment of a protein model to the template has a lower TM-score than the corresponding alignment of the X-ray structure. Thus, 20% to 36% of the templates (for 1 Å to 6 Å models correspondingly) detected in the docking of the X-ray structures because both TM-scores were > 0.4, become undetectable in the docking of models, because at least one TM-score dropped below 0.4 (light gray bars in Figure 3B). Interestingly, a significant amount of templates (22% for the 1 Å models, to 39% for the 6 Å models) is detected only in the models docking (dashed bars in Figure 3B). 95% of these templates have model/template TM-score 0.40 – 0.53, which means that their detection in the models docking is due to a small increase in the TM-score above the detection threshold due to “favorable” local structural variations in the models.
Shared templates (dark gray bars in Figure 3) yield model docking predictions with patterns of interface residue contacts similar to those in the X-ray predictions, irrespective of the monomers' accuracy (Figure 4). Local structural inaccuracies in the models of interacting proteins make some contacts disappear, or result in new contacts (average fnat values corresponding to 1 Å and 6 Å models in Figure 4B are 0.78 and 0.44 respectively, implying loss of 22 and 56% of the native contacts). Still, predominantly non-zero FSCij values suggest preservation of the docked monomers position with the increasing model inaccuracy. Such trend is similar for both good (acceptable and higher quality) and bad (incorrect) predictions with a slightly less pronounced effect for the latter (Figure S7), and with a fraction of the bad predictions (1.6% for the 1 Å models, to 6.1% for the 6 Å models) losing native contacts completely (minor peak in distributions at ~0, in Figure S7).
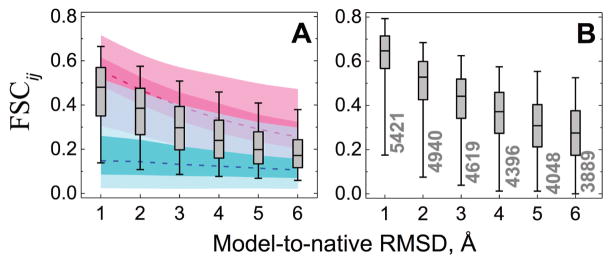
Figure 4. Comparison of free and template-based docking of models predictions with the docking of X-ray structures predictions in terms of fraction of shared contacts.
For each level of the model accuracy and each complex in the set, docking prediction of the X-ray structure with the maximum fraction of shared contacts FSCij (Eq. 2) was used for comparison with each of the top 1000 free docking of the models predictions. The resulting 165×1000 FSCij scores were plotted as gray box-and-whisker diagrams, separately for each distortion level (A). Box areas and whiskers contain 25 – 75 % and 5 – 95 % of data, respectively (outliers not shown). Lower bounds (blue) were estimated using 1000 randomly selected matches from the top 100,000 free docking of the models predictions. Upper bounds (red) were evaluated on a 1000-matches subset among 100,000 free docking of the models predictions with the maximum FSCij similarity to the top 1000 docking of the X-ray structures predictions. Darker and lighter areas of the upper and lower bounds correspond to boxes and whiskers respectively, and the dashed lines indicate medians. For the template-based docking (B), only pairs of the model and the X-ray predictions that share the same template (dark gray bars in Figure 3, and numbers at the whiskers in this figure) were considered. Upper and lower limits for the template-based docking were not estimated due to a statistically insufficient number of the docking predictions.
The templates' conservation and the models they produce are illustrated in Figure 5 for the two variable domains (chains L and H) in FV fragment of the anti-dansyl monoclonal antibody 1dlf. Immunoglobulins are widely represented in PDB, thus template-based docking of this structure results in a large pool of ~600 models. A tight cluster of the near-native solutions is preserved at all accuracy levels, whereas non-native matches have essentially random pattern with only a fraction of models shared between all accuracy levels.
Figure 5. Example of clustering in free and template-based docking.
Co-crystallized structures of the 1dlf chains H and L, along with their models at the six levels of accuracy from the Benchmark 2 were used in the free (left-hand panel) and the template-based (right-hand panel) docking. Top 1000 free and all template-based predictions are shown. Predicted matches are shown by yellow spheres, corresponding to the ligand (L chain) native interface center of mass. Magenta sphere corresponds to the native interface. The receptor structure (H chain) shown in cartoon is color-coded to reveal the location of distortions and their level. Distortions are measured as Cα-Cα distances calculated from RMSD-based superposition of the model onto the corresponding monomer from the co-crystallized complex.
In the free docking, the pool of initial models is much larger that in the template-based docking. Thus only the top 1000 solutions were selected for the scoring and the clustering. Since templates are not utilized in this method, we used a different approach to analyze the stability and conservation of the docking solutions.
Connectivity properties of similarity graphs constructed from the top or the randomly selected 1000 predictions were almost independent of the level of monomer distortion, albeit with substantial differences between these two groups of graphs (Figure S8). Pairwise comparison of distributions of cluster sizes for the six accuracy levels and the X-ray structures for each complex (in total, comparisons) indicated that only ~16% of the distribution pairs can be considered significantly different (comparison was done using two-sided Mann-Whitney U test34 at 0.05 significance level). This implies that the number and the size of clusters in top 1000 predictions do not vary significantly with the distortion level as well, albeit with some preference for the clusters originating from more distorted protein models to become less populated (169 distribution pairs with the clusters growing in size when distortion level increases, versus 389 opposite cases, as was identified by the one-sided Mann-Whitney U test).
However, in terms of the fraction of shared contacts, the free docking of models differs from the “native ensemble” (top 1000 X-ray free docking predictions) to a significantly larger extent, than in the template-based docking (Figure 4). The divergence of the model predictions from the native ensemble increases with the increase of the model inaccuracy (Figure 4A). The same trend is also observed for the upper bound of the average similarity (Figure 4A, red) indicating that even in the best case scenario local distortions in monomers allow only partial recovery of the residue contacts in the X-ray predictions. On the other hand, randomly selected docking models (Figure 4A, blue) share considerably less similarity to the native ensemble than the top 1000 predictions (box-and-whiskers in Figure 4A) for all model accuracy levels, implying preservation of some contacts from the native ensemble in all model predictions. Thus, local distortions in monomers substantially reduce the number of near-native solutions – Figure S3). However the clustering pattern remains almost unchanged (Figure S8).
A clustering example of the top 1000 free docking matches is shown in Figure 5A by the same variable fragment of the anti-dansyl monoclonal antibody. Most predictions are aggregated in the proximity of the large groove in the receptor (preserved in all models), formed by a concave β-sheet. The pool of the docking solutions in all cases is obviously non-random and some degree of similarity can be observed between docking of the co-crystallized X-ray structures and the models.
Template-based or free: which is preferable?
Protein docking methodologies are usually tested on unbound protein X-ray structures, with the challenge to accommodate the conformational difference from the bound protein. In this study, we challenged the docking programs much further since our protein models are, on average, significantly more different from the native bound structures, than the unbound X-ray structures. In the widely used protein-protein unbound X-ray docking Benchmark 435 only 24 out of 176 complexes (14%) are considered difficult, with I-RMSD in unbound/bound superposition > 2.2 Å. In comparison, in our Models Benchmarks 119 and 2,20 71% and 65% of the complexes, respectively, are that different from the bound X-ray structure. Thus, the unbound X-ray structures are easier to dock than the protein models (Figure S9).
In the free docking, the conformational deviation of the unbound X-ray structures can be mitigated by the low-resolution approach25 (albeit at the loss of atomic details in the docked complexes). Naturally, the low-resolution approach should help in the docking of protein models as well. Indeed, while the high-resolution docking outperforms the low-resolution one on the X-ray structures and on the models with small RMSDs from the native structures (Figure 6), starting from 2 Å RMSD (which roughly corresponds to the transition from “easy” to “difficult” unbound docking) the low-resolution docking systematically has higher success rate. Nevertheless, both the low- and the high-resolution docking have steeper decline of success rates with the increase of models' inaccuracy than the template-based docking (Figure 6). In our implementation, target/template similarity is assessed for the global fold, which determines the robustness of the docking solutions with respect to the local structural distortions in the protein models.
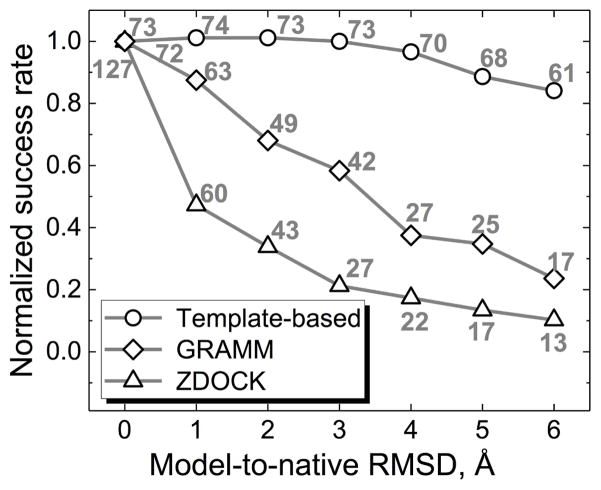
Figure 6. Normalized success rates for the template-based and the free docking.
The free docking at low resolution was performed by GRAMM, and at high resolution by ZDOCK 3.0.2.44 The complex was predicted successfully if one out of the top 10 predictions was correct (acceptable, medium or high quality). All success rates are normalized by the ones for the co-crystallized X-ray structures. The numbers above the data points show the absolute number of successful docking outcomes (out of 165 complexes in Benchmark 2).
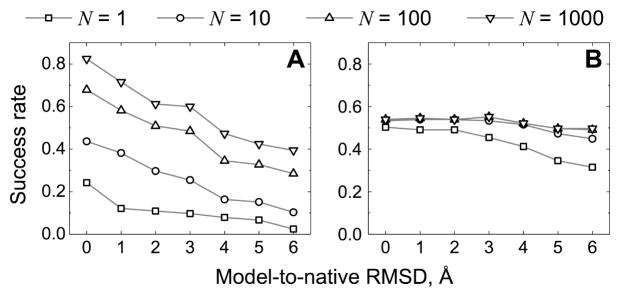
Interestingly, success rates of the free and the template-based dockings saturate differently with the increase of the number of considered top solutions (Figure 7). Rapid saturation of the template-based success rates indicates that the scoring scheme (see Methods) almost always finds the correct template among the top 10 detected templates (only 8 complexes have their good models ranks reduced to the top 1000 predictions at 6 Å accuracy). Moreover, ~60 % of the complexes retain a good model, although often in the lower quality category (larger N in the top N criterion), at the top of the list for all accuracy levels. Contrary to that, the free docking success rate consistently increases if more predictions are selected for the final analysis indicating a significant potential for improvement in the scoring of the initial scan stage models.
Figure 7. Docking success rates for different number of top solutions.
The successful prediction was defined as one correct structure (acceptable, medium or high quality) in the top N predictions. The rates are shown for the free (A) and the template-based docking (B).
Conclusions
We conducted systematic benchmark studies of the template-based and the free protein-protein docking methodologies on comprehensive sets of monomer protein models with a full array of accuracy levels. The results unambiguously show that the existing docking methodologies are applicable to protein models, even in case of relatively low protein structure accuracy. The template-based docking is significantly less sensitive to the distortions in protein models compared to the free docking. The template-based methodology yields model-model complexes with high degree of similarity to the docking predictions of the native X-ray structures, and its success rate is almost independent of the accuracy level (at the tested range). The results suggest that for protein models the use of the template-based docking is preferable provided a good template can be found. The hybrid (template-based and free) docking approach has been tested by our group in the “real case scenario” predictions of the joint CASP-CAPRI experiment,36 where all target proteins were bona fide models of different accuracy. Out of 25 target complexes, 11 were blindly predicted in at least acceptable docking accuracy category. The results show that the scoring scheme based on similarities of the global folds reliably finds good templates. However for some complexes (e.g., those with the alternative binding modes or in the twilight zone of target/template similarity) such scoring may not lead to the correct solution (similar conclusions were reached by Negroni et al.37). Thus, further improvement of the scoring would be useful in order to increase confidence in the model-model docking.
The free docking is essential for a number of important applications, including detection of transient complexes,38 modeling of protein association,39 and such. With the increase of the distortions in monomer models, the free docking performance significantly deteriorates. Still, the low-resolution in the free docking provides a degree of tolerance to local model distortions (success rates are non-negligible even at 4 – 6 Å distortion). However, to increase the docking reliability, the free docking scoring needs much greater improvement than the scoring for the template-based predictions.
The scoring for both the template-based and the free docking can be complemented by various constraints (e.g. automated literature search,40 evolutionary inferred residue-residue contacts,41,42 chemical shifts,43 etc.). With the continuous growth of publicly available information on protein interactions, the utility of such constrains will be increasing, expanding our abilities to reliably model the protein interactome.
Supplementary Material
Acknowledgments
This study was supported by NIH grant R01GM074255 and NSF grant DBI1262621. Calculations were conducted in part on ITTC computer cluster at The University of Kansas.
References
- 1.Kundrotas PJ, Zhu Z, Janin J, Vakser IA. Templates are available to model nearly all complexes of structurally characterized proteins. Proc Natl Acad Sci USA. 2012;109:9438–9441. doi: 10.1073/pnas.1200678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vakser IA. Low-resolution structural modeling of protein interactome. Curr Opin Struct Biol. 2013;23:198–205. doi: 10.1016/j.sbi.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vakser IA. Protein-protein docking: From interaction to interactome. Biophys J. 2014;107:1785–1793. doi: 10.1016/j.bpj.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosca R, Pons T, Ceol A, Valencia A, Aloy P. Towards a detailed atlas of protein–protein interactions. Curr Opin Struct Biol. 2013;23:929–940. doi: 10.1016/j.sbi.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Petrey D, Honig B. Structural bioinformatics of the interactome. Ann Rev Bioph. 2014;43:193–210. doi: 10.1146/annurev-biophys-051013-022726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrey D, Chen TS, Deng L, Garzon JI, Hwang H, Lasso G, Lee H, Silkov A, Honig B. Template-based prediction of protein function. Curr Opin Struct Biol. 2015;32:33–38. doi: 10.1016/j.sbi.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levitt M. Nature of the protein universe. Proc Natl Acad Sci USA. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrusier N, Mashiach E, Nussinov R, Wolfson HJ. Principles of flexible protein–protein docking. Proteins. 2008;73:271–289. doi: 10.1002/prot.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vakser IA, Matar OG, Lam CF. A systematic study of low-resolution recognition in protein-protein complexes. Proc Natl Acad Sci USA. 1999;96:8477–8482. doi: 10.1073/pnas.96.15.8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lensink MF, Wodak SJ. Docking, scoring, and affinity prediction in CAPRI. Proteins. 2013;81:2082–2095. doi: 10.1002/prot.24428. [DOI] [PubMed] [Google Scholar]
- 11.Vajda S, Vakser IA, Sternberg MJE, Janin J. Meeting report: Modeling of protein interactions in genomes. Proteins. 2002;47:444–446. doi: 10.1002/prot.10112. [DOI] [PubMed] [Google Scholar]
- 12.Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP) — round X. Proteins. 2014;82(Suppl 2):1–6. doi: 10.1002/prot.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skolnick J, Zhou H, Gao M. Are predicted protein structures of any value for binding site prediction and virtual ligand screening? Curr Opin Struct Biol. 2013;23:191–197. doi: 10.1016/j.sbi.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Dundas J, Kachalo S, Ouyang Z, Liang J. Accuracy of functional surfaces on comparatively modeled protein structures. J Struct Funct Genomics. 2011;12:97–107. doi: 10.1007/s10969-011-9109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kundrotas PJ, Vakser IA. Accuracy of protein-protein binding sites in high-throughput template-based modeling. PLoS Comp Biol. 2010;6:e1000727. doi: 10.1371/journal.pcbi.1000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodrigues JPGLM, Melquiond ASJ, Karaca E, Trellet M, van Dijk M, van Zundert GCP, Schmitz C, de Vries SJ, Bordogna A, Bonati L, Kastritis PL, Bonvin AMJJ. Defining the limits of homology modeling in information-driven protein docking. Proteins. 2013;81:2119–2128. doi: 10.1002/prot.24382. [DOI] [PubMed] [Google Scholar]
- 17.Maheshwari S, Brylinski M. Predicted binding site information improves model ranking in protein docking using experimental and computer-generated target structures. BMC Struct Biol. 2015;15:23. doi: 10.1186/s12900-015-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tovchigrechko A, Wells CA, Vakser IA. Docking of protein models. Protein Sci. 2002;11:1888–1896. doi: 10.1110/ps.4730102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anishchenko I, Kundrotas PJ, Tuzikov AV, Vakser IA. Protein models: The Grand Challenge of protein docking. Proteins. 2014;82:278–287. doi: 10.1002/prot.24385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anishchenko I, Kundrotas PJ, Tuzikov AV, Vakser IA. Protein models docking benchmark 2. Proteins. 2015;83:891–897. doi: 10.1002/prot.24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douguet D, Chen HC, Tovchigrechko A, Vakser IA. DOCKGROUND resource for studying protein-protein interfaces. Bioinformatics. 2006;22:2612–2618. doi: 10.1093/bioinformatics/btl447. [DOI] [PubMed] [Google Scholar]
- 22.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Molecular surface recognition: Determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vakser IA. Protein docking for low-resolution structures. Protein Eng. 1995;8:371–377. doi: 10.1093/protein/8.4.371. [DOI] [PubMed] [Google Scholar]
- 26.Miyazawa S, Jernigan RL. Self-consistent estimation of inter-residue protein contact energies based on an equilibrium mixture approximation of residues. Proteins. 1999;34(1):49–68. doi: 10.1002/(sici)1097-0134(19990101)34:1<49::aid-prot5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 27.Sinha R, Kundrotas PJ, Vakser IA. Docking by structural similarity at protein-protein interfaces. Proteins. 2010;78:3235–3241. doi: 10.1002/prot.22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anishchenko I, Kundrotas PJ, Tuzikov AV, Vakser IA. Structural templates for comparative protein docking. Proteins. 2015;83:1563–1570. doi: 10.1002/prot.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucl Acid Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 31.Lensink MF, Mendez R, Wodak SJ. Docking and scoring protein complexes: CAPRI 3rd Edition. Proteins. 2007;69:704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 32.Tovchigrechko A, Vakser IA. How common is the funnel-like energy landscape in protein-protein interactions? Protein Sci. 2001;10:1572–1583. doi: 10.1110/ps.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kundrotas PJ, Vakser IA. Protein-protein alternative binding modes do not overlap. Protein Sci. 2013;22:1141–1145. doi: 10.1002/pro.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mann HB, Whitney DR. On a test of whether one of 2 random variables is stochastically larger than the other. Ann Math Stat. 1947;18:50–60. [Google Scholar]
- 35.Hwang H, Vreven T, Janin J, Weng Z. Protein–protein docking benchmark version 4. 0. Proteins. 2010;78:3111–3114. doi: 10.1002/prot.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lensink MF, Velankar S, Kryshtafovych A, Huang SY, Schneidman-Duhovny D, Sali A, Segura J, Fernandez-Fuentes N, Viswanath S, Elber R, Grudinin S, Popov P, Neveu E, Lee H, Baek M, Park S, Heo L, Lee GR, Seok C, Qin S, Zhou HX, Ritchie DW, Maigret B, Devignes MD, Ghoorah A, Torchala M, Chaleil RA, Bates PA, Ben-Zeev E, Eisenstein M, Negi SS, Weng Z, Vreven T, Pierce BG, Borrman TM, Yu J, Ochsenbein F, Guerois R, Vangone A, Rodrigues JP, van Zundert G, Nellen M, Xue L, Karaca E, Melquiond AS, Visscher K, Kastritis PL, Bonvin AMJJ, Xu X, Qiu L, Yan C, Li J, Ma Z, Cheng J, Zou X, Shen Y, Peterson LX, Kim HR, Roy A, Han X, Esquivel-Rodriguez J, Kihara D, Yu X, Bruce NJ, Fuller JC, Wade RC, Anishchenko I, Kundrotas PJ, Vakser IA, Imai K, Yamada K, Oda T, Nakamura T, Tomii K, Pallara C, Romero-Durana M, Jimenez-Garcia B, Moal IH, Fernandez-Recio J, Joung JY, Kim JY, Joo K, Lee J, Kozakov D, Vajda S, Mottarella S, Hall DR, Beglov D, Mamonov A, Xia B, Bohnuud T, Del Carpio CA, Ichiishi E, Marze N, Kuroda D, Burman SSR, Gray JJ, Chermak E, Cavallo L, Oliva R, Tovchigrechko A, Wodak SJ. Prediction of homo- and hetero-protein complexes by ab-initio and template-based docking: A CASP-CAPRI experiment. Proteins. 2016 doi: 10.1002/prot.25007. [DOI] [Google Scholar]
- 37.Negroni J, Mosca R, Aloy P. Assessing the applicability of template-based protein docking in the twilight zone. Structure. 2014;22:1356–1362. doi: 10.1016/j.str.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Kozakov D, Li K, Hall DR, Beglov D, Zheng J, Vakili P, Schueler-Furman O, Paschalidis IC, Clore GM, Vajda S. Encounter complexes and dimensionality reduction in protein–protein association. eLife. 2014;3:e01370. doi: 10.7554/eLife.01370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou HX, Bates PA. Modeling protein association mechanisms and kinetics. Curr Opin Struct Biol. 2013;23:887–893. doi: 10.1016/j.sbi.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Badal VD, Kundrotas PJ, Vakser IA. Text mining for protein docking. PLoS Comp Biol. 2015;11:e1004630. doi: 10.1371/journal.pcbi.1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hopf TA, Scharfe CPI, Rodrigues JPGLM, Green AG, Kohlbacher O, Sander C, Bonvin AMJJ, Marks DS. Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife. 2014;3:e0343. doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue–residue interactions across protein interfaces using evolutionary information. eLife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stratmann D, Boelens R, Bonvin AMJJ. Quantitative use of chemical shifts for the modeling of protein complexes. Proteins. 2011;79:2662–2670. doi: 10.1002/prot.23090. [DOI] [PubMed] [Google Scholar]
- 44.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PloS One. 2011;6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.