Abstract
Late embryogenesis abundant (LEA) proteins are large and diverse group of polypeptides which were first identified during seed dehydration and then in vegetative plant tissues during different stress responses. Now, gene family members of LEA proteins have been detected in various organisms. However, there is no report for this protein family in watermelon and melon until this study. A total of 73 LEA genes from watermelon (ClLEA) and 61 LEA genes from melon (CmLEA) were identified in this comprehensive study. They were classified into four and three distinct clusters in watermelon and melon, respectively. There was a correlation between gene structure and motif composition among each LEA groups. Segmental duplication played an important role for LEA gene expansion in watermelon. Maximum gene ontology of LEA genes was observed with poplar LEA genes. For evaluation of tissue specific expression patterns of ClLEA and CmLEA genes, publicly available RNA-seq data were analyzed. The expression analysis of selected LEA genes in root and leaf tissues of drought-stressed watermelon and melon were examined using qRT-PCR. Among them, ClLEA-12-17-46 genes were quickly induced after drought application. Therefore, they might be considered as early response genes for water limitation conditions in watermelon. In addition, CmLEA-42-43 genes were found to be up-regulated in both tissues of melon under drought stress. Our results can open up new frontiers about understanding of functions of these important family members under normal developmental stages and stress conditions by bioinformatics and transcriptomic approaches.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-016-0405-8) contains supplementary material, which is available to authorized users.
Keywords: LEA gene family, Citrullus lanatus, Cucumis melo, Genome-wide analysis, Drought stress
Introduction
Late embryogenesis abundant (LEA) proteins were firstly identified in developing cotton seeds (Dure and Galau 1981) and then, abundance of this protein family was discovered in different organisms such as Brassica napus, Arabidopsis thaliana, Caenorhabditis elegans, Dictyostelium discoideum, etc. (Gal et al. 2004; Eichinger et al. 2005; Kikawada et al. 2006; Dalal et al. 2009; Denekamp et al. 2010). Especially most of LEA proteins are called as hydrophilins because of their high Arg/Lys, Glu, Ala, Thr and Gly amino acids contents (Battaglia et al. 2008). Analysis of LEA proteins revealed that this protein group can be divided into seven distinct classes named as LEA 1–5, dehydrin and SMP (Seed Maturation Protein) according to their different motif contents. However, their classification is still controversial and there are differences between authors (Dure et al. 1989; Hunault and Jaspard 2010). In this study, Pfam nomenclature was used for LEA protein classification.
Plant LEA proteins have many important functions in normal plant growth and in preserving cells from the detrimental effect of different stress conditions. For example, these proteins have been considered to prevent the inactivation of malate dehydrogenase, lactate dehydrogenase, catalase or citrate synthase enzymes under desiccation or cold conditions according to in vitro studies (Hara et al. 2001; Goyal et al. 2005; Reyes et al. 2008). Besides, membrane preservation with sugars and sequestration of ions were other roles of LEA proteins (Wolkers et al. 2001; Grelet et al. 2005; Liu et al. 2010). In addition, premature seed dehydration and maturation at the distal end of siliques were reported in LEA gene (ATEM6) knocked out Arabidopsis plants, which demonstrates the role of LEA genes in normal seed development and plant growth (Manfre et al. 2006).
This group of proteins were considered to be related with abiotic stress tolerance. As evidence, over expression of PM2 protein from soybean in Escherichia coli provided living of bacteria in high salt or extreme temperature conditions (Liu and Zheng 2005; Liu et al. 2010). Besides, Arabidopsis plants lack of one or two of three LEA 4 proteins were sensitive to water limitation conditions (Olvera-Carrillo et al. 2010). In addition, a LEA 3 like protein of Artemia franciscana revealed the increased survival capacity of Drosophila melanogaster cells under water deficiency conditions (Marunde et al. 2013). E. coli cells expressing OsLEA protein presented an increase resistance to cold, salinity, osmatic, heat and UV radiation conditions (He et al. 2012). Furthermore, some LEA proteins have a membrane stabilization function by association of anionic phospholipid vesicles under drought or cold conditions (Kosová et al. 2007; Tolleter et al. 2010).
There are a few studies subjected on LEA protein family by genome wide search. Up to date, studies, which make identification and characterization of LEA proteins in Arabidopsis, rice, soybean, tomato, potato, maize, poplar, sweet orange, cucumber, etc. were performed. In these studies, chromosomal location and gene duplications, cis-elements, phylogenetic relationships and conserved domains of LEA proteins were determined. Also, expression patterns of this gene family under abiotic stress conditions were evaluated using publically open RNA seq data and quantitative PCR results (Wang et al. 2007; Hundertmark and Hincha 2008; Li et al. 2011; Lan et al. 2013; Cao and Li 2015; Charfeddine et al. 2015; Pedrosa et al. 2015; Altunoglu et al. 2016; Li and Cao 2016).
Melon (Cucumis melo) and watermelon (Citrullus lanatus) are important crops and belong to Cucurbitaceae family, which includes cucumber (Cucumis sativus L.), squash (Cucurbita spp.), pumpkin (Cucurbita L. spp.) and gourd (Lagenaria Ser. spp., Luffa Mill. spp., and Cucurbita spp.). Production of watermelon is about 104 million Mg and it is in the top five of the most eaten fresh fruits around the world according to Food and Agriculture Organization (FAO) (http://faostat.fao.org). In addition, Turkey is the third largest producer of watermelon after China and Iran (Kumar and Wehner 2011). Besides, melon production is about 26 million tons worldwide according to FAO reports (http://faostat.fao.org), which has an economic value in some European and East Asian countries (Garcia-Mas et al. 2012).
Cucurbitaceae family is a useful model system for studying vascular biology and sex determination (Liu et al. 2004). In addition, whole genome sequences of melon and watermelon were reported in 2012 and 2013, respectively (Garcia-Mas et al. 2012; Guo et al. 2013). To our knowledge, LEA proteins of melon and watermelon have not been characterized yet. Therefore, we have performed genome wide analysis for these important family members and have identified 61 and 73 LEA gene members in melon and watermelon genomes, respectively. Then, bioinformatics analysis and molecular characterization of these LEA genes were performed. Besides, expression patterns were evaluated using RNA seq data and quantitative real time PCR analysis (qRT-PCR).
Materials and methods
Identification of LEA genes in melon and watermelon genomes
Three different applications were used for identification of LEA genes in Cucumis melo and Citrillus lanatus. LEA protein sequences of 14 plants (Arabidopsis thaliana, Gossypium hirsitum, Oryza sativa, Glycine max, Sorghum bicolor, Triticum aestivum, Triticum durum, Pisum sativum, Zea mays, Hordeum vulgare, Brassica napus, Medicago truncatula, Nicotiana tabacum, Vitis vinifera) were obtained by using LEAP database which is a data bank including LEA proteins from different organisms (http://forge.info.univ-angers.fr/~gh/Leadb/index.php?action=0&mode=0) (Hunault and Jaspard 2010).
These sequences were used in specification of cucumber and homologous peptides by making BLASTP search in open accessed PHYTOZOME v9.1 database which provides access to the sequenced and annotated green plant genomes (Goodstein et al. 2012). In addition to this, the database was scanned using LEA keywords. Further, Hidden Markov Model (HMM) profiles of LEA proteins in the Pfam database (http://pfam.sanger.ac.uk/) were compared with Cucumis sativus in the Phytozome database. Similar scanning of Cucumis sativus, Cucumis melo and Citrillus lanatus genomes EST sequences were conducted with TBLASTN in NCBI database for possible LEA proteins. By using decrease redundancy tool (web.expasy.org/decrease_redundancy), related sequences were added to the study and non-related sequences were removed from the study. Preserved areas of every sequence added to the study were also controlled by using SMART database which provides the identification and annotation of genetically mobile domains and the analysis of domain architectures (http://smart.emblheidelberg.de/) (Letunic et al. 2012) and Pfam database which is a large collection of protein families (http://pfam.sanger.ac.uk/).
Identification of chromosomal location and estimation of genomic distribution of LEA genes
Chromosomal locations of LEA genes were identified using Cucurbit Genomics database (http://www.icugi.org/cgi-bin/ICuGI/tool/blast.cgi) which allows search in genomes of Cucurbitaceae family members for watermelon.
Tandem and segmental duplications were identified with Plant Genome Duplication Database (PGDD) (Tang et al. 2008) which is a public database and provides to identify and catalog plant genes in terms of intragenome or cross-genome relationships. Briefly, top five matches ≤1e−05 were taken as potential anchors after BLASTP search against all potential peptide sequences of melon and watermelon LEA proteins. Collinear blocks were searched by MCScan and ≤1e−10 alignments were considered as meaningful matches (Tang et al. 2008; Du et al. 2013).
Sequence alignment, phylogenetic analysis and identification of preserved motifs
Amino acid sequences were loaded to MEGA6 (Tamura et al. 2013) program which enables to conduct statistical analysis of molecular evolution. Then, multiple sequence alignments were done by using ClustalW. Aligned file was used for constructing of phylogenetic tree by neighbor joining method with bootstrap analysis for 1000 iterations (Saitou and Nei 1987). Jones–Taylor–Thornton (JTT) substitution model was applied to phylogeny reconstruction and rates among sites were gamma distributed (G). Protein sequence motifs were identified by MEME motif search tool (http://meme.nbcr.net/meme3/meme.html) (Bailey and Elkan 1994) which allows to discover novel motifs in collections of unaligned nucleotide or protein sequences. Specified MEME motifs were scanned in InterPro database with InterProScan which enables functional analysis of proteins by classifying them into families and predicting domains and important sites (Quevillon et al. 2005).
Gene ontology analysis (GO)
Functional analysis of LEA proteins was performed by using Blast2GO (http://www.blast2go.com) program (Conesa and Götz 2008). Amino acid sequences of LEA proteins were loaded to Blast2GO program and three steps accomplished for functional analysis with this program (i) matching was conducted with the loaded sequences in the program (BLASTp) (ii) mapping associated with BLAST results was completed (Mapping) (iii) dump file related to sequences was created (Annotation). Briefly, with the program, determination of biological functions, cellular content and molecular functions which are the three categories of the GO categorization were provided.
Comparative physical mapping of watermelon LEA genes with other organisms
For the purpose of revealing orthologous relations between watermelon and other species including Arabidopsis, soybean, poplar, potato, grape and maize, peptide sequences were scanned in response to amino acid sequences of watermelon LEA proteins using BlastP program. Those having e-value ≤1e−5 and are at least 80% identical were considered as meaningful. Orthologous relations of LEA genes between watermelon and Arabidopsis, soybean, poplar, potato, grape and maize were viewed with MapChart program (Voorrips 2002).
Calculation of homologous and nonhomologous change rates
Beside the amino acid sequences of LEA genes coding duplicated protein, orthologous gene pairs between melon and watermelon and Arabidopsis, rice and soybean were also aligned by utilizing the ClustalW based multiple sequence alignment tools. CODEML (http://www.bork.embl.de/pal2nal/) program (Suyama et al. 2006) was used for calculating homologous (Ks) and nonhomologous (Ka) change rates through amino acid sequences of LEA proteins and their original complementary DNA sequences. Duplication and separation time of every LEA gene was calculated by using homologous mutation change of (million years ago, MYA) λ changes in response to every homologous area and year (T = Ks/2λ (λ = 6.5 × 10−9) (Lynch and Conery 2000; Yang et al. 2008).
In silico identification of miRNAs targeting LEA genes
Identification of miRNA controlled gene targets is important for understanding the miRNA functions. Plant miRNA database (http://bioinformatics.cau.edu.cn/PMRD/) was used for identification of miRNAs targeting melon and watermelon LEA genes. In order to obtain previously known plant miRNAs, miRBase v20.0 (http://www.mirbase.org/) program was utilized. All known plant miRNAs and melon, watermelon LEA gene transcripts were aligned together. Then, all melon and watermelon miRNAs were defined using internet based psRNA Target Server (http://plantgrn.noble.org/psRNATarget/). Using parameters described earlier by Zhang (2005), all known plant miRNAs and their potential target or targets were evaluated. These miRNA targets determined with computer method were analyzed by utilizing BLASTX scans in order to identify default gene homologous for the confirmation.
Estimation of predicated structures of ClLEA and CmLEA proteins
Melon and watermelon LEA genes were scanned in Protein Data Bank (PDB) which gives information about the 3D structures of proteins. In order to determine the best 3D structure of similar sequence, known three dimensional structures were used for BLASTP analysis (Berman et al. 2000). For the obtained information from Phyre2 program (Protein Homology/Analog/YRecognition Engine; http://www.sbg.bio.ic.ac.uk/phyre2), protein structure was estimated with homology modeling (Kelley et al. 2015).
Expression profiling of melon and watermelon LEA genes by using transcriptome data
For RNA-Seq analysis, all the Illimuna HiSeq readings were obtained from an open data bank archive (SRA, Sequence Read Archive). Entry numbers used in readings are as follows for melon; SRR411102, SRR411100, SRR411106, SRR411104, SRR1033647, SRR1033646, SRR2082958, SRR2082965, SRR2082865, SRR2082935, SRR2082943, SRR2082953, SRR2082831, SRR2082832, SRR2082790, SRR2082791, SRR2082796, SRR2082813 and for watermelon; SRR1724899, SRR1724900, SRR1724901, SRR1724902, SRR1724903, SRR1724943, WM-UR-1/SRR1001435, WM-UR-2/SRR1001436, WM-IM-1/SRR1001437, WM-IM-2/SRR1001438, WM-PM-1/SRR1001439, WM-PM-2/SRR1001440, WM-MA-1/SRR1001441, WM-MA-2/SRR1001442, SRR494474, SRR518988, SRR518988, SRR494479, SRR518992, SRR518993.
All readings were downloaded as raw sequence data in “sra” format and converted into “fastq” format. After removing of the readings at low quality [Phred quality (Q) score <20], all the clear readings were subjected to FastQC analysis in order to control their reading qualities in terms of sequence quality for every base, quality score for every base, nucleotide content for every base and sequence duplication level.
Plant samples, growing conditions and stress application
Melon and watermelon seeds were supplied from Monsanto Gıda ve Tarım Tic. Ltd. Şti (Antalya). Seed shells were removed and seeds were washed with distilled water three times. Afterwards, seeds were taken into plastic pots and grown inside the plant grow box in culture medium containing Hoagland solution (Hoagland and Arnon 1950) for 14 days at 400 μmol m−2 s−1 light intensity at 24 ± 2 °C and 16 h of light and 8 h of dark photoperiod. For drought stress, 10% polyethylene glycol 6000 (PEG-6000) was added to Hoagland solution. Considering morphological features of grown plants and taking parameters used by Baloglu et al. (2014) in the cucumber plant into account, at the zero, third, twelfth and twenty fourth hours of the stress application, samples were taken from stressed plants and control plants for RNA isolation. Stress applied plants and control plants were grown in the plant grow box at the same conditions. The zero hour was used as a control. Root and leaf samples of the grown plants were collected separately in order to use in the tissue specific expression analysis. Tissue samples taken by making biological sampling at three at a time were used in tissue specific gene expression analysis by being frozen with liquid nitrogen.
RNA isolation and quantitative real time PCR (qRT-PCR) analysis
Total plant RNA was isolated using Invitrogen Trizol reagent (Life Technologies Corporation, NY, USA). The quality and quantity of isolated RNA was checked using agarose gel electrophoresis and MultiscanGO nano-spectrophotometer (ThermoFisher Scientific, USA). Single and double stranded DNA fragments present in RNA samples were removed by DNase I enzyme (Thermo, Lithuania). cDNA synthesis was done using RevertAid First Strand cDNA Synthesis Kit according to manufacturers’ instructions (ThermoFisher Scientific, USA). For the qRT-PCR analysis, LEA genes in which it was seen an increase in gene expression were detected and gene specific primers were designed for those LEA genes which were showing increased expression using Primer 5 software program and a list of used primers is given in Supplementary Table 11. 18S rRNA gene was used as internal control gene in the study. Primer sequences of the used 18SrRNA gene is as follows (GenBank ID: X51542.1): 5-GTGACGGGTGACGGAGAATT-3 and 5-GACACTAATGCGCCCGGTAT-3 (Baloglu et al. 2014). Sampling was performed for three times for each step and each repetition, triple qRT-PCR test was done using SYBR Green master mix in Light Cycler 480 Real-Time PCR System (Roche Applied Science, Germany) according to manufacturers’ recommendations.
Taking 18SrRNA gene expression as reference, relative gene expressions were calculated. Proportioning gene expression levels to 18SrRNA gene, control and stress time periods were compared with each other. Calculating values of cycle threshold number (CT) for every uprising curve, calibration curves were formed. ΔCT and ΔΔCT values were calculated as ΔCT = CT sample − CT reference and ΔΔCT = ΔCT stress given sample − ΔCT control (in 0 h) and the difference at the expression level was determined as 2−ΔΔCt (Livak and Schmittgen 2001).
Statistical analysis
Statistical calculations and comparisons were per formed by using Minitab 17 package program. Student T test was used in the analysis of the differences between the stress given samples and control samples. If p value is <0.01; it is assumed that the expression difference in the relevant LEA gene is meaningful.
Results and discussion
Determination and characterization of LEA gene family members in melon and watermelon
LEA protein sequences of 14 plant from LEAP database were used for determination of melon and watermelon LEA genes. After detailed searches in Phytozome v9.1, Pfam and NCBI etc. databases, Melonomics database for melon and Cucurbit Genomics Database for watermelon were employed to find LEA sequences belong to these plants. Consequently, 73 LEA genes in watermelon (named as ClLEA-1 to ClLEA-73) and 61 LEA genes in melon (CmLEA-1 to CmLEA-61), mostly which were carrying conserved LEA domain, were determined. Protein lengths of identified LEA proteins changed between 64 and 464 amino acids in melon and between 64 and 1162 amino acids in watermelon. Also, molecular weights of these proteins varied between 6779.4 and 129,808.3 Da in watermelon. The smallest LEA protein was of 51.242 Da, whereas the biggest LEA protein was of 6795.4 Da in melon. Moreover, according to physicochemical analysis, 68 and 75% of LEA proteins were of a basic character in watermelon and melon, respectively. Most of the LEA2 and LEA1 group proteins were basic whereas most of the LEA4, LEA5, SMP and dehydrin group proteins were of an acidic character in watermelon and melon. LEA3 protein group composed of acidic and basic proteins in both plants. Our results were consistent with the results of Filiz et al. (2013) who found that LEA4, LEA5 and LEA6 group proteins were acidic and most of LEA proteins were basic (72.2%) in purple false brome. More information about results of used parameters for CmLEA and ClLEA proteins is available in Supplementary Table 1 and Supplementary Table 2.
Similar studies on genome wide identification of LEA family members in other plants revealed, 79 LEA genes in cucumber (Altunoglu et al.2016), 53 LEA genes in poplar (Lan et al. 2013), 51 LEA genes in Arabidopsis (Hundertmark and Hincha 2008), 36 LEA genes in soybean (Li et al. 2011), 34 LEA genes in rice (Wang et al. 2007), 32 LEA genes in maize (Li and Cao 2016), 29 LEA genes in potato (Charfeddine et al. 2015), 27 LEA genes in tomato (Cao and Li 2015) and 72 genes in sweet orange (Pedrosa et al. 2015). Accordingly, watermelon has the second highest number of LEA protein genes after cucumber enabling a lot more different types of searches in different databases and to include more plants for inquiry.
Chromosomal distribution and duplication of CmLEA and ClLEA genes
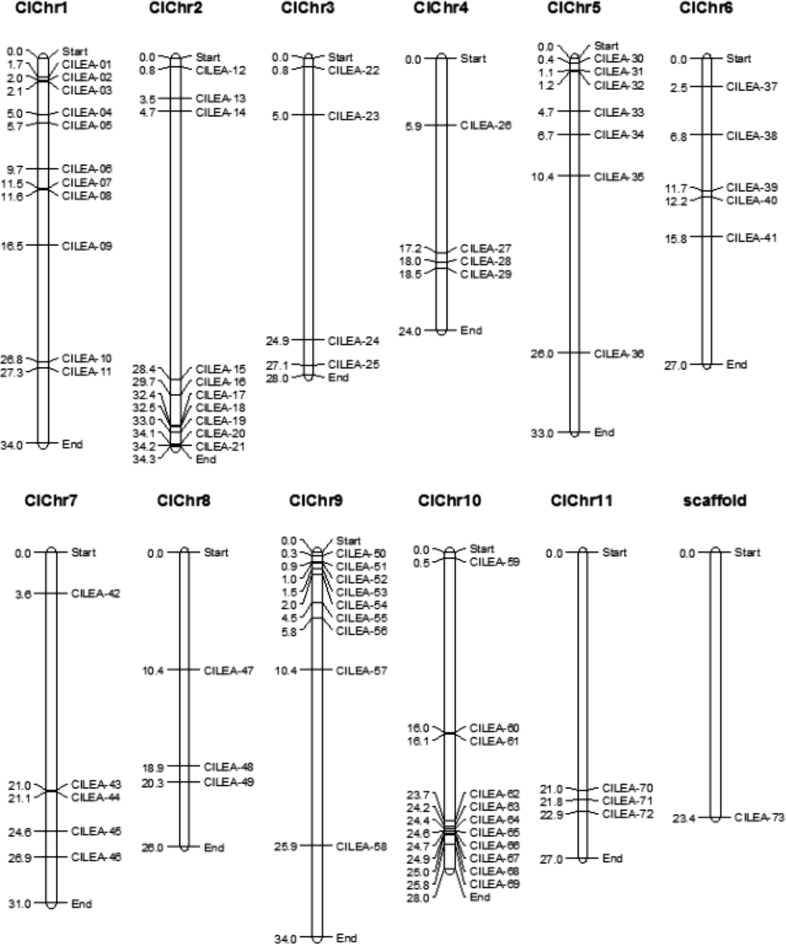
ClLEA genes were mapped on 11 watermelon chromosomes. Chromosome 1 and 10 had the most LEA genes with the number of 11 whereas chromosome 1 and 8 had a few LEA genes with the number of 3. ClLEA genes locating on chromosome 1, 5 and 9 were sited at the upper end of chromosomes but ClLEA genes locating on chromosome 2, 7, 10 and 11 were sited at the lower end of these chromosomes. Only ClLEA-73 gene couldn’t be mapped on any chromosome (Fig. 1). Chromosomal distributions of CmLEA genes could not be determined because locations of these genes were still in scaffold level.
Fig. 1.
Chromosomal distribution of 73 ClLEA genes. Physical locations of watermelon LEA genes were presented on 11 watermelon chromosomes numbered from ClChr1 to ClChr11
Tandem and segmental duplications events for ClLEA and CmLEA genes were evaluated because duplication of genes is one of the reason for occurrence of orthologous genes (Mehan et al. 2004). Twenty of CmLEA genes (32%) showed duplication events, however, we couldn’t elucidate the duplication type as a result of absence of exact chromosomal locations of CmLEA genes (Supplementary Table 4). Segmental duplications between LEA genes were determined in watermelon with the percentage of 31%. Seven ClLEA genes showed (9.5%) tandem duplication events. These tandem duplications located on chromosome 1 (ClLEA-6-9) and chromosome 10 (ClLEA-64-65-66-67-68) (Supplementary Table 3). According to tandem duplication analysis, frequencies were 70 and 30%, in cucumber and poplar, respectively. Our findings suggest that tandem duplication is not prevalent in watermelon as potato and rice. Besides, segmental duplication rates were 51% in tomato, 43% in Arabidopsis, 25% in maize, 20% in potato, 17% in Chinese plum and 15% in rice (Wang et al. 2007; Hundertmark and Hincha 2008; Lan et al. 2013; Cao and Li 2015; Charfeddine et al. 2015; Altunoglu et al. 2016; Li and Cao 2016; Du et al. 2013). Our results indicate that segmental duplication events may be one of the main reason for LEA gene family expansion in watermelon.
Phylogenetic distribution and identification of conserved motifs
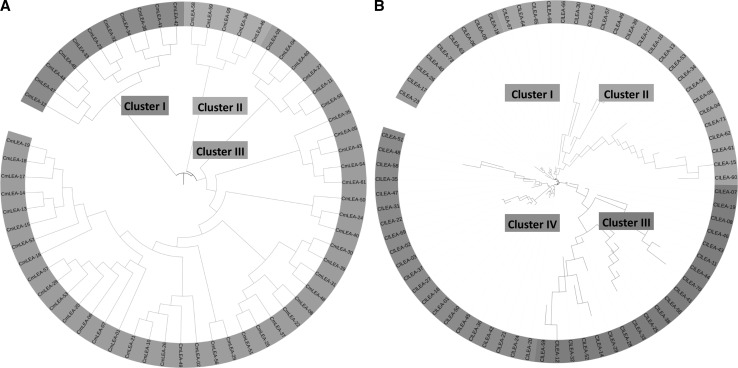
Phylogenetic analysis was implemented by Neighbour-Joining method with bootstrap analysis for 1000 repetitions for 61 melon and 73 watermelon LEA proteins to analyze evolutionary relationships between this groups of proteins. Jones-Taylor-Thornton (JTT) substitution model was used in MEGA v.6 software (Molecular Evolutionary Genetics Analysis) (Tamura et al. 2013) and rates among sites were gamma distributed (G). Four distinct clusters (Cluster I-IV) including 9, 23, 20 and 21 proteins were determined for watermelon LEA proteins, respectively (Fig. 2b). LEA 2 group proteins were dominant in watermelon according to Pfam nomenclature. LEA1 group proteins were located in Cluster I (ClLEA-26), Cluster II (ClLEA-62) and Cluster III (ClLEA-59). Three members of LEA3 group proteins (ClLEA-55-60-61) were in Cluster III whereas one member of this group (ClLEA-52) was located in Cluster III. In addition, only one member of LEA4 group protein (ClLEA-69) which located in Cluster IV was determined in watermelon. LEA5 (ClLEA-28-29), LEA6 (ClLEA-11-70) and SMP (ClLEA-25-33-38-56) group of proteins accumulated only in the Cluster III. Besides, dehydrin group of proteins were distributed in Cluster II (ClLEA-72), Cluster III (ClLEA-14-43-44) and Cluster IV (ClLEA-48) and LEA2 group of proteins were in all clusters mainly in Cluster IV.
Fig. 2.
Phylogenetic comparison of ClLEA and CmLEA genes. a Phylogenetic tree of melon LEA genes with three clusters. b Phylogenetic tree of watermelon LEA genes with four clusters
Three main clusters (Cluster I-III) were observed among melon LEA proteins, which comprised of 11, 5 and 45 proteins, respectively (Fig. 2a). LEA2 group was the dominant group between CmLEA proteins. SMP (CmLEA-23-32-33), LEA3 (CmLEA-34-38) and LEA4 (CmLEA-12) group proteins accumulated in Cluster I. LEA5 group proteins (CmLEA-58-59) were in Cluster II. Dehydrin group of proteins were located in Cluster II (CmLEA-36-46) and Cluster III (CmLEA-60). Cluster III contained only LEA2 group of proteins. LEA6 group proteins weren’t seen in melon.
Especially, SMP, LEA3 and LEA5 group proteins were distributed in a cluster whereas the dehydrin and LEA2 group proteins were allocated to different clusters in melon and watermelon. This kind of expansions may explain the different motif contents of LEA2 group proteins. According to phylogeny based analysis for LEA proteins in other plants, the highest number of LEA protein clusters were LEA4 group in Arabidopsis (Hundertmark and Hincha 2008), dehydrin group in tomato (Cao and Li 2015), LEA2 and LEA3 groups in maize (Li and Cao 2016), dehydrin and LEA2 groups in Chinese plum (Du et al. 2013), LEA2 group in cucumber (Altunoglu et al. 2016) and LEA4 group in poplar (Lan et al. 2013). LEA2 group was the mainly distributed group in watermelon, melon and cucumber which were in the Cucurbitacae family. LEA2 group proteins may more required than other group of LEA proteins for the specific functions in this important plant family.
Moreover, 17 pairs of segmentally and 5 pairs of tandemly duplicated watermelon LEA genes were classified in the same cluster in phylogenetic tree. As an example, ClLEA-16-27 and ClLEA-24-20 proteins which were segmentally duplicated and located in the fourth cluster and all of them belonged to LEA2 group proteins. Tandemly duplicated ClLEA-64-65 and ClLEA-65-68 proteins were in the second cluster. These motif and sequence conservations can be attributed to the reliability of phylogenetic tree and may be related to various of biological functions of this group of proteins.
Another phylogenetic tree which was containing all LEA proteins from cucumber, watermelon and melon was constructed (Altunoglu et al. 2016) (Supplementary Fig. 4). Six different clusters were observed among these LEA proteins from different origins. LEA2 group of proteins was the most dominant group in cluster 6. Other LEA groups (LEA1-6, SMP and dehydrin) dispersed to other clusters. Same LEA protein groups were determined in different clusters. This may be arisen from different motif contents of same LEA groups from different plants.
In addition, amino acid motif compositions of melon and watermelon LEA proteins were evaluated by MEME software. According to this analysis, 10 different motifs were determined for 67 ClLEA proteins in watermelon (Table 1; Supplementary Fig. 3b). LEA1 (ClLEA-26-59-62) and LEA4 (ClLEA-69) group proteins had only motif 6. All LEA3 (ClLEA-52-55-60-61) and LEA5 (ClLEA-28-29) group proteins included only motif 10. ClLEA-25-33-38-56 proteins all of which were in the SMP protein group had motif 5 whereas all of dehydrin group proteins had no motifs with the exception of ClLEA-48 which includes motif 6. LEA6 group proteins had no motifs like dehydrin group. LEA2 group proteins especially included motif 1, 2, 3 and 4 also motif 7 and 8 could be found in some members of LEA2 group.
Table 1.
Amino acid sequence composition of 10 different motifs observed in ClLEA proteins
Ten different motifs were found for 52 CmLEA proteins in melon using MEME software (Table 2; Supplementary Fig. 3a). Dehydrin and SMP group proteins had no motifs whereas LEA1, LEA4 group proteins and CmLEA-41-44-45-47 proteins had only motif 10. LEA2 group proteins included motif 1, 2, 3, 4 and 8 and some of LEA2 group proteins also had motif 5 and 7. Especially LEA1 and LEA4 protein groups had only one common motif, when LEA protein motif compositions of cucumber, watermelon and melon were compared (Altunoglu et al. 2016). This suggests that this group of LEA proteins may have similar function and role in these plants. In addition, dehydrin group proteins had no motifs in these plants in contrast to dehydrin proteins in Chinese plum and potato in which conserved motifs called K (a lysine-rich 15 amino acid sequence) and Y segments were reported before (Charfeddine et al. 2015; Du et al. 2013). This can be attributed to conserved structure of dehydrin group LEA proteins in the Cucurbitacea family.
Table 2.
Amino acid sequence composition of 10 different motifs observed in CmLEA proteins
Gene ontology analysis
Go slim analysis was utilized to determine possible molecular function, cellular localization and biological roles of LEA proteins in melon and watermelon by Blast2GO software (Fig. 3; Supplementary Table 5; Supplementary Table 6). Cellular and membrane localization ratios were higher between ClLEA and CmLEA proteins which may be compatible with their functions on protection of membranes and enzymes to maintain cellular functions under abiotic stress conditions (Goyal et al. 2005; Kosová et al. 2007; Reyes et al. 2008; Tolleter et al. 2010). In addition, our findings are relevant with the studies which reported that LEA proteins localized commonly in subcellular regions such as cytoplasm, mitochondria, nucleus and chloroplast in Arabidopsis, tomato and purple false brome (Hundertmark and Hincha 2008; Filiz et al. 2013; Cao and Li 2015). This may suggest that there is a strong relation between the subcellular localization and the role of the LEA protein in the cell.
Fig. 3.
Gene ontology analysis of a ClLEA and b CmLEA genes by Blast2Go program with three categories named as biological process, molecular function and cellular component
Binding to different molecules was the dominant activity for the action of ClLEA and CmLEA proteins as molecular function. Binding of LEA proteins to nucleic acids to protect cellular structures by constructing hydrogen network was reported, which is related to the roles of LEA proteins in stress tolerance (Wolkers et al. 2001; Shih et al. 2004). In addition, membrane stabilization of this protein family members by association with phospholipid vesicles and sugars were demonstrated in model membranes under dehydration conditions (Wolkers et al. 2001; Kosová et al. 2007; Liu et al. 2010; Tolleter et al. 2010). Binding activity as a molecular function of LEA proteins may be suggested the mode of action of these group of proteins when considering about these reported roles in stress tolerance.
In addition, response to stress is the most observed process when we analyzed biological roles of ClLEA and CmLEA proteins and this role was followed by the roles of LEA proteins in metabolic and cellular processes in melon and watermelon. These biological roles were consistent with the reported roles of LEA proteins such as membrane maintenance, enzyme and nucleic acid preservation and oxidant scavenging activity etc. to protect cell structures from the detrimental effects of abiotic stress factors (Hara et al. 2001; Wolkers et al. 2001; Shih et al. 2004; Goyal et al. 2005; Kosová et al. 2007; Reyes et al. 2008) Besides, reports about stress tolerance by overexpression of LEA genes in transgenic plants and bacteria can support the defined biological role of LEA proteins in our study (Puhakainen et al. 2004; Liu and Zheng 2005; Duan and Cai 2012).
Orthologous LEA genes between watermelon and other species
LEA genes which physically mapped on watermelon chromosomes and those of mapped on Arabidopsis, soybean, poplar, potato, grape and maize chromosomes were compared to evaluate orthologous relationships between them (Supplementary Fig. 5). Approximately 41% of ClLEA genes had orthologous genes with these plants. Maximum number of orthologous genes were with poplar with the ratio of 71%. These results are consistent with our group’s previous study which found that cucumber LEA genes distributed maximum orthology with LEA genes in poplar, which were on the fourth chromosome of poplar (Altunoglu et al. 2016). Detailed analysis revealed that the maximum orthologous relationships were observed between LEA genes located on the first chromosome of watermelon and LEA genes located on the fourth chromosome of poplar. According to these results, LEA genes which belong to Cucurbitacae family members distributed maximum orthology with the LEA genes accumulated on the fourth chromosome of poplar. These results can be useful for the determination of orthologous gene pairs between different organisms and for the evaluation of LEA gene family distributions. In addition, these findings suggest that gene duplication and inversion events may have important roles on expansion and organization of this gene family members in watermelon, Arabidopsis, rice, soybean, poplar, potato, grape and maize.
Duplication events and divergence rates of the ClLEA and CmLEA genes
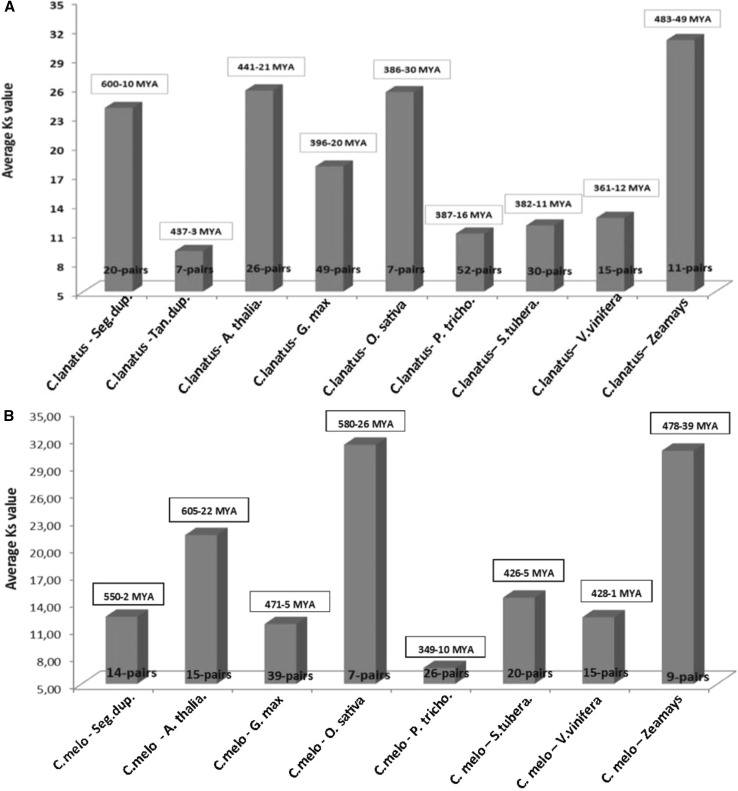
Tandem or segmental gene duplications and transposition events are the reason of the gene family broadening and generation (Cao and Shi 2012). We calculated the approximately dates of duplication events of ClLEA and CmLEA genes to understand broadening ranges of this gene family members. Thus, nonsynonymous (Ka) versus synonymous (Ks) substitutions (Ka/Ks) were estimated for LEA genes which showed duplication events in watermelon and melon. In addition, these calculations were repeated for orthologous LEA gene pairs between Arabidopsis, rice, soybean, grape, poplar, potato and maize in watermelon (Fig. 4a) and melon (Fig. 4b). The Ka/Ks ratios were between 0.0038 and 0.2503 for segmentally duplicated LEA genes and between 0.006 and 0.374 for tandemly duplicated LEA genes in watermelon (Supplementary Table 3). Analysis revealed that these segmental and tandem duplication events may occur approximately between 10 and 600 MYA (million years ago) and 3 and 437 MYA, respectively. In addition, the average Ka/Ks ratio was the maximum between watermelon and poplar with the ratio of 0.15. The earliest divergence time of these gene orthologous from other plants was with maize LEA genes with the average number of 237 MYA (Fig. 4a; Supplementary Table 7).
Fig. 4.
Estimation of duplication and divergence times of a ClLEA and b CmLEA genes using duplicated LEA genes of melon and watermelon and orthologous LEA gene pairs between watermelon–melon and Arabidopsis, soybean, rice, poplar, potato, grape and maize
Additionally, Ka/Ks ratios were 0.136 and divergence dates between duplicated LEA genes of melon varied from 2 to 550 MYA. Ka/Ks ratios of melon with Arabidopsis, soybean, rice, poplar, potato, grape and maize were with the average of 0.06, 0.081, 0.02, 0.07, 0.14, 6.65 and 0.03, respectively. The earliest divergence time was with rice with the average of 240 MYA (Fig. 4b; Supplementary Table 8).
According to these results, a vigorous purifying selection effected melon, watermelon and poplar. In addition, divergence times between cucumber and poplar were closer to date with the average of 78.4 MYA (Altunoglu et al. 2016) than other plants as melon and watermelon. The average of Ka/Ks ratio was 0.14 for tandem duplications in cucumber. All Ka/Ks ratios were observed as <1 in tandemly or segmentally duplicated LEA genes in Cucurbitaceae family members. These results may again show that LEA genes in Cucurbitacae family members exposed to a large purifying selection. In addition, poplar and Cucurbitacae family members may be closer genetically than other analyzed plants according to LEA gene analysis.
MicroRNA (miRNA) identification for CmLEA and ClLEA transcripts
MicroRNAs (miRNAs) are small, noncoding regulatory elements in organisms. Hence, they have important roles in gene regulation under stress conditions via destroying target gene transcripts in plants (Bartel 2004; Ambros and Chen 2007).
miRNAs which targeted CmLEA and ClLEA gene transcripts were identified by psRNA Target Server (http://plantgrn.noble.org/psRNATarget/). This server affords two important analyses: (1) reverse complementary matching is achieved between miRNA and target transcript by scoring scheme of miRU (Dai and Zhao 2011). The threshold of the score is maximum expectation for which cut off threshold was set as 3.0 in the study. (2) Target site accessibility analyses via calculating unpaired energy (UPE) which is necessary to open secondary structure around miRNA’s target site on mRNA. Connection of miRNA with target mRNA is going to be easier if needs low UPE.
Analyses revealed that 22 different ClLEA transcripts (ClLEA-06-10-11-13-14-15-17-19-20-33-38-39-42-45-46-52-58-63-64-67-72-73) were targeted by 24 different miRNAs in watermelon (Supplementary Table 9) and 21 different miRNAs targeted 21 different CmLEA transcripts (CmLEA-03-04-10-16-18-19-20-26-27-29-30-35-37-40-41-48-49-52-55-56-60) in melon (Supplementary Table 10). Mir854, mir414 and mir2673 were the common miRNAs and the most targeted gene was ClLEA-63 in watermelon. CmLEA-20-55 were the most abundant target transcripts and mir854, mir5021 and mir2629 were common miRNAs in melon. Proteins targeted by mir414 in Arabidopsis especially had roles in transcriptional regulation, protein modification, DNA repair or chromatin modification. In particular, mir414 mainly affects regulators of transcription such as bZIP transcription factors, WRKY, MYB, B3, scarecrow, heat shock proteins and TCP. In addition, this important miRNA had roles in posttranscriptional modifications of SNF2 transcriptional regulator, F-Box family proteins, SNF2 transcriptional regulator etc. (Eulgem et al. 2000; Gurley 2000; Jakoby et al. 2002; Suo et al. 2003; Zhang et al. 2003; Flaus et al. 2006; Guo et al. 2007; Guleria and Yadav 2011). This may be indicated that mir414 is very important in the regulation of plant growth. Besides, upregulated expression of another important miRNA family called miRNA854 were reported under water deficiency conditions in rice (Zhou et al. 2010). Mir854 family was present in both plants and animals and this suggests that these miRNAs have a common origin as regulators of basal transcriptional mechanisms (Arteaga-Vázquez et al. 2006). Additionally, a previous study reported that MYB transcription factor which had a key role in regulatory networks controlling development and in responses to stress factors. It was targeted by mir5021 in rosy periwinkle (Pani and Mahapatra 2013). Mir414 and mir854 were found the most abundant miRNAs in cucumber in an earlier study (Altunoglu et al. 2016). This suggested that LEA gene family members in Cucurbitacae family are especially targeted by miRNAs which contributed to the transcriptional regulation. In addition, this may be attributed to the important roles of LEA genes like transcription factors in these plants. Therefore, determination of miRNA targets of melon and watermelon LEA genes will be useful to elucidate the roles of these genes in melon and watermelon.
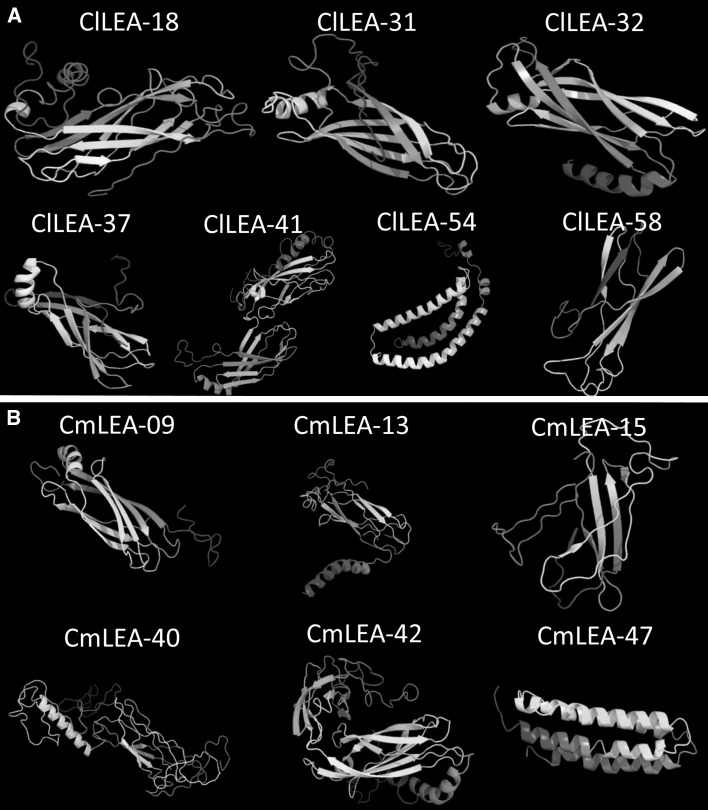
Estimation of predicated 3D-structures of ClLEA and CmLEA proteins
PDB was used by BLASTP search for homology modeling. Seven proteins in watermelon [ClLEA-18 (PDB ID:c1yycA), ClLEA-31 (PDB ID:d1xo8a), ClLEA-32 (PDB ID:d1hx6a1), ClLEA-37 (PDB ID:c1yycA), ClLEA-41 (PDB ID:d1xo8a), ClLEA-54 (PDB ID:c1xq8A), ClLEA-58 (PDB ID:c3butA)] (Fig. 5a) and six proteins in melon [CmLEA-09 (PDB ID:d1xo8a), CmLEA-13 (PDB ID:c1yycA), CmLEA-15 (PDB ID:c3butA), CmLEA-40 (PDB ID:d1xo8a), CmLEA-42 (PDB ID:d1xo8a), CmLEA-47 (PDB ID:d1 eq 1a)] (Fig. 5b) which showed high homology were chosen. Detection rates were used in Phyre2 tool with the selection of Hidden Markov Model (HMM) to predict protein structure (Kelley et al. 2015). Percentage of residues modelled were between 73 and 99 for watermelon and between 72 and 99 for melon with the >90% confidence interval. Interestingly, selected proteins from melon and watermelon were in LEA2 group. β sheets were the dominant structures between selected melon and watermelon LEA proteins. However, CmLEA-47 and ClLEA-54 proteins contained only α helix chains. Predicated structures of cucumber LEA proteins distributed structures with predominant β sheets (Altunoglu et al. 2016) and these findings were compatible with the predicated structures of LEA proteins in melon and watermelon. This can be attributed to the membership of these plants in the same family. In addition, a previous study suggested that LEA2 group proteins especially had two β sheets and one alpha helix structure, which was consistent with our results. This type of structure resembles the structure of fibronectin Type III residues on the surface of animal cells and this kind of configurations may be important in fluid loss to protect cellular structures from the effects of stress (Li and Cao 2016; Singh et al. 2005). Predicated three-dimensional structures of this proteins analysis may provide to elucidate molecular activity of them to understand their roles in the cell.
Fig. 5.
Predicated three dimensional structures of a seven LEA proteins of watermelon and b six proteins of melon with 90% confidence level
Genome-wide expression profiles of ClLEA and CmLEA genes and drought responsive LEA genes in watermelon and melon
SRA (Sequence Read Archive) data sets related to melon and watermelon were used to evaluate tissue specific expression patterns of CmLEA and ClLEA genes by RNA Seq approach. Hierarchically clustered heat map was constructed according to transcriptome data from different days of fruit development stages after pollination and phloem and vascular tissues (Supplementary Fig. 1). Expression levels of ClLEA-7-36-44 genes were upregulated in 10 days old white fruit stage while they were downregulated in 18, 28 and 34 days old fruit development stages. Besides, expression levels of ClLEA-10-13-16-17-23-29-31-38-43-45-46-50-53-63-70 genes showed a tendency to increase in 10 days old white fruit stage but expression levels of these genes decreased in other stages. Contrast to this, ClLEA-11-34-69 gene expressions showed a decrease in 10 days old white fruit stage while their gene expression levels increased in other fruit stages. In addition, ClLEA-7-31-32-36-40-43-44-47-50-51-52-54-55-73 gene expressions were upregulated in the 10th, 18th and 26th days of fruit development when compared with the expression levels of these genes in the 42nd and 52nd days of fruit development stages. In addition to this, transcriptome data from phloem and vascular tissues of watermelon was investigated. Expressions of ClLEA-7-8-10-16-23-26-31-32-36-37-42-54-59-72 genes were higher in phloem tissues than in vascular tissues.
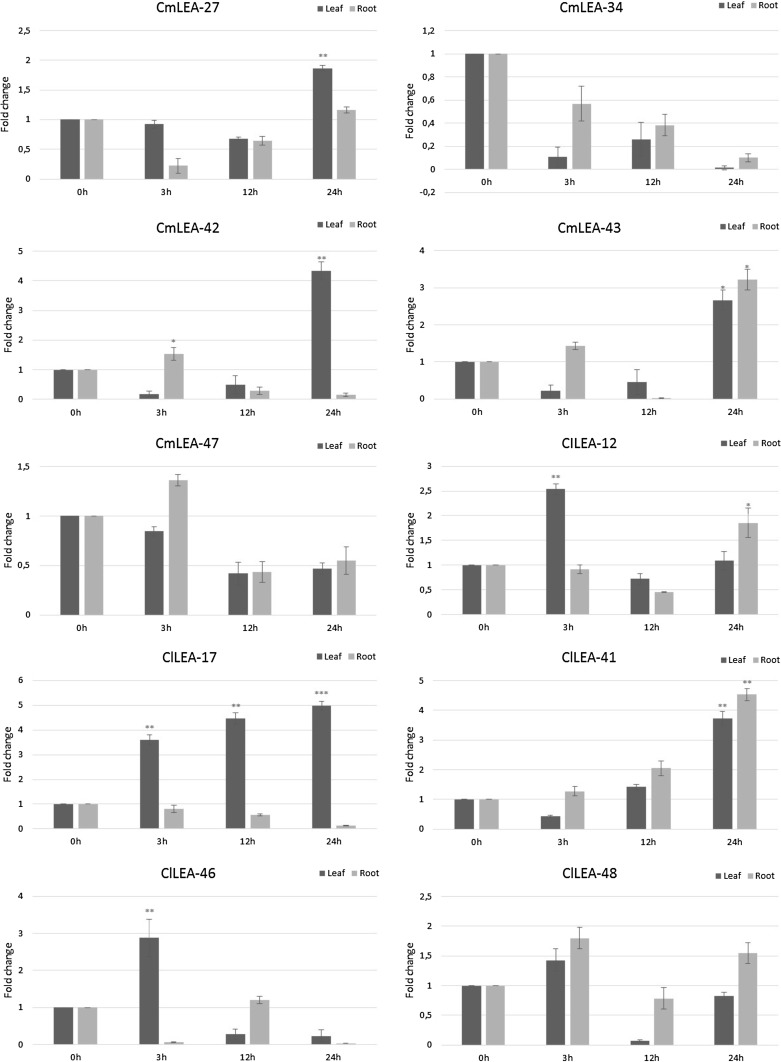
According to our qRT-PCR results, studied all LEA genes responded to drought in leaf tissues (Fig. 6). Especially ClLEA-12-17-46-48 genes expression levels were upregulated at the 3rd hour of drought stress application. ClLEA-17 gene was highly expressed in leaf tissues in all studied hours of water deficiency. Besides, expression levels of ClLEA-12-41-48 genes were high at the end of 24th hour of stress application in root tissues. In addition, ClLEA-41 was the only gene among the selected genes, whose expression level was upregulated in root and leaf tissues at the end of 24th hour of the stress treatment.
Fig. 6.
Expression patterns of selected ClLEA and CmLEA genes under water deficiency conditions in leaf (black) and root (grey) tissues (* means p < 0.01; ** means p < 0.05; *** means p < 0.001)
ClLEA-17 displayed an increase pattern in phloem and vascular tissues, 10th and 42nd days of fruit development and under drought stress conditions in our study when compared to the qRT-PCR data and the heat map. This may be attributed to its key predicated role in many metabolic processes and in water deficiency conditions. Same situation was observed for ClLEA-12 gene which plays important role in both metabolic processes and stress tolerance. In addition, ClLEA-41 gene had role in fruit development according to transcriptome data and upregulated under drought stress conditions. In addition, 26th LEA gene in Arabidopsis (At2g44060), which was the orthologous of ClLEA-41 gene, had high expression levels in non-seed tissues; also, it was induced by different stresses. Besides, 5th LEA gene (At1g20450) and 10th LEA gene (At1g76180) in Arabidopsis, which were orthologous of ClLEA-48 gene, were upregulated in different stress conditions. Especially, At1g20450 gene was induced by cold and salt stress conditions (Hundertmark and Hincha 2008). Our results are convenient with these findings, which suggests that ClLEA-41-48 genes and their orthologous in Arabidopsis were stress related genes in these plants. ClLEA-46 gene was related with fruit development while its expression was upregulated in vascular tissues and early in leaf cells under water deficiency. According to these findings, ClLEA-12-17-46 genes can be early response genes in drought stress.
Transcriptome data belonging to different melon varieties, fruit stages and salt stress conditions from SRA database were analyzed to evaluate expression patterns of LEA gene family members in melon (Supplementary Fig. 2). Gene expression levels of CmLEA-1-2-7-8-19-28-31-36-50-51-53-55-60 were upregulated in two melon varieties named as Cantalupo vedrantais and Piel de sapo pinonet when compared with expression levels of these proteins in Conomon SC and Piel de sapo-t111 melon varieties. In contrast, CmLEA-4-5-10-11-12-15-17-23-26-30-32-33-38-40-42-44-45-46-47-48-56-58-61 genes were highly expressed in Conomon SC and Piel de sapo-t111 varieties than in Cantalupo vedrantais and Piel de sapo pinonet varieties. Besides, gene expression levels of CmLEA-2-3-4-6-12-22-23-25-26-27-28-29-31-34-37-38-40-41-47-50-51-53-54-58 decreased while CmLEA-1-7-9-10-11-18-19-20-21-24-30-35-39-43-52-55-57-59-60 expression levels increased under salt stress conditions. CmLEA-43 gene was highly expressed in leaf and root tissues under drought stress in our study (Fig. 6). In addition, according to heat map its expression increased in normal vegetative tissues and under salt stress conditions. This gene may be one of the primary response genes to stress; also it may be important in normal cell functions. Expression levels of CmLEA-42 gene was the highest one in leaf and root tissues between analyzed LEA genes under drought stress. In addition, its expression was up regulated in yellow fruit stages when compared with the white fruit stages and a little increase was observed under salt stress based on heat map. 26th LEA gene in Arabidopsis (At2g44060), which was orthologous of CmLEA-42 gene, was upregulated by different stresses (Hundertmark and Hincha 2008). Besides, other gene orthologues of CmLEA-42 gene in maize [ZmLEA8 (GRMZ2G053637_T01) and ZmLEA10 (GRMZM2G352415_T01)] were upregulated in seed, innermost husk, silks or anthers according to transcriptome data (Li and Cao 2016). These results can support our results and it can be claimed that CmLEA-42 gene have a function in normal development stages besides its role in stress like function of CmLEA-43. However, CmLEA-34 gene was down regulated under water deficiency and salty conditions and compatible with transcriptome data, so this gene is not seemed to be related with stress.
Present study showed that some melon and watermelon LEA proteins have roles in protecting these plants from the effects of abiotic stresses while they have roles in normal cellular functions and fruit development stages which correlated with its abundance in maturing seeds (Galau et al. 1986; Manfre et al. 2006). Previous studies showed that gene expression of a LEA gene in citrus belong to LEA5 group highly increased in fruit than in leaves (Jeon et al. 2006) and also LEA5 gene expression were reported in cultured cells under freeze, water deficiency and salty conditions (Naot et al. 1995), which are relevant with our findings in melon and watermelon. Besides, expression of some LEA genes [STDHN1(YSK2) and StDHN25 (SK3)] in potato were reported in all tissues (Charfeddine et al. 2015), which may be related with their function in normal plant development stages, and these findings may support our results.
Present study provides a detailed molecular information about LEA gene family members in melon and watermelon. ClLEA-17 and CmLEA-42 genes seems to be important for trangenic interventions to determine their roles . Our results can open up new frontiers about understanding of functions of these important family members under normal and stress conditions by cloning and functional approaches.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Presentation of tissue specific expression of ClLEA genes by heatmap from different tissues of watermelon. Expression levels of genes were presented by colors from low (green) to high (red) expression pattern. (TIFF 7607 kb)
Tissue specific expression patterns of CmLEA genes by heatmap from different tissues of melon. Expression levels of genes were presented by colors from low (green) to high (red) expression pattern. (TIFF 4328 kb)
A total of 10 different motif clades with different colored boxes in melon (A) and watermelon (B) LEA proteins by MEME software. (TIFF 4075 kb)
Phylogenetic relationships between melon, watermelon and cucumber LEA proteins. (TIFF 2718 kb)
Comparative physical mapping of ClLEA genes with Arabidopsis, poplar, soybean, potato, grape and maize LEA genes. (TIFF 4326 kb)
Acknowledgements
This work was financially supported by The Scientific and Technological Research Council of Turkey (TUBITAK) with Grant Number 114O761.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-016-0405-8) contains supplementary material, which is available to authorized users.
References
- Altunoglu YC, Baloglu P, Yer EN, Pekol S, Baloglu MC. Identification and expression analysis of LEA gene family members in cucumber genome. Plant Growth Regul. 2016 [Google Scholar]
- Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134(9):1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- Arteaga-Vázquez M, Caballero-Pérez J, Vielle-Calzada JP. A family of microRNAs present in plants and animals. Plant Cell. 2006;18:3355–3369. doi: 10.1105/tpc.106.044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Proceedings of the second international conference on intelligent systems for molecular biology, AAAI Press, Menlo Park, CA, pp 28–36 [PubMed]
- Baloglu MC, Eldem V, Hajyzadeh M, Unver T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE. 2014;9(4):e96014. doi: 10.1371/journal.pone.0096014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Alejandra A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Li X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum) Planta. 2015;241(3):757–772. doi: 10.1007/s00425-014-2215-y. [DOI] [PubMed] [Google Scholar]
- Cao J, Shi F. Evolution of the RALF gene family in plants: gene duplication and selection patterns. Evol Bioinform. 2012;8:271–292. doi: 10.4137/EBO.S9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charfeddine S, Saïdi MN, Charfeddine M, Gargouri-Bouzid R. Genome wide identification and expression profiling of the late embryogenesis abundant genes in potato with emphasis on dehydrins. Mol Biol Rep. 2015;42(7):1163–1174. doi: 10.1007/s11033-015-3853-2. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal M, Tayal D, Chinnusamy V, Bansal KC. Abiotic stress and ABA-inducible group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J Biotechnol. 2009;139(2):137–145. doi: 10.1016/j.jbiotec.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Denekamp NY, Reinhardt R, Kube M, Lubzens E. Late embryogenesis abundant (LEA) proteins in nondesiccated, encysted, and diapausing embryos of rotifers. Biol Reprod. 2010;82(4):714–724. doi: 10.1095/biolreprod.109.081091. [DOI] [PubMed] [Google Scholar]
- Du D, Zhang Q, Cheng T, et al. Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Mol Biol Rep. 2013;40(2):1937–1946. doi: 10.1007/s11033-012-2250-3. [DOI] [PubMed] [Google Scholar]
- Duan J, Cai W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS ONE. 2012;7:e45117. doi: 10.1371/journal.pone.0045117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure LI, Galau GA. Developmental biochemistry of cotton seed embryogenesis and germination XIII. Regulation of biosynthesis of principal storage proteins. Plant Physiol. 1981;68(1):187–194. doi: 10.1104/pp.68.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L, Crouch M, Harada JJ, Ho T, Mundy J, Quatrano RS, Thomas TL, Sung ZR. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol. 1989;12:475–486. doi: 10.1007/BF00036962. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Filiz E, Ozyigit II, Tombuloglu H, Koc I. In silico comparative analysis of LEA (late embryogenesis abundant) proteins in Brachypodium distachyon L. Plant Omics. 2013;6(6):433. [Google Scholar]
- Flaus A, et al. Identification of multiple distinct SNF2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;43:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal TZ, Glazer I, Koltai H. An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett. 2004;577:21–26. doi: 10.1016/j.febslet.2004.09.049. [DOI] [PubMed] [Google Scholar]
- Galau GA, Hughes DW, Dure L. Abscisic acid induction of cloned cotton late embryogenesis abundant (LEA) messenger RNAs. Plant Mol Biol. 1986;7:155–170. doi: 10.1007/BF00021327. [DOI] [PubMed] [Google Scholar]
- Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, González VM, Alioto T, et al. The genome of melon (Cucumis melo L.) Proc Natl Acad Sci. 2012;109(29):11872–11877. doi: 10.1073/pnas.1205415109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokshar DS. Phytozome: a 14 comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J. 2005;388:151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelet J, Benamar A, Teyssier E, et al. Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol. 2005;137:157–167. doi: 10.1104/pp.104.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guleria P, Yadav SK. Identification of miR414 and expression analysis of conserved miRNAs from Stevia rebaudiana. Genom Proteom Bioinform. 2011;9(6):211–217. doi: 10.1016/S1672-0229(11)60024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, et al. Bioinformatic identification of microRNAs and their target genes from Solanum tuberosum expressed sequence tags. Chin Sci Bull. 2007;52:2380–2389. doi: 10.1007/s11434-007-0359-x. [DOI] [Google Scholar]
- Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H, Min J, et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet. 2013;45(1):51–58. doi: 10.1038/ng.2470. [DOI] [PubMed] [Google Scholar]
- Gurley WB. HSP101: a key component for the acquisition of thermotolerance in plants. Plant Cell. 2000;12:457–460. doi: 10.1105/tpc.12.4.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Terashima S, Kuboi T. Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J Plant Physiol. 2001;158:1333–1339. doi: 10.1078/0176-1617-00600. [DOI] [Google Scholar]
- He S, Tan L, Hu Z, Chen G, Wang G, Hu T. Molecular characterization and functional analysis by heterologous expression in E. coli under diverse abiotic stresses for OsLEA5, the atypical hydrophobic LEA protein from Oryza sativa L. Mol Genet Genom. 2012;287:39–54. doi: 10.1007/s00438-011-0660-x. [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ. 1950;347:1–32. [Google Scholar]
- Hunault G, Jaspard E. LEAPdb: a database for the late embryogenesis abundant proteins. BMC Genom. 2010;11:221. doi: 10.1186/1471-2164-11-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F, bZIP Research Group bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jeon OS, Kim CS, Lee SP, Kang SK, Kim CM, Kang BG, Hur Y, Kim IJ. Fruit ripening-related expression of a gene encoding group 5 late embryogenesis abundant protein in Citrus. J Plant Biol. 2006;49:403–408. doi: 10.1007/BF03178819. [DOI] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikawada T, Nakahara H, Kanamori Y, Iwata K, Watanabe M, McGee B, Tunnacli A, Okuda T. Dehydration induced expression of LEA proteins in an anhydrobiotic chironomid. Biochem Biophys Res Commun. 2006;348:56–61. doi: 10.1016/j.bbrc.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Kosová K, Vítámvás P, Prášil IT. The role of dehydrins in plant response to cold. Biol Plant. 2007;51(4):601–617. doi: 10.1007/s10535-007-0133-6. [DOI] [Google Scholar]
- Kumar R, Wehner TC. Breeding for high yield—a review. Cucurbit Genet Coop Rpt. 2011;33/34:41–42. [Google Scholar]
- Lan T, Gao J, Zeng QY. Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genet Genomes. 2013;9(1):253–264. doi: 10.1007/s11295-012-0551-2. [DOI] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cao J. Late Embryogenesis Abundant (LEA) gene family in maize: identification, evolution, and expression profiles. Plant Mol Biol Report. 2016;34(1):15–28. doi: 10.1007/s11105-015-0901-y. [DOI] [Google Scholar]
- Li L, Xu HL, Yang XL, Li YX, Hu YK. Genome-wide identification, classification and expression analysis of LEA gene family in soybean. Sci Agric Sin. 2011;44(19):3945–3954. [Google Scholar]
- Liu Y, Zheng Y. PM2, a group3 LEA protein from soybean, and its22-merrepeatingregion confersal tolerance in Escherichia coli. Biochem Biophys Res Commun. 2005;331:325–332. doi: 10.1016/j.bbrc.2005.03.165. [DOI] [PubMed] [Google Scholar]
- Liu SQ, Xu L, Jia ZQ, Xu Y, Yang Q, Fei ZJ, Lu XY, Chen HM, Huang SW. Genetic association of ethylene-insensitive-3-like sequence with the sex-determining M locus in cucumber (Cucumis sativus L.) Theor Appl Genet. 2004;117:927–933. doi: 10.1007/s00122-008-0832-1. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zheng Y, Zhang Y, Wang W, Li R. Soybean PM2 protein (LEA3) confers the tolerance of Escherichia coli and stabilization of enzyme activity under diverse stresses. Curr Microbiol. 2010;60:373–378. doi: 10.1007/s00284-009-9552-2. [DOI] [PubMed] [Google Scholar]
- Livak JK, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Manfre A, Lanni LM, Marcotte WR. The Arabidopsis group 1 late embryogenesis abundant protein ATEM6 is required for normal seed development. Plant Physiol. 2006;140:140–149. doi: 10.1104/pp.105.072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marunde MR, Samarajeewa DA, Anderson J, Li S, Hand SC, Menze MA. Improved tolerance to salt and water stress in Drosophila melanogaster cells conferred by late embryogenesis abundant protein. J Insect Physiol. 2013;59(4):377–386. doi: 10.1016/j.jinsphys.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Mehan MR, Freimer NB, Ophoff RA. A genome-wide survey of segmental duplications that mediate common human genetic variation of chromosomal architecture. Hum Genom. 2004;1(5):335–344. doi: 10.1186/1479-7364-1-5-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naot D, Ben-Hayyim G, Eshdat Y, Holland D. Drought, heat and salt stress induce the expression of a citrus homologue of an atypical late-embryogenesis Lea5 gene. Plant Mol Biol. 1995;27:619–622. doi: 10.1007/BF00019327. [DOI] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Campos F, Luis Reyes J, Garciarrubio A, Covarrubias AA. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010;154(1):373–390. doi: 10.1104/pp.110.158964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani A, Mahapatra RK. Computational identification of microRNAs and their targets in Catharanthus roseus expressed sequence tags. Genom Data. 2013;1:2–6. doi: 10.1016/j.gdata.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrosa AM, Martins CDPS, Gonçalve SLP, Costa MGC. Late embryogenesis abundant (LEA) constitutes a large and diverse family of proteins involved in development and abiotic stress responses in sweet orange (Citrus sinensis L. Osb.) PLoS ONE. 2015;10(12):e0145785. doi: 10.1371/journal.pone.0145785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhakainen T, Hess MW, Mäkelä P, Svensson J, Heino P, Palva ET. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol Biol. 2004;54:743–753. doi: 10.1023/B:PLAN.0000040903.66496.a4. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Campos F, Wei H, Arora R, Yang Y, Karlson DT, Covarrubias AA. Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ. 2008;31:1781–1790. doi: 10.1111/j.1365-3040.2008.01879.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shih MD, Lin SC, Hsieh JS, Tsou CH, Chow TY, Lin TP, Hsing YI. Gene cloning and characterization of a soybean (Glycine max L.) LEA protein, GmPM16. Plant Mol Biol. 2004;56(5):689–703. doi: 10.1007/s11103-004-4680-3. [DOI] [PubMed] [Google Scholar]
- Singh S, Cornilescu CC, Tyler RC, Cornilescu G, Tonelli M, Lee MS, Markley JL. Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stressrelated protein. Protein Sci. 2005;14(10):2601–2609. doi: 10.1110/ps.051579205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo J, et al. Identification of GhMYB109 encoding a R2R3MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.) Biochim Biophys Acta. 2003;1630:25–34. doi: 10.1016/j.bbaexp.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, et al. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Tolleter D, Hincha DK, Macherel D. A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. Biochim Biophys Acta. 2010;1798:1926–1933. doi: 10.1016/j.bbamem.2010.06.029. [DOI] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wang XS, Zhu HB, Jin GL, Liu HL, Wu WR, Zhu J. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.) Plant Sci. 2007;172(2):414–420. doi: 10.1016/j.plantsci.2006.10.004. [DOI] [Google Scholar]
- Wolkers WF, McCready S, Brandt WF, Lindsey GG, Hoekstra FA. Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim Biophys Acta. 2001;1544(1–2):196–206. doi: 10.1016/S0167-4838(00)00220-X. [DOI] [PubMed] [Google Scholar]
- Yang Z, Gu S, Wang X, Li W, Tang Z, et al. Molecular evolution of the cpp2 like gene family in plants: insights from comparative genomics of Arabidopsis and rice. J Mol Evol. 2008;67:266–277. doi: 10.1007/s00239-008-9143-z. [DOI] [PubMed] [Google Scholar]
- Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WS, et al. Interaction of wheat high-mobility-group proteins with four-way-junction DNA and characterization of the structure and expression of HMGA gene. Arch Biochem Biophys. 2003;409:357–366. doi: 10.1016/S0003-9861(02)00630-6. [DOI] [PubMed] [Google Scholar]
- Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot. 2010;61(15):4157–4168. doi: 10.1093/jxb/erq237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Presentation of tissue specific expression of ClLEA genes by heatmap from different tissues of watermelon. Expression levels of genes were presented by colors from low (green) to high (red) expression pattern. (TIFF 7607 kb)
Tissue specific expression patterns of CmLEA genes by heatmap from different tissues of melon. Expression levels of genes were presented by colors from low (green) to high (red) expression pattern. (TIFF 4328 kb)
A total of 10 different motif clades with different colored boxes in melon (A) and watermelon (B) LEA proteins by MEME software. (TIFF 4075 kb)
Phylogenetic relationships between melon, watermelon and cucumber LEA proteins. (TIFF 2718 kb)
Comparative physical mapping of ClLEA genes with Arabidopsis, poplar, soybean, potato, grape and maize LEA genes. (TIFF 4326 kb)