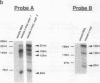
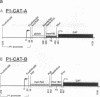
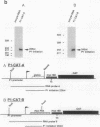
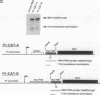
Abstract
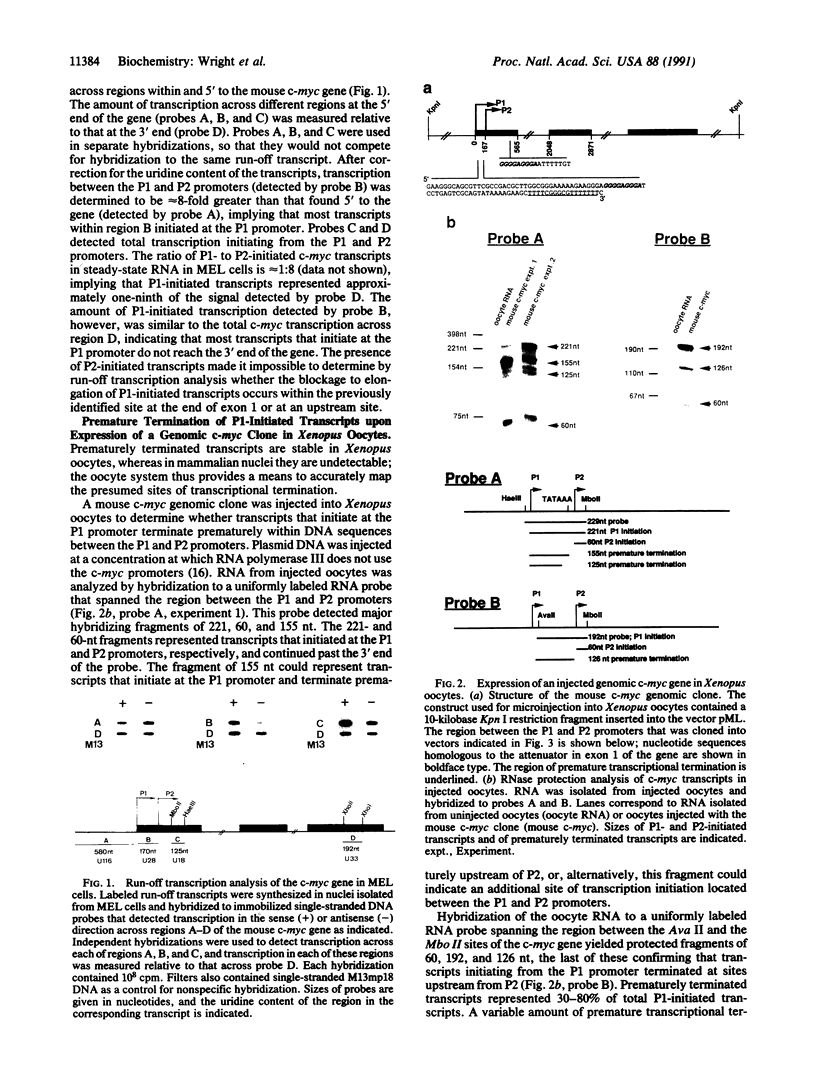
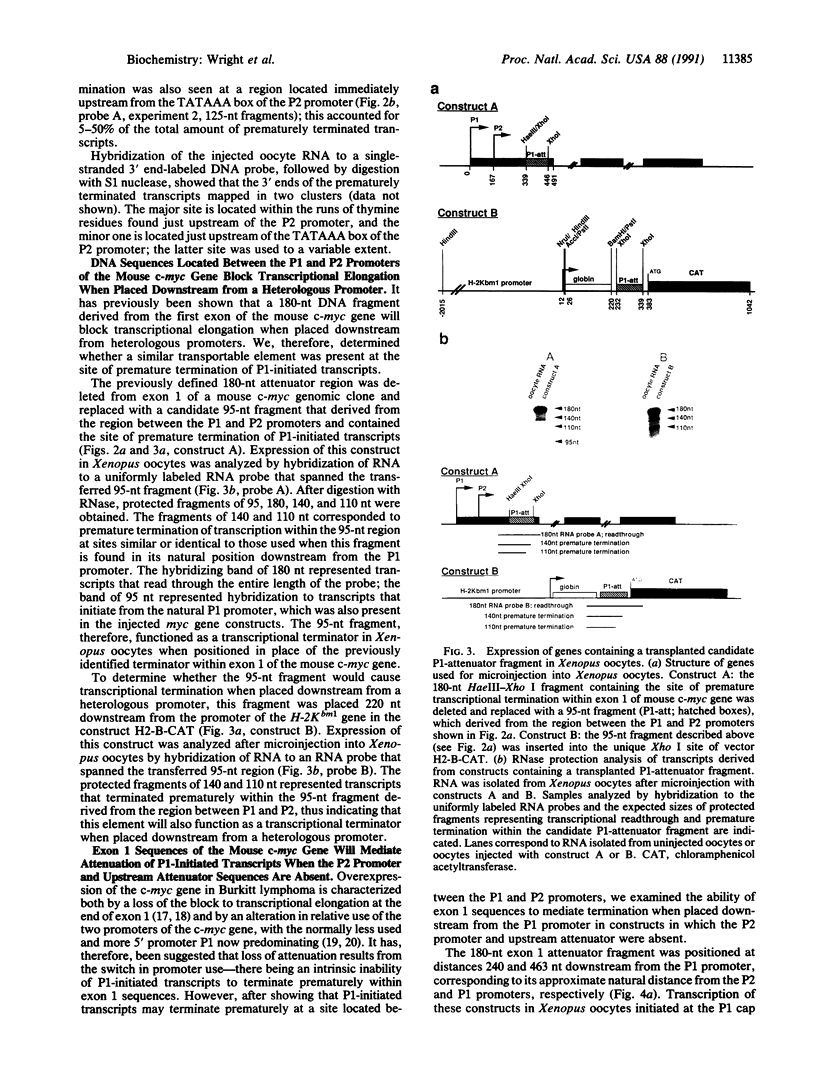
Modulation of transcriptional elongation within the c-myc gene is thought to play a major role in determining levels of c-myc mRNA in both normal and tumor cells. A discrete site of blockage to transcriptional elongation has previously been localized at the 3' end of exon 1 of the mouse and human c-myc genes. We here identify an additional site of transcriptional attenuation that is located between the P1 and P2 promoters of the c-myc gene and that mediates premature termination of transcripts initiating from the P1 promoter. A 95-nucleotide DNA fragment derived from this region prematurely terminated transcription when placed downstream from the promoter of the H-2Kbm1 gene and assayed by expression in Xenopus oocytes. We also show that the previously identified attenuation signal in exon 1 of the mouse c-myc gene can mediate premature termination of P1-initiated transcripts. Premature termination of P1-initiated transcripts presumably increases transcription from the downstream P2 promoter; aberrant regulation of this termination may explain the increased use of the P1 promoter that is characteristic of certain tumors in which myc is overexpressed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alitalo K., Schwab M. Oncogene amplification in tumor cells. Adv Cancer Res. 1986;47:235–281. doi: 10.1016/s0065-230x(08)60201-8. [DOI] [PubMed] [Google Scholar]
- Barik S., Ghosh B., Whalen W., Lazinski D., Das A. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell. 1987 Sep 11;50(6):885–899. doi: 10.1016/0092-8674(87)90515-0. [DOI] [PubMed] [Google Scholar]
- Bender T. P., Thompson C. B., Kuehl W. M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987 Sep 18;237(4821):1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Brown W. L., Groudine M. Accurate, TATA box-dependent polymerase III transcription from promoters of the c-myc gene in injected Xenopus oocytes. Genes Dev. 1989 Aug;3(8):1179–1189. doi: 10.1101/gad.3.8.1179. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Cesarman E., Dalla-Favera R., Bentley D., Groudine M. Mutations in the first exon are associated with altered transcription of c-myc in Burkitt lymphoma. Science. 1987 Nov 27;238(4831):1272–1275. doi: 10.1126/science.3685977. [DOI] [PubMed] [Google Scholar]
- Chen Z., Harless M. L., Wright D. A., Kellems R. E. Identification and characterization of transcriptional arrest sites in exon 1 of the human adenosine deaminase gene. Mol Cell Biol. 1990 Sep;10(9):4555–4564. doi: 10.1128/mcb.10.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Cory S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv Cancer Res. 1986;47:189–234. doi: 10.1016/s0065-230x(08)60200-6. [DOI] [PubMed] [Google Scholar]
- Cullen B. R., Lomedico P. T., Ju G. Transcriptional interference in avian retroviruses--implications for the promoter insertion model of leukaemogenesis. Nature. 1984 Jan 19;307(5948):241–245. doi: 10.1038/307241a0. [DOI] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C., Mechti N., Piechaczyk M., Lebleu B., Jeanteur P., Blanchard J. M. Increased rate of degradation of c-myc mRNA in interferon-treated Daudi cells. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4896–4899. doi: 10.1073/pnas.82.15.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Berger R., Polack A., Bornkamm G. W. Transcription of c-myc in human mononuclear cells is regulated by an elongation block. Oncogene. 1987;2(1):61–65. [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einat M., Resnitzky D., Kimchi A. Close link between reduction of c-myc expression by interferon and, G0/G1 arrest. Nature. 1985 Feb 14;313(6003):597–600. doi: 10.1038/313597a0. [DOI] [PubMed] [Google Scholar]
- Faus I., Richardson J. P. Structural and functional properties of the segments of lambda cro mRNA that interact with transcription termination factor Rho. J Mol Biol. 1990 Mar 5;212(1):53–66. doi: 10.1016/0022-2836(90)90304-5. [DOI] [PubMed] [Google Scholar]
- Fort P., Rech J., Vie A., Piechaczyk M., Bonnieu A., Jeanteur P., Blanchard J. M. Regulation of c-fos gene expression in hamster fibroblasts: initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res. 1987 Jul 24;15(14):5657–5667. doi: 10.1093/nar/15.14.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E., Steitz J. A. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989 Mar;8(3):851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E., Steitz J. A. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J. 1989 Mar;8(3):841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayhack E. J., Yang X. J., Lau L. F., Roberts J. W. Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell. 1985 Aug;42(1):259–269. doi: 10.1016/s0092-8674(85)80121-5. [DOI] [PubMed] [Google Scholar]
- Gurdon J. B., Wickens M. P. The use of Xenopus oocytes for the expression of cloned genes. Methods Enzymol. 1983;101:370–386. doi: 10.1016/0076-6879(83)01028-9. [DOI] [PubMed] [Google Scholar]
- Hann S. R., King M. W., Bentley D. L., Anderson C. W., Eisenman R. N. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell. 1988 Jan 29;52(2):185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- Hernandez N., Lucito R. Elements required for transcription initiation of the human U2 snRNA gene coincide with elements required for snRNA 3' end formation. EMBO J. 1988 Oct;7(10):3125–3134. doi: 10.1002/j.1460-2075.1988.tb03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz R. J., Li J., Greenblatt J. An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell. 1987 Nov 20;51(4):631–641. doi: 10.1016/0092-8674(87)90132-2. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Bartsch I., Grummt I. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature. 1990 Apr 5;344(6266):559–562. doi: 10.1038/344559a0. [DOI] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A DNA-binding protein is required for termination of transcription by RNA polymerase I in Xenopus laevis. Mol Cell Biol. 1990 Jun;10(6):2793–2800. doi: 10.1128/mcb.10.6.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. An RNA polymerase I termination site can stimulate the adjacent ribosomal gene promoter by two distinct mechanisms in Xenopus laevis. Genes Dev. 1990 Jul;4(7):1240–1251. doi: 10.1101/gad.4.7.1240. [DOI] [PubMed] [Google Scholar]
- Middleton K. M., Morgan G. T. Premature termination of transcription can be induced on an injected alpha-tubulin gene in Xenopus oocytes. Mol Cell Biol. 1990 Feb;10(2):727–735. doi: 10.1128/mcb.10.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H., Asselin C., Dufort D., Yang J. Q., Gupta K., Marcu K. B., Nepveu A. A cis-acting element in the promoter region of the murine c-myc gene is necessary for transcriptional block. Mol Cell Biol. 1989 Dec;9(12):5340–5349. doi: 10.1128/mcb.9.12.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Levine R. A., Campisi J., Greenberg M. E., Ziff E. B., Marcu K. B. Alternative modes of c-myc regulation in growth factor-stimulated and differentiating cells. Oncogene. 1987;1(3):243–250. [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B., Skoultchi A. I., Lachman H. M. Contributions of transcriptional and post-transcriptional mechanisms to the regulation of c-myc expression in mouse erythroleukemia cells. Genes Dev. 1987 Nov;1(9):938–945. doi: 10.1101/gad.1.9.938. [DOI] [PubMed] [Google Scholar]
- Pfleiderer C., Smid A., Bartsch I., Grummt I. An undecamer DNA sequence directs termination of human ribosomal gene transcription. Nucleic Acids Res. 1990 Aug 25;18(16):4727–4736. doi: 10.1093/nar/18.16.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986 Aug 7;322(6079):562–565. doi: 10.1038/322562a0. [DOI] [PubMed] [Google Scholar]
- Ramamurthy V., Maa M. C., Harless M. L., Wright D. A., Kellems R. E. Sequence requirements for transcriptional arrest in exon 1 of the murine adenosine deaminase gene. Mol Cell Biol. 1990 Apr;10(4):1484–1491. doi: 10.1128/mcb.10.4.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Amler L. C. Amplification of cellular oncogenes: a predictor of clinical outcome in human cancer. Genes Chromosomes Cancer. 1990 Jan;1(3):181–193. doi: 10.1002/gcc.2870010302. [DOI] [PubMed] [Google Scholar]
- Spandidos D. A., Paul J. Transfer of human globin genes to erythroleukemic mouse cells. EMBO J. 1982;1(1):15–20. doi: 10.1002/j.1460-2075.1982.tb01117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. A., LeStrange R. C., Novak U., Hayward W. S., Groudine M. The block to transcription elongation is promoter dependent in normal and Burkitt's lymphoma c-myc alleles. Genes Dev. 1990 Jan;4(1):75–88. doi: 10.1101/gad.4.1.75. [DOI] [PubMed] [Google Scholar]
- Taub R., Kelly K., Battey J., Latt S., Lenoir G. M., Tantravahi U., Tu Z., Leder P. A novel alteration in the structure of an activated c-myc gene in a variant t(2;8) Burkitt lymphoma. Cell. 1984 Jun;37(2):511–520. doi: 10.1016/0092-8674(84)90381-7. [DOI] [PubMed] [Google Scholar]
- Taub R., Moulding C., Battey J., Murphy W., Vasicek T., Lenoir G. M., Leder P. Activation and somatic mutation of the translocated c-myc gene in burkitt lymphoma cells. Cell. 1984 Feb;36(2):339–348. doi: 10.1016/0092-8674(84)90227-7. [DOI] [PubMed] [Google Scholar]
- Varmus H. E. The molecular genetics of cellular oncogenes. Annu Rev Genet. 1984;18:553–612. doi: 10.1146/annurev.ge.18.120184.003005. [DOI] [PubMed] [Google Scholar]
- Watson R. J. A transcriptional arrest mechanism involved in controlling constitutive levels of mouse c-myb mRNA. Oncogene. 1988 Mar;2(3):267–272. [PubMed] [Google Scholar]
- Wright S., Bishop J. M. DNA sequences that mediate attenuation of transcription from the mouse protooncogene myc. Proc Natl Acad Sci U S A. 1989 Jan;86(2):505–509. doi: 10.1073/pnas.86.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]