Abstract
Polymorphisms in miRNA and miRNA pathway genes have been previously associated with cancer risk and outcome, but have not been studied in esophageal adenocarcinoma outcomes. Here, we evaluate candidate miRNA pathway polymorphisms in esophageal adenocarcinoma prognosis and attempt to validate them in an independent cohort of esophageal adenocarcinoma patients. Among 231 esophageal adenocarcinoma patients of all stages/treatment plans, 38 candidate genetic polymorphisms (17 biogenesis, 9 miRNA targets, 5 pri‐miRNA, 7 pre‐miRNA) were genotyped and analyzed. Cox proportional hazard models adjusted for sociodemographic and clinicopathological covariates helped assess the association of genetic polymorphisms with overall survival (OS) and progression‐free survival (PFS). Significantly associated polymorphisms were then evaluated in an independent cohort of 137 esophageal adenocarcinoma patients. Among the 231 discovery cohort patients, 86% were male, median diagnosis age was 64 years, 34% were metastatic at diagnosis, and median OS and PFS were 20 and 12 months, respectively. GEMIN3 rs197412 (aHR = 1.37, 95%CI: [1.04–1.80]; P = 0.02), hsa‐mir‐124‐1 rs531564 (aHR = 0.60, 95% CI: [0.53–0.90]; P = 0.05), and KIAA0423 rs1053667 (aHR = 0.51, 95% CI: [0.28–0.96]; P = 0.04) were found associated with OS. Furthermore, GEMIN3 rs197412 (aHR = 1.33, 95% CI: [1.03–1.74]; P = 0.03) and KRT81 rs3660 (aHR = 1.29, 95% CI: [1.01–1.64]; P = 0.04) were found associated with PFS. Although none of these polymorphisms were significant in the second cohort, hsa‐mir‐124‐1 rs531564 and KIAA0423 rs1053667 had trends in the same direction; when both cohorts were combined together, GEMIN3 rs197412, hsa‐mir‐124‐1 rs531564, and KIAA0423 rs1053667 remained significantly associated with OS. We demonstrate the association of multiple miRNA pathway polymorphisms with esophageal adenocarcinoma prognosis in a discovery cohort of patients, which did not validate in a separate cohort but had consistent associations in the pooled cohort. Larger studies are required to confirm/validate the prognostic value of these polymorphisms in esophageal adenocarcinoma.
Keywords: Esophageal adenocarcinoma, miRNA pathways, polymorphisms, prognosis
Introduction
With a five‐fold increased incidence over the past three decades, esophageal adenocarcinoma is one of the most rapidly rising malignancies in the developed world 1. Advances in established treatment regimens including surgery, radiotherapy, and chemotherapy (including targeted therapies such as trastuzumab) have led to modest improvements in survival; approximately 35 months for localized disease, 15 months for locally advanced disease, and 6 months for metastatic disease 2, 3, 4. As one third of the patients with localized disease survive for more than 10 years, this suggests there is heterogeneity among patients 4. Molecular factors which may contribute to this variation are not yet fully understood, but may assist in prognostication and elucidation of therapeutic targets.
MicroRNA (miRNA) molecules are short noncoding RNA molecules that regulate mRNA stability. miRNAs are produced by cleavage of large primary precursors, known as pri‐miRNAs, into pre‐miRNAs. Further modification and subsequent cleavage yields mature miRNAs, which are capable of negatively regulating the expression of genes by binding to the 3′UTRs of the target mRNAs 5. miRNAs regulate/modulate the translation of hundreds of other genes in multiple genetic pathways, have been shown to modulate the transformation of cancer cells and are linked to the etiology, progression, and prognosis of cancer 5, 6, 7. In addition, expression profiles of these miRNA pathway genes have been linked to other (non‐esophageal adenocarcinoma) cancers 5. Specific to esophageal adenocarcinoma, altered tumoral miRNA expression profiles and cell‐free circulating microRNAs have been correlated with prognosis 8, 9.
Although rare, single‐nucleotide polymorphisms (SNPs) in miRNA and miRNA‐processing pathway genes, which may alter the expression, transcription, and processing of miRNA have also been linked with cancer‐related risk and outcomes in a variety of tumor subtypes including esophageal adenocarcinoma risk 5. However to date, the effects of polymorphisms in these pathways on esophageal adenocarcinoma prognosis have not been studied. As SNPs influencing cancer risk may also impact prognostication, analyzing previously identified polymorphisms in miRNA pathways associated with cancer risk in esophageal cancer may help to yield new prognostic biomarkers and possible therapeutic targets for esophageal adenocarcinoma 10.
We performed an evaluation of miRNA and miRNA pathway‐processing genes previously associated with risk of any cancer with esophageal adenocarcinoma prognosis. Our aims of the study are: (1) to identify miRNA and miRNA pathways polymorphisms associated with cancer risk that can serve as prognostic markers of esophageal adenocarcinoma; and (2) to evaluate any previously identified polymorphic prognostic relationships in cancer in a cohort of esophageal adenocarcinoma patients. This information may help to identify new biological pathways that may influence esophageal adenocarcinoma outcomes.
Materials and Methods
Study population
The study protocol was approved by the Research Ethics Board of the University Health Network (UHN), Toronto, Ontario, Canada. The study population consisted of patients with a histologically confirmed diagnosis of esophageal adenocarcinoma who were receiving care at Princess Margaret Cancer Centre‐University Health Network (Toronto, Ontario, Canada). Two separate cohorts of patients—(1) a discovery cohort and (2) a validation cohort—were created for this study from a molecular epidemiology study evaluating the association between germline SNPs, esophageal cancer risk, and prognosis. From May 2006 to August 2009, 231 consecutive patients were prospectively enrolled into the discovery cohort. Between August 2009 and January 2013, a second group of 137 consecutive patients were recruited for the validation cohort. The date separating these two datasets was based on the closure of the initial study on August 15, 2009.
Eligibility criteria
All patients recruited to our study required a histological diagnosis of esophageal or gastro‐esophageal junction adenocarcinoma, were at least 18 years of age at diagnosis, able to communicate in English language, and had no cognitive deficits that would affect ability to consent. The written consent consisted of completing a baseline study questionnaire for epidemiological data, obtaining blood sample collection for genotyping at study entry as well as access to hospital records for regular updates on their clinicopathological data and survival.
Baseline epidemiological data
The study questionnaire was derived from the Harvard Oncologic Molecular Epidemiological survey 11. This self‐reported questionnaire documented details on sociodemographics, education, occupation, smoking, alcohol consumption, height and weight, weight loss, and coexistent gastrointestinal problems (such as Barrett's esophagus and Helicobacter pylori infection) as well as performance status as measured by the patient‐reported Eastern Cooperative Oncology Group (ECOG) score. A positive smoking history was defined as a patient reporting consuming ≥100 cigarettes in their lifetime. Those with a positive smoking history were classified as current or ex‐smokers dependent on their current smoking status at diagnosis. Where relevant, the total number of pack years smoked was obtained through self‐reported number of cigarettes consumed/day and years smoked. Alcohol intake was also documented in terms of standard drinks consumed per week 12.
Follow‐up, endpoints, and assessment of clinical outcomes
All patients in the discovery cohort were followed up until June 2011, while those in the validation cohort were followed up until July 2014. Follow‐up of the discovery cohort was limited to June 2011 due to research ethics restrictions. The histological diagnosis, location/clinical stage of cancer, treatments received were obtained from the clinical records. For those who underwent surgical resection, successful surgery was defined as R0 margins (> 1 cm). Curative intent chemotherapy was defined as that given in the neoadjuvant or adjuvant settings, while radiotherapy was defined as that given with potentially curative intent to the primary tumor; palliative radiation therapy given to metastatic sites were excluded.
We selected two primary endpoints for this study, progression‐free survival (PFS) and overall survival (OS). PFS was defined as the interval between the date of diagnosis and the first date of disease recurrence, progression, or death. OS was defined as the interval between the date of diagnosis and the date of death. For patients lost to regular follow‐up, efforts were made to obtain information on their vital status from cancer registry records. Otherwise, they were censored for either outcome on the date of last follow‐up.
Candidate polymorphism selection
We performed a comprehensive literature search (NCBI PubMed) on previously published studies assessing SNPs in the miRNA pathway (until 2013). We specifically selected variants that had been assessed in types of esophageal cancer, and further included other variants where there was a putative association with cancer incidence and survival of any cancer type. A list of candidate SNPs was compiled, covering all four areas of the miRNA pathway: including pri‐miRNAs (let‐7f‐2, mir‐100, mir‐124‐1, mir‐219‐1, mir‐26a‐1, mir‐30‐a, mir‐30‐c, mir‐373), pre‐miRNAs (mir‐146a, mir‐196a‐2, mir‐492, mir‐499, mir‐604, mir‐608, mir‐631), genes involved in the biogenesis (i.e., cleavage and processing) of miRNAs (AGO1, AGO2, DGCR8, DICER, DROSHA, GEMIN3, GEMIN4, HIWI, RAN, XPO5), and genes containing miRNA target sequences (BMPR1P, CD14orf101, CD86, DAG1, GOLGA7, IL1A, KIAA0423, KRT81, LAMB3, RAN, RYR3, USP9X) 13, 14, 15, 16, 17, 18, 19, 20. Descriptions of the sequence variants and the respective pathways involved are provided in Table S1.
Genotyping
Genomic DNA was extracted from peripheral blood lymphocytes using Archive Pure DNA Blood Kits (5 PRIME, Inter Medico, ON, Canada) according to the manufacturer's recommendations. Genotyping was performed using the GoldenGate® Genotyping Assay (Illumina Inc. San Diego, CA) as per the manufacturer's protocol. Briefly, sequence variants were uploaded to Illumina's Assay Design Tool (ADT) (www.illumina.com) for probe design resulting in a custom panel of 384 matrix spots out of which 54 were allocated to miRNA sequence variants. All the sequence variants presenting a functionality score <0.4 and design ability rank <1, which is considered as a lower limit for genotyping success by the manufacturer, were discarded. A total of 5 μL of 50 ng/μL in 10 mmol/L Tris‐HCL pH 8.0, 1 mmol/L EDTA of genomic DNA underwent an allele‐specific oligonucleotide hybridization followed by extension and ligation. A universal polymerase chain reaction (PCR) step for all loci followed with primers labeled with either Cy3 (primer 1) or Cy2 (primer 2). The amplified products were then hybridized to a sentrix array matrix (SAM) and scanned using the Illumina Bead Array Reader (BAR) (Illumina Inc.). The resulting data were analyzed with Beadstudio v.3.0 using the default parameters. Only sequence variants with GenCall scores >0.25 were called and samples were discarded if call rates were below 85%.
Genotyping for significant SNPs identified in the training set was performed using SNaPShot analysis in the validation set. Multiplex PCR was performed in 25 μL of a reaction mixture with a final concentration of each component as: 4 ng/μL of genomic DNA, 0.2 μmol/L of each primer (nine pairs PCR primer mixture), 2.5 mmol/L of MgCl2, 0.2 mmol/L of each dNTP, and 0.04U/μL of Taq polymerase in 1x PCR buffer (KAPA2G Robust PCR Kit). After an initial 2 min denaturation at 95°C, 35 cycles of denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec, and extension at 72°C for 30 sec were followed by a final extension step at 72°C for 5 min in the thermal cycler (GeneAmp9700; Applied Biosystems Foster City, CA). The PCR product (4.0 μL) was incubated at 37°C for 30 min with 2U of Exonuclease I (New England BioLabs) and 2U of shrimp alkaline phosphatase (New England BioLabs). After a 15 min incubation to inactivate the enzyme at 85°C, 1 μL of enzyme‐purified PCR product was mixed with 5 μL of SNaPshot Multiplex Ready Reaction Mix (Applied Biosystems), 1 μL of 1 μmol/L nine extension primer mixture, and 3 μL of dH2O. This mixture was placed in the thermal cycler and underwent 25 cycles at 96°C for 10 sec, 50°C for 5 sec, and 60°C for 10 sec. When completed, 0.5U of shrimp alkaline phosphatase was added and the reaction mixture was incubated for 60 min at 37°C to stop nonspecific reaction of extension primers to reduce SnaPshot background. Before loading onto the ABI PRISM 310 Genetic Analyzer (Applied Biosystems), 12 μL of HiDi formamide (Applied Biosystems) was added to 1 μL of reaction mixture, and samples were heated to 95°C for 5 min. Analyses were performed with GeneScan 3.0 application software (Applied Biosystems). Table S2 illustrates the primers used for initial PCR amplifications and later SNaPshot analysis.
Statistical analysis
All statistical analyses were conducted on SAS 9.2. Descriptive statistics were used to assess frequencies of sociodemographics and clinicopathological variables for each cohort. For the discovery cohort, univariable analysis using Cox proportional hazard models were used to assess the association of each variable with OS and PFS. Baseline multivariable Cox proportional hazard models for each clinical outcome were created using a backward selection algorithm of all sociodemographic and clinicopathological variable significantly associated with each outcome (P < 0.10) with age also included in the selection algorithm as a clinically important predictor. Adjusted hazard ratios (aHR) were provided with 95% confidence interval (CI). For each genetic polymorphism, the association with each outcome was first assessed using Kaplan–Meier method (log‐rank test). Each polymorphism was then individually added into the baseline multivariable model created for each outcome (OS and PFS) and tested for significance using the Wald Test. We applied the additive model for genetic inheritance in the Cox proportional hazard models to increase the power for screening. Nominal significance level was set as P < 0.05.
For the validation cohort, each SNP identified as significantly associated with OS and PFS in the discovery cohort was evaluated for association with survival using the same multivariable model that had been developed in the training set. As a form of sensitivity analysis in the validation cohort, we also constructed an independent multivariable model using backward selection, as above, for univariable significant predictors associated with OS and/or PFS in the analysis of the validation cohort. In the validation cohort, each SNP associated with OS or PFS in the discovery cohort was reevaluated in this sensitivity model.
In addition, both the discovery and validation cohorts were combined together to assess the genetic associations using both the same multivariable model in the discovery cohort and sensitivity model from the validation cohort.
Results
Baseline sociodemographic and clinicopathological characteristics
Baseline sociodemographic and clinical characteristics of our discovery (n = 231) and validation (n = 137) cohorts can be found in Table 1. The mean and median follow‐up times were 31 and 20 months, respectively, for our discovery cohort and 22 and 17 months for our validation cohort. At the time of analysis, there were 147 (64%) deaths in the discovery cohort and 84 (61%) deaths in the validation cohort. Furthermore, patients with evidence of progressive disease who remained alive were 22 (10%) of the discovery cohort and 19 (14%) of the validation cohort. Median PFS was 12 months for the discovery cohort and 13 months for the validation cohort, while median OS was 20 months for the discovery cohort and 17 months for the validation cohort.
Table 1.
Summary of patient baseline sociodemographics, clinicopathological, and treatment characteristics of our esophageal adenocarcinoma discovery and validation cohorts
| Variable | Subgroup | Discovery Cohort | Validation Cohort | P Value |
|---|---|---|---|---|
| Sociodemographic variables | ||||
| Gender | Male | 86% | 85% | 0.88 |
| Age at diagnosis | Median (range) | 64 (29–88) | 62 (29–86) | 0.08 |
| Ethnicity | Caucasian | 91% | 91% | 1.00 |
| Occupation | White collar | 54% | 51% | 0.72 |
| Education | Any postsecondary | 52% | 50% | 0.82 |
| Marital status | Married or equivalent | 72% | 73% | 0.80 |
| BMI at diagnosis | Underweight (≤18.5) | 4% | 2% | 0.49 |
| Overweight (>25) | 48% | 54% | ||
| Smoking status | Current | 14% | 29% | 0.003 |
| Ex‐smoker | 56% | 43% | ||
| Pack years smoked | Median (range) | 13.5 (0–118) | 15 (0–180) | 0.25 |
| Alcohol use | Yes | 86% | 69% | < 0.001 |
| Years drinking | Median (range) | 41 (0–77) | 29 (0–70) | < 0.001 |
| Clinicopathological variables | ||||
| Barrett's esophagus | Yes | 19% | 15% | 0.34 |
| Heart burn | Yes | 78% | 72% | 0.27 |
| H. pylori | Yes | 4% | 4% | 1.00 |
| ECOG | 0/1+ | 21%/79% | 22%/78% | 0.89 |
| Weight loss | Median (range) | 5.4 (0–55.9) | 5.4 (0–34.4) | 0.36 |
| Tumor location | GEJ | 40% | 39% | 0.007 |
| Distal | 50% | 43% | ||
| Middle | 8% | 2% | ||
| Clinical stage overall | 1–3 | 66% | 71% | 0.33 |
| 4 | 34% | 29% | ||
| Overall treatment intent | Curative | 78% | 74% | 0.44 |
| Surgery attempted | Yes | 69% | 59% | 0.07 |
| Successful surgery | Yes | 62% | 53% | 0.12 |
| Radiation received | Yes | 59% | 60% | 0.91 |
| Chemotherapy | Adjuvant or NeoAdjuvant | 46% | 59% | 0.02 |
All values represent percentages of patients except for pack years smoked, years of alcohol drunk, weight loss and age where the median and range in brackets are given. P values compare characteristics between the discovery and validation cohorts.
GEJ, gastro‐esophageal junction; ECOG, Eastern Cooperative Oncology Group performance score; BMI, body mass index.
In both cohorts, the majority of patients were male, Caucasian, with a median age in the early 60s, were married, had a smoking history, and experienced heartburn symptoms. Furthermore, the majority had localized tumors, had not experienced a significant amount of weight loss, and were of good performance status. Alcohol consumption was more frequent in the discovery cohort, while ongoing smoking at diagnosis was more prevalent in the validation cohort. Furthermore, a relatively higher proportion of rare mid‐esophageal tumors were observed in the discovery cohort, while curative intent chemotherapy was more commonly given in the validation cohort.
Quality control of genetic data
Details of the original list of the selected polymorphisms can be found in Supplementary Table 1. The genotype information, MAF and genotypic frequency of the final listing of the 38 polymorphisms investigated in the discovery cohort of the study can be found in Table S2. Two SNPs (GOLGA7 rs11337, MIR30C1 rs16827546) were excluded due to MAF <5%, two SNPs (USP9X rs10463, hsa‐let‐7f‐2 rs17276588) were excluded as they are located on the X chromosome where time‐to‐event methodologic approaches have not been developed to take into account X inactivation, and five SNPs (DGCR8 rs3757, DROSHA rs10719, hsa‐mir‐100 rs1834306, hsa‐mir‐219‐1 rs213210, hsa‐mir‐26a‐1 rs7372209) were excluded for not being in Hardy–Weinberg Equilibrium (P < 0.05).
Association analysis of polymorphisms and cancer outcomes
Univariable and multivariable analysis of the association between baseline sociodemographic and clinicopathological parameters with OS and PFS is displayed in Table 2. The final multivariable model for OS was adjusted for weight loss, stage, and successful surgery, while the final multivariable model for PFS was adjusted for weight loss, stage, successful surgery, and occupation.
Table 2.
Univariate and Multivariate Results for our clinical base model for the outcomes of overall survival and progression‐free survival in our discovery cohort
| Variable | Comparison | Overall Survival (OS) | Progression‐free Survival (PFS) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Unadjusted results | Multivariate results | Unadjusted results | Multivariate results | ||||||
| HR (95% CI) | P Value | aHR (95% CI) | P Value | HR (95% CI) | P Value | aHR (95% CI) | P Value | ||
| Gender | Male vs. female | 1.71 (1.00–2.92) | 0.05 | – | – | 1.40 (0.88–2.24) | 0.15 | ||
| Age At Dx | Per 1 Year increase | 1.00 (0.98–1.02) | 0.67 | 1.00 (0.99–1.01) | 0.91 | ||||
| Ethnicity | Caucasian vs. non‐Caucasian | 0.80 (0.46–1.39) | 0.42 | 0.87 (0.51–1.49) | 0.62 | ||||
| Occupation | Industry vs. business | 1.43 (1.03–2.01) | 0.03 | – | – | 1.43 (1.05–1.95) | 0.03 | 1.47 (1.02–2.10) | 0.04 |
| Education | No postsecondary vs. postsecondary | 1.21 (0.73–1.70) | 0.26 | 1.25 (0.91–1.72) | 0.16 | ||||
| Marital Status | Married vs. single | 1.16 (0.78–1.73) | 0.46 | 1.27 (0.88–1.84) | 0.20 | ||||
| BMI | Overweight vs. normal | 1.76 (0.81–3.85) | 0.15 | 2.10 (0.96–4.57) | 0.06 | ||||
| Underweight vs. normal | 0.96 (0.68–1.36) | 0.83 | 1.04 (0.75–1.43) | 0.83 | |||||
| Smoking Status | Ex‐Smoker vs. never | 0.81 (0.57–1.15) | 0.23 | 0.81 (0.57–1.15) | 0.23 | ||||
| Current Smoker vs. never | 1.17 (0.74–1.84) | 0.50 | 1.17 (0.74–1.84) | 0.51 | |||||
| Pack Years | Per pack year increase | 1.00 (0.99–1.01) | 0.42 | 1.00 (0.99–1.01) | 0.57 | ||||
| Alcohol Use | Yes vs. No | 1.34 (0.82–2.19) | 0.24 | 1.34 (0.82–2.20) | 0.24 | ||||
| Years of EtOH | Per year increase | 0.99 (0.98–1.01) | 0.47 | 0.99 (0.98–1.00) | 0.21 | ||||
| Barrett's Esophagus | Yes vs. No | 0.45 (0.26–0.80) | 5.9E‐3 | – | – | 0.36 (0.21–0.61) | 2.0E‐4 | – | – |
| Heart Burn | Yes vs. No | 0.97 (0.64–1.46) | 0.89 | 1.05 (0.72–1.54) | 0.80 | ||||
| H pylori | Yes vs. No | 0.80 (0.35–1.81) | 0.59 | 0.80 (0.35–1.81) | 0.59 | ||||
| ECOG | 2–3 vs. 0–1 | 1.80 (1.05–3.07) | 0.03 | – | – | 1.80 (1.06–3.07) | 0.03 | ||
| Weight Loss | Per kg lost | 1.02 (1.01–1.04) | 5.2E‐3 | 1.02 (1.01–1.04) | 5.2E‐3 | 1.02 (1.01–1.04) | 7.0E‐4 | 1.02 (1.00–1.03) | 0.02 |
| Tumor Location | GEJ vs. distal third | 0.81 (0.57–1.14) | 0.22 | 0.94 (0.68–1.30) | 0.71 | ||||
| Middle vs. distal third | 1.70 (0.94–3.06) | 0.08 | 1.69 (0.97–2.92) | 0.06 | |||||
| Upper vs. distal third | 0.53 (0.07–3.85) | 0.54 | 0.45 (0.06–3.24) | 0.43 | |||||
| Clinical Stage | 4 vs. 1‐3 | 3.45 (2.45–4.87) | 1.6E‐12 | 1.59 (1.02–2.48) | 0.04 | 3.55 (2.56–4.90) | 1.9E‐14 | 2.10 (1.38–3.21) | 6.0E‐4 |
| Treatment Intent | Palliative vs. curative | 4.21 (2.92–6.08) | 1.5E‐14 | – | – | 4.61 (3.22–6.64) | 1.2E‐16 | – | – |
| Surgery | Successful vs. other | 0.25 (0.18–0.34) | 1.5E‐16 | 0.30 (0.19–0.48) | 2.1E‐4 | 0.27 (0.20–0.37) | 1.9E‐16 | 0.40 (0.27–0.61) | 2.0E‐5 |
| Radiation | Yes vs. No | 1.59 (1.13–2.23) | 8.0E‐3 | – | – | 1.54 (1.12–2.11) | 7.8E‐3 | – | – |
| Chemotherapy | Adj/NeoAdj vs. pall/none | 0.64 (0.46–0.89) | 8.7E‐3 | – | – | 0.71 (0.53–0.97) | 0.03 | – | – |
Backward selection of clinical variables significantly associated (P < 0.10) with each outcome with age included in the selection algorithm was conducted to create separate significant multivariate clinical base models for each outcome (P < 0.05). It is upon these base models that each genetic polymorphism was then evaluated upon for significance.
Dx, Diagnosis; EtOH, Alcohol; Adj, Adjuvant; NeoAdj, Neoadjuvant; Pall, Palliative; BMI, Body Mass Index.
Univariable and multivariable analysis results of our polymorphisms with OS and PFS for our discovery cohort can be found in Table 3. Univariable analysis identified five polymorphisms that were significantly associated with OS, namely biogenesis pathway gene polymorphisms GEMIN3 rs197412 and GEMIN4 rs3744741, miRNA target gene polymorphisms CD86 rs17281995 and KIAA0423 rs1053667, and pre‐miRNA polymorphism hsa‐mir‐492 rs2289030. In multivariable analysis, GEMIN3 rs197412 and KIAA0423 rs1053667 remained significantly associated with OS (aHR = 1.37, 95% CI: (1.04–1.80); P = 0.02 and aHR = 0.51, 95% CI: (0.28–0.96); P = 0.04, respectively). In addition, one pri‐miRNA polymorphisms was found to be significantly associated with OS in multivariable analysis that was originally not found associated with OS in univariable analysis—hsa‐mir‐124‐1 rs531564 (aHR = 0.60, 95% CI: (0.37–0.99); P = 0.05).
Table 3.
Significant multivariate associations results between the miRNA pathway polymorphisms and prognosis (OS and PFS) in esophageal adenocarcinoma in our discovery cohort
| Gene | RS Number | A1 | A2 | Overall Survival (OS) | Progression‐free Survival (PFS) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Unadjusted results | Multivariate results | Unadjusted results | Multivariate results | ||||||||
| HR (95% CI) | P Value | aHR (95% CI) | P Value | HR (95% CI) | P Value | aHR (95% CI) | P Value | ||||
| GEMIN3 | rs197412 | G | A | 1.34 (1.05–1.72) | 0.02 | 1.37 (1.04–1.80) | 0.02 | 1.31 (1.04–1.66) | 0.02 | 1.33 (1.03–1.74) | 0.03 |
| CD86 | rs17281995 | G | C | 1.50 (1.08–2.08) | 0.01 | 1.37 (0.98–1.94) | 0.06 | 1.35 (0.99–1.85) | 0.06 | 1.23 (0.88–1.73) | 0.23 |
| hsa–mir‐124‐1 | rs531564 | G | C | 0.85 (0.57–1.28) | 0.44 | 0.60 (0.37–0.99) | 0.05 | 0.98 (0.68–1.40) | 0.91 | 0.77 (0.50–1.19) | 0.24 |
| hsa‐mir‐492 | rs2289030 | G | C | 0.58 (0.33–1.00) | 0.05 | 0.82 (0.46–1.47) | 0.50 | 0.68 (0.42–1.10) | 0.12 | 0.94 (0.56–1.60) | 0.82 |
| KIAA0423 | rs1053667 | G | A | 0.57 (0.32–1.01) | 0.05 | 0.51 (0.28–0.96) | 0.04 | 0.72 (0.44–1.16) | 0.17 | 0.72 (0.42–1.23) | 0.22 |
| GEMIN4 | rs3744741 | A | G | 1.39 (1.01–1.91) | 0.04 | 1.25 (0.89–1.79) | 0.19 | 0.72 (0.44–1.16) | 0.18 | 0.72 (0.42–1.23) | 0.23 |
| KRT81 | rs3660 | C | G | 1.09 (0.86–1.38) | 0.47 | 1.20 (0.93–1.55) | 0.17 | 1.12 (0.90–1.39) | 0.32 | 1.29 (1.01–1.64) | 0.04 |
The multivariate analysis results were adjusted for weight loss, stage, and surgery for the OS baseline model and adjusted for weight loss, stage, surgery, and occupation for the PFS baseline model.
A1, Minor Allele; A2, Major Allele; aHR are per each risk (minor) allele (A1).
GEMIN3 rs197412 was found also to be associated with PFS in both univariable analysis and multivariable analysis (aHR = 1.33, 95% CI: (1.03–1.74); P = 0.03). One additional polymorphism was also significantly associated with PFS in multivariable analysis, but not in univariable analysis: KRT81 rs3660 (aHR = 1.29, 95% CI: (1.01–1.64); P = 0.04).
Upon evaluation of the significant polymorphisms found in multivariable analysis in our validation cohort, none of our identified polymorphisms were found significantly associated with their respective outcomes using either the multivariable modeling from the discovery cohort or with a sensitivity model from backward selection in the validation cohort (Table S3). KRT81 rs3660 was significantly associated with PFS, but showed opposite directionality (aHR = 0.62, 95% CI: (0.42–0.91); P = 0.02). The strongest identified nonsignificant trend that was consistent in directionality was hsa‐mir‐124‐1 rs531564 with OS (aHR = 0.72, 95% CI: (0.47–1.11); P = 0.13) (Table 3).
Upon combining both the discovery and validation cohorts, hsa‐mir‐124‐1 rs531564 (aHR = 0.72, 95% CI: (0.52–0.99); P = 0.05) remained significantly associated with OS using the discovery cohort model (Table 4). The Kaplan–Meier curves for hsa‐mir‐124‐1 rs531564 in the discovery, validation, and combined cohorts can be found in Figure 1. In addition, KIAA0423 rs1053667 was found significantly associated with OS in both the discovery cohort (aHR = 0.56, 95% CI: (0.32–0.97); P = 0.04) and sensitivity analysis model (aHR = 0.64, 95% CI: (0.41–0.99); P = 0.04) (Table 4, Table S4). GEMIN3 rs197412 was only found significantly associated with OS (aHR = 1.26, 95% CI: (1.03–1.55); P = 0.02) in the sensitivity analysis model (Table 4, Table S4). None of the originally identified polymorphisms were found significantly associated with PFS in the combined cohort.
Table 4.
Multivariate associations results between the miRNA pathway polymorphisms and overall survival in esophageal adenocarcinoma across all three esophageal adenocarcinoma cohorts (discovery, validation, and combined) among polymorphisms originally found to be significantly associated with overall survival in the discovery cohort
| Gene | RS Number | A1 | A2 | Discovery cohort results | Validation cohort results | Combined cohort results | |||
|---|---|---|---|---|---|---|---|---|---|
| aHR (95% CI) | P Value | aHR (95% CI) | P Value | aHR (95% CI) | P Value | ||||
| GEMIN3 | rs197412 | C | T | 1.37 (1.04–1.80) | 0.02 | 1.05 (0.72–1.54) | 0.80 | 1.19 (0.95–1.49) | 0.13 |
| hsa‐mir‐124‐1 | rs531564 | G | C | 0.60 (0.37–0.99) | 0.05 | 0.72 (0.47–1.11) | 0.13 | 0.72 (0.52–0.99) | 0.045 |
| KIAA0423 | rs1053667 | C | T | 0.51 (0.28–0.96) | 0.04 | 0.80 (0.34–1.86) | 0.60 | 0.56 (0.32–0.97) | 0.038 |
The multivariate analysis results were adjusted based upon the original model for the discovery cohort, which included weight loss, stage, and successful surgery.
A1, Minor Allele, A2, Major Allele. aHR are per each risk (minor) allele (A1).
Figure 1.
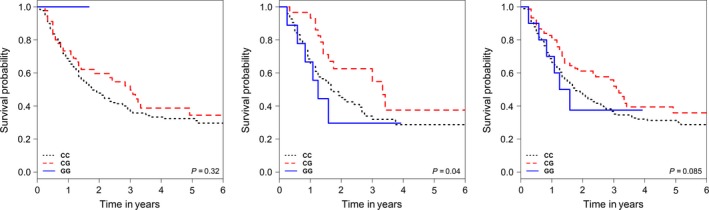
Kaplan–Meier Curves for overall survival with hsa‐mir‐124‐1 rs531564 in our discovery (left), validation (middle) and combined (right) cohort of patients with esophageal cancer.
As an exploratory analysis, we evaluated the combined effects of our two most consistently associated SNPs with overall survival (hsa‐mir‐124‐1 rs531564 and KIAA0423 rs1053667) in our combined patient (discovery and validation) cohort. As none of the patients carried more than two risk alleles in total, very few patients either were double heterozygotes (3%) or homozygous for the risk allele (3%), a comparison was done between patients who had at least one risk allele and those with no risk alleles. Patients carrying at least one variant allele were found to have reduced risk of OS (aHR = 0.59, 95% CI: (0.42–0.83); P = 0.002) and reduced risk of PFS (aHR1 = 0.71, 95% CI: (0.51–0.99); P = 0.043).
Discussion
Despite advances in therapy for esophageal adenocarcinoma, response rates and prognosis both remain poor and the need for new biomarkers and therapeutic targets is imperative. Given the diversity of pathways that are regulated by miRNA, polymorphisms in both miRNA and miRNA‐processing pathway genes may help to identify potential new targets for esophageal adenocarcinoma treatment. Here, by evaluating SNPs in miRNA and miRNA pathway genes previously associated with risk of development of cancer in esophageal adenocarcinoma prognosis, we have identified hsa‐mir‐124‐1 rs531564 as a relatively consistent predictor of overall survival whereby each variant allele contributed to a 30–40% decrease in mortality. Two additional polymorphisms were identified that may potentially be associated with OS in esophageal adenocarcinoma; namely KIAA0423 rs1053667 and GEMIN3 rs197412.
Previous studies have evaluated for prognostic biomarkers in esophageal adenocarcinoma in a variety of cancer‐related pathways including VEGF/angiogenesis, cell cycle pathways, cell free circulating microRNAs, DNA repair pathways, and a few studies have evaluated the role of SNPs in miRNA pathways in the risk of esophageal cancer and on esophageal squamous cell carcinoma prognosis, but no studies to date have evaluated the role of polymorphisms in miRNA pathway genes on esophageal adenocarcinoma prognosis 9, 21, 22, 23, 24, 25, 26, 27. This is the first study known to date, evaluating the potential for polymorphisms in miRNA pathways as prognostic markers in esophageal adenocarcinoma.
Pri‐mRNA hsa‐mir‐124‐1 rs531564 is a SNP that has previously been found associated with risk of development of cervical, colorectal, and esophageal squamous cell cancers 28, 29. Bioinformatics analyses have suggested that rs531564 may modulate the secondary structure of hsa‐mir124‐1 and alter the efficiency of the processing of pri‐miRNA‐124‐1, which can explain the association of different expression levels of mature miRNA‐124 with different alleles of this polymorphism 30. Furthermore, from the RegulomeDB database 31, data suggest that rs531564 is likely to affect the binding of a transcriptional factor called EZH2, which is involved in chromatin remodeling and gene silencing in cancer, and may alter the aggressiveness of tumors and their progression 32. miRNA‐124 has been described as a tumor suppressor, preventing development of a malignant phenotype in the cancer cell by the downregulation of several pathways including STAT3 signaling in colorectal cancer or EZH2 in gastric and hepatocellular cancers 33, 34, 35. In noncancer studies, mi‐RNA 124 has been suggested to have an immune modulatory role, as it has been found associated with experimental autoimmune encephalomyelitis (EAE), inflammatory bowel disease and hence may modulate the microenvironment by playing an immunosuppressive role to enhance tumorigenesis 36, 37, 38. Furthermore, miRNA‐124 has been found to inhibit ROCK, leading to reduction/modulation in the migration/invasion/aggressiveness of hepatocellular carcinomas, gliomas, and bladder cancer 35, 39, 40. Since the previously demonstrated effects of miRNA‐124 appears to be potentially pro‐tumorigenic in the tumor microenvironment but inhibitory to tumorigenesis in the cancer cell, further studies on the functional effects of the rs531564 in esophageal adenocarcinoma are necessary. Moreover, when searched in the Haploreg Database, no other SNPs were found to be highly linked with rs531564 41. This suggests that this SNP is likely to be the causative locus itself if its prognostic association is established in esophageal adenocarcinoma.
Among cancer studies, KIAA0423 rs1053667, a 3′ UTR polymorphism was found not associated with risk or OS in non‐Hodgkin's lymphoma or hepatocellular carcinoma and also not associated with prognosis in multiple myeloma patients undergoing autologous stem cell transplant 19, 42, 43, 44, 45. In addition, in a study evaluating SNP regulation of miRNA expression and colon cancer risk, rs1053667 was found associated with differential expression of its targeting miRNA, hsa‐miR‐19b‐3p in nontumor colonic tissues, but when comparing tumor versus nontumor tissue, the miRNA showed differential expression while rs1053667 was found not associated with risk of colon cancer 46. However, given the limited studies on the functional characteristics of KIAA0423, further genotype‐to‐phenotype analysis is required to better understand its function in carcinogenesis.
GEMIN3 rs197412 was also found consistently associated with OS in both the discovery and combined cohorts. GEMIN3 rs197412 was previously found to be associated with recurrence‐free survival in bladder cancer and overall survival in non‐Hodgkin's lymphoma 47, 48. GEMIN3 rs197412 was not associated with outcome in hepatocellular carcinoma and studies in colorectal cancer have yielded inconclusive results 49, 50, 51, 52. Genotype‐to‐phenotype analyses are required to better characterize the changes caused by this polymorphism (and the polymorphisms highly linked with them) on its gene product 47.
Landmark clinical trials reported within the past decade which have informed current clinical practice, have demonstrated only modest improvements (6–9%) in OS with peri‐operative chemotherapy for esophageal adenocarcinoma or a 26% improvement in overall survival with trastuzumab therapy for HER‐2‐positive advanced esophageal adenocarcinoma 53, 54. Thus, the need for new biomarkers in the prognostication and treatment of esophageal adenocarcinoma is acute. miRNA has the potential to regulate many cancer‐related pathways ranging from cell proliferation, invasion, and apoptosis (i.e., CDKs, Rb, E2F, and BCL‐2 family genes) and can provide insight into the diagnosis and treatment of cancer 55. Polymorphisms in miRNA can potentially modulate miRNA‐mRNA interaction and potentially create or destroy miRNA binding sites; while those in processing genes can influence the miRNA transcript either through altering transcription, processing, or maturation 5. By studying previously associated polymorphisms associated with either cancer risk or prognosis in other cancers, there is a possibility that the same polymorphisms may be able to predict clinical outcome in esophageal adenocarcinoma, yielding insights into new possible pathways to target for therapeutic agents 10, 56.
There were several limitations to this study. These include the self‐reported nature of the study questionnaire which could be affected by recall and social desirability bias. The relatively early stage of the cancer in most of our patients means that the impact of these polymorphisms on prognosis in advanced stage disease may be missed. Also, given that many of these SNPs identified are linked to other polymorphisms, we cannot ascertain if the biological effects seen are due to these polymorphisms or linked polymorphisms 41. Additionally, we have analyzed a set of polymorphisms previously associated with either cancer risk or prognosis in various cancers and further studies should attempt to identify new polymorphisms for analysis using methods including genome‐wide association studies or tagSNP approaches 23, 57. Furthermore, as this is a single center study analysis and our validation cohort was 50% the discovery cohort size with baseline demographic differences, further validation of this relationship in other esophageal adenocarcinoma cohorts and in other disease sites is warranted. However, this heterogeneity in sociodemographic and clinicopathological variables may help explain the differences seen between the results of our discovery and validation cohorts. Specifically, some of the factors that were different between the cohorts including smoking and alcohol status are known factors that can influence prognosis and may have influenced both the model selection and final results that were obtained 58, 59, 60. However, the heterogeneity in these sociodemographic and clinicopathological variables may support the robustness of the consistent associations between the discovery and combined cohorts that were identified.
In summary, this is the first study to evaluate the prognostic effects of miRNA pathways polymorphisms in a cohort of esophageal adenocarcinoma patients. We have identified multiple polymorphisms in miRNA pathway genes that were found associated with esophageal adenocarcinoma prognosis which was not validated in an independent cohort of esophageal adenocarcinoma patients but was found to have consistent relationships when both cohorts were combined. They were namely: hsa‐mir‐124‐1 rs531564, KIAA0423 rs1053667, and GEMIN3 rs197412. Future studies are needed to validate these identified relationships in other prospective studies of esophageal adenocarcinoma and evaluate their prognostic role in other cancer disease sites.
Disclaimers
The study sponsors had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript
Conflict of Interest
None declared.
Supporting information
Table S1. Summary of candidate polymorphisms belonging to mi‐RNA and mi‐RNA pathways genes selected for inclusion in our study on esophageal adenocarcinoma prognosis.
Table S2. Final listing of miRNA pathway polymorphisms investigated and their quality control metrics. A total of 47 polymorphisms were originally selected for investigation in the study and 38 polymorphisms were included in the final analysis. The specific genotype distribution frequency (percentages) is also listed.
Table S3. Results of our identified mi‐RNA pathway polymorphisms significantly associated with esophageal adenocarcinoma prognosis (OS and PFS) in our validation cohort.
Table S4. Results of the identified mi‐RNA pathway polymorphisms significantly associated with esophageal adenocarcinoma prognosis (OS and PFS) in the combined cohort.
Cancer Medicine 2017; 6(2):361–373
References
- 1. Brown, L. M. , Devesa S. S., and Chow W. H.. 2008. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J. Natl Cancer Inst. 100:1184–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Cutsem, E. , Van de Velde C., Roth A., Lordick F., Kohne C. H., Cascinu S., et al.; European Organisation for Research and Treatment of Cancer (EORTC)‐gastrointestinal cancer group . 2008. Expert opinion on management of gastric and gastro‐oesophageal junction adenocarcinoma on behalf of the European Organisation for Research and Treatment of Cancer (EORTC)‐gastrointestinal cancer group. Eur. J. Cancer 44:182–194. [DOI] [PubMed] [Google Scholar]
- 3. Bang, Y. J. , Van Cutsem E., Feyereislova A., Chung H. C., Shen L., Sawaki A., et al. 2010. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): a phase 3, open‐label, randomised controlled trial. Lancet 376:687–697. [DOI] [PubMed] [Google Scholar]
- 4. Dubecz, A. , Gall I., Solymosi N., Schweigert M., Peters J. H., Feith M., et al. 2012. Temporal trends in long‐term survival and cure rates in esophageal cancer: a SEER database analysis. J. Thorac. Oncol. 7:443–447. [DOI] [PubMed] [Google Scholar]
- 5. Ryan, B. M. , Robles A. I., and Harris C. C.. 2010. Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer 10:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carrington, J. C. , and Ambros V.. 2003. Role of microRNAs in plant and animal development. Science 301:336–338. [DOI] [PubMed] [Google Scholar]
- 7. Kumar, M. S. , Lu J., Mercer K. L., Golub T. R., and Jacks T.. 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39:673–677. [DOI] [PubMed] [Google Scholar]
- 8. Song, J. H. , and Meltzer S. J.. 2012. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology 143(35–47):e2. [DOI] [PubMed] [Google Scholar]
- 9. Zhai, R. , Wei Y., Su L., Liu G., Kulke M. H., Wain J. C., et al. 2015. Whole‐miRNome profiling identifies prognostic serum miRNAs in esophageal adenocarcinoma: the influence of Helicobacter pylori infection status. Carcinogenesis 36:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartman, M. , Loy E. Y., Ku C. S., and Chia K. S.. 2010. Molecular epidemiology and its current clinical use in cancer management. Lancet Oncol. 11:383–390. [DOI] [PubMed] [Google Scholar]
- 11. Zhou, W. , Liu G., Miller D. P., Thurston S. W., Xu L. L., Wain J. C., et al. 2002. Gene‐environment interaction for the ERCC2 polymorphisms and cumulative cigarette smoking exposure in lung cancer. Cancer Res. 62:1377–1381. [PubMed] [Google Scholar]
- 12. Skinner, H. A. , and Sheu W. J.. 1982. Reliability of alcohol use indices. The lifetime drinking history and the MAST. J. Stud. Alcohol 43:1157–1170. [DOI] [PubMed] [Google Scholar]
- 13. Ye, Y. , Wang K. K., Gu J., Yang H., Lin J., Ajani J. A., et al. 2008. Genetic variations in microRNA‐related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev. Res. (Phila.) 1:460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zou, H. Z. , and Zhao Y. Q.. 2013. Positive association between miR‐499A>G and hepatocellular carcinoma risk in a Chinese population. Asian Pac. J. Cancer Prev. 14:1769–1772. [DOI] [PubMed] [Google Scholar]
- 15. Zhou, X. , Chen X., Hu L., Han S., Qiang F., Wu Y., et al. 2010. Polymorphisms involved in the miR‐218‐LAMB3 pathway and susceptibility of cervical cancer, a case‐control study in Chinese women. Gynecol. Oncol. 117:287–290. [DOI] [PubMed] [Google Scholar]
- 16. Gao, Y. , He Y., Ding J., Wu K., Hu B., Liu Y., et al. 2009. An insertion/deletion polymorphism at miRNA‐122‐binding site in the interleukin‐1alpha 3′ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis 30:2064–2069. [DOI] [PubMed] [Google Scholar]
- 17. Saetrom, P. , Biesinger J., Li S. M., Smith D., Thomas L. F., Majzoub K., et al. 2009. A risk variant in an miR‐125b binding site in BMPR1B is associated with breast cancer pathogenesis. Cancer Res. 69:7459–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Landi, D. , Gemignani F., Naccarati A., Pardini B., Vodicka P., Vodickova L., et al. 2008. Polymorphisms within micro‐RNA‐binding sites and risk of sporadic colorectal cancer. Carcinogenesis 29:579–584. [DOI] [PubMed] [Google Scholar]
- 19. Yu, Z. , Li Z., Jolicoeur N., Zhang L., Fortin Y., Wang E., et al. 2007. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 35:4535–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yuan, L. , Chu H., Wang M., Gu X., Shi D., Ma L., et al. 2013. Genetic variation in DROSHA 3′UTR regulated by hsa‐miR‐27b is associated with bladder cancer risk. PLoS ONE 8:e81524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu, C. , Li M., Hu C., and Duan H.. 2014. Prognostic role of microRNA polymorphisms in patients with advanced esophageal squamous cell carcinoma receiving platinum‐based chemotherapy. Cancer Chemother. Pharmacol. 73:335–341. [DOI] [PubMed] [Google Scholar]
- 22. Umar, M. , Upadhyay R., Prakash G., Kumar S., Ghoshal U. C., and Mittal B.. 2013. Evaluation of common genetic variants in pre‐microRNA in susceptibility and prognosis of esophageal cancer. Mol. Carcinog. 52(Suppl 1):E10–E18. [DOI] [PubMed] [Google Scholar]
- 23. Eng, L. , Azad A. K., Qiu X., Kong Q. Q., Cheng D., Ying N., et al. 2015. Discovery and validation of vascular endothelial growth factor (VEGF) pathway polymorphisms in esophageal adenocarcinoma outcome. Carcinogenesis 36:956–962. [DOI] [PubMed] [Google Scholar]
- 24. Eng, L. , Azad A. K., Habbous S., Pang V., Xu W., Maitland‐van der Zee A. H., et al. 2012. Vascular endothelial growth factor pathway polymorphisms as prognostic and pharmacogenetic factors in cancer: a systematic review and meta‐analysis. Clin. Cancer Res. 18:4526–4537. [DOI] [PubMed] [Google Scholar]
- 25. Renouf, D. J. , Zhai R., Sun B., Xu W., Cheung W. Y., Heist R. S., et al. 2013. Association of MDM2 T309G and p53 Arg72Pro polymorphisms and gastroesophageal reflux disease with survival in esophageal adenocarcinoma. J. Gastroenterol. Hepatol. 28:1482–1488. [DOI] [PubMed] [Google Scholar]
- 26. Cescon, D. W. , Bradbury P. A., Asomaning K., Hopkins J., Zhai R., Zhou W., et al. 2009. p53 Arg72Pro and MDM2 T309G polymorphisms, histology, and esophageal cancer prognosis. Clin. Cancer Res. 15:3103–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang, L. , Wang C., Sun C., Xu Y., Ding Z., Zhang X., et al. 2014. The impact of pri‐miR‐218 rs11134527 on the risk and prognosis of patients with esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 7:6206–6212. [PMC free article] [PubMed] [Google Scholar]
- 28. Gao, X. R. , Wang H. P., Zhang S. L., Wang M. X., and Zhu Z. S.. 2015. Pri‐miR‐124 rs531564 polymorphism and colorectal cancer risk. Sci. Rep. 5:14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li, W. J. , Wang Y., Gong Y., Tu C., Feng T. B., and Qi C. J.. 2015. MicroRNA‐124 rs531564 polymorphism and cancer risk: a meta‐analysis. Asian Pac. J. Cancer Prev. 16:7905–7909. [DOI] [PubMed] [Google Scholar]
- 30. Qi, L. , Hu Y., Zhan Y., Wang J., Wang B. B., Xia H. F., et al. 2012. A SNP site in pri‐miR‐124 changes mature miR‐124 expression but no contribution to Alzheimer's disease in a Mongolian population. Neurosci. Lett. 515:1–6. [DOI] [PubMed] [Google Scholar]
- 31. Boyle, A. P. , Hong E. L., Hariharan M., Cheng Y., Schaub M. A., Kasowski M., et al. 2012. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Volkel, P. , Dupret B., Le Bourhis X., and Angrand P. O.. 2015. Diverse involvement of EZH2 in cancer epigenetics. Am. J. Transl. Res. 7:175–193. [PMC free article] [PubMed] [Google Scholar]
- 33. Ohigashi, Y. , Sho M., Yamada Y., Tsurui Y., Hamada K., Ikeda N., et al. 2005. Clinical significance of programmed death‐1 ligand‐1 and programmed death‐1 ligand‐2 expression in human esophageal cancer. Clin. Cancer Res. 11:2947–2953. [DOI] [PubMed] [Google Scholar]
- 34. Xie, L. , Zhang Z., Tan Z., He R., Zeng X., Xie Y., et al. 2014. MicroRNA‐124 inhibits proliferation and induces apoptosis by directly repressing EZH2 in gastric cancer. Mol. Cell. Biochem. 392:153–159. [DOI] [PubMed] [Google Scholar]
- 35. Zheng, F. , Liao Y. J., Cai M. Y., Liu Y. H., Liu T. H., Chen S. P., et al. 2012. The putative tumour suppressor microRNA‐124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 61:278–289. [DOI] [PubMed] [Google Scholar]
- 36. Ponomarev, E. D. , Veremeyko T., Barteneva N., Krichevsky A. M., and Weiner H. L.. 2011. MicroRNA‐124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP‐alpha‐PU.1 pathway. Nat. Med. 17:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koukos, G. , Polytarchou C., Kaplan J. L., Morley‐Fletcher A., Gras‐Miralles B., Kokkotou E., et al. 2013. MicroRNA‐124 regulates STAT3 expression and is down‐regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology 145(842–52):e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang, J. , Huang X., Xiao J., Yang Y., Zhou Y., Wang X., et al. 2014. Pri‐miR‐124 rs531564 and pri‐miR‐34b/c rs4938723 polymorphisms are associated with decreased risk of esophageal squamous cell carcinoma in Chinese populations. PLoS ONE 9:e100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. An, L. , Liu Y., Wu A., and Guan Y.. 2013. microRNA‐124 inhibits migration and invasion by down‐regulating ROCK1 in glioma. PLoS ONE 8:e69478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu, X. , Li S., Lin Y., Chen H., Hu Z., Mao Y., et al. 2013. MicroRNA‐124‐3p inhibits cell migration and invasion in bladder cancer cells by targeting ROCK1. J. Transl. Med. 11:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ward, L. D. , and Kellis M.. 2012. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40:D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang, B. , Liu C., Diao L., Wang C., and Guo Z.. 2014. A polymorphism at the microRNA binding site in the 3′ untranslated region of C14orf101 is associated with non‐Hodgkin lymphoma overall survival. Cancer Genet. 207:141–146. [DOI] [PubMed] [Google Scholar]
- 43. Peng, C. , Guo Z., Wu X., and Zhang X. L.. 2015. A polymorphism at the microRNA binding site in the 3′ untranslated region of RYR3 is associated with outcome in hepatocellular carcinoma. Onco. Targets Ther. 8:2075–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xie, Y. , Diao L., Zhang L., Liu C., Xu Z., and Liu S.. 2014. A miR‐SNP of the KRT81 gene is associated with the prognosis of non‐Hodgkin's lymphoma. Gene 539:198–202. [DOI] [PubMed] [Google Scholar]
- 45. de Larrea, C. F. , Navarro A., Tejero R., Tovar N., Diaz T., Cibeira M. T., et al. 2012. Impact of MiRSNPs on survival and progression in patients with multiple myeloma undergoing autologous stem cell transplantation. Clin. Cancer Res. 18:3697–3704. [DOI] [PubMed] [Google Scholar]
- 46. Mullany, L. E. , Wolff R. K., Herrick J. S., Buas M. F., and Slattery M. L.. 2015. SNP regulation of microRNA expression and subsequent colon cancer risk. PLoS ONE 10:e0143894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ke, H. L. , Chen M., Ye Y., Hildebrandt M. A., Wu W. J., Wei H., et al. 2013. Genetic variations in micro‐RNA biogenesis genes and clinical outcomes in non‐muscle‐invasive bladder cancer. Carcinogenesis 34:1006–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao, Y. , Diao L., Li H., and Guo Z.. 2015. Single nucleotide polymorphisms of microRNA processing genes and outcome of non‐Hodgkin's lymphoma. Onco. Targets Ther. 8:1735–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu, S. , An J., Lin J., Liu Y., Bao L., Zhang W., et al. 2014. Single nucleotide polymorphisms of microRNA processing machinery genes and outcome of hepatocellular carcinoma. PLoS ONE 9:e92791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin, M. , Gu J., Eng C., Ellis L. M., Hildebrandt M. A., Lin J., et al. 2012. Genetic polymorphisms in MicroRNA‐related genes as predictors of clinical outcomes in colorectal adenocarcinoma patients. Clin. Cancer Res. 18:3982–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee, H. C. , Kim J. G., Chae Y. S., Sohn S. K., Kang B. W., Moon J. H., et al. 2010. Prognostic impact of microRNA‐related gene polymorphisms on survival of patients with colorectal cancer. J. Cancer Res. Clin. Oncol. 136:1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Savas, S. , Xu J., Werdyani S., Shestopaloff K., Dicks E., Green J., et al. 2015. A Survival Association Study of 102 Polymorphisms Previously Associated with Survival Outcomes in Colorectal Cancer. Biomed. Res. Int. 2015:968743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Allum, W. H. , Stenning S. P., Bancewicz J., Clark P. I., and Langley R. E.. 2009. Long‐term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J. Clin. Oncol. 27:5062–5067. [DOI] [PubMed] [Google Scholar]
- 54. Cunningham, D. , Allum W. H., Stenning S. P., Thompson J. N., Van de Velde C. J., Nicolson M., et al.; MAGIC Trial P . 2006. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N. Engl. J. Med. 355:11–20. [DOI] [PubMed] [Google Scholar]
- 55. Sharma, P. , and Sharma R.. 2015. miRNA‐mRNA crosstalk in esophageal cancer: From diagnosis to therapy. Crit. Rev. Oncol. Hematol. 96:449–462. [DOI] [PubMed] [Google Scholar]
- 56. Azad, A. K. , Qiu X., Boyd K., Kuang Q., Emami M., Perera N., et al. 2014. A genetic sequence variant (GSV) at susceptibility loci of 5p15.33 (TERT‐CLPTM1L) is associated with survival outcome in locally advanced and metastatic non‐small‐cell lung cancer (NSCLC). Lung Cancer 84:289–294. [DOI] [PubMed] [Google Scholar]
- 57. Eng, L. , and Liu G.. 2013. VEGF pathway polymorphisms as prognostic and pharmacogenetic factors in cancer: a 2013 update. Pharmacogenomics 14:1659–1667. [DOI] [PubMed] [Google Scholar]
- 58. Thrift, A. P. , Nagle C. M., Fahey P. P., Russell A., Smithers B. M., Watson D. I., et al.; Australian Cancer Study Clinical Follow‐Up Study . 2012. The influence of prediagnostic demographic and lifestyle factors on esophageal squamous cell carcinoma survival. Int. J. Cancer 131: E759–E768. [DOI] [PubMed] [Google Scholar]
- 59. Druesne‐Pecollo, N. , Keita Y., Touvier M., Chan D. S., Norat T., Hercberg S., et al. 2014. Alcohol drinking and second primary cancer risk in patients with upper aerodigestive tract cancers: a systematic review and meta‐analysis of observational studies. Cancer Epidemiol. Biomarkers Prev. 23:324–331. [DOI] [PubMed] [Google Scholar]
- 60. Wright, C. D. , Kucharczuk J. C., O'Brien S. M., Grab J. D., and Allen M. S.; Society of Thoracic Surgeons General Thoracic Surgery Database . 2009. Predictors of major morbidity and mortality after esophagectomy for esophageal cancer: a Society of Thoracic Surgeons General Thoracic Surgery Database risk adjustment model. J. Thorac. Cardiovasc. Surg. 137: 587–595; discussion 596 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of candidate polymorphisms belonging to mi‐RNA and mi‐RNA pathways genes selected for inclusion in our study on esophageal adenocarcinoma prognosis.
Table S2. Final listing of miRNA pathway polymorphisms investigated and their quality control metrics. A total of 47 polymorphisms were originally selected for investigation in the study and 38 polymorphisms were included in the final analysis. The specific genotype distribution frequency (percentages) is also listed.
Table S3. Results of our identified mi‐RNA pathway polymorphisms significantly associated with esophageal adenocarcinoma prognosis (OS and PFS) in our validation cohort.
Table S4. Results of the identified mi‐RNA pathway polymorphisms significantly associated with esophageal adenocarcinoma prognosis (OS and PFS) in the combined cohort.


