Abstract
The heavily used protein-protein docking server ClusPro performs three computational steps as follows: (1) rigid body docking, (2) RMSD based clustering of the 1000 lowest energy structures, and (3) the removal of steric clashes by energy minimization. In response to challenges encountered in recent CAPRI targets, we added three new options to ClusPro. These are (1) accounting for Small Angle X-ray Scattering (SAXS) data in docking; (2) considering pairwise interaction data as restraints; and (3) enabling discrimination between biological and crystallographic dimers. In addition, we have developed an extremely fast docking algorithm based on 5D rotational manifold FFT, and an algorithm for docking flexible peptides that include known sequence motifs. We feel that these developments will further improve the utility of ClusPro. However, CAPRI emphasized several shortcomings of the current server, including the problem of selecting the right energy parameters among the five options provided, and the problem of selecting the best models among the 10 generated for each parameter set. In addition, results convinced us that further development is needed for docking homology models. Finally we discuss the difficulties we have encountered when attempting to develop a refinement algorithm that would be computationally efficient enough for inclusion in a heavily used server.
Keywords: protein-protein docking, SAXS data, docking with distance restraints, dimer classification, peptide-protein docking, structure refinement, scoring function
INTRODUCTION
The protein-protein docking server ClusPro performs three computational steps as follows: (1) rigid body docking by sampling billions of conformations, (2) RMSD based clustering of the 1000 lowest energy structures generated to find the largest clusters that will represent the most likely models of the complex, and (3) removal of steric clashes using energy minimization. The server was introduced in 2004,1,2 but at that time we did not have our own docking program, and used two of the best programs available, DOT by the Ten Eyck group3 and ZDOCK4 by the Weng lab.4 We considered the 20,000 lowest energy structures generated by DOT, and re-scored the conformations to retain 1500 structures with the best electrostatic energies and 500 structures with the best desolvation energies.1,2 ZDOCK provided 2000 structures selected using a combined scoring function, and hence we kept them without rescoring. Our main contribution that substantially improved the predictions has been the clustering of the 2000 conformations on the basis of their pairwise interface root mean-square deviation (IRMSD) values using 9Å as the clustering radius, and selecting the centers of the most populated low energy clusters as the models of the complex.5 The biophysical meaning of clustering is isolating low energy basins of the energy landscape that are both deep and broad.6 More recently we have shown that the size of each cluster is proportional to its probability, and hence retaining the largest clusters we can capture the most likely states of the associating proteins.7 Selecting models based on clustering rather than simply considering the lowest energy conformations remains an important part of the ClusPro server. While model selection based on cluster size is not widely adopted by the docking community, we note that a similar clustering step was implemented in the protein structure prediction program Rosetta.8 ClusPro was the only automated docking server in rounds 3–5 of CAPRI.9 It produced the 9th best results among all predictor groups,9 and we have been participating in CAPRI ever since.10–12
In 2006 we introduced PIPER,13 an FFT based docking program that was able to include a pairwise potential as part of its scoring function. The pairwise potential was based on the approach called DARS (Decoys As the Reference State).14 The novelty of DARS was that we generated a large decoy set of docked conformations using only shape complementarity as the scoring function (i.e., without any account for the atom types), and used the frequencies of interacting atom pairs in these decoy structures as the reference state. Thus, developing the potential we compared the frequency of contacts between two specific atom types in X-ray structures of protein complexes to the frequency of contacts in the decoys that are devoid of specific interactions. PIPER with DARS was implemented in the new server ClusPro 2.0, which clusters the top 1000 structures without any filtering.15 The server substantially increased the number of near-native complex structures found for enzyme-inhibitor and “other “ type of complexes in the protein benchmark set, but results for antibody-antigen pairs remained much poorer.13 However, in 2012 we have introduced a special DARS-type potential for this type of interactions, which provided some improvement.16
The last CAPRI evaluation meeting in April 2016 focused on the results of rounds 28–29 and 31–35 that included targets 59–67 and 95–105, respectively. Most targets in these rounds were challenging for ClusPro, as they included 8 protein-peptide complexes, 7 complexes considered difficult, and one target classified as very difficult. In spite, or possibly because, of this high level of general difficulty, ClusPro was the best performer among servers (TABLE I), and the 3rd best overall performer in terms of the number of targets for which acceptable and better predictions were submitted (TABLE II, note that for 2016 evaluation, following official assessment, T60–T64 where counted as 3 targets). As shown in TABLE II the server is almost competitive with the best human predictor groups in terms of such targets. However, the results of CAPRI also show that docking in general and ClusPro in particular need substantial further development. In fact, although CAPRI allows for the submission of ten models, acceptable predictions have been obtained only about half of the targets.
Table I.
Server performance based on the last three CAPRI evaluation meetings
| CAPRI evaluation meeting year and number of targets | ||||||
|---|---|---|---|---|---|---|
| 2009, 12 Targets | 2013, 14 Targets | 2016, 20 Targets | ||||
| Rank | Server | Successa | Server | Successa | Server | Successa |
| 1 | ClusPro (Vajda/Kozakov) |
5/1***/3** | ClusPro (Vajda/Kozakov) |
6/4** | ClusPro (Vajda/Kozakov) |
9/3** |
| 2 | HADDOCK (Bonvin) |
4/1***/1** | HADDOCK (Bonvin) |
4/1***/1** | PIE-DOCK (Elber) |
6/2** |
| 3 | GRAMM-X (Vakser) |
2/2** | SWARMDOCK (Bates) |
4/1** | LzerD (Kihara) |
4/1***/3** |
| 4 | SKE-DOCK (Umeyama) |
2/1** | PIE-DOCK (Elber) |
3/1** | HADDOCK (Bonvin) |
4/2** |
As defined by CAPRI evaluators, ***, **, and * denote high, medium, and acceptable accuracy submissions
Table II.
Predictor group performance based on the last three CAPRI evaluation meetings
| CAPRI evaluation meeting, year and targets | ||||||
|---|---|---|---|---|---|---|
| 2009, 12 Targets | 2013, 14 Targets | 2016, 20 Targets | ||||
| Rank | Group | Successa | Group | Successa | Group | Successa,b |
| 1 | Vajda/Kozakov | 6/4***/2** | Bonvin | 9/1***/3** | Guerois | 10/1***/8** |
| 2 | Zacharias | 6/4***/1** | Bates | 8/2** | Zacharias | 10/3***/2** |
| 3 | Zou | 6/3***/2** | Vakser | 7/1*** | ClusPro | 9/3** |
| 4 | Eisenstein | 6/3***/1** | Kozakov/Vajda | 6/2***/3** | Kozakov/Vajda | 8/3***/2** |
| 5 | Wolfson | 6/3***/1** | Shen | 6/1***/3** | Seok | 8/3***/2** |
| 6 | Weng | 6/2***/2** | Fernandez- Recio |
6/1***/3** | Fernandez-Recio | 7/1***/3** |
| 7 | Zhou | 6/2***/2** | ClusPro | 6/4** | Zou | 7/1***/2** |
| 8 | Bonvin | 6/1***/4** | Zou | 6/1***/2** | Weng | 6/1***/4** |
| 9 | ClusPro | 5/1***/3** | Zacharias | 6/1*** | Vakser | 6/2***/2** |
| 10 | Fernandez-Recio | 5/2** | Eisenstein | 5/1***/2** | Bates | 6/3** |
As defined by CAPRI evaluators, ***, **, and * denote high, medium, and acceptable accuracy submissions.
For 2016 joint protein-protein and protein-peptide docking performance is shown. Targets T60–T64 are counted as three targets, following official CAPRI evaluation
The goal of this paper is to describe three types of docking related problems. First, we consider the ones we have solved by developing new algorithms. Three of these algorithms have already been implemented as new options in ClusPro, and the remaining two will be added to the server in the near future. Second, we discuss problems that are difficult and still need further work, and finally problems that so far we have failed to solve in spite of substantial efforts – thus, the good, the bad, and the ugly. All work described here has been done during the last year, and was heavily influenced by the challenges posed by CAPRI targets. Unfortunately, in most cases the development was actually finished only after the submission deadline for the particular target, and hence some of the new methods could not be used in time. However, we show here some of the results indicating how the new methods would improve our predictions.
RESULTS AND DISCUSSION
Accounting for Small Angle X-ray Scattering (SAXS) data
SAXS experiments yield a one-dimensional scattering profile, containing information about the shape and size of the molecule17. The amount of this information is much lower than the one that can be obtained by X-ray crystallography, and on its own does not provide atom-level resolution. Rather than searching for complex structures for the best fit to SAXS data, in ClusPro we use the SAXS information as a filter, and focus on the regions of the configurational space containing the structures that are compatible with the scattering results, but otherwise perform docking as usual.18 This approach has the advantage that we avoid “overfitting” to the SAXS data, and hence the docking results will not get worse even in cases where the SAXS experiment provides very limited additional information. The use of SAXS experimental data as restraints has been already added as an advanced option to the server.18
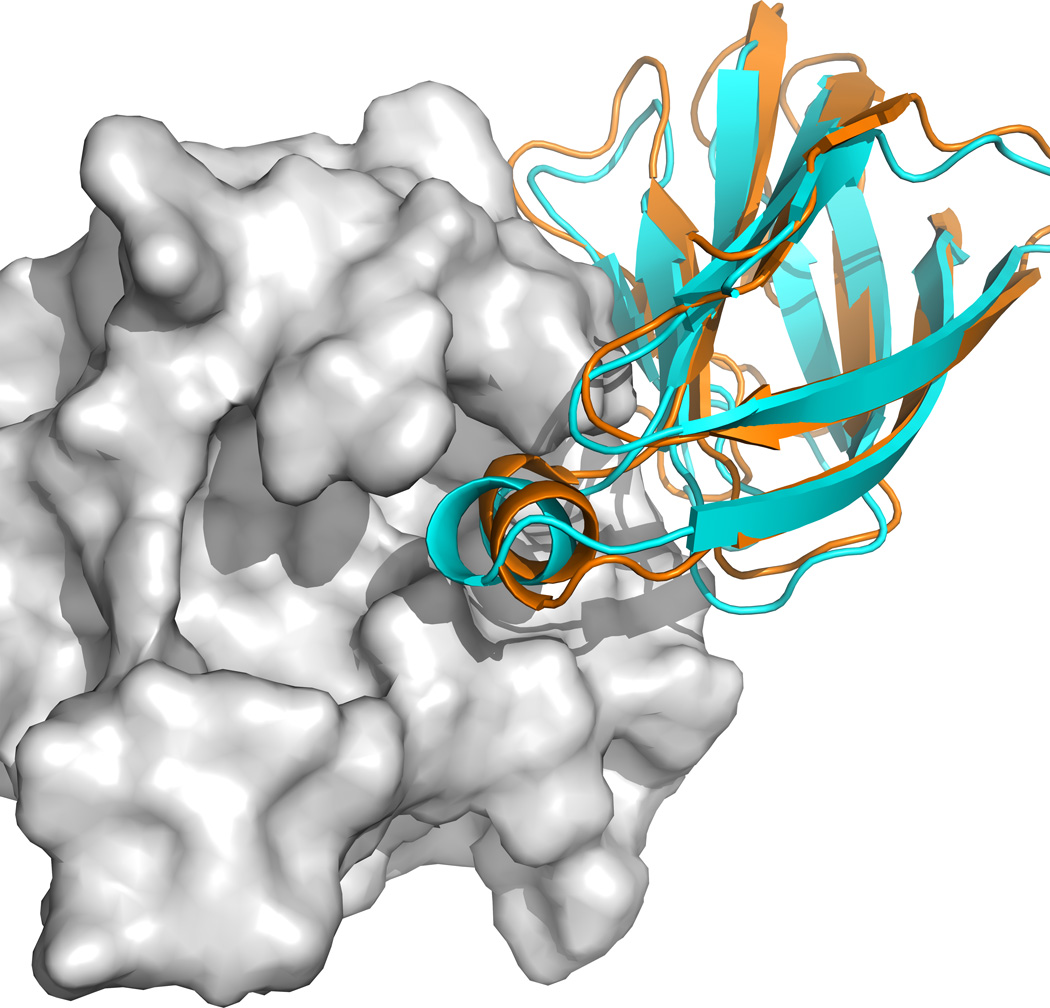
The development of a method to account for SAXS data was directly motivated by target 58 in Round 27 of CAPRI. The challenge was determining the complex between the salmon cold-active goose-type lysozyme and the E. coli PliG lysozyme inhibitor by constructing and docking a homology model of the lysozyme. SAXS data on the complex were provided to facilitate the docking.19 At the time of the challenge, we did not have the SAXS option. In addition, we selected a non-optimal template, and had no acceptable solutions for this target. Later we understood that, in spite of the moderate sequence identity of 57.8%, the best template to obtain correct conformation of a loop of the salmon lysozyme, interacting with the inhibitor, was the structure of black swan goose lysozyme (PDB ID 1GBS), and built the model using the MODELLER v9.020 program based on this template. Aromatic residues (Tyr, Phe, and Trp) that were not present in the template were placed in the most probable non-clashing rotamer positions. Other side chains that were not present in the template were not modeled, since placing them generally does not improve docking results. Docking to the resulting homology model ClusPro provided a near-native model even without the use of the SAXS restraints, but it was the center of the 6th largest cluster. Essentially the same near-native model was obtained when accounting for the SAXS data (see cyan cartoon in Figure 1), but now it was the center of the 3rd largest cluster. The X-ray structures of the bound ligand and the receptor in the complex (PDB ID 4G9S) are shown, respectively, as orange cartoon and grey surface in Figure 1.
Figure 1.
Docking the E. coli PliG lysozyme inhibitor to a homology model of the salmon goose-type lysozyme using SAXS data as restraints as given in Target 58 of Round 27 of CAPRI. A near-native model (shown as cyan cartoon) was obtained as the center of the 3rd largest cluster. The X-ray structures of the bound ligand and the receptor are shown, respectively, as orange cartoon and grey surface.
Accounting for pairwise distance restraints
The second enhancement, added to ClusPro in 2016, was the option of considering pairwise interaction data as restraints.21 This was an important but challenging development. Such pairwise restraints can be derived, e.g., from NMR Nuclear Overhauser Effect (NOE) experiments or by chemical crosslinking, and it is clear that accounting for them is needed in many applications. Docking with pairwise distance restraints is fundamental to HADDOCK,22 a heavily used protein-protein docking server. HADDOCK represents pairwise restraints by additional terms in its scoring function. However, this approach is computationally very inefficient with the fast Fourier transform (FFT) correlation approach used by PIPER. The problem is that each pairwise restraint in the scoring function requires a new correlation function term, and thus an additional Fourier transform, thereby reducing the numerical efficiency of the method. Since PIPER globally and systematically samples the entire conformational space of two interacting proteins on a dense grid, we accounted for restraints directly by selecting low energy solutions that also satisfy the restraints, rather then considering additional terms in the scoring function.21 Thus, similarly to using SAXS restraints, the scoring function is not affected, and hence the server can also be used with restraints that provide very limited information. Accounting for restraints was added to ClusPro as an advanced option.
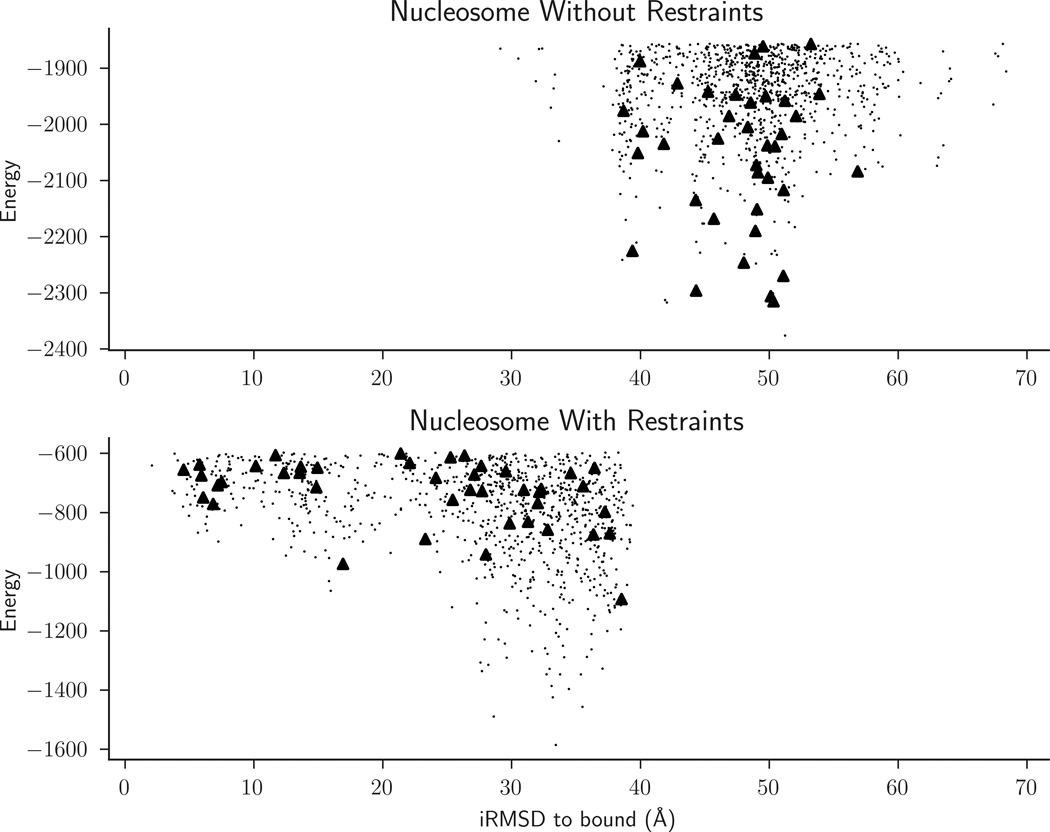
Considering pairwise interactions was motivated by CAPRI target 95, which required docking the PRC1 ubiquitylation module to the nucleosome. The nucleosome is a large complex of 8 protein chains with a total of 751 amino acid residues, plus two DNA chains 145 nucleotides each.23 The docking of PRC1 ubiquitylation module, which itself is a complex of three chains with 359 residues, is clearly a very difficult problem, and ClusPro did not produce any acceptable submission. However, evidence available from the literature provided pairwise distance restraints that we have later used to facilitate the docking.21 Catalytic competency of the PRC1 requires an interaction between Lys119 of the nucleosome histone H2A and Cys85 of the UbcH5c subunit of the PRC1 ubiquitylation module, resulting in a distance restraint that had to be satisfied between these two residues.24 The required range, 0 to 8Å, was fairly large, because these residues were located in flexible tail regions of the proteins. In addition, known mutation data suggested that Lys97 and Arg98 of PRC1 interact with the histone in the nucleosome, resulting in a second restraint group with multiple restraints, from Lys97 to the set of surface residues on the histone. In this second group we only required one of the restraints to be satisfied, since we did not know which of the residues on the surface of the histone interacted with Lys97. The best structure without restraints had the IRMSD of 38.68 Å, whereas docking using this restraint set produced a near-native cluster ranked 2 with the IRMSD of 4.53 Å (Figure 2).
Figure 2.
IRMSD versus energy plots for docking the PRC1 ubiquitylation module to the nucleosome as required in CAPRI Target 95. The energy of each docked structure is shown as a dot, and the cluster centers are shown as open triangles. The energy is in kcal/mol as provided by the PIPER program. Docking with restraints drastically shifts the distribution to the left, with the IRMSD of the best cluster center moving from 38.68 Å to 4.53 Å, and the latter was the 2nd largest cluster. Note that without restraints the lowest energy 1000 structures retained by ClusPro completely miss the near-native region.
Discrimination between biological and crystallographic homo-oligomers
The third problem we have recently considered is related to the recent CAPRI-CASP challenges that required the predictions of homodimers of protein chains that could be modeled using templates from the Protein Data Bank. Results summarized by Lensink et al.25 showed that the prediction of homodimer assemblies by homology modeling techniques and docking calculations was quite successful for targets featuring large enough subunit interfaces resulting in stable associations. However, targets with ambiguous or inaccurate oligomeric state assignments, often featuring crystal contact-sized interfaces, represented a confounding factor. For those, a much poorer prediction performance was achieved, while nonetheless often providing helpful clues on the correct oligomeric state of the protein. In complete agreement with this observation we have added the option of discriminating between biological and crystallographic dimers to ClusPro.26 The method is based on docking the protein (one subunit of the dimer) to itself to exhaustively sample the interaction energy landscape. If a substantial number of low energy docked poses cluster in a narrow vicinity of the structure of the dimer, then one can assume that there is a well-defined free energy well around the native state, which makes the interaction stable, and the dimer is most likely biological. In contrast, if the interaction sites in the docked poses do not form a large enough cluster around the native structure, then it is unlikely that the subunits form a stable biological dimer, and we most likely have a crystallographic dimer. The number of near-native structures is used to estimate the probability of a dimer being biological. Currently ClusPro examines only the stability of a given interface rather than generating all putative quaternary structures as accomplished by the PISA27 and EPPIC28 servers. Thus, unless already in the PDB file, the symmetry mates have to be generated, which can be accomplished using, say, PISA or the appropriate tools in PyMOL. However, the dimer classification option complements the information provided by PISA27 and EPPIC,28 which is particularly useful when results from the two established servers contradict to each other.
TABLE III shows author determined the and predicted oligomeric state of the targets in CAPRI Round 30 that have now structures in PDB and have been considered “easy” by Lensink et al.25 As shown, for all these targets the predictions by both PISA and EPPIC agree well with the author’s assignment. Notice that T79 has no author assigned state and the PISA prediction is uncertain as suggested by the server.27 In the case of EPPIC we adopt “bio” and “xtal” to denote biological and crystallographic association as used by the server,28 and thus all targets are considered biological. The last column in TABLE III shows that that the probability of forming a biological dimer, as predicted by ClusPro, is larger than 50% for all these targets. No attempts have been made here to study whether the dimers further dimerize, resulting in tetramers.
Table III.
Oligomeric state assignment for easy homodimer targets in CAPRI Round 30
| Target | PDB ID |
Protein name | Author assigned |
PISA | EPPIC | Probability of dimer by ClusPro |
|---|---|---|---|---|---|---|
| T69 | 4q34 | Putative esterase | dimer | dimer | bio | 0.79 |
| T71 | 4oju | Leucine rich repeat protein | tetramer | tetramer | bio | 0.83 |
| T73 | 4qhz | Putative glycosyl hydrolase | tetramer | tetramer | bio | 0.82 |
| T75 | 4q9a | Putative GDSL-like lipase | dimer | dimer | bio | 0.84 |
| T78 | 4qvu | Hypothetical protein | tetramer | tetramer | bio | 0.83 |
| T79 | 5a49 | LOTUS domain of OSKAR | n/a | tetramera | bio | 1.00 |
| T80 | 4piw | Sugar aminotransferase | dimer | dimer | bio | 0.77 |
| T81 | 4ojk | cGMP-dependent protein kinase |
tetramer | tetramer | bio | 0.83 |
| T85 | 4wji | Cyclohexadienyl dehydrogenase |
dimer | dimer | bio | 0.79 |
| T87 | 4wbt | Histidinol-phosphate aminotransferase |
dimer | dimer | bio | 0.78 |
| T90 | 4xau | Ats13 | dimer | dimer | bio | 0.69 |
| T91 | 4urj | Human Bj-Tsa-9 | monomer | dimer | bio | 1 |
| T92 | 4w66 | Glutathione S-transferase domain |
dimer | dimer | bio | 0.7 |
| T94 | 4w9r | APC103154 | dimer | dimer | bio | 0.7 |
Uncertain ensemble assignment by PISA
As noted by Lensink et al., some other targets were difficult or problematic. PDB structures are now available for six of these targets, and their more detailed analyses are reported in TABLE IV. The main problem with T68 (T0759) was that the crystal structure contains an artificial N-terminal peptide representing a His-tag that was used for protein purification.25 The N-terminal segments of neighboring subunits, which contain the artificial peptide, associate to form the largest interface between the subunits in the crystal (1150Å). With this peptide as part of the structure, ClusPro predicts that forming a biological dimer has 81% probability, although both PISA and EPPIC consider the protein to be monomeric. However, without the His-tag the ClusPro predicted probability of biological association is less than 50% for all interfaces, in agreement with the other two methods. T70 is a dimer by the author, but a tetramer by PISA, and analysis of all interfaces using ClusPro confirms this latter assignment.26 Target T72 is a dimer by both the authors and PISA but a monomer by EPPIC, and CluPro predicts that the protein forms a weak dimer (59% probability of being biological). Finally, ClusPro shows that targets T74, T77, and T86 are actually monomers, contradicting to the predictions by PISA, but in agreement with EPPIC predictions. These last cases emphasize that to have a third prediction method, in addition to PISA and EPPIC, can be very useful.
Table IV.
Oligomeric state assignment for difficult or problematic homodimer targets in CAPRI Round 30
| Target | PDB ID |
Chains | Interface Area, Å2 |
Author assigned |
PISA | EPPIC | Probability of dimer by ClusPro |
|---|---|---|---|---|---|---|---|
| T68 | 4q28 | monomer | monomer | monomer | |||
| (A:D with His tag) | 1150 | xtal | (0.81) | ||||
| A:D without His | 258 | xtal | 0.42 | ||||
| A:B | 861 | xtal | 0.21 | ||||
| T70 | 4pwu | dimer | tetramer | xtal | tetramer | ||
| A:B | 562 | xtal | 0.97 | ||||
| A:C | 196 | xtal | 0.11 | ||||
| AB:AB | 1036 | xtal | 0.75 | ||||
| T72 | 4q69 | A:B | 1119 | dimer | dimer | xtal | 0.59 |
| T74 | 4qb7 | monomer | tetramer | xtal | monomer | ||
| A:Y | 521 | xtal | 0.12 | ||||
| A:U | 492 | xtal | 0.11 | ||||
| T77 | 4qdy | A:A | 1600 | dimer | dimer | xtal | 0.13 |
| T86 | 4u13 | 680 | dimer | dimer? | xtal | 0.12 |
The good: Two additional problems we have solved
The fourth development, already published but not yet implemented in ClusPro, is a new algorithm and software to perform docking by employing fast generalized Fourier transforms on 5D rotational manifolds.29 While we started to work on such algorithms several years ago,30 the need for further speed-up of the FFT based docking was emphasized in this round of CAPRI, since several targets, including T59, involved docking of ensembles of structures obtained by Nuclear Magnetic Resonance (NMR). Dealing with such ensembles may require docking hundreds of multiple rigid body conformations, which significantly increases the computational costs of the docking, and would make server submissions within 48 hours extremely difficult. The new algorithm, based on generalizing the FFT correlation approach to the rotational group, is an order of magnitude faster than the classical Cartesian FFT approach implemented in PIPER, while providing very similar accuracy.29 An additional and very significant advantage of the generalized FFT approach is that adding extra correlation function terms to the energy function leads only to very minor increases in the computational efforts required. The new approach will be soon added to the server.
The fifth development we have completed but neither published nor added to ClusPro is the docking of flexible peptides to proteins. As mentioned, in latest rounds of CAPRI several targets required this type of calculation, leading to novel methodology based on the observation that we usually dock peptides that contain a sequence motif – i.e., a sequence of amino acids that frequently occur in protein-peptide interactions. For example, SH3 domains typically recognize proline-rich PXXP peptides, where P is fixed as the amino acid proline and X represents any amino acid. The key idea of the new algorithm is that protein fragments that include such a frequently occurring sequence motif also have some recurrent structure, and hence extracting and clustering fragments from the PDB with the motif provides a limited number of putative peptide structures in their protein environments. Given a specific target protein, the most likely structure and location can be relatively well identified by docking the extracted structures to the target protein. The method has been validated by applications to the established benchmark sets of protein-peptide interactions,31 and will be soon available as part of ClusPro.
The bad: Difficult problems that need solution
Here we describe three problems that were extensively studied during the last year, but are not yet solved, in spite of some progress. The first problem is selecting the relative weights of the different contributions to the energy expression used as the scoring function in docking. The PIPER interaction energy between two proteins is given by the expression of the form E = w1Erep + w2Eattr + w3Eelec + w4EDARS, where Erep and Eattr denote the repulsive and attractive contributions to the van der Waals interaction energy, and Eelec is an electrostatic energy term. EDARS is the DARS14 potential that primarily represents desolvation contributions13, and the coefficients w1, w2, w3, and w4 define the weights of the corresponding terms. After some trial and error we decided that by default ClusPro generates four sets of models using four different sets of energy coefficients that we call (1) balanced, (2) electrostatic-favored, (3) hydrophobic-favored, and (4) van der Waals + electrostatics. The balanced option works generally well for enzyme-inhibitor complexes, whereas options (2) and (3) are suggested for complexes where the association is primarily driven by electrostatic and hydrophobic interactions, respectively. The fourth option, van der Waals + electrostatics, means that w4=0, i.e., assumes that using the DARS potential, parameterized on a particular set of complexes, would make the predictions worse. We also have a fifth option that generates structures using three different coefficient sets and retains 500 conformations from each docking calculation to give 1500 conformations. This last option generally improves results for complexes that are in the “others” category of the protein docking benchmark set.32–35
Docking of the protein pairs in the protein-protein benchmark32–35 has shown that none of the above generic sets of weights works well for every protein, and the number of near-native structures generated substantially depends on selecting the right parameters. However, the server is currently unable to perform automated selection of the best scoring function, and we leave the task to the user, who may or may not have information facilitating such decision, Thus, a well defined challenge calls for the development of criteria that, based on the properties of interacting protein, would be able to suggest the most appropriate values of energy parameters. It is not clear how to solve this problem, and we envision the use of machine learning approaches. Another possibility is using adaptive docking that changes the weighting coefficients based on the outcome of an initial docking that provides information on the properties of the putative interface.
The second problem we have extensively studied is finding the models closest to the native structure among the many generated by the docking. After selecting the energy coefficients, ClusPro users face the problem of selecting one of the 10 (or up to 30, if required) models, each represented by the center of a highly populated cluster of low energy conformations. The problem of selecting the right cluster and thus the most likely docked structure is not yet solved. Fortunately many users of the server were able to reduce this uncertainty. Survey of the more than 400 articles that discuss models generated by ClusPro shows that in many applications the models were validated by experimental techniques, including site-directed mutagenesis with NMR, calorimetry, FRET, or surface plasmon resonance, cross-linking, various spectroscopic methods, X-ray scattering, electron self-exchange reaction, radiolytic protein footprinting with mass spectrometry, hydrogen/deuterium exchange, or intermolecular Nuclear Overhauser Effect restraints. Nevertheless, it would be desirable to provide computational tools for the selection of the clusters that are most likely near-native.
The third problem we have studied is the docking of homology models. A number of latest CAPRI targets required the docking of such models rather than separately crystallized components of the complex. Success in these cases strongly depends on the choice of the template and on the method of homology modeling. Examples included targets 96 and 97, in which the challenge was predicting the interaction of eGFP with two different artificial alpha repeat proteins. Although the sequence identity of the proteins to a known template was very high (80%), ClusPro was able to obtain acceptable quality model only for T96 but not for T97. This led us to realization that we have to develop special approaches to the docking of models. In consultation with CAPRI community, we have developed a web-based tool that generalizes the protein docking benchmark32–35 by including the need for homology modeling. Accordingly, the new benchmark provides lists of potential template structures that can be used to model the component proteins of the complexes in the benchmark set.36 If desired, the user can exclude the templates that could be used to obtain the entire complex by template based docking, since such templates were generally not available for the CAPRI targets.
The ugly: Problems we failed to solve
As shown in the last column of TABLE II, the main difference between the results by ClusPro and the best human predictor groups in the latest rounds of CAPRI is that while the server obtained almost the same number of acceptable predictions, it produced no high accuracy and much fewer medium accuracy submissions. Thus, in many cases the server was able to identify near-native clusters within 10Å IRMSD, but was not able to move these solutions closer to the X-ray structure. This is not surprising, since apart from local energy minimization to remove steric clashes, ClusPro does not have a refinement stage. In fact, we were not able to find an effective and computationally efficient refinement tool to include as part of our heavily used server. This is not for the lack of trying: during the last two years we have developed and tested several refinement algorithms. In terms of computational efficiency, the most promising approach was local resampling of near-native clusters on a denser grid.37 The method has been tested on 49 complexes using structures obtained by docking unbound protein pairs. The re-sampling increased the number of near-native structures for 31 of the 49 complexes, in 19 cases by more than 50%, whereas the number of near-native structures substantially decreased only in 5 cases.37 Since we generally had more near-native structure than in the initial docking, we clustered these with a smaller clustering radius, and expected to find some cluster centers closer to the native structure. However, this was the case only for a few of the proteins studied, and generally the distances of clusters centers from the X-ray structure did not substantially change.
Due to the failure of refinement by resampling we focused our efforts on the established Monte Carlo minimization (MCM) approach. Earlier we have employed MCM for eliminating about half of the non-native clusters in an approach we called stability analysis,38 but the simulations were computationally too expensive to be used in a server for refinement. Therefore our primary goal was to increase the efficiency of the MCM method, which was achieved by introducing three algorithmic innovations. First, we have developed a novel distributed algorithm for adjusting side chain conformations in the MCM steps.39 Second, we have used a pre-calculated structure-dependent rotamer library that had relatively few rotamers for most side chains.40 Third, we have developed a manifold based local energy minimization algorithm.41 The method was based on a new representation of the search space, and increased the efficiency of local energy minimization within the steps of the MCM method.41 These innovations enabled us to test refinement by MCM for a substantial number of complexes.39 Unfortunately we have found the refinement algorithm not very useful. The IRMSD from the X-ray structure generally improved for complexes that already had low IRMSD cluster centers. However, no improvement was observed for complexes that had no near-native clusters to begin with. The results in Table II clearly show this shortcoming. The three medium accuracy structures provided by ClusPro were refined to high accuracy by our “human” predictor team using the refinement algorithm, and two of the acceptable models were refined to medium accuracy (although for one target we lost the acceptable prediction due to “human” error). However, the refinement did not yield any acceptable model for targets that had no acceptable ClusPro results. We have determined that the main determinant of successful refinement has been whether the scoring function worked for the particular complex: if the energy function had false positive minima with lower energy than the minima around the native structure, then no refinement was able to improve the conformations. Refining already good accuracy predictions provided limited gain, and considering the high computational costs we decided not to add a refinement stage to the ClusPro server apart from the local minimization of the Charmm energy. While the energy minimization removes steric clashes, it does not change the IRMSD to any noticeable degree.
CONCLUSIONS
Our protein-protein docking server ClusPro has been used by the research community beyond our wildest expectations. By June 2016 the server had over 17,000 users (among them over 7,000 registered, which is not required), and completed more than 172,000 docking calculations, currently adding about 3,000 per month. Models built by ClusPro have been reported in over 400 publications. These statistics demonstrate that there is definitive need for protein-protein docking. Here we describe successful, as well as less successful, recent developments toward improving the server that were largely motivated by the challenges we have encountered when working on CAPRI targets. Successful development resulted in three options that were not previously available in ClusPro. These are (1) accounting for Small Angle X-ray Scattering (SAXS) data in docking; (2) the option of considering pairwise interaction data as restraints; and (3) enabling discrimination between biological and crystallographic dimers. In addition, we have developed an extremely fast docking algorithm based on 5D rotational manifold FFT, and an algorithm for docking flexible peptides that include known sequence motifs. The two new algorithms will also be part of the server. We feel that all these developments will further improve the utility of ClusPro. However, CAPRI also emphasized several shortcomings of the current server. In particular, we describe the problem of selecting the right energy parameters among the five options provided by ClusPro, and the problem of selecting the best models among the 10 generated for each of the five parameter sets. In addition, the results convinced us that we need specialized tools for docking homology models, and constructed a benchmark set to promote the development of such tools. Finally we described our efforts toward developing a refinement algorithm that would be computationally efficient enough for inclusion in a heavily used server, and recognize that so far this problem remains unsolved, reducing the server’s ability of producing high or medium accuracy predictions. Nevertheless, as shown in this paper, in the server category ClusPro achieved the best performance for all the last three CAPRI evaluation meetings, and was getting close to the performance of the best human groups. While acceptable or better solutions were submitted only about half of the targets, this indicates that the rigid body assumption is adequate for a large fraction of proteins, and that the global systematic search is very useful if no information on the interaction site can be used. We are also convinced that model selection based on the size of low energy clusters rather than based on energy values improves docking results and prides added robustness to the method.
Acknowledgments
Grant sponsor: NIH/NIGMS; Grant numbers: R35 GM118078 and R01 GM093147; Grant sponsor: National Science Foundation; Grant number: DBI 1458509 and AF 1645512
REFERENCES
- 1.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 3.Mandell JG, Roberts VA, Pique ME, Kotlovyi V, Mitchell JC, Nelson E, Tsigelny I, Ten Eyck LF. Protein docking using continuum electrostatics and geometric fit. Protein Eng. 2001;14:105–113. doi: 10.1093/protein/14.2.105. [DOI] [PubMed] [Google Scholar]
- 4.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 5.Kozakov D, Clodfelter KH, Vajda S, Camacho CJ. Optimal clustering for detecting near-native conformations in protein docking. Biophys J. 2005;89:867–875. doi: 10.1529/biophysj.104.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vajda S, Hall DR, Kozakov D. Sampling and scoring: A marriage made in heaven. Proteins. 2013;81:1874–1884. doi: 10.1002/prot.24343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, Vajda S. How good is automated protein docking? Proteins. 2013;81:2159–2166. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schueler-Furman O, Baker D. Conserved residue clustering and protein structure prediction. Proteins. 2003;52:225–235. doi: 10.1002/prot.10365. [DOI] [PubMed] [Google Scholar]
- 9.Mendez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3–5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 10.Lensink MF, Mendez R, Wodak SJ. Docking and scoring protein complexes: CAPRI 3rd Edition. Proteins. 2007;69:704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 11.Lensink MF, Wodak SJ. Docking and scoring protein interactions: CAPRI 2009. Proteins. 2010;78:3073–3084. doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- 12.Lensink MF, Wodak SJ. Docking, scoring, and affinity prediction in CAPRI. Proteins. 2013;81:2082–2095. doi: 10.1002/prot.24428. [DOI] [PubMed] [Google Scholar]
- 13.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 14.Chuang GY, Kozakov D, Brenke R, Comeau SR, Vajda S. DARS (Decoys As the Reference State) potentials for protein-protein docking. Biophys J. 2008;95:4217–4227. doi: 10.1529/biophysj.108.135814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comeau SR, Kozakov D, Brenke R, Shen Y, Beglov D, Vajda S. ClusPro: performance in CAPRI rounds 6–11 and the new server. Proteins. 2007;69:781–785. doi: 10.1002/prot.21795. [DOI] [PubMed] [Google Scholar]
- 16.Brenke R, Hall DR, Chuang GY, Comeau SR, Bohnuud T, Beglov D, Schueler-Furman O, Vajda S, Kozakov D. Application of asymmetric statistical potentials to antibody-protein docking. Bioinformatics. 2012;28:2608–2614. doi: 10.1093/bioinformatics/bts493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang S. Methods for SAXS-Based Structure Determination of Biomolecular Complexes. Advanced materials. 2014;26:7902–7910. doi: 10.1002/adma.201304475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia B, Mamonov A, Leysen S, Allen KN, Strelkov SV, Paschalidis IC, Vajda S, Kozakov D. Accounting for observed small angle X-ray scattering profile in the protein-protein docking server cluspro. J Comput Chem. 2015;36:1568–1572. doi: 10.1002/jcc.23952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leysen S, Vanderkelen L, Weeks SD, Michiels CW, Strelkov SV. Structural basis of bacterial defense against g-type lysozyme-based innate immunity. Cell Mol Life Sci. 2013;70:1113–1122. doi: 10.1007/s00018-012-1184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 21.Xia B, Vajda S, Kozakov D. Accounting for pairwise distance restraints in FFT-based protein-protein docking. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 23.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. Embo J. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lensink MF, Velankar S, Kryshtafovych A, Huang SY, Schneidman-Duhovny D, Sali A, Segura J, Fernandez-Fuentes N, Viswanath S, Elber R, Grudinin S, Popov P, Neveu E, Lee H, Baek M, Park S, Heo L, Rie Lee G, Seok C, Qin S, Zhou HX, Ritchie DW, Maigret B, Devignes MD, Ghoorah A, Torchala M, Chaleil RA, Bates PA, Ben-Zeev E, Eisenstein M, Negi SS, Weng Z, Vreven T, Pierce BG, Borrman TM, Yu J, Ochsenbein F, Guerois R, Vangone A, Rodrigues JP, van Zundert G, Nellen M, Xue L, Karaca E, Melquiond AS, Visscher K, Kastritis PL, Bonvin AM, Xu X, Qiu L, Yan C, Li J, Ma Z, Cheng J, Zou X, Shen Y, Peterson LX, Kim HR, Roy A, Han X, Esquivel-Rodriguez J, Kihara D, Yu X, Bruce NJ, Fuller JC, Wade RC, Anishchenko I, Kundrotas PJ, Vakser IA, Imai K, Yamada K, Oda T, Nakamura T, Tomii K, Pallara C, Romero-Durana M, Jimenez-Garcia B, Moal IH, Fernandez-Recio J, Joung JY, Kim JY, Joo K, Lee J, Kozakov D, Vajda S, Mottarella S, Hall DR, Beglov D, Mamonov A, Xia B, Bohnuud T, Del Carpio CA, Ichiishi E, Marze N, Kuroda D, Roy Burman SS, Gray JJ, Chermak E, Cavallo L, Oliva R, Tovchigrechko A, Wodak SJ. Prediction of homo- and hetero-protein complexes by protein docking and template-based modeling: a CASP-CAPRI experiment. Proteins. 2016 doi: 10.1002/prot.25007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yueh C, Hall DR, Xia B, Padhorny D, Kozakov D, Vajda S. ClusPro-DC: Dimer classification by the ClusPro server for protein-protein docking. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Duarte JM, Srebniak A, Scharer MA, Capitani G. Protein interface classification by evolutionary analysis. BMC Bioinformatics. 2012;13:334. doi: 10.1186/1471-2105-13-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padhorny D, Kazennov A, Zerbe BS, Porter KA, Xia B, Mottarella SE, Kholodov Y, Ritchie DW, Vajda S, Kozakov D. Protein-protein docking by fast generalized Fourier transforms on 5D rotational manifolds. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1603929113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie DW, Kozakov D, Vajda S. Accelerating and focusing protein-protein docking correlations using multi-dimensional rotational FFT generating functions. Bioinformatics. 2008;24:1865–1873. doi: 10.1093/bioinformatics/btn334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.London N, Raveh B, Cohen E, Fathi G, Schueler-Furman O. Rosetta FlexPepDock web server--high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011;39:W249–W253. doi: 10.1093/nar/gkr431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R, Mintseris J, Janin J, Weng Z. A protein-protein docking benchmark. Proteins. 2003;52:88–91. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- 33.Mintseris J, Wiehe K, Pierce B, Anderson R, Chen R, Janin J, Weng Z. Protein-Protein Docking Benchmark 2.0: an update. Proteins. 2005;60:214–216. doi: 10.1002/prot.20560. [DOI] [PubMed] [Google Scholar]
- 34.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein-protein docking benchmark version 3.0. Proteins. 2008;73:705–709. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang H, Vreven T, Janin J, Weng Z. Protein-protein docking benchmark version 4.0. Proteins. 2010;78:3111–3114. doi: 10.1002/prot.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohnuud T, Luo L, Wodak SJ, Bonvin AM, Weng Z, Vajda S, Schueler-Furman O, Kozakov D. A benchmark testing ground for integrating homology modeling and protein docking. Proteins. 2016 doi: 10.1002/prot.25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mamonov AB, Moghadasi M, Mirzaei H, Zarbafian S, Grove LE, Bohnuud T, Vakili P, Ch Paschalidis I, Vajda S, Kozakov D. Focused grid-based resampling for protein docking and mapping. J Comput Chem. 2016 doi: 10.1002/jcc.24273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozakov D, Schueler-Furman O, Vajda S. Discrimination of near-native structures in protein-protein docking by testing the stability of local minima. Proteins. 2008;72:993–1004. doi: 10.1002/prot.21997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moghadasi M, Mirzaei H, Mamonov A, Vakili P, Vajda S, Paschalidis I, Kozakov D. The impact of side-chain packing on protein docking refinement. J Chem Inf Model. 2015;55:872–881. doi: 10.1021/ci500380a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beglov D, Hall DR, Brenke R, Shapovalov MV, Dunbrack RL, Jr, Kozakov D, Vajda S. Minimal ensembles of side chain conformers for modeling protein-protein interactions. Proteins. 2012;80:591–601. doi: 10.1002/prot.23222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirzaei H, Zarbafian S, Villar E, Mottarella S, Beglov D, Vajda S, Paschalidis IC, Vakili P, Kozakov D. Energy Minimization on Manifolds for Docking Flexible Molecules. J Chem Theory Comput. 2015;11:1063–1076. doi: 10.1021/ct500155t. [DOI] [PMC free article] [PubMed] [Google Scholar]