Abstract
Abiotic stress and climate change is the major concern for plant growth and crop yield. Abiotic stresses lead to enhanced accumulation of reactive oxygen species (ROS) consequently resulting in cellular damage and major losses in crop yield. One of the major scavengers of ROS is ascorbate (AA) which acts as first line of defense against external oxidants. An enzyme named ascorbate oxidase (AAO) is known to oxidize AA and deleteriously affect the plant system in response to stress. Genome-wide analysis of AAO gene family has led to the identification of five, three, seven, four, and six AAO genes in Oryza sativa, Arabidopsis, Glycine max, Zea mays, and Sorghum bicolor genomes, respectively. Expression profiling of these genes was carried out in response to various abiotic stresses and during various stages of vegetative and reproductive development using publicly available microarray database. Expression analysis in Oryza sativa revealed tissue specific expression of AAO genes wherein few members were exclusively expressed in either root or shoot. These genes were found to be regulated by both developmental cues as well as diverse stress conditions. The qRT-PCR analysis in response to salinity and drought stress in rice shoots revealed OsAAO2 to be the most stress responsive gene. On the other hand, OsAAO3 and OsAAO4 genes showed enhanced expression in roots under salinity/drought stresses. This study provides lead about important stress responsive AAO genes in various crop plants, which could be used to engineer climate resilient crop plants.
Keywords: abiotic stress, ascorbate oxidase, genome-wide analysis, reactive oxygen species (ROS), qRT-PCR
Introduction
Reactive oxygen species (ROS) are unavoidable consequence of aerobic metabolism. ROS are formed as a byproduct of various metabolic pathways present in different cellular compartments in plants (Foyer and Harbinson, 1994; Foyer, 1997; Sanmartin et al., 2003; del Río et al., 2006; Blokhina and Fagerstedt, 2010). Environmental stresses such as salinity, drought, chilling, metal toxicity, and UV-B radiations can intensify generation of ROS in plants by disturbing cellular homeostasis (Shah et al., 2001; Mittler, 2002; Sharma and Dubey, 2005, 2007; Hu et al., 2008; Maheshwari and Dubey, 2009; Tanou et al., 2009; Mishra et al., 2011; Srivastava and Dubey, 2011). ROS also act as secondary messengers in variety of cellular processes including environmental stresses (Desikan et al., 2001; Neill et al., 2002; Yan et al., 2007). Whether ROS act as signaling molecules or damaging molecules depends upon fine balance between ROS production and ROS scavenging. In spite of the fact that ROS are involved in signaling, they are also known to cause cellular damage (Fridovich, 1998). Increase in production of ROS during environmental stress can cause threat to cells by causing peroxidation of lipids, damage to nucleic acids, enzyme inhibition, activation of programmed cell death (PCD) pathway and ultimately causing death of the cells (Shah et al., 2001; Mittler, 2002; Verma and Dubey, 2003; Meriga et al., 2004; Sharma and Dubey, 2005; Mishra et al., 2011; Srivastava and Dubey, 2011). As ROS show multifunctional roles, it is important for the cell to control the level of ROS tightly to avoid any oxidative injury and not to get rid of them completely.
Scavenging and detoxification of ROS is achieved by antioxidant system including various enzymatic and non-enzymatic antioxidants (Noctor and Foyer, 1998). One of the non-enzymatic antioxidants vitamin C or L-ascorbic acid or ascorbate (AA; the anion of ascorbic acid) is the most abundant antioxidant found in photosynthetic tissues and plays a key role in defense against oxidative stress (Foyer et al., 1983; Smirnoff, 2000). Over 90% of the AA is present in the cytoplasm, but a considerable amount is also exported and localized in the apoplast. It is believed that apoplastic AA acts as first line of defense against the external oxidants such as ozone, SO2, and NO2 (Plöchl et al., 2000; Barnes et al., 2002).
In the apoplast, an enzyme named AAO (a glycoprotein which belongs to the blue copper oxidase enzyme family) oxidizes AA into MDA by release of an electron (Smirnoff, 2000). Unlike ascorbate peroxidase (APX) this released electron is not utilized to reduce H2O2 (Raven, 2000), rather it is accepted by O2 that gets reduced to H2O. MDA being an unstable radical undergoes rapid disproportionation to yield dehydroascorbate (DHA) and AA. The DHA can be recycled back to AA through AA-glutathione cycle (AA-GSH cycle) and MDA radical can also be recycled back to AA by the activity of an enzyme NAD(P)-dependent MDAR (Smirnoff, 2000). Transport of DHA in exchange of AA from apoplast to symplast is thought to occur via AA-DHA antiporter (or plasma membrane AA-DHA carrier), this is done to ensure continuous flux of reducing power to the cell wall (Horemans et al., 2000). Changes in AA-DHA ratio in apoplast is regulated and plays an important role in transition of cell from division to elongation state (Córdoba and González-Reyes, 1994; Kato and Esaka, 1999).
The expression of AAO is modulated by complex transcriptional and translational controls (Esaka et al., 1992), with transcript levels induced by growth promoters, e.g., auxin, (Pignocchi et al., 2003); jasmonates, (Sanmartin, 2002), and reduced by growth suppressors, e.g., salicylic acid, (Sanmartin, 2002; Pignocchi et al., 2003). The expression of AAO is induced under the influence of light and repressed in dark and this diurnal pattern of regulation is independent of circadian rhythm in Nicotiana tabacum (Pignocchi et al., 2003). Expression of AAO is also high in roots and young fruits (Pignocchi et al., 2003; Sanmartin et al., 2007).
Diverse roles have been ascribed to AAO enzyme in plants. AAO has been shown to maintain AA in its oxidized form which is necessary for cells to undergo mitosis (Kerk and Feldman, 1995). However, it cannot induce proliferation in non-competent cells (Citterio et al., 1994). Moreover, it is widely believed that AAO plays a critical role in cell elongation evident by its extracellular localization and high activity in rapidly expanding tissues (Esaka et al., 1992; Mosery and Kanellis, 1994; Ohkawa et al., 1994; Kato and Esaka, 1999). Tobacco (Nicotiana tabacum) Bright Yellow-2 protoplasts overexpressing AAO cDNA of pumpkin (Cucurbita pepo) shows cell elongation more rapidly than the other untransformed controls (Kato and Esaka, 2000). The overexpression of AAO in tobacco (Sanmartin et al., 2003) reduces stomatal aperture, consequently reducing rates of leaf water loss upon detachment, and a higher apoplast DHA content than the wild type. Transgenic tomato plants with suppressed AAO expression showed increased accumulation of AA in fruits (Zhang et al., 2011), also increased fruit yield was seen in wild type plants, where assimilates became limiting factor due to removal of leaves (Garchery et al., 2013). Enhancing AAO expression could be a possible strategy in down regulating oxygen diffusion in root nodules containing nitrogen-fixing bacteria, as well as during symbiosis with arbuscular mycorrhizal fungi (Balestrini et al., 2012). AAO overexpression was shown to delay dark induced senescence due to increase in antioxidant enzyme activity and induction in the expression of AA recycling genes such as APX and glutathione reductase (GR) (Fotopoulos and Kanellis, 2013). T-DNA mutant and antisense suppression of AAO gene in tobacco also leads to delayed flowering time and shorter stem length during the vegetative growth stage (Yamamoto et al., 2005).
Although, AAO genes have been implicated in varied cellular responses but they have not been explored with respect to their transcriptional modulation under stress conditions. Additionally, there are only few reports regarding their orthologs and paralogs in diverse plant genera. In the present report, genome-wide analysis for AAO genes has been carried out in Oryza sativa, Arabidopsis thaliana, Glycine max, Zea mays, and Sorghum bicolor genomes, which indicated the presence of five AAO genes in Oryza sativa, three AAO genes in Arabidopsis thaliana, seven AAO genes in Glycine max, four AAO genes in Zea mays, and six AAO genes in Sorghum bicolor genomes. Expression profiling of these genes based on publicly available microarray data has also been carried out, which indicate AAO are differentially regulated in response to various abiotic stresses and developmental cues. The temporal expression pattern of AAO genes in root and shoot tissue of rice seedlings in absence and presence of abiotic stress with the help of qRT-PCR was analyzed to comment about their transcriptional regulation in crop plant rice. The expression analysis of AAO genes in developmental stages and in response to various abiotic stress helped in identification of most stress responsive AAO genes in the five crop plants. Since AAO genes have been implicated in stomatal aperture reduction, increase in antioxidant activity and regulation by jasmonates as well as salicylic acid in previous studies, directed targeting of these stress responsive genes could be harnessed in engineering climate resilient crop plants.
Materials and Methods
In silico Identification of Ascorbate Oxidase Family Members
Putative AAO gene members in Oryza sativa and Arabidopsis thaliana were identified using protein profiles of AAO from Pfam database1 using HMMER 3.0 software2 against genome browser database TIGR Rice 6.13 and TAIR4. Arabidopsis gene identifier At5g21100 was also utilized to verify all the putative AAO proteins by searching against the annotated proteins in the whole rice and Arabidopsis genomes. The protein sequence of At5g21100 was obtained from ‘The Arabidopsis Information Resource (TAIR),’ and sequence based homology search was under taken to retrieve AAO proteins from rice (TIGR) and Arabidopsis (TAIR) genomes using BLASTp search tool and gene search tool.
In order to search AAO genes in Glycine max, Zea mays, and Sorghum bicolor, the protein sequences of known AAOs with annotation score three or above were searched in UniProtKB database5. Sequences of six such proteins (Q40588.1, P29162, P14133.1, P37064.1, P24792.2, and Q00624.1) were retrieved and aligned through Clustal Omega6 and retained for analysis in Stockholm output format. Consensus sequence from these proteins was retrieved from EMBOSS CONS7 by providing the aligned protein sequence file. Thereafter, the entire proteome sequence was downloaded from the respective databases of Glycine max8, Zea mays9, and Sorghum bicolor10. A BLAST compatible protein database was created on our local computer from the downloaded proteome sequences and masked to remove simple internal repeats using SAGE. Standalone BLAST+ (Camacho et al., 2009) and HMMER 3.1b22 were used in Linux platform on local machine for searching putative AAO genes in Glycine max, Zea mays, and Sorghum bicolor. The aligned sequences of pre-known AAOs obtained from UniProtKB in Stockholm format was used as query for Psi-BLAST against the masked proteome databases, separately for each species. An e-value cut-off of ±1e – 05 was taken as search threshold; redundant entries were removed from resultant list of proteins in order to create a non-redundant dataset. To further refine the search, JACK HMMER was used for iterative searching via Hidden Markov Model where consensus protein sequence of known AAOs obtained from EMBOSS CONS was used as query against the non-redundant set of protein sequences obtained from psi-BLAST. In all three plants, bit score threshold was kept 500 for HMMER search. Subsequently, the presence of three copper oxidase domains in each of the identified putative AAOs was determined through online database Pfam11 and SMART12.
Genomic Distribution of Ascorbate Oxidase Genes on Different Chromosomes
Chromosomal locations of AAO genes in Arabidopsis thaliana were determined using chromosomal map tool of Arabidopsis information resource (TAIR13), and chromosomal location of AAO genes in rice were determined using the Oryza sativa genome browser3 along with the Ensembl genome browser14, while chromosomal location of AAO genes in Glycine max, Zea mays, and Sorghum bicolor was determined by Ensembl genome browser14 and mapped on their respective chromosomes. Gene duplication and their presence on duplicated chromosomal segments were also investigated. Nucleotide sequences of putative AAO genes in all five plants were separately aligned by Clustal Omega and a neighbor-hood joining phylogenetic tree was made without distance corrections to determine the percent identity. AAO genes with sequence similarity of 90% or above were considered segmentally duplicated and the series of putative genes found near each other, without any other genes in between were considered tandem duplicated genes. The relative positions of AAO genes and segmental duplications are shown on their respective chromosomes. Orthologs among the putative AAO genes in all five plants were analyzed by online tool ORCAN15. Genes that fall in in same orthologous groups were determined based on orthology prediction value of more than 75%. For nomenclature of AAO gene family in respective species, the ascorbate oxidase genes were named “AAO” with a prefix “Os,” “At,” “Gm,” “Zm,” and “Sb” for Oryza sativa, Arabidopsis thaliana, Glycine max, Zea mays, and Sorghum bicolor, respectively, and a suffix number to indicate the different members and their order of occurrence on the respective chromosomes.
Phylogeny and Divergence of Ascorbate Oxidase Gene Family
Sequences of AAO proteins in Arabidopsis thaliana and Oryza sativa were obtained from their respective databases (TIGR and TAIR). Sequences of previously defined AAO proteins from Nicotiana tabacum (Q40588.1, P29162), Cucumis sativus (P14133.1), Cucurbita pepo (P37064.1), Cucurbita maxima (P24792.2), and Brassica napus (Q00624.1) were obtained from the database UniProtKB. AAO members of Triticum aestivum, Hordeum vulgare, and Brassica rapa were deduced using the same methodology followed for Glycine max, Zea mays, and Sorghum bicolor. These sequences were aligned by Clustal Omega and subjected to phylogenetic analysis by MEGA 7 (Kumar et al., 2015). Phylogenetic tree was constructed using neighbor-joining method with Poisson substitution model and complete deletion for gaps/missing data with 1000 Bootstrap replications. Alignment data is given in Supplementary Figure S1.
Domain Search and Conserved Motif Identification in Ascorbate Oxidase Proteins
Multiple EM for Motif Elicitation (MEME) program (Bailey et al., 2006) was used for de novo motif detection of AAO proteins from Oryza sativa, Arabidopsis thaliana, Glycine max, Zea mays, and Sorghum bicolor with the parameters of minimum width six and maximum width of fifty amino acids. The maximum number of motifs to be searched were kept ten. Each de novo detected motif was further subjected for search in Interpro database16 to find resemblance with known domains. Consensus sequence was also separately scanned in Interpro database to find the domains present in pre-identified AAOs.
Expression Analysis Using Microarray Data
The microarray data for the expression of AAO gene family members in rice during abiotic stress conditions such as cold, drought, and salt stress was retrieved from genevestigator17. The dataset obtained corresponds to 7-day-old IR64 rice seedlings subjected to various abiotic stress conditions.
The microarray data for Arabidopsis AAO genes under various abiotic stress conditions, such as cold, oxidative, drought, salt, and osmotic stresses was taken from AtGenExpress18. The datasets obtained were corresponding to different time points of stress, viz., 0.5, 1, 3, 6, 12, and 24 h for root and shoot tissues.
The abiotic stress microarray data of AAO gene family members in Zea mays was retrieved form genevestigator17. The dataset obtained corresponds to cold and drought stress. The data was retrieved for shoot samples of different varieties of Zea mays such as B73, OH43, and MO17 from 14-day-old seedlings subjected to cold stress of 5°C for 16 h. Data for drought stress was obtained from B73 variety of Zea mays wherein 4–5 days old seedlings germinated in distilled water soaked paper rolls were transferred to paper rolls soaked in PEG8000 solution with a water potential of -0.2 and -0.8 M. Pa for a period of 6 and 24 h. Data corresponds to different anatomical parts such as whole radicle, radicle tip, radicle elongation zone, stele and cortex of stress treated seedlings. Expression of AAO in anatomical parts such as shoot, tassel, leaf blade, ear, foliar leaf, and caryopsis were also analyzed in drought stressed plants at various developmental stages like seedling, stem elongation, inflorescence and anthesis. Zea mays (B73) seedlings were mostly grown till V8 developmental stage with optimal irrigation, then not irrigated completely for “n” number of days wherein number of days represents different developmental stages, such as 11, 18, 27, and 32 days represents V12 stage, V14 stage, V16 stage, and R1 stage, respectively.
Data was also retrieved for broad developmental stages of Oryza sativa, Arabidopsis thaliana, Zea mays, Glycine max, and Sorghum bicolor from genevestigator17. Heatmap was generated using the log2 signal values for dataset pertaining to stress samples of rice and Zea mays while mean normalized values were used for Arabidopsis to generate heatmap using MeV software package (Eisen et al., 1998).
Plant Material and Stress Treatment for qRT-PCR Analysis
The seedlings of IR64 rice were grown under standard growth conditions in the growth chamber at 28 ± 2°C with the photoperiod of 16 h and humidity of 70–80%. The seeds were sterilized with 1% Bavistin for 20 min and allowed to germinate in hydroponic system. The germinated seeds were then supplied with yoshida media (Yoshida et al., 1971). After 10 days, rice seedlings were exposed to salinity stress (200 mM NaCl dissolved in yoshida media) and drought stress (seedlings removed from hydroponics followed by desiccation on a tissue paper towel) for a time-period of 1 and 24 h whereas untreated seedlings were used as control.
Real-Time PCR
Total RNA was isolated from shoot and root tissue of control and stressed rice plants using IRIS kit (Bangalore, Genei) as per the manufacturer’s protocol. RNase free DNase I (Fermentas Life Sciences, USA) enzyme was used to get rid of the genomic DNA contamination in RNA samples. First strand cDNA synthesis was carried out using Maxima first strand cDNA synthesis kit for qPCR-RT (Fermentas Life Sciences, USA). Primers for real-time PCR analysis of AAO genes in rice were designed using NCBI primer BLAST for a product length ranging between 70 and 120 bp. The sequences for these primers are listed in Supplementary Table S1 and their binding sites are highlighted in Supplementary Figure S2. Rice β-actin and eIF-4α (eukaryotic initiation factor-4α) genes were used as reference genes for the normalization of real-time data. The PCR mixture contained 2.5 μl first strand cDNA (10 times diluted), 5 μl of 2X SYBR green PCR master mix (Fermentas Life Sciences, USA), and 2 μM of each gene-specific primer in a final volume of 10 μl. Negative template controls (NTC) were also performed for each of the primer pair. The real-time PCRs were performed employing ViiA7TM real-time PCR machine (Applied Biosystems, USA). All the PCRs were performed under the following conditions: 10 min at 95°C, and 40 cycles of 15 s at 95°C, 30 s at 60°C and melt curve with single reaction cycle with following conditions 95°C for 15 s, 60°C for 1 min and dissociation at 95°C for 15 s. Three biological replicates were analyzed for each sample. The relative expression ratio was calculated using delta Ct value method (Livak and Schmittgen, 2001).
Results
Ascorbate Oxidase Gene Members Constitute a Small Family
In the present study, we have employed bioinformatics tools to carry genome wide analysis of AAO genes present in Oryza sativa, Arabidopsis thaliana, Glycine max, Zea mays, and Sorghum bicolor. Genome wide search revealed that AAO genes in all the five-plant species under taken in this study constitute a multigene family. Five putative AAO genes in Oryza sativa, three in Arabidopsis thaliana, seven in Glycine max, four in Zea mays, and six in Sorghum bicolor were identified, details of which are given in Table 1. AAO genes identified in all the genera analyzed here encode for respective AAO proteins with no incidence of alternative splicing, barring At5g21105 (AtAAO3) gene in Arabidopsis which has three alternative spliced forms, named At5g21105.1, At5g21105.2, and At5g21105.3 and GLYMA20G12230 (GmAAO7) in Glycine max with two alternative spliced forms, named GLYMA20G12230.2 and GLYMA20G12230.3.
Table 1.
List of putative AAO genes in Oryza sativa, Arabidopsis thaliana, Glycine max, Zea mays, and Sorghum bicolor along with their, splice forms, nucleotide lengths, polypeptide length, CDS coordinates, chromosome number (bp base pair, aa amino acid).
| Gene | Locus identifier | Splice forms | Nucleotide length (bp) | Polypeptide length (aa) | CDS coordinates 5′–3′ | Chromosome no. | |
|---|---|---|---|---|---|---|---|
| Oryza sativa | OsAAO1 | LOC_Os06g37080 | 1 | 1746 | 582 | 21893247–21898264 | VI |
| OsAAO2 | LOC_Os06g37150 | 1 | 1902 | 634 | 21951199–21956670 | VI | |
| OsAAO3 | LOC_Os07g02810 | 1 | 1638 | 546 | 1055140–1058131 | VII | |
| OsAAO4 | LOC_Os09g20090 | 1 | 1734 | 578 | 12035268–12029912 | IX | |
| OsAAO5 | LOC_Os09g32952 | 1 | 1725 | 575 | 19662808–19665268 | IX | |
| Arabidopsis thaliana | AtAAO1 | At4g39830 | 1 | 2415 | 582 | 18478929–18481343 | IV |
| AtAAO2 | At5g21100 | 1 | 2745 | 573 | 7168184–7170928 | V | |
| AtAAO3 | At5g21105 | 3 | 5155 | 588 | 7172687–7177841 | V | |
| Glycine max | GmAAO1 | GLYMA05G33470 | 1 | 2113 | 577 | 38106684–38109689 | V |
| GmAAO2 | GLYMA08G14730 | 1 | 1962 | 576 | 10724343–10727358 | VIII | |
| GmAAO3 | GLYMA13G03650 | 1 | 2182 | 576 | 3663820–3672157 | XIII | |
| GmAAO4 | GLYMA14G04530 | 1 | 2182 | 581 | 3114029–3120767 | XIV | |
| GmAAO5 | GLYMA20G12150 | 1 | 2118 | 575 | 17069647–17077629 | XX | |
| GmAAO6 | GLYMA20G12220 | 1 | 2112 | 584 | 17148230–17152571 | XX | |
| GmAAO7 | GLYMA20G12230 | 2 | 1572 | 523 | 17209500–17214632 | XX | |
| Zea mays | ZmAAO1 | GRMZM2G064106 | 1 | 1997 | 514 | 102651781–102655212 | VII |
| ZmAAO2 | GRMZM2G163535 | 1 | 1829 | 574 | 102761977–102774858 | VII | |
| ZmAAO3 | GRMZM2G386170 | 1 | 1794 | 593 | 27419887–27421680 | VIII | |
| ZmAAO4 | GRMZM2G141376 | 1 | 2269 | 580 | 80614913–80620082 | IX | |
| Sorghum bicolor | SbAAO1 | Sb02g023140 | 1 | 1713 | 570 | 56601683–56603523 | II |
| SbAAO2 | Sb02g023150 | 1 | 2090 | 571 | 56649618–56655081 | II | |
| SbAAO3 | Sb02g023160 | 1 | 1740 | 579 | 56657530–56661262 | II | |
| SbAAO4 | Sb03g001450 | 1 | 1972 | 587 | 1309944–1311915 | III | |
| SbAAO5 | Sb10g022430 | 1 | 2000 | 578 | 50151435–50156051 | X | |
| SbAAO6 | Sb10g022440 | 1 | 1917 | 538 | 50171334–50174340 | X |
Chromosomal localization analysis showed that AAO members were dispersed on few chromosomes, i.e., three in Oryza sativa, two in Arabidopsis thaliana, three in Zea mays, five in Glycine max, and three in Sorghum bicolor. A scaled representation of all AAO genes on their respective chromosomes has been shown in Figure 1. OsAAO1 and OsAAO2 were located on chromosome VI, OsAAO3 was present on chromosome VII and OsAAO4 and OsAAO5 were present on chromosome IX (Figure 1A). In case of Arabidopsis, AtAAO1 was located on chromosome IV while AtAAO2 and AtAAO3 were located on chromosome V (Figure 1B). In Zea mays ZmAAO1, ZmAAO2 were located on chromosome VII and ZmAAO3 and ZmAAO4 were located on chromosome VIII and IX, respectively (Figure 1C). In Glycine max GmAAO1, GmAAO2, GmAAO3, GmAAO4 were located on chromosome V, VIII, XIII, XIV, respectively, and GmAAO5, GmAAO6, and GmAAO7 were located on chromosome XX (Figure 1D). In Sorghum bicolor, SbAAO1, SbAAO2, SbAAO3 were located on chromosome II, SbAAO4 was located on chromosome III and SbAAO5, SbAAO6 were located on chromosome X, respectively (Figure 1E).
FIGURE 1.
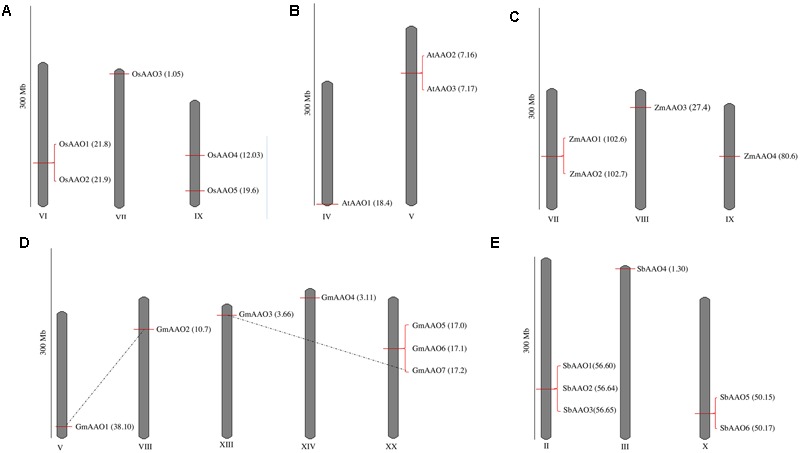
Chromosomal distribution of AAO genes in diverse genera. Chromosomal distribution of AAO genes in (A) rice, (B) Arabidopsis, (C) Zea mays, (D) Glycine max, and (E) Sorghum bicolor. Only the chromosomes having ascorbate oxidase (AAO) genes are shown. The scale is in (300 Mb). The chromosomal position (Mb) depicted in parenthesis along with gene name for each AAO is also marked as horizontal bar on the chromosome. Pairs of segmentally duplicated genes are shown by a dotted line.
Few of these gene members were present in close proximity to each other on the same chromosome suggesting a possible incidence of duplication events. Analysis for segmental duplications revealed that GmAAO1 on chromosome V could be segmentally duplicated on chromosome VIII as GmAAO2. GmAAO3 on chromosome XIII could be segmentally duplicated on chromosome XX as GmAAO7, as these genes share more than 90% sequence identity at nucleotide level. On the other hand, GmAAO5, GmAAO6, and GmAAO7 might have undergone tandem duplication as they appear as continuous cluster on the same chromosome. The sequence identity at the nucleotide level between GmAAO5 and GmAAO6, GmAAO6 and GmAAO7, and GmAAO5 and GmAAO7 genes is 85, 92, and 90%, respectively. Similarly, in Sorghum bicolor SbAAO1, SbAAO2, and SbAAO3 genes on chromosome II and SbAAO5 and SbAAO6 genes on chromosome X may represent tandem duplication due to their presence in close proximity to each other. The level of sequence identity between SbAAO1 and SbAAO2, SbAAO2 and SbAAO3, and SbAAO1 and SbAAO3 genes is 66, 87, and 86%, respectively. AAO genes in Oryza sativa, Arabidopsis thaliana, and Zea mays show no incidence of segmental duplication, instead OsAAO1 and OsAAO2 genes on chromosome VI, AtAAO2 and AtAAO3 genes on chromosome V, and ZmAAO1 and ZmAAO2 on chromosome VII show a possible incidence of tandem duplication.
The orthologous AAO genes were also found among the five-plant species. OsAAO1 gene in rice was predicted to be orthologous to AtAAO3, GmAAO3, ZmAAO4, and SbAAO5. OsAAO4 was predicted to be orthologous to AtAAO1, GmAAO1, ZmAAO1, and SbAAO2. OsAAO2 was predicted to be orthologous to SbAAO6 while OsAAO5 was predicted to be orthologous to ZmAAO3 and SbAAO4. ZmAAO2 in Zea mays was predicted to be orthologous to SbAAO3. No orthologs could be found for OsAAO3 from Oryza sativa, AtAAO2 from Arabidopsis thaliana, SbAAO1 from Sorghum bicolor, GmAAO2, GmAAO4, GmAAO5, GmAAO6, and GmAAO7 from Glycine max.
Ascorbate Oxidase Genes Are Highly Conserved across Diverse Genera
In order to investigate evolutionary relationship, a total of 53 AAO protein sequences from 13 different plant species were subjected to phylogenetic analysis. An unrooted tree was constructed by neighbor joining from the alignment of full-length protein sequences. The unrooted tree showed three distinct clades (Figure 2).
FIGURE 2.
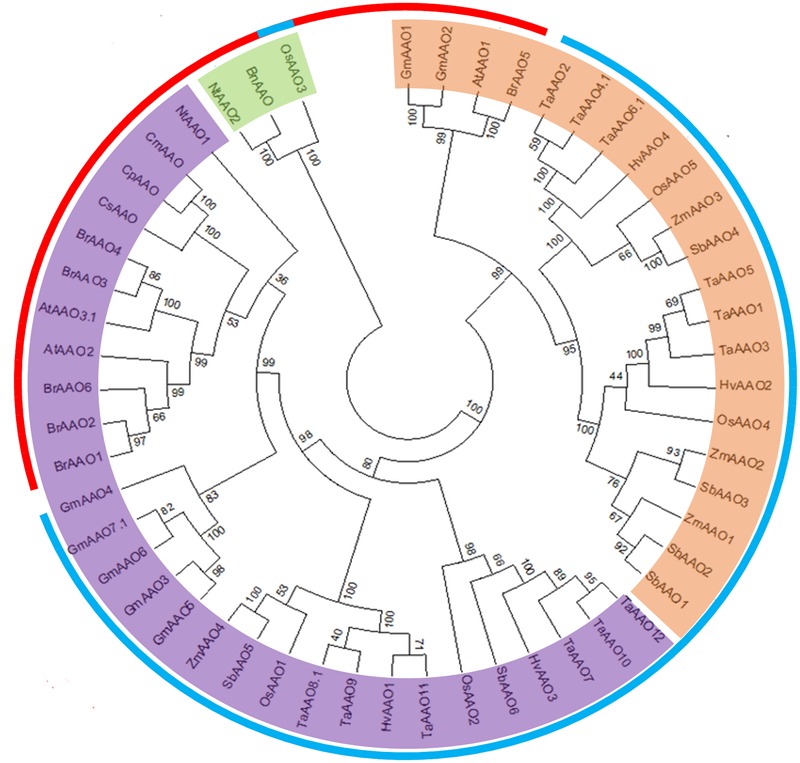
Phylogenetic analysis of AAO from diverse genera. An unrooted parsimonious tree of AAO proteins showing different clades. Clade I is marked in orange color while, clade II and clade III are marked in purple and green color, respectively. The outer circle denotes monocot (blue color) and dicot (red color) plant species taken for phylogenetic study. The tree was plotted using Mega7 software.
It was observed that clade I contained AAO family members mainly from monocot plants like Oryza sativa, Zea mays, Sorghum bicolor, Triticum aestivum, and Hordeum vulgare. However, several AAOs from these monocot species were also found in clade II and clade III. Most of AAO family members from dicot plants like Arabidopsis thaliana, Glycine max, Brassica rapa, Nicotiana tabacum, and Brassica napus were exclusively present in clade II and clade III except a small cluster of AtAAO1, GmAAO1, GmAAO2, and BrAAO5 which were the part of clade I. Although based on the signature sequence OsAAO3 has been classified as an AAO, but as per an unrooted phylogenetic tree, it was an outlier forming a third clade, clustered together with pre-known BnAAO from Brassica napus and NtAAO2 from Nicotiana tabacum, which have diverged substantially in terms of their protein sequence as compared to other AAO proteins.
All three clades contain the proteins from both dicots and monocots, suggesting that the divergence of AAO gene family might have occurred before the split of dicot and monocots during course of plant evolution. Additionally, it was also observed that the genes which are segmentally duplicated tend to be clustered together in the phylogenetic tree, further supporting the possibility of a duplication event.
Identification of Conserved Motifs in Ascorbate Oxidase Proteins
Scanning of consensus sequence of previously defined AAOs in Interpro identified three domains of cupredoxin family present across the entire length of its protein sequence. Each cupredoxin family domain consisted of either multicopper oxidase type 1, type 2, or type 3 domains. Multicopper oxidase conserved sites and copper binding sites were also present in sequence of multicopper oxidase type 2 domain. Pattern of domains is depicted in Figure 3A.
FIGURE 3.
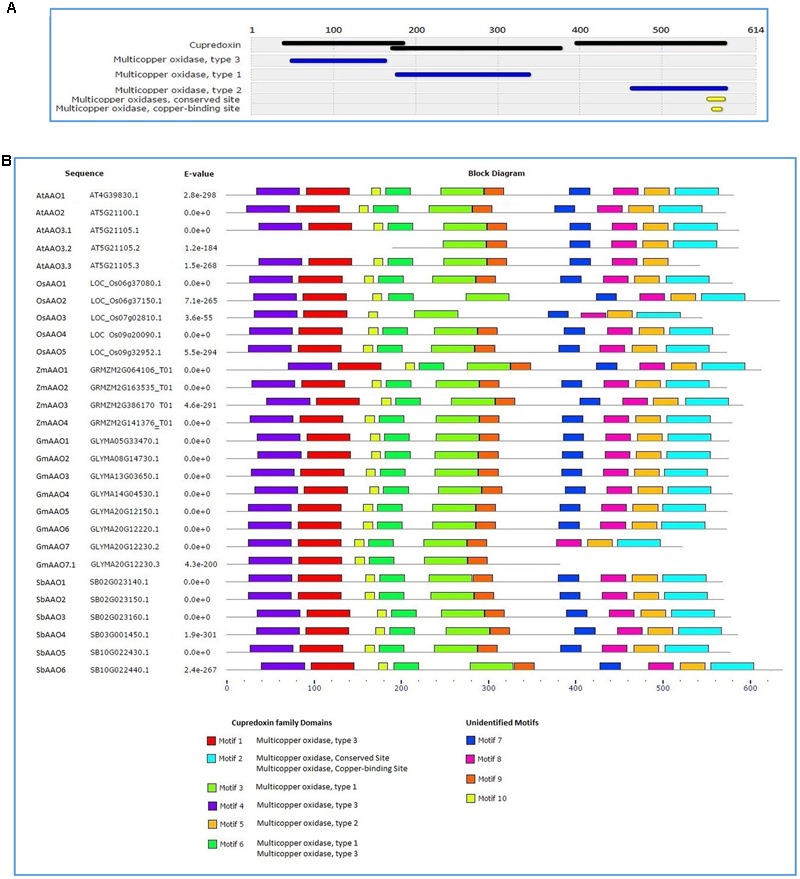
Conserved motif identification in AAO proteins from diverse plant genera (A) Interpro analysis revealed three types of domains as a part of cupredoxin family in previously defined AAO proteins. (B) De novo motif identification of AAO proteins; motifs 1, 2, 3, 4, 5, and 6 show resemblance to cupredoxin family and motifs 7, 8, 9, and 10 are unidentified motifs.
When MEME motif search tool was employed to identify the conserved motifs in AAOs identified in the present study, 10 discrete motifs were found in all putative AAOs except a spliced form of AtAAO3 and GmAAO7 (Figure 3B). Further, individual search of the motifs obtained from MEME in the Interpro database revealed that six motifs correspond to known domains present in copper binding AAOs while remaining four motifs did not resemble with any known domain in Interpro database. Motifs 1 and 4 were identified as the part of multicopper oxidase type 3 domain while motif 5 and motif 3 were the part of multicopper oxidase type 2 and type 1, respectively. Motif 2 displayed similarity to the multicopper oxidase conserved site and copper binding site while motif 6 resembled the sequence shared by multicopper oxidase type 1 and type 3 domains present in known AAOs (Figures 3A,B).
Ascorbate Oxidase Genes Are Differentially Regulated under Stress Conditions
Analysis of microarray data in response to stress revealed AAO genes to be differentially regulated by various abiotic stresses. In rice seedlings, expression of OsAAO1, OsAAO2, and OsAAO3 was downregulated in cold stress. In salt stress, expression of OsAAO2 was downregulated while in drought stress, expression of OsAAO2, OsAAO3, and OsAAO5 was downregulated (Figure 4A).
FIGURE 4.
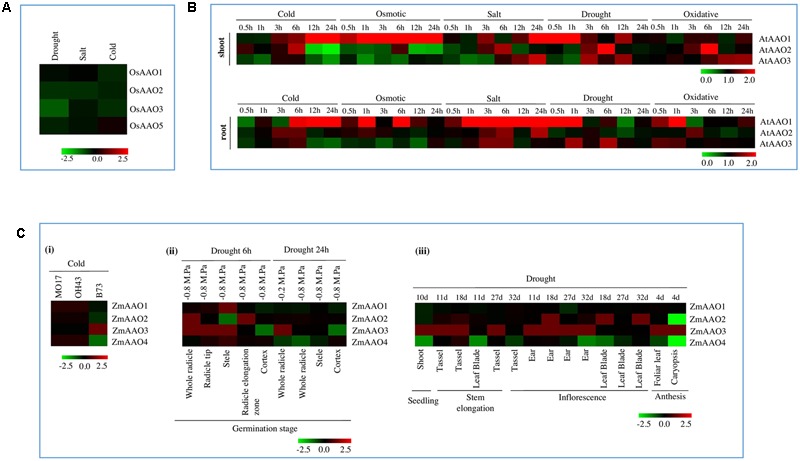
Stress regulated expression of AAO genes. (A) Heatmap analysis of AAO genes OsAAO1, OsAAO2, OsAAO3, and OsAAO5 from rice in 7 days old seedlings. Heatmap is based on the microarray data depicting the expression profile of AAO genes under abiotic stress conditions such as, drought, salt, and cold (B) Heatmap analysis of AAO genes AtAAO1, AtAAO2, and AtAAO3 from 18 days old Arabidopsis seedlings. Microarray data depicting expression of AAO gene under various abiotic stress such as osmotic, cold, salt, drought, and oxidative stress. The dataset obtained correspond to shoot and root tissue at different time point of stress such as 0.5, 1, 3, 6, 12, and 24 h with respect to control. (C) Heatmap analysis of AAO genes ZmAAO1, ZmAAO2, ZmAAO3, and ZmAAO4 in response to abiotic stress. Heatmap is based on the microarray data depicting expression of AAO genes (i) under cold stress on different varieties of Zea mays (MO17, B73, and OH43), (ii) under drought stress of 6 and 24 h on different anatomical parts of germination stage with water potential ranging from -0.2 M. Pa to -0.8 M. Pa, (iii) in response to drought stress on different anatomical parts harvested from various developmental stages such as seedling stage, stem elongation, inflorescence and anthesis stage. The color bar in all figures represents the expression values, green color representing down regulation, black no change in expression and red signifies highest level of expression.
Microarray data analyzed for Arabidopsis also revealed AAO members to be differentially regulated at different time points in response to various abiotic stresses (Figure 4B). In cold stress, expression of AtAAO1 gradually increased from early to late duration of stress in both shoot and root tissue. AtAAO2 was briefly induced at 3 and 6 h of cold stress in shoot as well as root. Whereas, its expression was downregulated during late duration of stress in shoot and 0.5 and 12 h stress in root. AtAAO3 expression was slightly upregulated in 3 and 6 h of stress in root and downregulated at early and late duration of cold stress in both shoot and root.
Osmotic stress lead to major upheavals in transcript abundance of AtAAO1 gene in both root and shoot tissue. Expression of AtAAO2 was mostly downregulated except at 6 h time point in shoot and at 1 h time point in root, where it was slightly induced. On the other hand, AtAAO3 expression was downregulated at 3 and 12 h of stress in root and also at 1 h till 6 h of stress in root.
AtAAO genes were highly upregulated under salt stress in both shoot and root tissue. In shoot, expression of AtAAO1 was high except at 0.5, 1, and 6 h of stress. Whereas, in root expression of AtAAO1 was always high under all time-points of stress barring 0.5 h. In shoot, expression of AtAAO2 was induced at early durations of stress and expression of AtAAO3 was induced during late duration of stress. Whereas, in root, expression of AtAAO2 was upregulated at both early and late duration of stress and expression of AtAAO3 was upregulated only in early durations of salt stress.
In drought stress, expression of all AtAAO genes were mostly up regulated in shoot tissue, except AtAAO2 and AtAAO3, which were downregulated at early duration of stress. In root tissue, expression of AtAAO1 was mostly upregulated except at 12 h of drought stress. Expression of AtAAO2 was mostly downregulated in root tissue, whereas, expression of AtAAO3 was briefly induced 1 and 6 h of stress.
AtAAO genes mostly displayed high abundance under oxidative stress barring AtAAO1 and AtAAO2. AtAAO1 was downregulated at 1 and 12 h of stress in shoot and 3 h of stress in root. Whereas, AtAAO2 was slightly downregulated during late duration of stress in both shoot and root.
All four AAO genes in different varieties of Zea mays MO17 and OH43 showed no change in expression in response to cold stress, except B73 variety, where expression of ZmAAO4, ZmAAO2 was downregulated and ZmAAO3 was upregulated (Figure 4Ci). This clearly reflects upon a genotype specific regulation of AAO genes in Zea mays. In drought stress, ZmAAO1 and ZmAAO4 showed slight down regulation or no change in expression in different anatomical parts of various developmental stage such as germination stage, seedling stage, stem elongation, inflorescence stage and anthesis except of 6 h time point in stele where ZmAAO1 and ZmAAO4 showed slight up regulation (Figures 4Cii,iii). Expression of ZmAAO2 was upregulated in whole radicle and radicle elongation zone stage and downregulated in stele at 6 h time point and caryopsis stage of anthesis, under drought stress. ZmAAO3 was most stress responsive among all the four AAOs of Zea mays and its expression was mostly seen to be upregulated in cold and drought stress. Abiotic stress microarray data for Sorghum bicolor and Glycine max was not available on genevestigator, therefore it could not be included in the study.
Ascorbate Oxidase Gene Are Differentially Regulated under Various Developmental Stages
Since AAO genes are directly involved in managing ROS levels in plants, they might play a role in plant growth and development. Therefore, microarray data was also analyzed with respect to broad developmental stages of Oryza sativa, Arabidopsis thaliana, Glycine max, Zea mays, and Sorghum bicolor (Figure 5), to see the effect of development on transcript accumulation of AAO genes in plants. In Arabidopsis, AtAAO1 expression was moderate in all developmental stages whereas expression of AtAAO2 and AtAAO3 was mostly moderate to high in all stages of development (such as germination, seedling, young rosette, developed rosette, bolting, young flower, developed flower and flower and silique) except for the last two stages of mature silique and senescence (Figure 5A).
FIGURE 5.
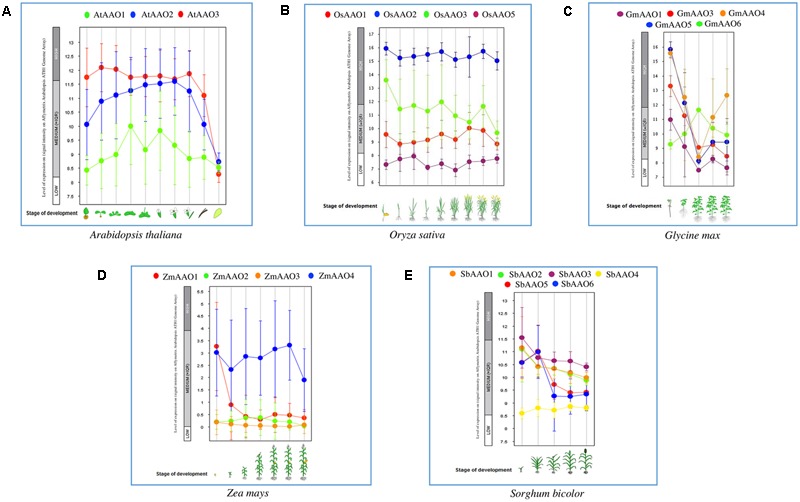
Developmental stage specific expression of AAO genes. Scatter plot displays the expression of AAO genes across various stages of development in (A) Oryza sativa, (B) Arabidopsis thaliana, (C) Glycine max, (D) Zea mays, and (E) Sorghum bicolor. Expression of different genes are represented by different color dots.
All the rice genes showed higher relative expression at the seedling and flowering stage which are also considered as most stress sensitive stages (Figure 5B). OsAAO2 and OsAAO3 maintained a high expression in all the developmental stages while OsAAO5 and OsAAO1 were low expressers. Data was not available for OsAAO4. Vegetative and tillering stages showed relatively low abundance for all the transcripts (Figure 5B).
In Glycine max (Figure 5C), expression of GmAAO3, GmAAO4, and GmAAO5 was high and that of GmAAO1 and GmAAO6 was moderate in germination stage. During shoot growth expression of GmAAO4 and GmAAO5 was maintained high whereas expression of GmAAO3, GmAAO6, and GmAAO1 was moderate. In the flowering stage, expression of all AAOs dropped except for GmAAO6 whose expression slightly increased. For fruit formation and bean development expression of all AAOs was low except for GmAAO4 whose expression slightly increased.
In Zea mays (Figure 5D) expression of all AAO genes in germination, seedling, stem elongation, inflorescence, anthesis, fruit formation and dough stage was slightly less except for ZmAAO1 and ZmAAO4. Expression of ZmAAO1 was moderate only in germination stage, whereas expression of ZmAAO4 was moderate in all stages of plant development. Expression of ZmAAO4 was highest among all AAOs.
In germination stage of Sorghum bicolor (Figure 5E) SbAAO3 expression was slightly high while expression of SbAAO1, SbAAO2, SbAAO5, and SbAAO6 was moderate. In all other developmental stages such as stem elongation, booting, flowering and dough stage, expression of all SbAAO genes was moderate except for SbAAO4, which was a low expresser among other SbAAOs.
RT-PCR and qRT-PCR Analysis Showed Shoot/Root Specific Expression of Ascorbate Oxidase Genes
PCR amplification using cDNA as template was carried out to determine tissue specific (root and shoot) expression of AAO genes. Separate PCR reaction was set for all five AAO genes where pooled first strand cDNA of control and stress sample from root and shoot tissue was used as a template. It was observed that OsAAO2 expressed specifically in shoot tissue while OsAAO1, OsAAO3, and OsAAO4 genes expressed in the shoot as well as the root tissue. Unlike the other AAO genes, OsAAO5 neither expressed in the shoot nor the root tissue (Figure 6A). However, there is a possibility that OsAAO5 expresses in other developmental stage or tissue sample of a rice plant or the expression of OsAAO5 might be significantly low to be detected as observed in the microarray based expression analysis wherein OsAAO5 displayed very low expression across various developmental stages in this study.
FIGURE 6.
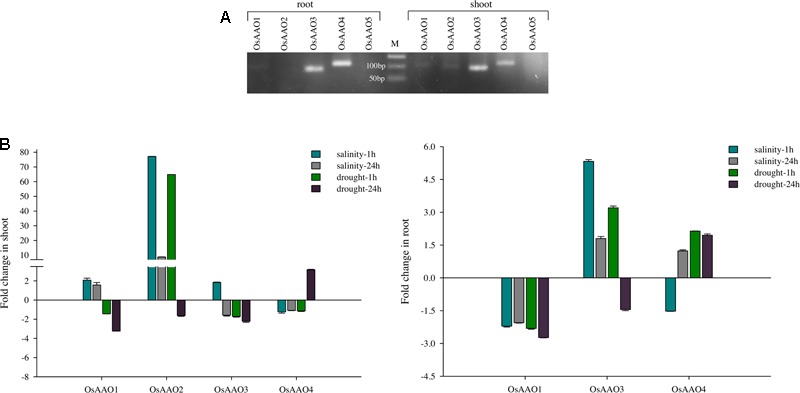
Expression pattern of AAO genes in rice based on RT-PCR and qRT-PCR analysis. (A) EtBr stained agarose gel depicting the expression of AAO genes in root and shoot tissue of rice seedling. Individual lanes show amplicons corresponding to OsAAO1–OsAAO5 amplified from rice root and shoot tissue using gene specific real-time PCR primers. M corresponds to 50 bp ladder. (B) Histogram representing fold change of OsAAO1, OsAAO2, OsAAO3, and OsAAO4 in 1 and 24 h stress treated shoot and root tissue of rice seedling based on qRT-PCR analysis. OsAAO5 could not be amplified hence it was not included in real-time analysis. Real-time PCR was done with cDNA template synthesized from shoot and root tissue of 10 days old control or stressed (salinity 200 mM NaCl and drought) rice seedlings.
The relative transcript level of OsAAO1, OsAAO2, OsAAO3, and OsAAO4 was determined in response to salinity and drought stress in shoot and root tissue of rice seedlings. OsAAO5 gene could not be amplified from both shoot and root cDNA used in this study; therefore, it is not included in the expression analysis. The real-time data reveals differential tissue specific stress inducibility of the members of AAO gene family in Oryza sativa IR64. OsAAO2 was found to be specifically expressed in shoot and it was upregulated under both drought and salinity stress treatment except in tissue subjected to late durations of drought stress (24 h) where it was downregulated. Expression of OsAAO1 was upregulated in shoot tissue in response to both early and late durations of salinity stress while it was downregulated at both time points of drought stress. OsAAO3 and OsAAO4 genes were upregulated only in response to early duration of salinity stress and late duration of drought stress, respectively. Both the genes were downregulated in shoots in response to all the other stress conditions analyzed in this study (Figure 6B). OsAAO1 was downregulated in root tissue under both early and late duration of salinity and drought stress. OsAAO3 gene showed enhanced expression in roots under both salinity (early and late duration) and drought (early duration) while it was downregulated during late durations of drought stress. OsAAO4 showed enhanced expression under drought stress conditions as well as in late duration (24 h) of salinity (Figure 6B).
Discussion
Availability of whole genome sequences of model and important food crops has served as a useful platform to carry out comprehensive analysis of gene families, e.g., study of glyoxalase gene family in rice, Arabidopsis (Mustafiz et al., 2011), soybean (Ghosh and Islam, 2016), analysis of CBS domains containing proteins (CDCPs) in Arabidopsis and rice, (Kushwaha et al., 2009), analysis of F-box protein, auxin-responsive SAUR gene family and homeobox genes in rice (Jain et al., 2006, 2007, 2008), analysis of CaHsp20 gene family in pepper (Guo et al., 2015), study of AAAP (amino acid/auxin permease) gene family in maize (Sheng et al., 2014) and analysis of PIN auxin transporter gene family in soybean (Wang et al., 2015) etc. In the present study, we attempted a genome-wide analysis of AAO gene family in Oryza sativa, Arabidopsis thaliana, Glycine max, Zea mays, and Sorghum bicolor and as well as analysis of their transcript abundance specific to abiotic stress and developmental stages. AAO genes form a small gene family in plants. AAO multigene families identified in the five-plant species under study were found to have variable number of members, viz., five in Oryza sativa, three in Arabidopsis thaliana, seven in Glycine max, four in Zea mays, and six in Sorghum bicolor. Variation in gene number of AAO among the five-plant species could be due to variation in number of duplication events, for example, AAO genes in soybean show more duplication events and constitutes a larger AAO gene family as compared to Arabidopsis, Zea mays, and Oryza sativa. Gene family size variation is a common phenomenon in plant genera which could be attributed to gene duplication, deletion, pseudogenization, and/or functional diversification (Zhang et al., 2010). AAOs are best described from plants (Kues and Ruhl, 2011), however, much is not known about their precise biological function. Genes for putative AAOs have also been reported from some fungi (Hoegger et al., 2006). Plant AAOs have been implicated in oxygen homeostasis and ROS balancing (De Tullio et al., 2004; Semchuk et al., 2009), various stress reactions (Sanmartin et al., 2003; Caputo et al., 2010), defense (Barbehenn et al., 2008), growth and cell wall formation (Ros-Barceló et al., 2006; Díaz-Vivancos et al., 2010), and signaling (Pignocchi et al., 2003; Fotopoulos et al., 2008). Based on the previous studies, it has been established that they belong to multiple copper oxidase family proteins. To further comment upon the type of copper oxidase domain, AAO proteins were investigated for the type of copper binding domains.
Similar de novo identified motifs are present and conserved across all of the putative AAOs considered in this study. Some of these motifs correspond to one of the three conserved domains (multicopper oxidase types 1, 2, and 3) identified in pre-defined AAO proteins. The presence and similar pattern of the conserved domains/motifs in all putative AAOs suggests the high structural similarity among the AAO gene family members found in Oryza sativa, Arabidopsis thaliana, Glycine max, Zea mays, and Sorghum bicolor. Furthermore, the resemblance of de novo identified motifs in putative AAOs with that of known domains in pre-identified AAOs suggests that all AAO genes taken in this study are functionally similar and can be active AAOs. Although these gene members showed known copper binding motifs, some unreported motifs were also found in these proteins.
It has been postulated that the three-domain multi-copper blue proteins, such as AAO, have evolved by a single domain addition to the two-domain protein. Alternatively, it has been suggested that AAO could have evolved from a six-domain protein by replacing domains 3 through 5 with a short linker (Nakamura et al., 2003). These postulations are further strengthened by the presence of a small stretch of non-conserved linker like sequence between multicopper oxidase domain types 1 and 2 in pre-defined AAOs as well as between motifs 7 and 9 in all putative AAOs depicted in Figures 3A,B.
An important aspect of response to stress occurs at transcriptional level, which alters gene expression (Tester and Davenport, 2003). In several previous studies, AAO activity and expression are closely co related with light, salicylic acid, auxin, and jasmonates (Sanmartin, 2002). Light driven ROS production detrimentally affects the redox balance of photosynthetic tissues and also the overall plant growth and development (Foyer and Shigeoka, 2011). AA acts as one of the most important non-enzymatic, water soluble antioxidant in plants to metabolize ROS (Foyer et al., 1983; Smirnoff, 2000). However, AAO leads to oxidation of AA and therefore preventing the detoxification of ROS (Smirnoff, 2000). These findings suggest AAO to be involved in negative regulation of stress response. However, no direct correlation has been established between environmental perturbations and AAO activity or transcriptional activity. For this reason, we wanted to study if AAO genes are differentially regulated in response to abiotic stress, viz., salinity and drought conditions. Expression pattern of AAO genes retrieved from publically available microarray data and revalidated through RT-PCR as well as qRT-PCR suggested these genes to be strongly stress responsive showing a genotype/genus and tissue specific temporally regulated expression. OsAAO2 is one of the most stress responsive gene in shoot tissue of rice while OsAAO3 and OsAAO4 show high expression in root tissue in response to salinity and drought stress, as analyzed by qRT-PCR. AtAAO1 was highly upregulated in shoot and root in response to abiotic stress and AtAAO3 displayed high expression in various developmental stages. In Zea mays, ZmAAO3 is the most stress responsive gene as seen in both cold and drought stress. OsAAO2, OsAAO3 and OsAAO4 from Oryza sativa, AtAAO1 and AtAAO3 from Arabidopsis and ZmAAO3 from Zea mays can serve as a good candidate for raising stress tolerant transgenic crops.
Based on the previous reports as depicted in Figure 7A, AA works in close coordination with GSH in ASA-GSH cycle (Foyer and Halliwell, 1976) and water–water cycle (Asada, 1999) to metabolize ROS. Therefore, the redox state of AA is of utmost importance to prevent ROS induced cell injury. However, the cell wall localized enzyme AAO leads to oxidation of AA, to form unstable MDHA in turn reducing the AA/DHA ratio (Smirnoff, 2000) hence affecting the detoxification of ROS leading to more damage to plants (Figure 7A). Additionally, overexpression of AAO in tobacco reduces capacity of plant to scavenge ROS in leaf apoplast due to oxidation of AA in this compartment consequently leading to enhanced sensitivity to ozone (Sanmartin et al., 2003). On the other hand, if the activity of AAO is suppressed, the availability of AA in its reduced form is increased. This increased level of AA in its reduced form helps scavenge more ROS as compared to wild type plants and plants show stress tolerant phenotype (Figure 7B). It has also been reported in tobacco and Arabidopsis that if the AAO gene expression is suppressed, it grants resistance to oxidative damage brought about by methyl viologen or H2O2 (Yamamoto et al., 2005). Furthermore, in the same study it was also shown that AA/DHA ratio was higher in case of antisense-AAO tobacco plants and Arabidopsis T-DNA AAO mutant plant than those of wild type plants during growth under salt stress conditions, whereas overexpressing of AAO in tobacco leads to greater damage of transgenic plants as compared to wild type plants (Yamamoto et al., 2005). In plants, such as tobacco and Arabidopsis it has been seen, that the level of H2O2 decreased on suppressing the activity of AAO gene. It was also seen in previous studies that antisense suppression of the AAO gene in tobacco enhanced percentage germination and increased seed yield at high salinity conditions (Yamamoto et al., 2005). These findings suggest that the targeted knock down of OsAAO2, OsAAO3, and OsAAO4 in rice which have been found to show steep changes in stress responsive transcript accumulation can be a possible strategy to engineer stress tolerance in rice. It would be interesting to study the changes in redox state of AA in engineered plants and its effect on the level of ROS and damage caused by them.
FIGURE 7.
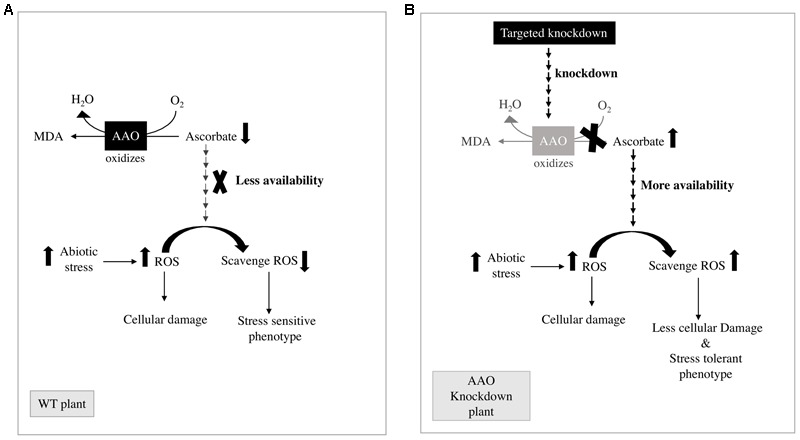
Mechanistic details of possible pathway of AAO action in regulating stress phenotype of plants. (A) Reduced AA availability due to the oxidizing activity of AAO leads to stress sensitive phenotype in WT plants. (B) Enhanced AA availability due to the knock down of AAO gene leads to stress tolerant phenotype in AAO knock down plants.
Author Contributions
The idea, concept, design of experiments and manuscript preparation are done by AM. SK has done the bioinformatics work and contributed in preparing the manuscript. RB has done the wet lab experiments and also contributed in manuscript preparation and KS has contributed in bioinformatics work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding from Science and Engineering Research Board (SERB) and Department of Science and Technology (DST), India and South Asian University (SAU) are duly acknowledged.
Abbreviations
- AA
ascorbate
- AAO
ascorbate oxidase
- MDA
monodehydroascorbate
- MDAR
monodehydroascorbate reductase
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00198/full#supplementary-material
References
- Asada K. (1999). The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 601–639. 10.1146/annurev.arplant.50.1.601 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Williams N., Misleh C., Li W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34 W369–W373. 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini R., Ott T., Güther M., Bonfante P., Udvardi M. K., De Tullio M. C. (2012). Ascorbate oxidase: the unexpected involvement of a “wasteful enzyme” in the symbioses with nitrogen-fixing bacteria and arbuscular mycorrhizal fungi. Plant Physiol. Biochem. 59 71–79. 10.1016/j.plaphy.2012.07.006 [DOI] [PubMed] [Google Scholar]
- Barbehenn R. V., Jaros A., Yip L., Tran L., Kanellis A. K., Constabel C. P. (2008). Evaluating ascorbate oxidase as a plant defense against leaf-chewing insects using transgenic poplar. J. Chem. Ecol. 34 1331–1340. 10.1007/s10886-008-9539-7 [DOI] [PubMed] [Google Scholar]
- Barnes J., Zheng Y., Lyons T. (2002). “Plant resistance to ozone: the role of ascorbate,” in Air pollution and Plant Biotechnology eds Omasa K., Saji H., Youssefian S., Kondo N. (Tokyo: Springer; ) 235–252. [Google Scholar]
- Blokhina O., Fagerstedt K. V. (2010). Reactive oxygen species and nitric oxide in plant mitochondria: origin and redundant regulatory systems. Physiol. Plant. 138 447–462. 10.1111/j.1399-3054.2009.01340.x [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo E., Ceglie V., Lippolis M., La Rocca N., De Tullio M. C. (2010). Identification of a NaCl-induced ascorbate oxidase activity in Chaetomorpha linum suggests a novel mechanism of adaptation to increased salinity. Environ. Exp. Bot. 69 63–67. 10.1016/j.envexpbot.2010.02.008 [DOI] [Google Scholar]
- Citterio S., Sgorbati S., Scippa S., Sparvoli E. (1994). Ascorbic acid effect on the onset of cell proliferation in pea root. Physiol. Plant. 92 601–607. 10.1034/j.1399-3054.1994.920409.x [DOI] [Google Scholar]
- Córdoba F., González-Reyes J. A. (1994). Ascorbate and plant cell growth. J. Bioenerg. Biomembr. 26 399–405. [DOI] [PubMed] [Google Scholar]
- De Tullio M. C., Liso R., Arrigoni O. (2004). Ascorbic acid oxidase: an enzyme in search of a role. Biol. Plant. 48 161–166. 10.1023/B:BIOP.0000033439.34635.a6 [DOI] [Google Scholar]
- del Río L. A., Sandalio L. M., Corpas F. J., Palma J. M., Barroso J. B. (2006). Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiol. 141 330–335. 10.1104/pp.106.078204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R., A-H-Mackerness S., Hancock J. T., Neill S. J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127 159–172. 10.1104/pp.127.1.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Vivancos P., Barba-Espín G., Clemente-Moreno M. J., Hernández J. A. (2010). Characterization of the antioxidant system during the vegetative development of pea plants. Biol. Plant. 54 76–82. 10.1007/s10535-010-0011-5 [DOI] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95 14863–14868. 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaka M., Fujisawa K., Goto M., Kisu Y. (1992). Regulation of ascorbate oxidase expression in pumpkin by auxin and copper. Plant Physiol. 100 231–237. 10.1104/pp.100.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotopoulos V., De Tullio M. C., Barnes J., Kanellis A. K. (2008). Altered stomatal dynamics in ascorbate oxidase over-expressing tobacco plants suggest a role for dehydroascorbate signalling. J. Exp. Bot. 59 729–737. 10.1093/jxb/erm359 [DOI] [PubMed] [Google Scholar]
- Fotopoulos V., Kanellis A. K. (2013). Altered apoplastic ascorbate redox state in tobacco plants via ascorbate oxidase overexpression results in delayed dark-induced senescence in detached leaves. Plant Physiol. Biochem. 73 154–160. 10.1016/j.plaphy.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Foyer C., Rowell J., Walker D. (1983). Measurement of the ascorbate content of spinach leaf protoplasts and chloroplasts during illumination. Planta 157 239–244. 10.1007/BF00405188 [DOI] [PubMed] [Google Scholar]
- Foyer C. H. (1997). Oxygen Metabolism and Electron Transport in Photosynthesis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 587–621. [Google Scholar]
- Foyer C. H., Halliwell B. (1976). The presence of glutathione and glutathione reductase in chloroplasts: a proposed role in ascorbic acid metabolism. Planta 133 21–25. 10.1007/BF00386001 [DOI] [PubMed] [Google Scholar]
- Foyer C. H., Harbinson J. (1994). “Oxygen metabolism and the regulation of photosynthetic electron transport,” in Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants eds Foyer C. H., Mullineaux P. M. (Londres: CRC press; ) 42. [Google Scholar]
- Foyer C. H., Shigeoka S. (2011). Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 155 93–100. 10.1104/pp.110.166181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich I. (1998). Oxygen toxicity: a radical explanation. J. Exp. Biol. 201(Pt 8) 1203–1209. 10.1242/jeb.037663 [DOI] [PubMed] [Google Scholar]
- Garchery C., Gest N., Do P. T., Alhagdow M., Baldet P., Menard G., et al. (2013). A diminution in ascorbate oxidase activity affects carbon allocation and improves yield in tomato under water deficit. Plant Cell Environ. 36 159–175. 10.1111/j.1365-3040.2012.02564.x [DOI] [PubMed] [Google Scholar]
- Ghosh A., Islam T. (2016). Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 16:87 10.1186/s12870-016-0773-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Liu J. H., Lu J. P., Zhai Y. F., Wang H., Gong Z. H., et al. (2015). Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 6:806 10.3389/fpls.2015.00806.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegger P. J., Kilaru S., James T. Y., Thacker J. R., Kües U. (2006). Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 273 2308–2326. 10.1111/j.1742-4658.2006.05247.x [DOI] [PubMed] [Google Scholar]
- Horemans N., Foyer C. H., Asard H. (2000). Transport and action of ascorbate at the plant plasma membrane. Trends Plant Sci. 5 263–267. 10.1016/S1360-1385(00)01649-6 [DOI] [PubMed] [Google Scholar]
- Hu W. H., Song X. S., Shi K., Xia X. J., Zhou Y. H., Yu J. Q. (2008). Changes in electron transport, superoxide dismutase and ascorbate peroxidase isoenzymes in chloroplasts and mitochondria of cucumber leaves as influenced by chilling. Photosynthetica 46 581–588. 10.1007/s11099-008-0098-5 [DOI] [Google Scholar]
- Jain M., Nijhawan A., Arora R., Agarwal P., Ray S., Sharma P., et al. (2007). F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Tyagi A. K., Khurana J. P. (2006). Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88 360–371. [DOI] [PubMed] [Google Scholar]
- Jain M., Tyagi A. K., Khurana J. P. (2008). Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J. 275 2845–2861. 10.1111/j.1742-4658.2008.06424.x [DOI] [PubMed] [Google Scholar]
- Kato N., Esaka M. (1999). Changes in ascorbate oxidase gene expression and ascorbate levels in cell division and cell elongation in tobacco cells. Physiol. Plant. 105 321–329. 10.1034/j.1399-3054.1999.105218.x [DOI] [Google Scholar]
- Kato N., Esaka M. (2000). Expansion of transgenic tobacco protoplasts expressing pumpkin ascorbate oxidase is more rapid than that of wild-type protoplasts. Planta 210 1018–1022. 10.1007/s004250050712 [DOI] [PubMed] [Google Scholar]
- Kerk N. M., Feldman L. J. (1995). A biochemical model for the initiation and maintenance of the quiescent center: implications for organization of root meristems. Development 2833 2825–2833. [Google Scholar]
- Kues U., Ruhl M. (2011). Multiple multi-copper oxidase gene families in basidiomycetes - what for? Curr. Genomics 12 72–94. 10.2174/138920211795564377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2015). MEGA7: molecular evolutionary genetics analysis version 7.0. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha H. R., Singh A. K., Sopory S. K., Singla-Pareek S. L., Pareek A. (2009). Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics 10:200 10.1186/1471-2164-10-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Maheshwari R., Dubey R. S. (2009). Nickel-induced oxidative stress and the role of antioxidant defence in rice seedlings. Plant Growth Regul. 59 37–49. 10.1007/s10725-009-9386-8 [DOI] [Google Scholar]
- Meriga B., Reddy B. K., Rao K. R., Reddy L. A., Kavi Kishor P. B. (2004). Aluminium-induced production of oxygen radicals, lipid peroxidation and DNA damage in seedlings of rice (Oryza sativa). J. Plant Physiol. 161 63–68. 10.1078/0176-1617-01156 [DOI] [PubMed] [Google Scholar]
- Mishra S., Jha A. B., Dubey R. S. (2011). Arsenite treatment induces oxidative stress, upregulates antioxidant system, and causes phytochelatin synthesis in rice seedlings. Protoplasma 248 565–577. 10.1007/s00709-010-0210-0 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7 405–410. 10.1016/S1360-1385(02)02312-9 [DOI] [PubMed] [Google Scholar]
- Mosery O., Kanellis A. K. (1994). Ascorbate oxidase of Cucumis melo L. var. reticulatus: purification, characterization and antibody production. J. Exp. Bot. 45 717–724. 10.1093/jxb/45.6.717 [DOI] [Google Scholar]
- Mustafiz A., Singh A. K., Pareek A., Sopory S. K., Singla-Pareek S. L. (2011). Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics 11 293–305. 10.1007/s10142-010-0203-2 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Kawabata T., Yura K., Go N. (2003). Novel types of two-domain multi-copper oxidases: Possible missing links in the evolution. FEBS Lett. 553 239–244. 10.1016/S0014-5793(03)01000-7 [DOI] [PubMed] [Google Scholar]
- Neill S., Desikan R., Hancock J. (2002). Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 5 388–395. 10.1016/S1369-5266(02)00282-0 [DOI] [PubMed] [Google Scholar]
- Noctor G., Foyer C. H. (1998). Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 249–279. 10.1146/annurev.arplant.49.1.249 [DOI] [PubMed] [Google Scholar]
- Ohkawa J., Ohya T., Ito T., Nozawa H., Nishi Y., Okada N., et al. (1994). Structure of the genomic DNA encoding cucumber ascorbate oxidase and its expression in transgenic plants. Plant Cell Rep. 13 481–488. 10.1007/BF00232941 [DOI] [PubMed] [Google Scholar]
- Pignocchi C., Fletcher J. M., Wilkinson J. E., Barnes J. D., Foyer C. H., Yamamoto A., et al. (2003). The function of ascorbate oxidase in tobacco. Plant Physiol. 132 1631–1641. 10.1104/pp.103.022798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plöchl M., Lyons T., Ollerenshaw J., Barnes J. (2000). Simulating ozone detoxification in the leaf apoplast through the direct reaction with ascorbate. Planta 210 454–467. 10.1007/PL00008153 [DOI] [PubMed] [Google Scholar]
- Raven E. L. (2000). Peroxidase-catalyzed oxidation of ascorbate. Structural, spectroscopic and mechanistic correlations in ascorbate peroxidase. Subcell. Biochem. 35 317–349. 10.1007/0-306-46828-X_10 [DOI] [PubMed] [Google Scholar]
- Ros-Barceló A., Gómez-Ros L. V., Ferrer M. A., Hernández J. A. (2006). The apoplastic antioxidant enzymatic system in the wood-forming tissues of trees. Trees 20 145–156. 10.1007/s00468-005-0020-8 [DOI] [Google Scholar]
- Sanmartin M. (2002). Regulation of Melon Ascorbate Oxidase Gene Expression and Effect of its Modification in Transgenic Tobacco and Melon Plants. PhD thesis, University of Valencia; Spain. [Google Scholar]
- Sanmartin M., Drogoudi P. A. M. D., Lyons T., Pateraki I., Barnes J., Kanellis A. K. (2003). Over-expression of ascorbate oxidase in the apoplast of transgenic tobacco results in altered ascorbate and glutathione redox states and increased sensitivity to ozone. Planta 216 918–928. 10.1007/s00425-002-0944-9 [DOI] [PubMed] [Google Scholar]
- Sanmartin M., Pateraki I., Chatzopoulou F., Kanellis A. K. (2007). Differential expression of the ascorbate oxidase multigene family during fruit development and in response to stress. Planta 225 873–885. 10.1007/s00425-006-0399-5 [DOI] [PubMed] [Google Scholar]
- Semchuk N. M., Lushchak O. V., Falk J., Krupinska K., Lushchak V. I. (2009). Inactivation of genes, encoding tocopherol biosynthetic pathway enzymes, results in oxidative stress in outdoor grown Arabidopsis thaliana. Plant Physiol. Biochem. 47 384–390. 10.1016/j.plaphy.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Shah K., Kumar R. G., Verma S., Dubey R. S. (2001). Effect of cadmium on lipid peroxidation, superoxide anion generation and activities of antioxidant enzymes in growing rice seedlings. Plant Sci. 161 1135–1144. 10.1016/S0168-9452(01)00517-9 [DOI] [Google Scholar]
- Sharma P., Dubey R. S. (2005). Drought induces oxidative stress and enhances the activities of antioxidant enzymes in growing rice seedlings. Plant Growth Regul. 46 209–221. 10.1007/s10725-005-0002-2 [DOI] [Google Scholar]
- Sharma P., Dubey R. S. (2007). Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminum. Plant Cell Rep. 26 2027–2038. 10.1007/s00299-007-0416-6 [DOI] [PubMed] [Google Scholar]
- Sheng L., Deng L., Yan H., Zhao Y., Dong Q., Li O., et al. (2014). A genome-wide analysis of the AAAP gene family in maize. J. Proteomics Bioinform. 7 023–033. 10.4172/jpb.1000299 [DOI] [Google Scholar]
- Smirnoff N. (2000). Ascorbic acid: Metabolism and functions of a multi-facetted molecule. Curr. Opin. Plant Biol. 3 229–235. 10.1016/S1369-5266(00)00069-8 [DOI] [PubMed] [Google Scholar]
- Srivastava S., Dubey R. S. (2011). Manganese-excess induces oxidative stress, lowers the pool of antioxidants and elevates activities of key antioxidative enzymes in rice seedlings. Plant Growth Regul. 64 1–16. 10.1007/s10725-010-9526-1 [DOI] [Google Scholar]
- Tanou G., Molassiotis A., Diamantidis G. (2009). Induction of reactive oxygen species and necrotic death-like destruction in strawberry leaves by salinity. Environ. Exp. Bot. 65 270–281. 10.1016/j.envexpbot.2008.09.005 [DOI] [Google Scholar]
- Tester M., Davenport R. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 91 503–527. 10.1093/aob/mcg058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Dubey R. S. (2003). Lead toxicity induces lipid peroxidation and alters the activities of antioxidant enzymes in growing rice plants. Plant Sci. 164 645–655. 10.1016/S0168-9452(03)00022-0 [DOI] [Google Scholar]
- Wang Y., Chai C., Valliyodan B., Maupin C., Annen B., Nguyen H. T. (2015). Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max). BMC Genomics 16:951 10.1186/s12864-015-2149-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Bhuiyan M. N. H., Waditee R., Tanaka Y., Esaka M., Oba K., et al. (2005). Suppressed expression of the apoplastic ascorbate oxidase gene increases salt tolerance in tobacco and Arabidopsis plants. J. Exp. Bot. 56 1785–1796. 10.1093/jxb/eri167 [DOI] [PubMed] [Google Scholar]
- Yan J., Tsuichihara N., Etoh T., Iwai S. (2007). Reactive oxygen species and nitric oxide are involved in ABA inhibition of stomatal opening. Plant Cell Environ. 30 1320–1325. 10.1111/j.1365-3040.2007.01711.x [DOI] [PubMed] [Google Scholar]
- Yoshida S., Forno D. A., Cock J. H. (1971). Laboratory Manual for Physiological Studies of Rice. Los Baños, CA: Int. Rice Res. Inst. 10.1007/978-3-642-22067-8_9 [DOI] [Google Scholar]
- Zhang M., Wu Y. H., Lee M. K., Liu Y. H., Rong Y., Santos T. S., et al. (2010). Numbers of genes in the NBS and RLK families vary by more than four-fold within a plant species and are regulated by multiple factors. Nucleic Acids Res. 38 6513–6525. 10.1093/nar/gkq524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li H., Shu W., Zhang C., Zhang W., Ye Z. (2011). Suppressed expression of ascorbate oxidase gene promotes ascorbic acid accumulation in tomato fruit. Plant Mol. Biol. Rep. 29 638–645. 10.1007/s11105-010-0271-4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


