Abstract
Plastid DNA markers sequencing and DNA fingerprinting approaches were used and compared for resolving molecular phylogeny of closely related, previously unexplored Amorphophallus species of India. The utility of individual plastid markers namely rbcL, matK, trnH–psbA, trnLC–trnLD, their combined dataset and two fingerprinting techniques viz. RAPD and ISSR were tested for their efficacy to resolves Amorphophallus species into three sections specific clades namely Rhaphiophallus, Conophallus and Amorphophallus. In the present study, sequences of these four plastid DNA regions as well as RAPD and ISSR profiles of 16 Amorphophallus species together with six varieties of two species were generated and analyzed. Maximum likelihood and Bayesian Inference based construction of phylogenetic trees indicated that among the four plastid DNA regions tested individually and their combined dataset, rbcL was found best suited for resolving closely related Amorphophallus species into section specific clades. When analyzed individually, rbcL exhibited better discrimination ability than matK, trnH–psbA, trnLC–trnLD and combination of all four tested plastid markers. Among two fingerprinting techniques used, the resolution of Amorphophallus species using RAPD was better than ISSR and combination of RAPD +ISSR and in congruence with resolution based on rbcL.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-016-0400-0) contains supplementary material, which is available to authorized users.
Keywords: Amorphophallus, Molecular phylogeny, DNA finger printing, rbcL, matK, Maximum likelihood, Bayesian inference
Introduction
Genus Amorphophallus Blume ex Decne belongs to the family Araceae and comprises of perennial herbaceous plants that are mainly adapted to grow in the shady and mountainous areas. This genus is represented by 200 species distributed in the tropical Africa, Madagascar, tropical and subtropical Asia, the Malay Archipelago, Melanesia and Australia (Mayo et al. 1997). The genus has economic potential as starch and other carbohydrate yielding plants. Some of the species are used as food and vegetables while others have ornamental potentials. With the merger of Amorphophallus sect. synantherias into section Rhaphiophallus (Sivadasan 1989), the genus at present has 10 sections, of which 3 sections viz. Candarum Engl., Conophallus (Schott) Engl. and Rhaphiophallus (Schott) Engl. are represented in India. Recently section Candarum has been revised and renamed as section Amorphophallus (Jaleel et al. 2014). In India, these sections are represented by 18 species and 7 varieties of which 12 species and 3 varieties are endemic to the country. Identification and discrimination of Amorphophallus species is difficult due to closely related morphological characters. Because of the timing of emergence of inflorescences and their relatively short active period of existence, the taxonomic identification of Amorphophallus species becomes difficult. Moreover, close similarity of morphological characters of the leaves of many Amorphophallus species makes their identification with vegetative specimens difficult or impossible (Jaleel et al. 2011).
Previously, several studies investigated the phylogenetic relationships within the genus Amorphophallus. Recently, Grob et al. (2002, 2004) used molecular data for phylogenetic reconstruction and attempted to interpret morphological character evolution in Amorphophallus based on a combined nuclear and plastid phylogeny. This study based on matK and trnL intron sequences indicated that Amorphophallus can be divided into five well supported clades (Grob et al. 2002). Also, Sedayu et al. (2010) studied the morphological character evolution of Amorphophallus based on a combined phylogenetic analysis of trnL, rbcL and LEAFY second intron sequences. Three major clades were identified which reflected the biogeographical distribution of Amorphophallus and some of the clades were supported by morphological characters. The study conducted by Grob et al. (2002) included only three Amorphophallus species from India namely A. commutatus, A. margaritifer and A. smithsonianus whereas the study by Sedayu et al. (2010) included six species from India namely A. commutatus, A. hirsutus, A. hohenackeri, A. konkanensis, A. longiconnectivus and A. margaritifer. The molecular phylogeny of remaining Amorphophallus species from India is still not known.
It is generally agreed that a multilocus approach based on plastid (‘chloroplast’) data is currently the most effective strategy for species identification and species recognition in plants (Chase et al. 2005, 2007a, b; Kress et al. 2005, Kress and Erickson 2007; Newmaster et al. 2006, 2008; Cowan et al. 2006). Beside multilocus DNA barcoding approach, molecular marker based fingerprinting techniques can also be used to study the similarity of genome of the different species distributed in same geological area (Kumar et al. 2008). Molecular markers such as randomly amplified polymorphic DNA (RAPD) and Inter Simple Sequence Repeats (ISSR) have been successfully utilized for phylogenetic studies of many plant species (Fang et al. 1997; Joshi et al. 2000; Rajesh et al. 2003; Arnau et al. 2003).
In this report, we reconstructed the molecular phylogeny of Amorphophallus species from India using four different plastid gene regions sequences viz. rbcL, matK, trnH–psbA, trnLC–trnLD and two fingerprinting markers namely RAPD and ISSR. Molecular phylogeny was reconstructed to infer species relationship and to test if molecular phylogeny resolves Amorphophallus species into three section specific clades namely Rhaphiophallus, Conophallus and Amorphophallus. We evaluated the sequences of these plastid gene regions individually as well as their combined data for reconstruction of phylogenetic trees using for maximum likelihood (ML) and Bayesian Inference (BI) analyses and compared their efficacies with RAPD and ISSR. The objective of the present study was to assess if the resolution of closely related Amorphophallus species of India into three sections is also supported by molecular phylogeny. This was achieved by using (1) multi-locus plastid gene regions sequencing approach (2) DNA fingerprinting method, and (3) and comparing the performance of both approaches.
Materials and methods
Sampling
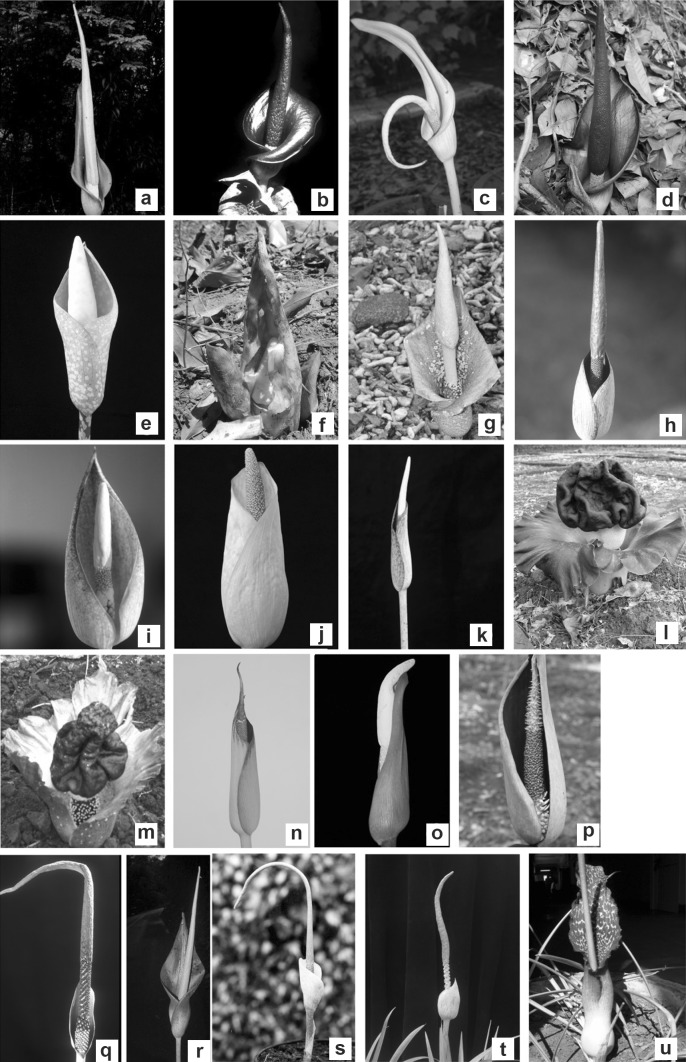
Tubers or plantlets of 16 Amorphophallus species together with six varieties of two species were initially collected from different locations in India and maintained in Lead Botanical Garden of Department of Botany, Shivaji University, Kolhapur, India. Our specimen included ten species from section Rhaphiophallus, four species together with four varieties of Amorphophallus commutatus from section Conophallus, two species together with two varieties of Amorphophallus paeoniifolius from section Amorphophallus, and Sauromatum venosum as an out-group (Table 1). Specimens were taxonomically identified by studying floral characters (Fig. 1), using basic floras, monographs; papers and herbarium were submitted at Dept. of Botany, Shivaji University, Kolhapur (SUK), India.
Table 1.
Collection localities and sections of Amorphophallus species and varieties
| Sr. no. | Name of species | Vouchers specimens | Sections | Locality |
|---|---|---|---|---|
| 1 | A. commutatus var. commutatus | ARG-19 | Conophallus | Amba, Maharashtra, India. |
| 2 | A. commutatus var. anmodensis | ARG-21 | Conophallus | Anmod Ghat, Goa, India |
| 3 | A. commutatus var. wayanadensis | ARG-20 | Conophallus | Mulshi, Maharashtra, India |
| 4 | A. commutatus var. anshiensis | ARG-22 | Conophallus | Anshi National Park, Karnataka, India |
| 5 | A. bulbifer | ARG-18 | Conophallus | Jog fall, Karnataka, India |
| 6 | A. hirsutus | ARG-35 | Amorphophallus | Andaman Island, India |
| 7 | A. oncophyllus | ARG-36 | Conophallus | Andaman Island, India |
| 8 | A. konkanensis | ARG-12 | Rhaphiophallus | Belgaum, Karnataka, India |
| 9 | A. hohenackeri | ARG-16 | Rhaphiophallus | Kaddan pedika, Kerala, India |
| 10 | A. margaritifer | ARG-8 | Rhaphiophallus | Parasnath, Jharkhand, India |
| 11 | Amorphophallus species | ARG-24 | Rhaphiophallus | Gondia, Maharashtra, India |
| 12 | A. paeoniifolius var. campanulatus | ARG-34 | Amorphophallus | Botanical Garden. Shivaji Uinversity, India |
| 13 | A. paeoniifolius var. paeoniifolius | ARG-37 | Amorphophallus | Botanical Garden. Shivaji Uinversity, India |
| 14 | A. bhandarensis | ARG-38 | Rhaphiophallus | Bhandara, Tumsar, Maharashtra, India |
| 15 | A. bonaccordensis | ARG-13 | Rhaphiophallus | Bonaccord, Kerala, India |
| 16 | A. longiconnectivus | ARG-3 | Rhaphiophallus | Khandva, Madhya Pradesh, India |
| 17 | A. mysorensis | ARG-33 | Rhaphiophallus | Billigirirangan Hills., Karnataka, India |
| 18 | A. nicolsonianus | ARG-15 | Conophallus | Bonaccord, Kerala, India |
| 19 | A. smithsonianus | ARG-14 | Rhaphiophallus | Bonaccord, Kerala, India |
| 20 | A. sylvaticus | ARG-40 | Rhaphiophallus | Pillathikkuppam, Tamil Nadu, India. |
| 21 | Sauromatum venosum | ARG-39 | Outgroup | Botanical Garden. Shivaji Uinversity, India |
Fig. 1.
Kinds of inflorescence/flower in 16 Amorphophallus species and six varieties of India included in this study: A. commutatus var. commutatus (a) A. commutatus var. anmodensis (b), A. commutatus var. wayanadensis (c), A. commutatus var. anshiensis (d), A. bulbifer (e), A. hirsutus (f), A. oncophyllus (g), A. konkanensis (h), A. hohenackeri (i), A. margaritifer (j), Amorphophallus species (k), A. paeoniifolius var. campanulatus (l), A. paeoniifolius var. paeoniifolius (m), A. bhandarensis (n), A. bonaccordensis (o), A. longiconnectivus (p), A. mysorensis (q), A. nicolsonianus (r), A. smithsonianus (s), A. sylvaticus (t), Sauromatum venosum-outgroup (u)
DNA extraction, PCR amplification and sequencing
Total genomic DNA from each species under study was isolated from approximately 0.5 g of fresh leaf material using CTAB method (Doyle and Doyle 1987). The extracted DNAs were checked for their purity on agarose gel (1%) and quantified spectrophotometrically. Prior to PCR amplification of barcoding, RAPD and ISSR markers, working stock (20 ng/µl) of each of genomic DNA was prepared and used. The PCR amplifications of various barcode regions were performed with optimized annealing temperatures (Ta) depending upon the primers used (Table 2). DNA was amplified in 50 µl reaction mixtures containing 2U of high fidelity Taq DNA polymerase (Merck Bioscience Pvt. Ltd, Bangalore, India) with 1 X Buffer A, 2.5 mM MgCl2 (Merck Bioscience Pvt. Ltd, Bangalore, India), 400 µM dNTPs, 400 µM of each primer and, 40 ng of template DNA. PCR conditions were as follows: initial denaturation at 96 °C for 5 min., followed by 35 cycles of denaturation at 96 °C for 1 min., annealing at optimized temperature for 1 min, extension at 72 °C for 1 min, and final extension at 72 °C for 10 min. Successful amplification was checked on 1% agarose gel, amplified products were purified using PCR purification Kit (Sigma Aldrich, India). Purified PCR products were sequenced bidirectionally on ABI’s 3500×L genetic analyzer using a BigDye Terminator v3.1 cycle sequencing kit (Invitrogen, Carlsbad, USA) using standard conditions.
Table 2.
List of primer pairs and polymerase chain reaction (PCR) annealing temperatures of candidate barcodes markers used
| Locus | Primer name | Sequence | Annealing temperature | References |
|---|---|---|---|---|
| rbcL | rbcL 1F | ATGTCACCACAAACAGAAAC | 46 | Fay et al. (1998) |
| rbcL 724R | TCGCATGTACCTGCAGTAGC | |||
| rbcL aF | ATGTCACCACAAACAGAGACTAAAGC | 48 | Levin et al. (2003), Kress et al. (2009) | |
| rbcL aR | GTAAAATCAAGTCCACCRCG | |||
| matK | mat K 390 | CGATCTATTCATTCAATATTC | 48 | Cuénoud et al. (2002) |
| matK 1326 | TCTAGCACACGAAAGTCGAAGT | |||
| matK F | ACCCAGTCCATCTGGAAATCTTGGTTC | 50 | Fofana et al. (1997) | |
| matK R | CGTACAGTACTTTTGTGTTTACGAG | |||
| ITS | ITS 8F | AACAAGGTTTCCGTAGGTGA | 58 | Wen and Zimmer (1996) |
| ITS 9R | TATGCTTAAAYTCAGCGGGT | |||
| ITS 10F | CGAACACGTTACAATACCG | 55 | ||
| ITS 11R | ACCACTTGTCGTGACGTCC | |||
| trnLC–trnLD | trnL C | CGAAATCGGTAGACGCTACG | 48.5 | Taberlet et al. (1991) |
| trnL D | GGGGATAGAGGGACTTGAAC | |||
| trnH–psbA | psbA | GTTATGCATGAACGTAATGCTC | 55 | Sang et al. (1997) |
| trnH | CGCGCATGGTGGATTCACAATCC | Tate and Simpson (2003) |
Phylogenetic reconstructions and analyses
Sequences were analyzed, edited and assembled using ChromasPro software (http://www.technelysium.com.au/ChromasPro.html). For phylogenetic analysis, multiple sequence alignments of sequences were performed with ClustalW (Thompson et al. 1994) and edited manually using DAMBE (Xia and Xie, 2001) to obtain an unambiguous sequence alignment. For the analyses of combined datasets, sequences were concatenated using DAMBE. Interspecific genetic divergences was calculated using Kimura 2-Parameter (K2P) distances in MEGA 5.0 following the instruction of the CBOL for distance calculations (Tamura et al. 2011).
Maximum likelihood (ML) analyses for individual and combined datasets of rbcL, matK, trnH–psbA, trnLC–trnLD were carried out in RAxML (Stamatakis, 2006, 2014) using the application raxmlGUI 1.5b1(Silvestro and Michalak, 2011). The clade support was estimated using ML + rapid bootstrap with 1000 replicates based on a maximum likelihood analysis with the model GTRGAMMA using RAxML version 8.0.0 (Stamatakis 2014).
Bayesian inference (BI) of phylogeny using Monte Carlo Markov chains (Yang and Rannala, 1997) was performed with MrBayes v. 3.2 (Ronquist et al. 2012). Individual and combined datasets of rbcL, matK, trnH–psbA, trnLC–trnLD were tested for the appropriate model of nucleotide evolution with MrModelTest 2.3 (Nylander, 2004). The optimal model was implemented according to the Akaike information criterion and then implemented for the analyses (Posada and Buckley 2004). In BI analyses, 1,000,000 generations of two independent Monte Carlo Markov chains with equal rates until convergence was achieved, implemented as ‘stoprule’ when average deviation of split frequencies was below 0.01, were carried out. Trees were sampled every 100 generations and initial 10% trees before stationarity were discarded as burn-in (set to 0.1). Plot of the generation versus log probability was generated by giving the “sump” command and used for estimation of stationarity of the analyses. Convergence between runs was reported by MrBayes using ‘‘mcmcdiagn’’ command. The majority-rule consensus trees with posterior probabilities (PP) values were constructed using remaining trees. PP values between 0.8 and 0.89 were considered as moderate support whereas PP values over 0.9 were considered as high support.
Nucleotide accession numbers
Sequences generated in this study have been submitted to NCBI’s GeneBank under following accession numbers. rbcL: KM093835–KM093855, matK: KM113324–KM113344, psbA–trnH: KM113366–KM113372, trnLC–D: KM113345–KM113365.
RAPD and ISSR analyses
Initially, a total of 25 decamer RAPD primers from Bangalore Genei Kit (Merck Bioscience Pvt. Ltd, Bangalore, India) and 50 ISSR primers (UBC primer set No. 9, University of British Columbia, Canada) were screened for RAPD and ISSR analyses respectively. In order to select ISSR primers that could give good amplification and suitable annealing temperatures, a preliminary screening was carried out using gradient annealing temperatures (Ta). The PCR amplification reaction (25 µl) consisted of 40 ng of DNA, 1X PCR buffer A (10 mM Tris pH 9.0, 50 mM KCl, 1.5 mM MgCl2), 100 µM of each of the four dNTPs, 0.4 µM of RAPD or ISSR primer and 1U of Taq DNA polymerase (Merck Bioscience Pvt. Ltd, Bangalore, India). PCR amplifications were performed in C1000 Touch Thermal Cycler (Biorad, India) with an initial denaturation at 94 °C for 3 min followed by 45 cycles at 94 °C for 45 s, 36 °C for 30 s and 72 °C for 2 min with a final extension at 72 °C for 7 min. Regardless of the marker system used, all the PCR amplifications included a negative control (no DNA) to avoid erroneous interpretations. The amplified PCR products were separated on 1.8% agarose gel in 1X TAE buffer by electrophoresis at 100 V for 3 h., stained with ethidium bromide and documented on G:Box gel imaging system (Syngene Bioimaging Pvt. Ltd, India). To ensure reproducibility, DNA amplified profiles with ISSR and RAPD markers were tested for repeatability at least thrice. This was done by repeating the PCR reactions on two different thermal cyclers. The generated band profiles were considered reproducible and scorable only after observing and comparing them in three separate amplifications for each primer. Only clear and intense bands were considered and scored while faint bands were not scored for the analysis.
Statistical analysis
For each primer, the presence (1) or absence (0) of polymorphic bands in each species profile was scored using GeneTool image analysis software (Syngene Bioimaging Pvt. Ltd, India) and set in a binary matrix. Genetic similarities were calculated using Jaccard similarity coefficient for RAPD and ISSR polymorphisms individually as well as together. The Mantel test of significance was determined to measure the goodness of fit between the similarity matrices produced with the two marker systems. Dendrograms were constructed using the un-weighted pair-group method with an arithmetic average (UPGMA) using the NTSYS PC version 2.11 (Applied Biostatistics Inc, Setauket, USA).
Results
PCR amplification and sequence analyses
To evaluate the utility of different plastid DNA markers for assessing the phylogenetic relationships among the Amorphophallus species of India, we initially tested different plastid regions such as rbcL, matK, trnH–psbA, trnLC–trnLD, rpoB and rpoC and nuclear locus ITS. The efficiency of PCR amplification of ITS, rpoB and rpoC were less than 40% (Data not shown), therefore were not used further. The most successful PCR rate (100%) was observed for rbcL, matK, trnH–psbA, trnLC–trnLD. Except trnH-psbA, substantial sequence length differences were not detected in these three markers and PCR products were sequenced without complications.
For individual marker regions, aligned sequence lengths was 524 for rbcL, 724 for matK, 144 for trnH–psbA, and 529 for trnLC–trnLD (Table 3). Among the four loci analyzed, rbcL was found most conserved (93.12%) in Amorphophallus species and trnH-psbA was least conserved (22.22%). Locus trnH-psbA was also noted with highest percentage of Parsimony Informative (PI, 66.66%) characters and maximum mean interspecific K2P distance (0.58). When sequence of all four studied loci were concatenated, it resulted aligned length of 2014 bp with 65.59% conserved sites, 11.71% PI characters and interspecific K2P distance of 0.05.
Table 3.
Properties of the four plastid DNA loci and their combinations (the outgroup sequence is not included in the calculation)
| Locci and their combinations | Align length (bp) | No. of variable sites | No. of conserved sites | No. of par-informative sites | No. of singleton sites | Mean interspecific K2P distance |
|---|---|---|---|---|---|---|
| rbcL | 524 | 27 (5.15) | 488 (93.12) | 13 (2.48) | 14 (2.67) | 0.011 (0.002–0.034) |
| matK | 724 | 201 (27.76) | 455 (62.84) | 68 (9.39) | 122 (16.85) | 0.046 (0.018–0.097) |
| trnH–psbA | 144 | 110 (76.38) | 32 (22.22) | 96 (66.66) | 14 (9.72) | 0.585 (0.069–1.153) |
| trnLC–trnLD | 529 | 101 (19.09) | 378 (71.45) | 59 (11.15) | 42 (7.93) | 0.069 (0.003–0.126) |
| rbcL + matK + trnH–psbA + trnLC–trnLD | 2014 | 437 (21.69) | 1321 (65.59) | 236 (11.71) | 190 (9.43) | 0.051 (0.009–0.067) |
Figures in the parenthesis for no. of variable sites, no. of conserved sites and no. of Parsimony informative sites indicate the percentage of aligned length; Figures in the paranthesis for Mean K2P interspecific distance indicate range of K2P interspecific distance in 16 Amorphophallus species and six varieties
Maximum likelihood analyses
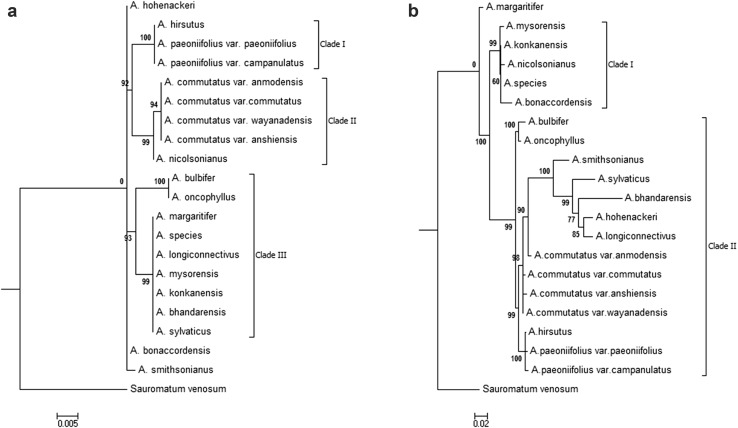
The majority rule consensus trees with bootstrap support values resulting from the ML analyses based on rbcL, matK, trnH–psbA, trnLC–trnLD and combined dataset are shown in Fig. 2 and Supplementary Figure S1. The ML analysis based on rbcL marker resolved tested taxa in three strongly supported section specific clades I (BS 100%), clade II (BS 99%) and clade III (BS 93%) (Fig. 2a). In clade I, resolved topology strongly (BS 100%) supported the monophyly of all three tested members of section Amorphophallus. Clade II comprised of monophyletic group of four tested varieties of A. commutatus of section Conophallus (BS 94%) whereas remaining two members namely A. bulbifer and A. oncophyllus were sister (BS 100%) to clade III. In Clade III, seven of the eleven species of section Rhaphiophallus were resolved as monophyletic with 99% BS and remaining four were resolved either as paraphyletic or polyphyletic (Fig. 2a).
Fig. 2.
Maximum likelihood majority rule consensus trees (RAxML) showing the relationships among Amorphophallus species of India based on a rbcL sequences and b combined dataset of rbcL, matK, TrnH–psbA and TrnL C–D sequences. ML bootstrap values are shown next to the node
Section specific clades were not resolved in ML tree based on matK markers and tree was highly polytomous with low bootstrap supports (Fig. S1a). ML tree based on trnH–psbA resolved the taxa in two clades and the topologies in both of these clades were polytomic and none resolved tested taxa into the section specific clades (Fig. S1b). Clade I was strongly supported with BS 100% and comprised of all of the tested species of sections Conophallus and Amorphophallus, whereas poorly supported (BS 41%) clade II comprised of species of section Rhaphiophallus. The members of section Amorphophallus were found embedded in the clade I that was composed of most of the members of section Conophallus. The monophyly of tested section was not supported in both these clades. The topology of ML tree based on trnLC–trnLD was similar to that obtained in ML tree based on matK. Although species were resolved in two clades, they were polytomy and none of these clades was section specific (Fig. S1c).
Similar to resolutions and tree topologies resulted from ML analysis of individual markers, resolutions and tree topology resulted from combined data was not in congruence with the rbcL. Unlike on the rbcL phylogeny, the resolution of relationships among Amorphophallus species based on combined data were poor. None of the two clades supported monophyly of tested sections and none resolved species into section specific clades. In clade II (BS 90%), five of the ten members of section Rhaphiophallus formed an unresolved polytomy together with members of section Conophallus whereas remaining members formed clade I (Fig. 2b).
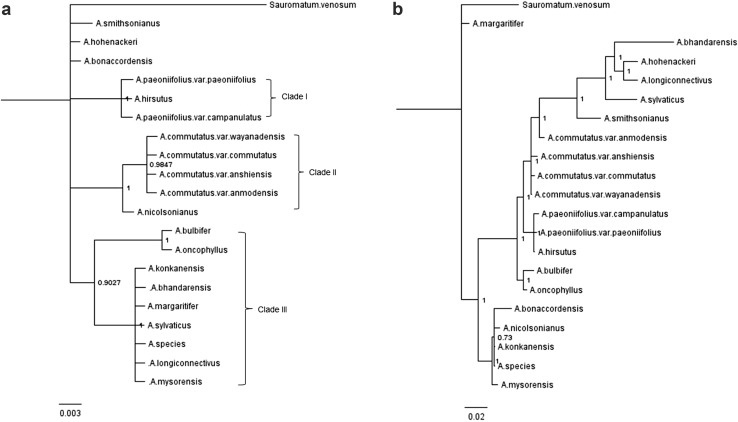
BI analysis
The Bayesian 50% majority rule consensus trees with posterior probability values resulting from the BI analyses based on individual marker and their combined dataset are shown in Fig. 3 and Supplementary Figure S3.
Fig. 3.
The Bayesian 50% majority rule consensus trees showing the relationships among Amorphophallus species of India based on a rbcL sequences and b combined dataset of rbcL, matK, TrnH–psbA and TrnL C–D sequences. The Bayesian posterior probability values are shown next to the node
When we carried out BI analysis, resolution of the species relationship and tree topology based on rbcL were similar to that obtained in ML analysis (Figs. 2a, 3a). Like ML analysis, BI analysis also resolved tested taxa into in three section specific clades viz. clade I, II and III that were strongly supported with higher posterior probabilities (PP 1, 1, and 0.9 respectively) (Fig. 3a). The clade I comprised of all three tested members of section Amorphophallus and their monophyly was strongly supported (PP 1). Clade II comprised of four tested varieties of A. commutatus of section Conophallus as monophyletic group with strong support (PP 1). Remaining two members of section Conophallus namely A. bulbifer and A. oncophyllus were embedded in clade III as sister to monophyletic group of seven of the ten species of section Rhaphiophallus (PP 1) (Fig. 3a).
The species resolutions and tree topologies in BI trees based on matK was better than ML tree and resolved tested species into three clades that were in congruence with ML and BI trees based on rbcL (Figs. S2a, 2a, 3a). The BI analysis of trnH–psbA and trnLC–trnLD were also similar to those obtained in ML trees (Fig. S2 b, c, Fig. S1 b, c). In BI trees of these three markers, section specific, monophyletic clades were not resolved and showed high polytomy with low supports of posterior probabilities.
When BI analysis was performed for the combined dataset of all four tested markers, species resolution and tree topology were comparable and in congruence with ML tree of combined dataset. Similar to ML tree, BI tree also resolved tested taxa in two clades (Fig. 3b). Major Clade I (PP 1) comprised of most of the species of all three section while minor clade II (PP 1) comprised of five species of section Rhaphiophallus. Similar to ML tree, section specific monophyly of tested species was not supported in either of these two clades and none resolved species into section specific clades (Fig. 3b).
Resolution based on DNA fingerprinting approaches
Resolution based on RAPD
To study the suitability of fingerprinting techniques for resolving molecular phylogeny of Amorphophallus species, we initially screened 25 decamer RAPD primers and 50 ISSR primers. Out of 25 RAPD primers tested, nine primers produced amplification products all of which revealed polymorphic fingerprint patterns in tested Amorphophallus species. These nine RAPD primers produced a total of 162 reproducible and scorable bands, 126 (77%) were polymorphic with an average of 14 polymorphic bands per primer. The total number of bands per amplification varied from 13 to 23 in the molecular size range of 77–1507 bp. Maximum 23 bands were produced by primer RPi 4 whereas maximum 18 polymorphic bands were produced by RPi 1 and RPi 4. Using RAPD, the percentage of polymorphism ranged from 64.79% (RPi 6) to 85% (RPi 2) with an average polymorphism of 77.30% per primer (Table 4).
Table 4.
RAPD and ISSR primers and their characteristics
| No | Name | Annealing temperature used (Ta) | No. of Bands | Band size (bp) | No. of polymorphic bands | % polymorphism |
|---|---|---|---|---|---|---|
| RAPD | ||||||
| 1 | RPi 1 | 38.0 | 22 | 119–1507 | 18 | 81.18 |
| 2 | RPi 2 | 39.0 | 20 | 85–660 | 17 | 85.00 |
| 3 | RPi 3 | 38.5 | 21 | 85–117 | 17 | 80.95 |
| 4 | RPi 4 | 38.0 | 23 | 313–1082 | 18 | 78.36 |
| 5 | RPi 5 | 40.0 | 16 | 530–1340 | 13 | 81.25 |
| 6 | RPi 6 | 39.0 | 17 | 450–920 | 11 | 64.79 |
| 7 | RPi 7 | 40.0 | 13 | 480–1340 | 10 | 76.92 |
| 8 | RPi 9 | 38.5 | 14 | 77–682 | 11 | 78.57 |
| 9 | RPi 10 | 38.0 | 16 | 80–387 | 11 | 68.75 |
| ISSR | ||||||
| 1 | UBC 2 | 44.6 | 11 | 170–2900 | 9 | 81.81 |
| 2 | UBC 3 | 44.6 | 14 | 340–2050 | 8 | 57.14 |
| 3 | UBC 4 | 44.6 | 8 | 300–1950 | 7 | 87.50 |
| 4 | UBC 5 | 49.2 | 10 | 450–2370 | 8 | 80.00 |
| 5 | UBC 6 | 49.2 | 9 | 250–3010 | 7 | 77.77 |
| 6 | UBC 18 | 59.8 | 12 | 300–1810 | 9 | 75.00 |
| 7 | UBC 22 | 55.0 | 13 | 1060–2240 | 10 | 76.92 |
| 8 | UBC 26 | 60.0 | 13 | 200–3200 | 11 | 84.61 |
| 9 | UBC 27 | 60.0 | 9 | 600–1790 | 6 | 66.66 |
| 10 | UBC 36 | 59.8 | 14 | 470–2740 | 11 | 78.57 |
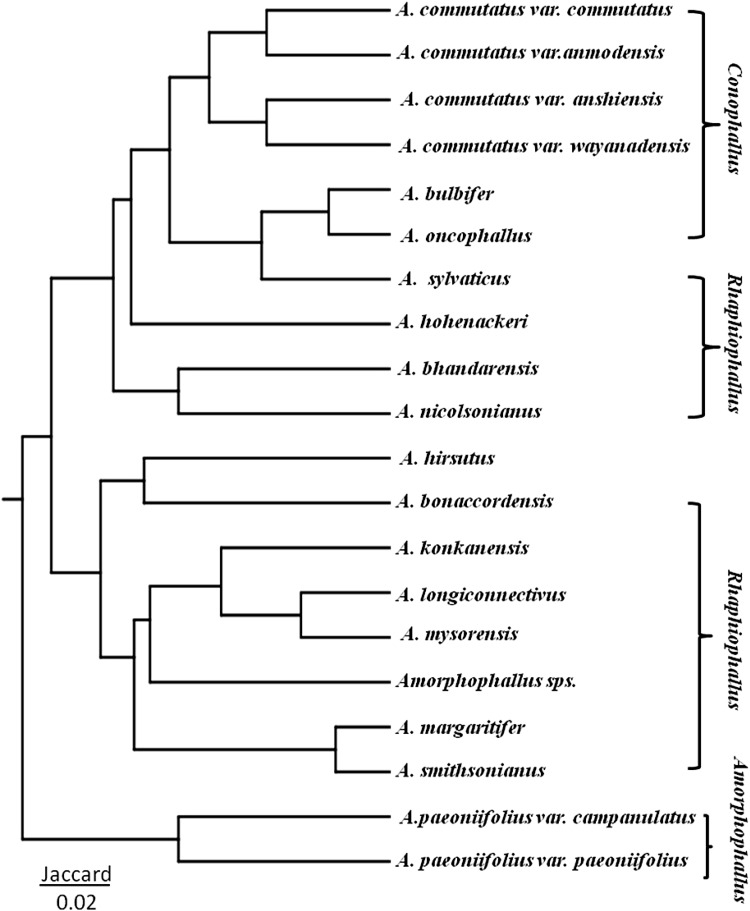
The genetic similarity based on Jaccard coefficients among 16 Amorphophallus species and six varieties was used to construct a dendrogram (Fig. 4) by UPGMA method. The genetic distance among the Amorphophallus species ranged from 0.11 to 0.84. The dendrogram clustered 16 Amorphophallus species and six varieties into clusters that were in congruent with the morphological characters. All of the three species and four varieties A. commutatus of section Conophallus were clustered together. However, this resolved group of Conophallus also contained A. sylvaticus, a member of Rhaphiophallus, whereas eight of the remaining nine species of Rhaphiophallus were clustered together in two groups. With the exception of A. hirsutus that was clustered with species of section Rhaphiophallus, remaining two varieties of A. paeoniifolius of section Amorphophallus were also found clustered together (Fig. 4).
Fig. 4.
Species dendrogram of 16 Amorphophallus species and six varieties based on 126 polymorphic bands from RAPD profile
Resolution based on ISSR
Of the 50 ISSR primers screened, 16 primers resulted amplification of which ten primers generated polymorphic banding patterns. Out of 101 bands (10.1 per primer), 86 were polymorphic indicating an average of 8.6 polymorphic bands per primer. With tested ISSR primers, the number of bands amplified per primer ranged from eight (UBC 4) to 14 (UBC 3 and 36) with band size between 170 bp and 3.2 kb. The percentage of polymorphism ranged from 57.14% (UBC 3) to 87.50% (UBC 4) with an average polymorphism of 76.59% across the Amorphophallus species studied. The genetic distance based on Jaccard coefficients ranged from 0.15 to 0.71. UPGMA dendrogram based on ISSR profile did not resolve Amorphophallus species into section specific clusters congruent with the sections proposed by Sivadasan (1989) (Fig. S3). This clearly indicated that the DNA based marker ISSR alone is not suitable for resolution of Amorphophallus species.
Resolution based on combined data of RAPD and ISSR
The NJ analysis and UPGMA dendrogram of the combined 126 RAPD and 86 ISSR bands for 16 species six varieties clustered together all of the three species of section of Amorphophallus while 9 of the ten species of section Rhaphiophallus were also clustered together. The species of section Conophallus did not clustered together (Fig. S4). The resolution based on combined data of RAPD and ISSR also could not resolve Amorphophallus species completely into three proposed section. In comparison, the resolution based on combined data of RAPD and ISSR was better than the ISSR alone but not better that RAPD alone.
Discussion
In the present study, the molecular phylogeny of Amorphophallus species of India was reconstructed using plastid gene regions’ sequences and two DNA fingerprinting markers namely RAPD and ISSR. We tested the performances of sequences of four plant loci viz. rbcL, matK, trnH–psbA, trnLC–trnLD for the resolution of Amorphophallus species into three Indian sections namely Rhaphiophallus, Conophallus, and Amorphophallus. In addition, the utility of two DNA finger printing techniques namely RAPD and ISSR for species resolution was also tested. Our study included most of the previously unexplored species of Amorphophallus occurring in India. Because of rare and endangered status of some of Amorphophallus species, we could not include multiple specimen and accessions. For the selection of suitable locus regions, criteria of efficiency of reproducible amplification and ease of sequencing were applied. The efficiency of PCR amplification, which is an important index to evaluate the candidate barcode, was determined to select the suitable candidate barcoding marker. In comparison to rbcL, matK, trnH–psbA and trnLC–trnLD, the efficiency of PCR amplification of ITS, rpoB and rpoC was low (<40%), therefore were not used further. Unlike two barcode markers namely matK and rbcL that have already been established and recommended for plant barcoding (CBOL 2009), the use of ITS2 is still debated for its potential use as a DNA barcode in different plant groups. In our study, the use of ITS specific primers was consistently resulting lower amplification efficiency and amplification of multiple bands.
Despite of our attempts to amplify other barcode regions, successful amplifications were achieved only for rbcL, matK, trnH–psbA and trnLC–trnLD. For these loci, amplification and sequencing success was 100%. Among the tested markers, substantial difference in the length of amplified product was not observed for rbcL, matK and trnLC–trnLD, however, despite of 100% amplification efficiency of trnH–psbA, the length of amplified product varied in the range of 140–200 bp. Similar variation in amplified length of trnH–psbA was demonstrated in Umbelliferae and in Araliaceae (Degtjareva et al. 2012). The ease of amplification of rbcL and matK in Amorphophallus species was congruent with those reported by Grob et al. (2002, 2004) and Sedayu et al. (2010). The performance of these four loci was analyzed individually as well as in combinations of four loci.
Locus rbcL as one of the core plant plastid barcode regions has been recommended as universal and was found easy to be sequenced and aligned in group of plants such as ferns, mosses and angiosperms (Hasebe et al. 1995; Hollingsworth et al. 2009; Liu et al. 2011). Despites of some doubts in few plant groups about the universality and applicability, another plastid coding region matK has also been recommended as one of the core plant barcode by CBOL (Cuénoud et al. 2002; Chase et al. 2007a, b). In the view of universality and applicability of rbcL and matK, we anticipated good resolution of phylogeny of Amorphophallus species using these two barcode markers. However, in our study, rbcL as a single locus was found most conserved than other three loci analyzed. It also contained low percentages of PI sites, singleton sites and lowest interspecific K2P distance. Similar observation wherein rbcL possessed very low inter-specific variation in closely related species was made by few workers (Newmaster et al. 2008; Kress et al. 2005). Among four loci tested, rbcL was found most suitable for resolving Amorphophallus species than matK, trnH–psbA and trnLC–trnLD. Surprisingly, in the order of their efficacy for resolution of tested species, the performance of small intergenic spacer region trnH–psbA was found better suited than widely used plastid barcode marker matK. trnH–psbA was also noted with the highest percentage of variable sites, PI sites, interspecific K2P distance. Using matK and trnLC–trnLD, tree topology and relationships among the Amorphophallus species in ML and BI trees were not fully resolved. In comparison to ML and BI trees based on matK, trnH–psbA and trnLC–trnLD, the ML and BI trees based on rbcL clearly resolved Amorphophallus species into three clades which were strongly supported with boorstrap, posterior probabilities values and morphological characters. As expected, four tested varieties of A. commutatus of section Conophallus were resolved as monophyletic in ML and BI tress based on rbcL. Two members of Conophallus namely A. oncophyllus and A. bulbifer always grouped together as monophyletic with strong support and morphological characters. Also the species of section Amorphophallus namely A. hirsutus and two varieties of A. paeoniifolius namely var. paeoniifolius and campanulatus were recovered as monophyletic with strong support and morphological characters.
The monophyly of species of sections Rhaphiophallus, Conophallus and Amorphophallus was not fully supported and section specific clades were not recovered when ML and BI analyses were performed on combined datasets as well. In comparison, the efficacy and performance of rbcL alone was found better than matK, trnH–psbA and trnLC–trnLD and combined dataset for species resolution and recovery of section specific clades. The lower efficacy and poor performances of matK, trnH–psbA and trnLC–trnLD was also reflected in efficacy and performances of combined dataset.
In the view of our observation that individual locci matK, trnH–psbA and trnLC–trnLD were not able to resolve Amorphophallus species into clades supported by morphological characters, we further carried out ML and BI analyses on concatenated sequences of various combinations of two and three loci and found that their performances and efficiencies were also poor in terms of complete resolution and recovery of tested taxa into section specific clades (Data not shown). The Indian section Rhaphiophallus was founded by Engler and it is still in use (Grob et al. 2002). In our analysis, the species forming the clade of Rhaphiophallus were supported by morphological characters such the presence of stipitate spadix that contains staminodes between the female and the male zone, large stigma and undifferentiated spathe, whereas the species forming clade of Conophallus were supported by characters like globose or disciform tuber, with globose or rhizomatus offsets, spadix longer than spathe. Species forming clade of Amorphophallus had globose or depressed tuber with distinct annular root-scar, highly variable appendix and psilate pollen.
In previous analysis by Grob et al. (2002), two species namely A. smithsonianus and A. margaritifer of Rhaphiophallus section were not placed together. In congruence with this observation, we also observed that these two species were also not placed together, instead they were found polyphyletic in most of our ML and BI analyses. Based on these results, our study demonstrates and recommends that rbcL should also be considered better suited locus for resolving molecular phylogeny of closely related species of genus Amorphophallus of India. In terms of the resolving power, trnH-psbA had ranked second to rbcL and was found polymorphic, consequently suitable for resolving closely related Amorphophallus species. However, none of these tested loci completely resolved Amorphophallus species into Rhaphiophallus, Conophallus, and Amorphophallus section specific clades. The use of matK, trnH–psbA and trnLC–D as a single locus and multi-locus marker approach based on combination of two and three loci were not much suitable for resolution.
Recently, several workers have used RAPD and ISSR markers for the study of phylogeny and species discrimination. RAPD was successfully used for the study of relationships among Verbascum species (Muazaz et al. 2014) and combination of ISSR and RAPD was used to study genetic relationships in Coffea and Indian Psilanthus species (Kumar et al. 2008). In the view of these reports, we have also accessed the utility of RAPD and ISSR for resolving the phylogeny of Amorphophallus species. Since RAPD markers are known to suffer from a lack of reproducibility, we checked the consistency of the electrophoretic patterns and the polymorphism detected by repeating every PCR reaction twice on two different thermal cyclers. Phylogenetic dendrogarm based on RAPD profile clustered together all of the species of section Conophallus and species of section Rhaphiophallus were grouped in two clusters. In our study, to resolve the phylogeny of Amorphophallus species using DNA fingerprinting technique, RAPD alone performed better than ISSR and combination of RAPD and ISSR, but was not better than plastid gene region sequencing and analysis approach. In comparison, plastid gene region sequencing approach using rbcL was found most suitable for resolving molecular phylogeny and relationship among the Amorphophallus species of India.
Earlier studies of molecular phylogeny of Amorphophallus species by Grob et al. (2002, 2004) and Sedayu et al. (2010) included only five Amorphophallus species each from India. Our study differs from these two reports in having studied most of the previously unexplored Indian species. Moreover, as plastid gene region sequencing approach, we have compared the performance of single and combinations of four barcode loci for resolution of phylogeny. In addition, the utility and performance of ISSR and RAPD fingerprinting technique has also been compared with plastid DNA region sequencing approach.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by Department of Biotechnology, Govt. of India and was carried out as a part of SUK-DBT IPLS program sanctioned to Department of Biotechnology, Shivaji University, Kolhapur, India. Dr. Kiran D. Pawar gratefully acknowledges SUK-DBT IPLS program for funding his tenure as Assistant Professor. Mr. Avinash Gholave acknowledges SUK-DBT IPLS program for Junior Research fellowship. Prof. V. A. Bapat is thankful to Indian National Science Academy, New Delhi, India for senior scientist fellowship. Prof. S. P. Govindwar and Mr. Swapnil Patil are gratefully acknowledged for providing sequencing facility and help in sequencing experiments, respectively. Authors are also grateful to Mr. Mudasir Dar for his help in sequencing efforts.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Avinash R. Gholave and Kiran D. Pawar have contributed equally to this work and shared first author.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-016-0400-0) contains supplementary material, which is available to authorized users.
References
- Arnau G, Lallemand J, Bourgoin M. Fats and reliable strawberry cultivar identification using ISSR amplification. Euphytica. 2003;129:69–79. doi: 10.1023/A:1021509206584. [DOI] [Google Scholar]
- CBOL, Plant Working Group A DNA barcode for land plants. Proc Natl Acad Sci USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haidar N, Phi VS. Land plants and DNA barcodes: short-term and long-term goals. Trans R Soc B. 2005;360:1889–1895. doi: 10.1098/rstb.2005.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW, Cowan RS, Hollingsworth PM, et al. A proposal for a standardized protocol to barcode all land plants. Taxon. 2007;56:295–299. [Google Scholar]
- Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madrinan S, et al. A proposal for a standardised protocol to barcode all land plants. Taxon. 2007;56:295–299. [Google Scholar]
- Cowan RS, Chase MW, Kress WJ, Savolainen V. 300,000 species to identify: problems, progress, and prospects in DNA barcoding of land plants. Taxon. 2006;55:611–616. doi: 10.2307/25065638. [DOI] [Google Scholar]
- Cuénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, et al. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB and matK DNA sequences. Am J Bot. 2002;89:132–144. doi: 10.3732/ajb.89.1.132. [DOI] [PubMed] [Google Scholar]
- Degtjareva GV, Logacheva MD, Samigullin TH, Terentieva E, Valiejo-Roman CM. Organization of chloroplast psbA_trnH intergenic spacer in dicotyledonous angiosperms of the family umbelliferae. Biochemistry. 2012;77:1056–1064. doi: 10.1134/S0006297912090131. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Fang DQ, Roose ML, Krueger RR, Fedrici CT. Fingerprinting trifoliate orange germplasm accession with isozymes, RFLPs and inter simple sequence repeat markers. Theor Appl Genet. 1997;95:211–219. doi: 10.1007/s001220050550. [DOI] [Google Scholar]
- Fay MF, Bayer C, Alverson WS, de Bruijn AY, Chase MW. Plastid rbcL sequence data indicate a close affinity between Diegodendron and Bixa. Taxon. 1998;47:43–50. doi: 10.2307/1224017. [DOI] [Google Scholar]
- Fofana B, Harvengt L, Baudoin JP, du Jardin P. New primers for the polymerase chain amplification of cpDNA intergenic spacers in Phaseolus phylogeny. Belg J Bot. 1997;129:118–122. [Google Scholar]
- Grob GBJ, Gravendeel B, Eurlings MCM, Hetterscheid WLA. Phylogeny of the tribe Thomsoniae (Araceae) based on Chloroplast matK and trnL intron sequences. Syst Bot. 2002;27:453–467. [Google Scholar]
- Grob GBJ, Gravendeel B, Eurlings MCM. Potential phylogenetic utility of the nuclear floricaula/leafy second intron: comparison with three chloroplast DNA regions in Amorphophallus (Araceae) Mol Phylogenet Evol. 2004;30:13–23. doi: 10.1016/S1055-7903(03)00183-0. [DOI] [PubMed] [Google Scholar]
- Hasebe M, Wolfe PG, Pryer KM. Fern phylogeny based on rbcL nucleotide sequences. Am Fern J. 1995;85:134–181. doi: 10.2307/1547807. [DOI] [Google Scholar]
- Hollingsworth ML, Clark AA, Forrest LL, et al. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour. 2009;9:439–457. doi: 10.1111/j.1755-0998.2008.02439.x. [DOI] [PubMed] [Google Scholar]
- Jaleel A, Sivadasan M, Alfarhan AH, Thomas J, Alatar AA. Revision of Amorphophallus blume ex decne. Sect. Rhaphiophallus (schott) engl. (araceae) in India. Bangladesh J Plant Taxon. 2011;18:1–26. doi: 10.3329/bjpt.v18i1.7835. [DOI] [Google Scholar]
- Jaleel A, Sivadasan M, Alfarhan AH, Thomas J, Alatar AA. Revision of Amorphophallus blume ex decne. Sect. Amorphophallus. (Araceae) in India. J Plant Taxon. 2014;21(2):105–120. [Google Scholar]
- Joshi SP, Gupta VS, Aggrawal RK, et al. Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus Oryza. Theor Appl Genet. 2000;100:1311–1320. doi: 10.1007/s001220051440. [DOI] [Google Scholar]
- Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE. 2007;2:e508. doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, et al. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. PNAS. 2009;106:18621–18626. doi: 10.1073/pnas.0909820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SA, Sudisha J, Srimath HL. Genetic relation of coffea and Indian Psilanthus species as revealed through RAPD and ISSR markers. Int J Integr Biol. 2008;3:150–158. [Google Scholar]
- Levin RA, Wagner WL, Hoch PC, Nepokroeff M, Pires JC, Zimmer EA, et al. Family-level relationships of Onaagraceae based on chloroplast rbcL and ndhf data. Am J Bot. 2003;90:107–115. doi: 10.3732/ajb.90.1.107. [DOI] [PubMed] [Google Scholar]
- Liu J, Möller M, Gao LM, Zhang DQ, Li DZ. DNA barcoding for the discrimination of Eurasian yews (Taxus L, Taxaceae), and the discovery of cryptic species. Mol Ecol Resour. 2011;11:89–100. doi: 10.1111/j.1755-0998.2010.02907.x. [DOI] [PubMed] [Google Scholar]
- Mayo SJ, Bogner J, Boyce PC. The genera of Araceae. Kew: Royal Botanic Gardens; 1997. [Google Scholar]
- Muazaz AH, Jawad MM, Nantawan K, Hazim AJ, Athiya AM, Talib AK, Sahapat B, Theerakulpisut P. Genetic diversity and relationships among Verbascum species in Iraq by RAPD-PCR technique. Glob J Mol Evol Genom. 2014;2:63–74. [Google Scholar]
- Newmaster SG, Fazekas AJ, Ragupathy S. DNA barcoding in the land plants: evaluation of rbcL in a multigene tiered approach. Can J Bot. 2006;84:335–341. doi: 10.1139/b06-047. [DOI] [Google Scholar]
- Newmaster SG, Fazekas AJ, Steeves RAD, Janovec J. Testing candidate plant barcode regions in the Myristicaceae. Mol Ecol Resour. 2008;8:480–490. doi: 10.1111/j.1471-8286.2007.02002.x. [DOI] [PubMed] [Google Scholar]
- Nylander JA. MrModeltest v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Rajesh PN, Sant VJ, Gupta VS, et al. Genetic relationship among annual and perennial wild species of Cicer using ISSR polymorphism. Euphytica. 2003;129:15–23. doi: 10.1023/A:1021567821141. [DOI] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres D, Dar-ling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesianphylogenetic inference and model choice across a largemodel space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution and biogeography of Paeonia (Paeoniaceae) Am J Bot. 1997;84:1120–1136. doi: 10.2307/2446155. [DOI] [PubMed] [Google Scholar]
- Sedayu A, Eurlings MCM, Gravendeel B, Hetterscheid WLA. Morphological character evolution of Amorphophallus (Araceae) based on a combined phylogenetic analysis of trnL, rbcL and Leafy second intron sequences. Bot Stud. 2010;51:473–490. [Google Scholar]
- Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Organ Divers Evol. 2011;12(4):335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- Sivadasan M. Amorphophallus smithsonianus (Araceae), a new species from India and a note on A. sect. Synantherias. Willdenowia. 1989;18:435–440. [Google Scholar]
- Stamatakis A. Raxml-vi-hpc: maximum likelihood based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogeneticanalysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Mol Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JA, Simpson BB. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst Bot. 2003;28:723–737. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Zimmer EA. Phylogeny and biogeography of Panax L. (the Ginseng genus, araliaceae): inferences from ITS sequences of nuclear ribosomal DNA. Mol Phylogenet Evol. 1996;6:167–177. doi: 10.1006/mpev.1996.0069. [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Rannala B. Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo method. Mol Biol Evol. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.