Summary
Bacterial RNA polymerase-promoter open complexes can exist in a range of states in which the leading edge of the enzyme moves but the trailing edge does not, a phenomenon we refer to as “open complex scrunching”. Here we describe how open complex scrunching can determine the position of the transcription start site for some promoters, modulate the level of expression, and potentially could be targeted by factors to regulate transcription. We suggest that open complex scrunching at the extraordinarily active ribosomal RNA promoters might have evolved to initiate transcription at an unusual position relative to the core promoter elements in order to maximize the rate of promoter escape.
Keywords: promoter, promoter escape, RNA polymerase, scrunching, transcription initiation, transcription start site selection
Introduction
Transcription, translation, and replication each go through initiation, elongation, and termination phases. There is a lot of information about each of these steps in bacterial transcription, but our understanding of the transitions between the various steps in the transcription cycle is in its infancy. Furthermore, why transcription initiates at different positions at different promoters has long been a mystery. Some recent publications from our group have provided insights into these problems and more generally about the role of specific promoter sequences in the mechanism of transcription initiation [1, 2].
Transcription initiation is a multistep process that involves initial binding of RNA polymerase (RNAP) to double-stranded promoter DNA followed by a series of conformational changes of the complex that ultimately result in selection of a transcription start site and RNA formation [3-6]. The standard view of the transcription initiation reaction is that promoter DNA initially interacts with RNAP to form a “closed complex”. This is followed by a series of intermediates that culminate in the formation of an “open complex” in which approximately one turn of DNA is melted, and the template DNA strand is placed into the active center of RNAP. The open complex is then competent to bind NTPs and synthesize an RNA transcript.
Every promoter has its own kinetic properties, leading to its own individual activity. Ultimately, promoter sequence determines the extent of interaction with a series of different RNAP sub-domains (see Box 1), and the combined effects of these interactions result in promoter strengths that vary over at least three orders of magnitude [3]. Not only the overall promoter activity but also the rates of formation and decay of each initiation intermediate are functions of the DNA sequence. Although sequences closer to consensus generally form open complexes faster, the rate of open complex formation does not always correlate with the amount of RNA transcript production. For example, promoters with perfect -10 and -35 elements for binding to RNAP, spaced the perfect distance apart, have lower activity than promoters that deviate slightly from the perfect sequence elements with optimal spacing between the elements[11].
Box 1.
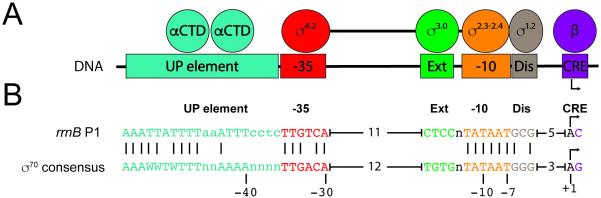
Modular elements in promoters make sequence-specific interactions with distinct regions of the RNAP σ subunit or with core RNAP (α2ββ’ω). The primary determinants of promoter strength are the -35 and -10 elements which bind σ region 4 (σ4) and σ region 2 (σ2), respectively (Fig. 1) [6]. Several additional promoter elements can also interact sequence-specifically with RNAP, thereby affecting the kinetics of the initiation reaction and influencing the strength of the promoter. These include the UP element (which binds the carboxyl-terminal domain of the α subunit of RNAP, αCTD) [7], the extended -10 element (which binds σ3.0)[8]; the discriminator element (which binds σ1.2) [9]; and the core recognition element (CRE; which binds the β subunit of RNAP)[10]. The strength of each of these interactions ultimately determines the kinetic properties of the promoter.
To understand the molecular interactions responsible for promoter escape, the transition between open complex formation and transcription elongation has been examined in more detail. Synthesis of the first 8-15 nt of RNA occurs without release of promoter contacts to RNAP, resulting in formation of a “scrunched” complex [12, 13]. Energy stored in this scrunched complex either allows the promoter-bound RNAP to go back to the open complex, releasing the nascent RNA as an “abortive” transcript, or the stored energy causes RNAP’s contacts with the promoter to break, allowing RNAP to escape from the promoter to form a productive elongation complex.
This article focuses on a phenomenon we call “open complex scrunching”. We discuss how open complex scrunching can affect the selection of the transcription start site (TSS), and we hypothesize that some promoters, namely those responsible for making ribosomal RNA, use this mechanism to improve the efficiency of promoter escape. We then discuss the potential regulatory role of open complex scrunching and start site selection more broadly in bacteria and eukaryotes.
Scrunching occurs during initial transcription
The term scrunching was initially used to describe the change in DNA conformation during initial transcription (Fig. 2A) [12-14]. During this step, some promoter interactions with RNAP that were established in the initial binding step are retained. The trailing edge of RNAP is stationary on promoter DNA, while the leading edge of RNAP moves downstream, 1 bp for each nt added to the growing RNA. Movement of the leading edge of RNAP without corresponding movement of the trailing edge results in expansion of the number of single-stranded nucleotides in the complex (i.e. the transcription bubble) to form a “scrunched” complex. These scrunched complexes are unstable and provide the driving force for either breaking promoter contacts and escaping from the promoter or for relaxing the complex back to an open state and releasing the RNA to form what is called an abortive RNA. During initial transcription, scrunching is driven by nucleotide addition. Below, we describe evidence for formation of a scrunched state not only in initiating complexes, but also before nucleotide addition, i.e. in the open complex at certain promoters.
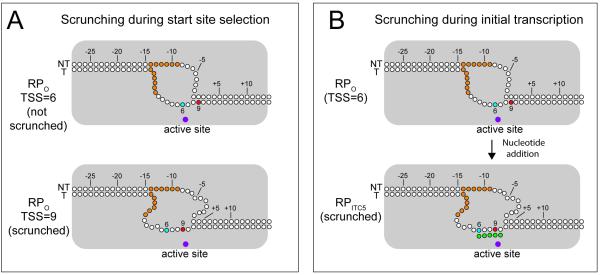
Figure 2.
Scrunching during transcription start site selection and initial transcription. RNAP is gray. Promoter DNA nucleotides (nt) are drawn as circles. The numbering is from wild-type rrnB P1. +1 is 9 bp downstream from the -10 hexamer. The -10 hexamer is shown as filled orange circles. Positions of two alternative transcription start sites are indicated as filled circles on the template strand, blue for the TSS 6 bp downstream from the end of the -10 element and red for the wild-type TSS 9 bp from the -10 element. The active site Mg2+ is shown as a filled purple circle. A: Scrunching during TSS selection. Two different forms of the open complex are shown. Top, non-scrunched complex results in a TSS 6 bp downstream from the -10 hexamer [1, 21]. Bottom, 3 nt of DNA are pulled into RNAP past the active site, resulting in a scrunched open complex and a TSS 9 bp downstream from the -10 hexamer. B: Scrunching during initial transcription. The proposed change in the DNA path during the transition from RPO (top) at an rrnB P1 promoter variant with a TSS of +6 with respect to the end of the -10 hexamer [21], to an initial transcribing complex with a 5 nt RNA (RPITC5, bottom). RNA nt are shown as filled green circles. Similar structural changes occur during initial transcription and during TSS selection. In each case, interconversion between the scrunched and unscrunched forms is likely to occur.
Evidence that scrunching can occur prior to nucleotide addition and is a determinant of the TSS
Transcription initiates most frequently 7 nt downstream from the end of the -10 promoter element, but it can start in a window 6-9 nt downstream, depending on the promoter [15-19]. Variability in transcription start site (TSS) selection suggested that the template strand DNA can be positioned in several different configurations in the open complex [20]. To determine the mechanism of variability in TSS selection, we took a similar approach in each of two studies: we identified promoter sequences that affected TSS selection and determined how the promoter DNA was positioned in complexes with different TSSs. In one study, we analyzed the ribosomal RNA promoter rrnB P1, which has an unusual TSS position 9 bp downstream from the -10 element [1]. We mapped transcription start sites from a library of promoter mutants to identify mutations that shifted the start site. Intriguingly, mutations in several discrete locations in the promoter affected TSS selection. Substitutions in the region immediately downstream of the -10 hexamer, the discriminator element, increased interactions with σ1.2, substitutions in the extended -10 element increased interactions with σ3.0, and several 1 bp insertions in the spacer between the -10 and -35 elements each resulted in an upstream shift of the TSS. To determine if these mutations affected the positioning of DNA in open complexes, we introduced the crosslinkable amino acid p-benzoyl-L-phenylalanine (Bpa) at 14 different positions in RNAP and mapped the locations of the resulting crosslinks to DNA at nt resolution [21]. We also mapped the position of the template strand of the promoter relative to the active site by replacing the active-site Mg2+ with Fe2+ to generate hydroxyl radicals that cleaved promoter DNA.
Together, the data indicated that the upstream part of the transcription bubble, as well as the double-stranded DNA upstream, were positioned identically on each of the promoters tested, regardless of the TSS. However, the downstream part of the bubble, as well as the double-stranded DNA downstream, were pulled into RNAP, past the active site in the wild-type rrnB P1 complex, correlating with an increased distance of the TSS from the -10 element. These data suggested that rRNA promoters form a “scrunched open complex”, similar to the scrunched complex that forms at most promoters following RNA synthesis. rRNA promoters thereby start further downstream than standard promoters.
In a second study (with the Nickels and Ebright labs), we generated transcripts from a library of promoter templates containing more than one million variants of the 10 bp sequence immediately downstream from the -10 element [2]. By mapping the TSS from this transcript library, we showed that the discriminator element sequence affects the position of the TSS and that purine-rich discriminator sequences favor TSSs further upstream of those formed by promoters with pyrimidine-rich discriminators. We then used the crosslinking approach to map the trailing and leading edges of the open complexes formed by RNAP and the more than one million promoter variants. The trailing-edge position did not change with discriminator sequence or TSS, but the leading edge position and TSS correlated with the discriminator element sequence. Since movement of the leading edge without movement of the trailing edge is one of the defining hallmarks of scrunching, we concluded that TSS selection is a function of DNA scrunching.
Together, the two studies showed that the transcription bubble in open complexes formed by different promoter sequences is not identical, that some promoters favor scrunching more than others, and that open complex scrunching is dependent on weak discriminator element interactions with σ1.2, weak extended -10 promoter element interactions with σ3.0, and with non-consensus -10 / -35 element spacing. Another study showed that the Core Recognition Element (CRE; see Box 1) can also affect TSS selection; it was proposed that the effects of the CRE also resulted from open complex scrunching [22].
Does open complex scrunching facilitate escape at ribosomal RNA promoters?
In E. coli, each of the 7 ribosomal RNA P1 promoters initiates transcription 9 bp downstream from the -10 element [1]. Furthermore, although rrn P1 TSS position has not been examined extensively in other species and the rRNA promoter sequences themselves have diverged greatly, transcription still initiates 9 bp downstream from the -10 element in species other than E. coli [23]. Conservation of TSS position while TSS region sequence identity has diverged suggests that initiating from a scrunched open complex has been selected for at rRNA promoters in evolution, at least in fast-growing γ-proteobacteria. Why have rRNA promoters evolved to initiate transcription from a scrunched state?
During fast growth, transcription of rRNA can account for up to 70% of the total transcription in the cell [24], and rRNA promoters have to be extremely efficient. It has been proposed that rRNA promoters approach the theoretical limit for initiation rate during fast growth [3]. This has been visualized in so-called “Miller spreads” generated from exponentially growing E. coli, in which it appears that RNAP molecules completely cover rRNA genes with essentially no gaps between them [25]. Several properties of rRNA promoters account for their fast transcription initiation rates. First, rRNA promoters recruit RNA polymerase very effectively because they have excellent UP elements [7], they are stimulated by interactions of their α-subunit C-terminal domains with the transcription factor Fis [26], and they have near consensus -35 elements and consensus -10 elements. However, rRNA promoters differ from consensus in their spacing between the -35 and -10 elements, and they have poor discriminator and extended -10 element sequences. We suggest that they compensate for these deviations from consensus in part by having evolved an extremely efficient mechanism for promoter escape.
Most promoter complexes go through multiple rounds of transcription initiation, in each round relaxing back to the open complex and releasing a short RNA transcript [27]. These released products are termed abortive RNAs. Escape from this cycle of binding and abortive product formation is rate-limiting at some promoters. In contrast, rRNA promoters bypass the abortive cycling step, escaping from the promoter without making abortive products [1].
Consistent with our hypothesis, we found that neither the wild-type rRNA promoter nor a variant that also initiated 9 nt downstream of the -10 element made abortive RNAs, whereas variants that initiated 6 nt downstream of the -10 element produced abortive products [1]. Thus, it appears that escape from the promoter without production of abortive RNAs correlates with initiation further downstream of the -10 hexamer in a scrunched state.
We hypothesize that initiating transcription from a scrunched open complex facilitates rapid RNA synthesis by “priming” the promoters for escape. Whereas scrunching in most promoter complexes is driven by RNA synthesis and results in formation of a “stressed intermediate” required for promoter escape, we propose that promoters that form scrunched open complexes can enter the intermediate state without the associated stress. Entry into this scrunched state, like entry into the other states on the pathway to formation of standard open complexes, is driven by binding free energy and does not require energy derived from nucleotide addition.
If promoter efficiency is the goal, why haven’t rRNA promoters just evolved to have better -35 hexamers, extended -10 elements, discriminator sequences, or better spacing between the -10 and -35 hexamers? Presumably evolution could have picked some other sequence combinations that maximized activity without impeding escape. We suggest another explanation, however.
Protein synthesis is the major consumer of energy in the cell. Cells have to down-regulate the rate of rRNA synthesis (and thus ribosome synthesis) when nutritional resources are scarce and growth slows down so as not to over-invest their resources in ribosome synthesis. They do this in two ways: (1) by producing increased concentrations of ppGpp, a non-canonical nucleotide that binds directly to RNAP and reduces the synthesis of rRNA (and some 400 other transcripts) [28], and (2) by reducing the concentrations of the NTPs needed for initiation at the 7 rrn P1 promoters, ATP and GTP, to inhibit rRNA promoter activity [29].
ppGpp inhibits rRNA promoters by binding to two sites on E. coli RNAP, reducing the lifetime of the open complex and thereby shifting the equilibrium between the closed and open complex back towards the closed complex [30-31]. Only promoters that are already rate-limited at this kinetic step by virtue of forming relatively unstable open complexes are subject to the inhibitory effects of ppGpp [32]. The same weak interactions of rRNA promoters with σ1.2, and σ3.0, along with their non-optimal -10 and -35 element spacing, that result in open complex scrunching also make these promoters susceptible to the inhibitory effects of high levels of ppGpp and reduced levels of NTPs. rRNA promoters are more sensitive to low initiating NTP concentrations than most promoters because they require high NTP concentrations to drive the closed complex – open complex equilibrium towards the open complex [29]. We suggest that rRNA promoters have evolved sequences that result in open complex scrunching as a means of facilitating promoter escape while at the same time employing non-consensus interactions with RNAP that allow regulation by high ppGpp and low initiating NTP concentration.
Is transcription start site selection regulated through effects on scrunching?
We have argued above that rRNA promoters have evolved to use open complex scrunching as a means of facilitating promoter escape for high activity without compromising regulatory capacity. That is, the 9 bp spacing between the -10 hexamer and the TSS can be considered a consequence of selection for a promoter with kinetic properties that result in rapid escape and at the same time regulation by small molecules.
However, there are other cases where changes in TSS more directly affect the level of gene expression. For example, Turnbough and coworkers found that changes in nucleotide concentrations shift the start sites of the pyrC and pyrD promoters in E. coli, thereby regulating pyrimidine biosynthesis. The TSS-dependent 5′ ends of these RNAs result in alternative secondary structures that are translated with different efficiencies [33]. Changes in the 5′ ends of RNAs can also result in alternative RNA structures that either favor or disfavor transcription attenuation or affect RNA stability.
Several studies in eukaryotic cells have also found that mRNAs derived from genes with alternative transcription start sites can be differentially terminated or translated [34-38]. In these cases, the start sites can vary over 100s of bp, instead of the 3-4 bp observed with bacterial promoters. The mechanism by which eukaryotic RNA polymerases select different transcription start sites as well as the sequence determinants of TSS selection are still unclear, but these results highlight the role that alternative RNA 5′ ends could play in modulating gene expression in all organisms.
Conclusion
Although changes in TSS can have consequences for expression of the final product, the frequency of such regulation by alternative TSS selection remains unclear. Global changes in TSSs have been mapped under a few different environmental or nutritional conditions in E. coli [39-41] and in a range of bacterial species. Other than the concentrations of the initiating NTP [33] and small RNA products referred to as nano RNAs [40-41], trans-acting factors affecting TSS selection have not yet been reported. However, the finding that open complex scrunching is dependent on promoter sequence and can cause shifts in TSS provides a mechanistic basis for how such a regulatory factor could potentially alter TSS selection.
How could a regulatory factor affect open complex scrunching? Since movement of the clamp domain of RNAP is thought to open the main channel in an early transcription initiation intermediate and to re-close the channel in a late intermediate [42], we speculate that a factor that acted on the clamp might facilitate open complex scrunching by allowing more DNA to be accommodated within the main channel. Closing of the channel in the late stages of open complex formation would then decrease the available room in the active site channel, inhibit open complex scrunching, and tighten RNAP-promoter interactions.
Figure 1.
Promoter elements and RNA polymerase regions responsible for recognition of each promoter element. A: Regions of core (αCTD and β) and σ70 that make sequence-specific interactions with promoter DNA are indicated above the DNA elements with which they interact. The transcription start site (TSS) is shown as a bent arrow. αCTD (carboxyl-terminal domain of α subunit; -35 (-35 hexamer), Ext (extended -10 element), -10 (-10 hexamer), Dis (discriminator element), CRE (core recognition element). B: rrnB P1 and consensus sequence for σ70-dependent promoters. rrnB P1 bases identical to the consensus sequence are indicated. Promoter elements are colored as in (A). W=A or T, n= any base. Spacing between -35 and extended -10 and between discriminator element and TSS are indicated by brackets. The number of bp separating elements is indicated.
Acknowledgements
We thank Wilma Ross and Bryce Nickels for comments on the manuscript. Work in the Gourse lab is supported by a grant from the National Institutes of Health R37 GM37048.
Abbreviations
- RNAP
RNA polymerase
- TSS
transcription start site
References
- 1.Winkelman J, Chandrangsu P, Ross W, Gourse R. Open complex scrunching before nucleotide addition accounts for the unusual transcription start site of E. coli ribosomal RNA promoters. Proc Natl Acad Sci USA. 2016;113:E1787–1795. doi: 10.1073/pnas.1522159113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winkelman J, Vvedenskaya I, Zhang Y, Zhang Y, et al. Multiplexed protein-DNA cross-linking: Scrunching in transcription start site selection. Science. 2016;351:1090–3. doi: 10.1126/science.aad6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Record M, Reznikoff W, Craig M, McQuade K, et al. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, et al., editors. ASM Press; Washington, DC: 1996. pp. 792–820. [Google Scholar]
- 4.Browning D, Busby S. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol. 2016;14:638–50. doi: 10.1038/nrmicro.2016.103. [DOI] [PubMed] [Google Scholar]
- 5.Ruff E, Record M, Artsimovitch I. Initial events in bacterial transcription initiation. Biomolecules. 2015;5:1035–62. doi: 10.3390/biom5021035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haugen S, Ross W, Gourse R. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–19. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross W, Gosink K, Salomon J, Igarashi K, et al. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–13. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 8.Barne K, Brown J, Busby S, Minchin S. Region 2.5 of the Escherichia coli RNA polymerase sigma 70 subunit is responsible for the recognition of the ‘extended-10′ motif at promoters. EMBO J. 1997;16:4034–40. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haugen S, Berkmen M, Ross W, Gaal T, et al. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–82. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Feng Y, Chatterjee S, Tuske S, et al. Structural basis of transcription initiation. Science. 2012;338:1076–80. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graña D, Gardella T, Susskind M. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988;120:319–27. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Revyakin R, Liu C, Ebright R, Strick T. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–43. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapanidis A, Margeat E, Ho S, Kortkhonjia E, Weiss S, et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–7. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheetham D, Jeruzalmi D, Steitz T. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–3. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- 15.Jeong W, Kang C. Start site selection at lacUV5 promoter affected by the sequence context around the initiation sites. Nucleic Acids Res. 1994;22:4667–72. doi: 10.1093/nar/22.22.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis D, Adhya S. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol Microbiol. 2004;54:692–701. doi: 10.1111/j.1365-2958.2004.04318.x. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Turnbough C. Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J Bacteriol. 1994;176:2938–45. doi: 10.1128/jb.176.10.2938-2945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vvedenskaya I, Zhang Y, Goldman S, Valenti A, et al. Massively systematic transcript readout (MASTER): transcription start site selection, transcriptional slippage, and transcript yields. Mol Cell. 2015;60:953–65. doi: 10.1016/j.molcel.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker K, Osuna R. Factors affecting start site selection at the Escherichia coli fis promoter. J Bacteriol. 2002;184:4783–91. doi: 10.1128/JB.184.17.4783-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robb N, Cordes T, Hwang L, Gryte K, et al. The transcription bubble of the RNA polymerase-promoter open complex exhibits conformational heterogeneity and millisecond-scale dynamics: implications for transcription start-site selection. J Mol Biol. 2013;425:875–85. doi: 10.1016/j.jmb.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkelman J, Winkelman B, Boyce J, Maloney M, et al. Crosslink mapping at amino acid-base resolution reveals the path of scrunched DNA in initial transcribing complexes. Mol Cell. 2015;59:768–80. doi: 10.1016/j.molcel.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vvedenskaya I, Vahedian-Movahed H, Zhang Y, Taylor D, et al. Interactions between RNA polymerase and the core recognition element are a determinant of transcription start site selection. Proc Natl Acad Sci USA. 2016;113:E2899–2905. doi: 10.1073/pnas.1603271113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aiyar S, Gaal T, Gourse RL. rRNA promoter activity in the fast-growing bacterium Vibrio natriegens. J Bacteriol. 2002;184:1349–58. doi: 10.1128/JB.184.5.1349-1358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bremer H, Dennis P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. pp. 1553–1569. [Google Scholar]
- 25.French S, Miller O. Transcription mapping of the Escherichia coli chromosome by electron microscopy. J Bacteriol. 1989;171:4207–16. doi: 10.1128/jb.171.8.4207-4216.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross W, Thompson J, Newlands J, Gourse R. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–42. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu L. Promoter clearance and escape in prokaryotes. Biochem Biophys Acta. 2002;1577:191–207. doi: 10.1016/s0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 28.Durfee T, Hansen A, Zhi H, Blattner F, et al. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–96. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaal T, Bartlett M, Ross W, Turnbough C, et al. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–7. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 30.Ross W, Vrentas C, Sanchez-Vazquez P, Gaal T, et al. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–9. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross W, Sanchez-Vazquez P, Chen A, Lee J, et al. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol Cell. 2016;62:811–23. doi: 10.1016/j.molcel.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barker M, Gaal T, Josaitis C, Gourse R. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–88. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 33.Turnbough C, Switzer R. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuehner J, Brow D. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell. 2008;31:201–11. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 35.Thiebaut M, Colin J, Neil H, Jacquier A, et al. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell. 2008;31:671–82. doi: 10.1016/j.molcel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Arribere J, Gilbert W. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res. 2013;23:977–87. doi: 10.1101/gr.150342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malabat C, Feuerbach F, Ma L, Saveanu C, Jacquier A. Quality control of transcription start site selection by nonsense-mediated-mRNA decay. eLife. 2015;4:e06722. doi: 10.7554/eLife.06722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Floor S, Doudna J. Tunable protein synthesis by transcript isoforms in human cells. eLife. 2016;5:e10921. doi: 10.7554/eLife.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomason M, Bischler T, Eisenbart S, Forstner K, et al. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol. 2015;197:18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Druzhinin S, Tran N, Skalenko K, Goldman S, et al. A conserved pattern of primer-dependent transcription initiation in Escherichia coli and Vibrio cholera revealed by 5′ RNA-seq. PLoS Genet. 2015;11:e1005348. doi: 10.1371/journal.pgen.1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vvedenskaya I, Sharp J, Goldman S, Kanabar P, et al. Growth phase-dependent control of transcription start site selection and gene expression by nanoRNAs. Genes Dev. 2012;26:1498–507. doi: 10.1101/gad.192732.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty A, Wang D, Ebright Y, Korlann Y, et al. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–5. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]