Dear Editor,
I am Dr. Ji-Hong Wu, from the Department of Ophthalmology, Eye & ENT Hospital of Fudan University, China. I write to present a case report of retinitis pigmentosa (RP) caused by novel digenic heterozygous mutations in a Chinese family.
RP, the most frequently described inherited retinal dystrophies (IRD), shows a prevalence of 1/3500 to 1/5000 worldwide, but statistics from the Beijing Eye Research Center show that the incidence of RP in China is approximately 1 in 1000[1]. RP commences with progressive night blindness, followed by gradual loss of peripheral visual fields (VF) and complete blindness in the later stage[2]. RP can be transmitted via autosomal dominant (24%), autosomal recessive (41%), and X-linked (22%)[3]. The remaining 12% of cases are presumed to cause by non-genetic factors, non-Mendelian inheritance (for example, mitochondrial or de novo mutations) or complex inheritance (digenic or polygenic inheritance)[4].
To date, more than 60 genes have been associated with RP. EYS is considered to be one of the most prevalent mutated genes worldwide[5]. The EYS gene contains 43 exons and spanning over 2000 bp within the RP25 locus (6q12). EYS encodes a protein of 3165 amino acids, which is localized to the outer segment of the photoreceptor cell layer[6]. EYS mutations demonstrate great genotypic and phenotypic varieties, and are one of the primary major gene responsible for recessive RP[7]. The low-density lipoprotein (LDL)-related receptor-5 (LRP5), a member of the LDL receptor superfamily, is a co-receptor for Wnt ligands involved in the wingless (Wnt) signaling pathway, which is essential for the development of vascular endothelial cells, Müller cells and retinal interneurons[8]. Loss-of-function mutations in LRP5 can cause familial exudative vitreoretinopathy (FEVR) in humans in the way of recessive RP[9], but no mutations have been identified in RP.
Diverse diagnostic strategies are used in RP cases. Custom-designed re-sequencing microarrays is an effective alternative for the detection of novel mutations, although this approach is limited to a known set of genes. Sanger sequencing is an indispensable and the most reliable method for identifying causative gene mutations, although it is not affordable due to the great genetic heterogeneity of RP. However, most of these limitations can now be overcome with next-generation sequencing (NGS), which has become widely used and facilitated the discovery of many causative genes and gene variants of complex traits[10]. In this study, we conducted a high-throughput sequence capture microarray with 99.67% coverage of all exons and combined with NGS to identify the possible causal genes in a Chinese family with RP, in which the cause of the disease had not been determined yet.
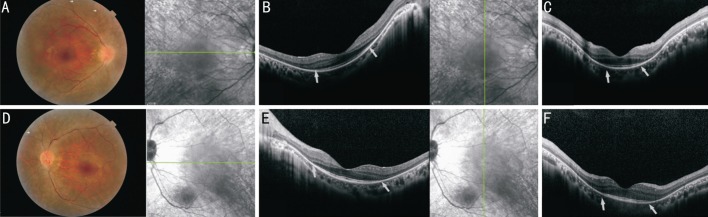
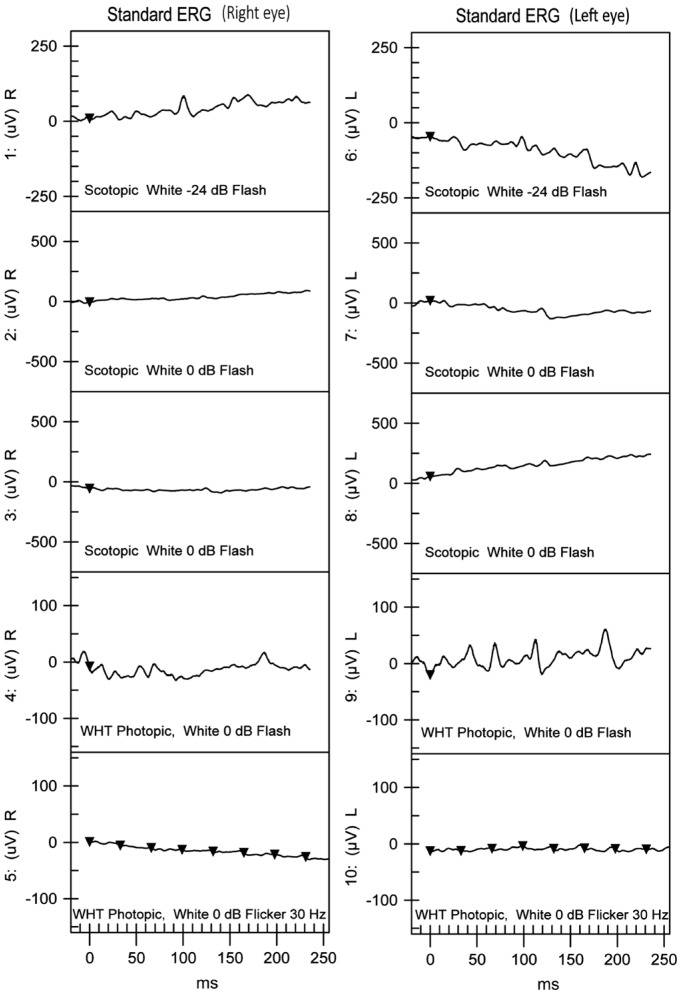
Our study involved a Chinese family consisting of the proband and his unaffected parents. The proband, a 30-year-old man, received a diagnosis of RP on the basis of clinical symptoms, such as decreased vision, night blindness, red-green blindness and the results of a comprehensive ocular examination, as described elsewhere[11]. Fundus examination revealed attenuated vessels and bone spicule-shaped pigment deposits in the peripheral retina in both eyes (Figure 1); spectral-domain optical coherence tomography (SD-OCT) line scans of both eyes through the central maculae showed a profound loss of photoreceptor layer structure in the retina. The remaining photoreceptors and the transitional zone were only seen in the macular zone (Figure 1); No detectable cone or rod responses were recorded by electroretinography (ERG) in the patient (Figure 2). All of the retinal phenotypes of the patient were typical for RP without any other ocular abnormalities. His parents reported no major issues with vision, and there was no family history of ocular or systemic diseases.
Figure 1. Fundus photographs and SD-OCT scans of the right and left eyes of the proband.
Fundus photography (A: right eye; D: left eye) in the proband exhibited attenuated vessels and bone spicule-shaped pigment deposits in the peripheral retina (marked by white arrow) in both eyes. SD-OCT scans along the horizontal (B: right eye; E: left eye) and vertical (C: right eye; F: left eye) meridian of the central retina highlighted the remaining photoreceptors and the transitional zone (marked by white arrow heads).
Figure 2. ERG results of the right and left eyes of the proband. Both dark-adapted and light-adapted responses were non-detectable.
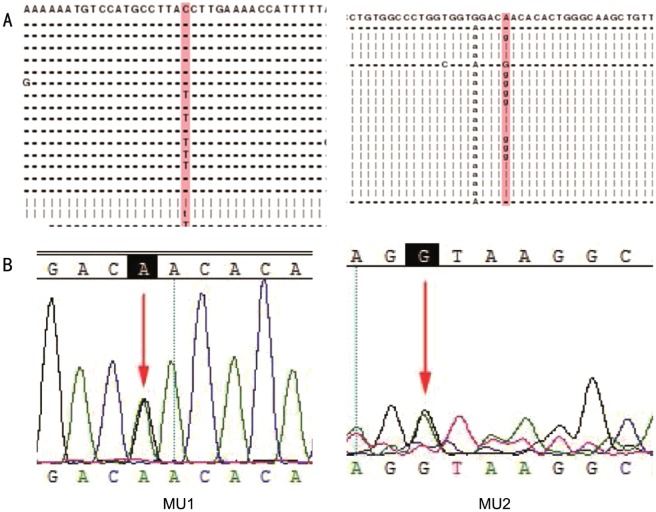
Genomic DNA from the family was extracted from the peripheral blood using a standard method[12]. The index patient was analyzed firs. We performed a targeted NGS approach on the patient. The NGS strategy included 129 target genes, spanning a length of 495 953 bp, with 191.5X depth and 99.65% coverage of the targeted region (Table 1). Fifteen heterozygous mutations and two homozygous mutations, including five novel and eleven known mutations, were identified. After subjecting the results to a pipeline of database filtering, including the dbSNP137, HapMap Project, 1000 Genome Project, YH database and Exome Variant Server databases, a novel nonsense mutation c.7723+1G>A in the EYS gene and a formerly reported missense mutation c.3361A>G in the LRP5 gene[9] were identified as potentially causative mutations for RP (Figure 3, Table 2). Sanger sequencing for these two mutations in his parents showed that his mother carried the EYS c.7723+1G>A mutation in the heterozygous state, and his father carried the LRP5 c.3361 A>G mutation in the heterozygous state.
Table 1. Overview of data production.
| Gene | NCBI reference sequence | Nucleotide change | Amino acid substitution | Chromosomal location | Gene subregion | Allele status |
| EYS | NM_001142800 | c.7723+1G>A | - | Chr6:6449797 | IN39 | Het |
| LRP5 | NM_002335 | c.3361 A>G | p.Asn1121Asp | Chr11:68192694 | EX15/CDS1 | Het |
Figure 3. Mutations identified in the EYS and LRP5 genes.
A: Protein alignment for the mutations identified in EYS (left) and LRP5 (right) gene; B: Sequencing results of the novel mutation in the EYE gene (MU1) and LRP5 gene (MU2). Arrows indicate the position of the mutated nucleotide.
Table 2. Mutations identified in the present study.
| No. of target genes | 129 |
| Target gene length (bp) | 495 953 |
| Coverage of target region | 99.67% |
| Mean depth of target region (X) | 264.26 |
| Fraction of target covered ≥30 X (%) | 96.80% |
Het: Heterozygous.
The advent of NGS has significantly increased the possibilities of identification of many rare disease genes, including genes for retinal dystrophies. NGS is a high-throughput tool which is capable of sequencing large gene pools efficiently and precisely, so it provides large data sets. Therefore, NGS has become a powerful and cogent tool for elucidating thorough mutation profiles for heterogeneous diseases. The clinical and genetic complexity of RP makes the accurate diagnosis of some cases possible only through NGS approaches. In this study, we comprehensively screened 129 genes involved in common inherited nonsyndromic eye diseases and successfully identified 2 potentially causative mutations for RP: EYS c.7723+1G>A and LRP5 c.3361 A>G.
The types of EYS mutations identified in RP include missense, nonsense, microdeletions and insertions, 5′UTR variations, and copy number variations, such as midsize genomic rearrangements[13]. The onset ages of RP caused by EYS varied greatly from 6- to 62-year-old[14]. In the present study, we detected a novel splicing mutation of EYS c.7723+1G>A in the proband in a heterozygous state. The potential pathogenicity of the filtered variants was then interpreted according to the existing and proposed American College of Medical Genetics and Genomics guidelines. Result shows that EYS c.7723+1G>A is splice site mutation, which may change the splicing way of the RNA precursor, resulting in abnormal protein coding. However, his unaffected mother is also in a heterozygous state, implying that heterozygous mutation of EYS c.7723+1G>A alone may not be sufficient to cause RP.
Mutations in the gene LRP5 have been shown to be responsible for both FEVR and osteoporosis-pseudoglioma syndrome, a disease that is characterized by blindness and loss of bone mass[15]. Heterozygous mutation of LRP5 c.3361 A>G has been reported in FEVR but not in RP. However, this previous study only tested the coding exons and adjacent intronic regions of the FZD4 and LRP5 genes. Thus, mutations in the intronic or regulatory regions could not be detected, and we cannot rule out the possibility of the existence of other mutations at work. In our study, we found that the mutation was present in both the proband and his unaffected father in heterozygous status, suggesting that single heterozygous mutation of LRP5 c.3361 A>G might not be sufficient to cause RP or FEVR.
Our data suggest a possible digenic form of inheritance for RP, whereby the co-existence of EYS c.7723+1G>A and LRP5 c.3361A>G heterozygous mutations can result in RP. Because EYS c.7723+1G>A is a splicing mutation and LRP5 c.3361A>G is missense mutation, we hypothesize this result can be explained by the fact that the LRP5 c.3361A>G mutation affects only part of the mRNA, and the normal amount of LRP5 protein is sufficient to sustain proper functioning of the cells. The EYS splicing mutation may affect pre-mRNA splicing and mRNA stability and translation efficiency; as a result, normal LRP5 protein is decreased, whereas abnormal protein accumulates and eventually leads to RP. However, there may be other unknown mechanisms responsible for the phenotype observed. This study may provide for the first time evidence for the digenic or polygenic nature of this disease, which further extended our current understanding of the genetic basis of RP, although more samples are needed to further understand the mutation spectrum of RP.
In summary, this is the first report that digenic heterozygous mutations of EYS c.7723+1G>A and LRP5 c.3361A>G can cause RP in a Chinese individual. These results enhance our current understanding of the genetic basis of RP and provide helpful clues for designing future studies to further investigate genetic factors associated with familial RP.
Acknowledgments
We would like to thank the patient and his parents for their collaboration. We are grateful to the technical staff at Eye and ENT Hospital of Fudan University for their assistance and BGI-Shenzhen for technical support.
Foundations: Supported by National Natural Science Foundation of China (No.81470623; No.81470624; No.81470625); National Key Basic Research Program of China (No.2013CB967503).
Conflicts of Interest: Gao FJ, None; Zhang SH, None; Chen JY, None; Xu GZ, None; Wu JH, None.
REFERENCES
- 1.Xu L, Hu L, Ma K, Li J, Jonas JB. Prevalence of retinitis pigmentosa in urban and rural adult Chinese: The Beijing Eye Study. Eur J Ophthalmol. 2006;16(6):865–866. doi: 10.1177/112067210601600614. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y, Tian D, Lee J, Zeng J, Zhang H, Chen S, Guo H, Xiong Z, Xia K, Hu Z, Luo J. Clinical and genetic identification of a large chinese family with autosomal dominant retinitis pigmentosa. Ophthalmic Genetics. 2015;36(1):64–69. doi: 10.3109/13816810.2013.809458. [DOI] [PubMed] [Google Scholar]
- 3.Birch DG, Locke KG, Felius J, Klein M, Wheaton DK, Hoffman DR, Hood DC. Rates of decline in regions of the visual field defined by frequency-domain optical coherence tomography in patients with RPGR-mediated X-linked retinitis pigmentosa. Ophthalmology. 2015;122(4):833–839. doi: 10.1016/j.ophtha.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010;11(4):273–284. doi: 10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- 5.Pieras JI, Barragan I, Borrego S, Audo I, Gonzalez-Del Pozo M, Bernal S, Baiget M, Zeitz C, Bhattacharya SS, Antinolo G. Copy-number variations in EYS: a significant event in the appearance of arRP. Invest Ophthalmol Vis Sci. 2011;52(8):5625–5631. doi: 10.1167/iovs.11-7292. [DOI] [PubMed] [Google Scholar]
- 6.Abd El-Aziz MM, Barragan I, O'Driscoll CA, Goodstadt L, Prigmore E, Borrego S, Mena M, Pieras JI, El-Ashry MF, Abu Safieh L, Shah A, Cheetham ME, Carter NP, Chakarova C, Ponting CP, Bhattacharya SS, Antinolo G. EYS, encoding an ortholog of Drosophila spacemaker, is mutated in autosomal recessive retinitis pigmentosa. Nat Genet. 2008;40(11):1285–1287. doi: 10.1038/ng.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosono K, Ishigami C, Takahashi M, Park DH, Hirami Y, Nakanishi H, Ueno S, Yokoi T, Hikoya A, Fujita T, Zhao Y, Nishina S, Shin JP, Kim IT, Yamamoto S, Azuma N, Terasaki H, Sato M, Kondo M, Minoshima S, Hotta Y. Two novel mutations in the EYS gene are possible major causes of autosomal recessive retinitis pigmentosa in the Japanese population. PLoS One. 2012;7(2):e31036. doi: 10.1371/journal.pone.0031036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407(6803):530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Li S, Xiao X, Wang P, Guo X, Zhang Q. Identification of FZD4 and LRP5 mutations in 11 of 49 families with familial exudative vitreoretinopathy. Mol Vis. 2012;18:2438–2446. [PMC free article] [PubMed] [Google Scholar]
- 10.Rangachari D, VanderLaan PA, Le X, Folch E, Kent MS, Gangadharan SP, Majid A, Haspel RL, Joseph LJ, Huberman MS, Costa DB. Experience with targeted next generation sequencing for the care of lung cancer: insights into promises and limitations of genomic oncology in day-to-day practice. Cancer Treat Commun. 2015;4:174–181. doi: 10.1016/j.ctrc.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Liu XX, Sheng XL, Gao X, Zhang XM, Li ZL, Li H, Liu Y, Rong W, Zhao K, Zhao C. Targeted next-generation sequencing reveals novel EYS mutations in Chinese families with autosomal recessive retinitis pigmentosa. Sci Rep. 2015;5:8927. doi: 10.1038/srep08927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma X, Guan L, Wu W, Zhang Y, Zheng W, Gao YT, Long J, Wu N, Wu L, Xiang Y, Xu B, Shen M, Chen Y, Wang Y, Yin Y, Li Y, Xu H, Xu X, Li Y. Whole-exome sequencing identifies OR2W3 mutation as a cause of autosomal dominant retinitis pigmentosa. Sci Rep. 2015;5:9236. doi: 10.1038/srep09236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonilha VL, Rayborn ME, Bell BA, Marino MJ, Pauer GJ, Beight CD, Chiang J, Traboulsi EI, Hollyfield JG, Hagstrom SA. Histopathological comparison of eyes from patients with autosomal recessive retinitis pigmentosa caused by novel EYS mutations. Grafe Arch Clin Exp. 2015;253(2):295–305. doi: 10.1007/s00417-014-2868-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Littink KW, van den Born LI, Koenekoop RK, Collin RW, Zonneveld MN, Blokland EA, Khan H, Theelen T, Hoyng CB, Cremers FP, den Hollander AI, Klevering BJ. Mutations in the EYS gene account for approximately 5% of autosomal recessive retinitis pigmentosa and cause a fairly homogeneous phenotype. Ophthalmology. 2010;117(10):2026–2033. doi: 10.1016/j.ophtha.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 15.Qin M, Hayashi H, Oshima K, Tahira T, Hayashi K, Kondo H. Complexity of the genotype-phenotype correlation in familial exudative vitreoretinopathy with mutations in the LRP5 and/or FZD4 genes. Hum Mutat. 2005;26(2):104–112. doi: 10.1002/humu.20191. [DOI] [PubMed] [Google Scholar]