Abstract
The mitochondrial transcription initiation machinery in humans consists of three proteins: the RNA polymerase (POLRMT) and two accessory factors, transcription factors A and B2 (TFAM and TFB2M, respectively). This machinery is required for the expression of mitochondrial DNA and the biogenesis of the oxidative phosphorylation system. Previous experiments suggested that TFB2M is required for promoter melting, but conclusive experimental proof for this effect has not been presented. Moreover, the role of TFB2M in promoter unwinding has not been discriminated from that of TFAM. Here we used potassium permanganate footprinting, DNase I footprinting, and in vitro transcription from the mitochondrial light-strand promoter to study the role of TFB2M in transcription initiation. We demonstrate that a complex composed of TFAM and POLRMT was readily formed at the promoter but alone was insufficient for promoter melting, which only occurred when TFB2M joined the complex. We also show that mismatch bubble templates could circumvent the requirement of TFB2M, but TFAM was still required for efficient initiation. Our findings support a model in which TFAM first recruits POLRMT to the promoter, followed by TFB2M binding and induction of promoter melting.
Keywords: mitochondrial DNA (mtDNA), RNA polymerase, transcription, transcription initiation factor, transcription promoter, mitochondria, transcription factor
Introduction
Human mitochondria contain a small circular genome that encodes 22 tRNA, 2 rRNA, and 13 proteins required for oxidative phosphorylation. Transcription of this genome is initiated from two promoters, the heavy- and light-strand promoters (HSP2 and LSP, respectively), and generates polycistronic transcripts that are processed to produce individual RNA molecules (1).
Mitochondrial transcription is carried out by the RNA polymerase POLRMT, which is structurally related to the single-subunit RNA polymerase of bacteriophage T7 (T7 RNAP) (2, 3). In contrast to its phage counterpart, POLRMT cannot initiate promoter-specific transcription on its own, but needs the assistance of accessory factors (4, 5). The human system requires two accessory proteins, transcription factors A (TFAM) and B2 (TFB2M) (1). POLRMT contains a large N-terminal extension (NTE) in addition to the T7 RNAP-like core of the enzyme. This NTE enhances promoter specificity by inhibiting transcription initiation from random DNA sequences (6).
TFAM is a high-mobility group box (HMG) protein that binds sequence-specifically to regions upstream of mitochondrial transcription start sites (7–10). TFAM bound at this position introduces a sharp 180° bend in promoter DNA, and this structural change is required for transcription initiation (11, 12). TFAM recruits POLRMT to mitochondrial promoters, and mutations that impair sequence-specific binding and the precise distance to the transcription start site also impair transcription initiation (6, 13, 14). In addition to the function as a transcription factor, TFAM also binds DNA in a nonspecific manner, and it has been shown to completely cover mtDNA. In this way, TFAM functions as an mtDNA packaging factor that wraps the genome into the compact nucleoid structures (8, 15, 16).
TFB2M enters the transcription initiation complex after POLRMT has been recruited to the promoter by TFAM, and triggers initiation of transcription (6, 14). TFB2M interacts with POLRMT and functions as a transient component of the catalytic site during transcription initiation, where it is placed in direct proximity of the priming substrate in the process of transcription initiation (17). After transcription has been initiated, TFB2M probably dissociates from POLRMT when the enzyme enters the elongation phase (18). TFB2M has been suggested to be required for promoter melting, but this has never been directly demonstrated (17).
POLRMT contributes actively to promoter recognition by sequence-specific interactions in the vicinity of the transcription start site, but possibly also by recognition of a region upstream of the TFAM binding site (6, 14, 19, 20). DNase I footprinting and crosslinking experiments have shown that TFAM is absolutely required to initiate the formation of a pre-initiation complex. In the absence of TFAM, neither POLRMT nor the combination of POLRMT and TFB2M binds to promoter DNA in a sequence-specific manner (6, 14, 19). In the presence of TFAM, POLRMT on its own is efficiently recruited to the promoter, whereas TFB2M is only recruited when both TFAM and POLRMT are present (6). These and other findings have led us to propose a model for transcription initiation where TFAM recruits POLRMT to the promoter, aided by POLRMT-DNA interactions. Formation of this pre-initiation complex releases the negative effect of the NTE of POLRMT and opens up the polymerase for TFB2M binding and transcription initiation (6).
In yeast, the mitochondrial RNA polymerase Rpo41 and the TFB2M homologue Mtf1 are the only factors required for transcription initiation (21). Interestingly, dependence on Mtf1 can be circumvented by the introduction of a mismatch region over the transcription start site, or by the use of negatively supercoiled templates for transcription (22). When a similar mismatch bubble template is used in the human transcription system, transcription is independent of both TFB2M and TFAM (17). This observation is difficult to reconcile with the sequential model for the assembly of the mitochondrial pre-initiation complex, which states that TFAM is required to recruit POLRMT to the promoter region.
In the current work, we have specifically analyzed the role of TFB2M in the process of mitochondrial transcription initiation. We have used DNase I and potassium permanganate footprinting to study promoter interactions and unwinding. We have also performed in vitro transcription with mismatch bubble oligonucleotide templates and supercoiled plasmid templates to elucidate the requirement of transcription factors for promoter melting. Our analysis helps to distinguish the roles of TFAM and TFB2M at the promoter, and provides further evidence for the sequential model of transcription initiation in human mitochondria.
Results
Transcription Start Site Melting Is Strictly Dependent on TFB2M
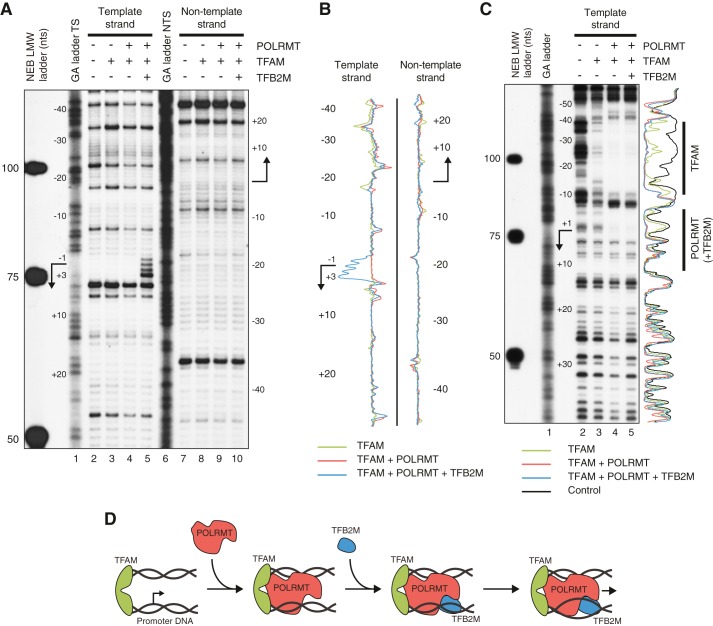
To monitor structural changes in the promoter region during transcription initiation, we used potassium permanganate (KMnO4) footprinting (Fig. 1, A and B). Permanganate primarily oxidizes thymidine residues, and the reaction has a strong preference for nucleotides within single-stranded regions. DNA is next cleaved with piperidine at the modified bases (23–25). In parallel to the permanganate footprinting, we performed DNase I footprinting to monitor binding of the mitochondrial transcription factors to the promoter (Fig. 1C). Both sets of reactions were performed using the same buffer and salt conditions, which allowed us to directly compare promoter binding events with effects on promoter structure. For the analysis, we used a P32-labeled PCR fragment covering the LSP promoter region. The permanganate footprinting templates showed some background cleavage in the absence of proteins (Fig. 1A, lanes 2 and 7). This effect could be due to intrinsic permanganate-sensitive sites induced by breathing in the DNA. The background signals do not however interfere with the interpretation of the data. The addition of TFAM created a DNase I footprint from positions −15 to −40 relative to the transcription start site, corresponding to the previously characterized TFAM binding site (Fig. 1C, lane 3) (9). On their own, neither TFB2M nor POLRMT binds to LSP (19). The addition of POLRMT together with TFAM caused an even stronger footprint over the −15 to −40 region. The presence of POLRMT also promoted changes in DNase I sensitivity around position −45 and strong protection over the transcription start site (−4 to +5), indicating correct positioning of the polymerase (Fig. 1C, lane 4). These changes were in agreement with previous studies, which had shown that TFAM recruits POLRMT to the transcription start site (6, 19). In parallel with the DNase I experiments, we monitored permanganate sensitivity. The addition of TFAM did not change permanganate sensitivity, demonstrating that TFAM on its own is unable to unwind promoter sequences (Fig. 1A, lanes 3 and 8, and Fig. 1B). Furthermore, no changes in the permanganate sensitivity were observed when POLRMT was added (Fig. 1A, lanes 4 and 9, and Fig. 1B).
FIGURE 1.
DNA footprinting reveals that TFB2M is the promoter melting-inducing factor in human mitochondria. A, potassium permanganate footprinting of the LSP region template strand (TS, lanes 2–5) and non-template strand (NTS, lanes 7–10), in the absence or presence of transcription proteins as indicated. Indicated markers are New England Biolabs Low Molecular Weight Marker (NEB LMW) and strand-specific GA ladders (lanes 1 and 6, Maxam-Gilbert G+A sequencing reaction, see “Experimental Procedures”). The positions on the DNA in relation to the LSP start site (arrow) are indicated. B, densitometric scanning profiles of the autoradiographs in A (where background signals, lane 2 for the template strand and lane 7 for the non-template strand, are subtracted) are shown in green for lanes 3 and 8 (TFAM), red for lanes 4 and 9 (TFAM and POLRMT), and blue for lanes 5 and 10 (TFAM, POLRMT and TFB2M). C, DNase I footprinting of the LSP region (template strand labeled) in the absence or presence of transcription proteins as indicated. The positions on the DNA in relation to the LSP start site (arrow) are indicated as well as the footprint positions of TFAM and POLRMT/TFB2M. For indicated markers, see the description of panel A. Densitometric scanning profiles of the autoradiographs are shown in black for lane 2 (absence of protein), green for lane 3 (TFAM), red for lane 4 (TFAM and POLRMT), and blue for lane 5 (TFAM, POLRMT and TFB2M). D, the sequential model for transcription initiation with TFB2M induced melting of the promoter.
When TFB2M was added in addition to TFAM and POLRMT, we observed no obvious effect on DNase I protection (Fig. 1C, lane 5). However, TFB2M induced a permanganate-sensitive region at positions −1 to +3 relative to the transcription start site on the template strand (Fig. 1A, lane 5, and Fig. 1B), indicating that the addition of TFB2M causes DNA unwinding in this region. Apart from the bases at −1 to +3, the closest thymidines on the template strand are located at positions −7 and +5. No melting was observed on the non-template strand, where the closest thymidines are found at −8 and +6 (Fig. 1A, lane 10, and Fig. 1B).
Our results demonstrate that the promoter-bound TFAM-POLRMT complex is unable to melt the dsDNA promoter region, but requires TFB2M to catalyze this reaction. We also conclude that the start site melting is seen at positions −1 to +3 and does not propagate farther than positions −6 and +4.
A 7-bp Pre-melted Bubble Template Causes Non-sequence-specific Initiation of Transcription
In previous in vitro transcription experiments, a 7-bp mismatch region over the LSP transcription start site circumvented the requirement of both TFAM and TFB2M (17). We found this result puzzling, because our data suggested that TFAM was needed to recruit POLRMT in a step preceding start site unwinding, whereas the actual unwinding also required TFB2M (the sequential model for transcription initiation is summarized in Fig. 1D). We reasoned that if the current transcription initiation complex formation model is correct, it should be possible to find a mismatch template that can circumvent the requirement for TFB2M, but still depend on TFAM for recruitment of POLRMT and transcription initiation. To investigate this possibility, we used a 62-mer DNA template strand (−42 to +20) that includes the TFAM binding site and the LSP promoter, annealed to either WT or mismatch non-template strands. As expected, transcription from the double-stranded WT template (Fig. 2A, template LWT) was strictly dependent on TFAM and TFB2M (Fig. 2B, lanes 1–4) and gave two clear run-off products (LSP transcription can be initiated from two different positions, +1 and +2). We also examined a template with the same sized mismatch bubble (−3 to +4) as used in earlier published experiments (17) (Fig. 2A, template LP1). Transcription from this LP1 template was indeed independent of both TFAM and TFB2M; however, on this template, POLRMT did not produce LSP-specific run-off products, but instead caused the formation of multiple shorter transcripts (Fig. 2B, lanes 5–8). POLRMT uses ATP as the initiating nucleotide, and we have previously demonstrated that it can initiate non-sequence-specifically from ssDNA-containing thymidines (26). Most likely, the observed products produced with the 7-bp mismatch bubble were the result of initiation from thymidines in the mismatch template, e.g. at positions +3 and +5. We hypothesized that the 7-bp mismatch bubble template displays a sufficiently long ssDNA stretch to allow POLRMT to initiate transcription in a non-sequence-specific manner. When the same sized mismatch template was used in a previous study, this effect was not observed because short RNA primers were used in combination with the bubble template and such primers will direct transcription initiation to the correct start site (17).
FIGURE 2.
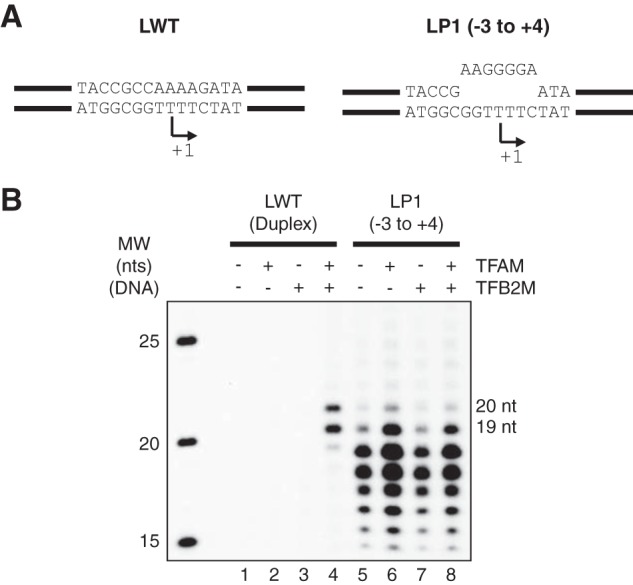
A 7-bp pre-melted (mismatch) bubble fails to produce promoter-specific transcription. A, graphic representation of the two templates used for in vitro transcription. LWT is composed of two complementary oligonucleotides, whereas LP1 contains a 7-bp mismatch region. B, in vitro transcription from duplex and mismatch oligonucleotide templates as described under “Experimental Procedures.” Reactions with duplex LSP template (LWT, lanes 1–4) or LSP 7-bp mismatch template (LP1, mismatch −3 to +4, lanes 5–8) are shown; complete oligonucleotide sequences are found under “Experimental Procedures.” POLRMT was added to all reactions, and TFAM and TFB2M were added as indicated. The expected LSP run-off products of 19 and 20 nt are indicated. MW, molecular weight marker.
Specific Bypass of TFB2M by Pre-melted Templates
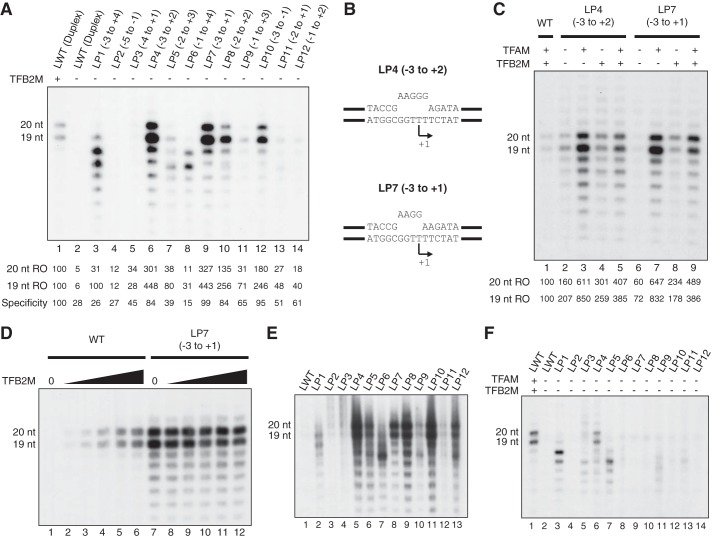
We decided to create a series of shorter mismatch bubble templates in an attempt to find an unwound region that could specifically circumvent the requirement for TFB2M, and thus lead to transcription initiation from the same sites observed in vivo, i.e. positions +1 and +2. To this end, we screened 11 new templates, each with a 3–5-bp mismatch, for TFB2M-independent, LSP-specific transcription (Fig. 3A). We found that 4 out of 11 templates displayed LSP-specific transcription patterns in the absence of TFB2M: LP4 (mismatch −3 to +2), LP7 (mismatch −3 to +1), LP8 (mismatch −2 to +2), and LP10 (mismatch −3 to −1), with specific run-off products at 19 and 20 nucleotides (nt) (Fig. 3A, lanes 6, 9, 10, and 12). All four templates were highly active in the absence of TFB2M and were transcribed at higher levels than the WT control transcribed by the complete transcription machinery (Fig. 3A, compare lane 1 with lanes 6, 9, 10, and 12). Furthermore, the fraction of 19- and 20-nt products produced by correct transcription initiation, relative to all transcription products observed in the same lane, was nearly as high for these four bubble templates as what was observed for the WT LSP construct (84, 99, 84, and 95% of the WT levels for LP4, LP7, LP8, and LP10, respectively, Fig. 3A).
FIGURE 3.
Screen for TFB2M-independent, TFAM-dependent, LSP templates. A, screen for TFB2M-independent LSP templates. In vitro transcription from WT duplex LSP template (lanes 1 and 2) and mismatch templates with the indicated mismatch region (templates LP1-LP12, lanes 3–14) are shown; the complete oligonucleotide sequences are found under “Experimental Procedures.” POLRMT and TFAM were added to all reactions, and the duplex reaction in lane 1 also contained TFB2M. The expected LSP run-off (RO) products of 19 and 20 nt are indicated. Quantifications below each lane show the intensity of the 19- and 20-nt run-off products for each template relative to the band intensities in lane 1 (WT LSP duplex). We also indicate the specificity of the reaction, i.e. the fraction of correct transcription initiation events (19- and 20-nt run-off products) relative to the total lane intensity. For comparison, the indicated number is relative to the specificity in lane 1, which is set to 100. B, graphic representation of templates LP4 and LP7 used for further in vitro transcription (in panel C). C, screen for TFAM dependence of TFB2M-independent LSP templates. In vitro transcription with template LP4 (lanes 2–5) and LP7 (lanes 6–9) was performed, and WT duplex LSP template was used as control (lane 1). POLRMT was added to all reactions, and TFAM and TFB2M were added as indicated. The expected LSP run-off products of 19 and 20 nt are indicated. Quantifications show the intensity of the 19- and 20-nt run-off products for each template relative to the band intensities in lane 1 (WT LSP duplex). D, in vitro transcription from the WT (lanes 1–6) and LP7 (lanes 7–12) templates in the absence of TFB2M (lanes 1 and 7) or in the presence of increasing amounts of TFB2M (lanes 2–6 and 8–12, 0.125, 0.25, 0.5, 1, and 2 pmol, respectively). The expected LSP run-off products of 19 and 20 nt are indicated. E, transcription from WT duplex (lane 1) and mismatch bubble templates (LP1–12, lanes 2–13) in the presence of POLRMT, TFAM, and TFB2M. Complete oligonucleotide sequences are found under “Experimental Procedures.” F, POLRMT transcription from WT duplex (lanes 1 and 2) and mismatch bubble templates (LP1–12, lanes 3–14) in the absence of TFAM and TFB2M (lanes 2–14). TFAM and TFB2M were added to the reaction in lane 1 as a positive control. Complete oligonucleotide sequences are found under “Experimental Procedures.”
We continued analyzing the two templates that produced the highest levels of correctly initiated transcripts in the absence of TFB2M, i.e. LP4, with a mismatch between −3 to +2, and LP7, with a mismatch between −3 and +1 (Fig. 3B). These two templates were tested in reactions with different combinations of transcription factors. Again, both templates displayed TFB2M-independent, LSP-specific transcription (Fig. 3C, lanes 3 and 7). In the absence of TFAM, LSP-specific transcription was reduced on both templates (Fig. 3C, lanes 2 and 4 for LP4 and lanes 6 and 8 for LP7). The TFB2M-independent, TFAM-dependent pattern was most obvious with the LP7 template (Fig. 3C lanes 6–9). In addition, TFB2M-independent transcription on the LP4 and the LP7 templates was markedly higher than that from a WT duplex template (Fig. 3C, compare lane 1 with lanes 3 and 7).
When TFB2M was added in addition to TFAM, transcription was slightly inhibited from both the LP4 and the LP7 template (Fig. 3C, compare lane 3 with lane 5 and lane 7 with lane 9). This could indicate an effect similar to that seen in yeast, where the presence of Mtf1 inhibits run-off transcription from a mismatch bubble template (22, 27). To further investigate the effect of TFB2M on the human system, we used the WT and LP7 templates for in vitro transcription and compared the effects of increasing TFB2M concentrations (Fig. 3D). As expected, we observed increased transcription with higher TFB2M concentrations using the WT template, leveling out when the POLRMT:TFB2M ratio exceeded 1:1 (Fig. 3D, lanes 5 and 6). In contrast, the highest levels of bubble template transcription were seen in the absence of TFB2M, and increasing concentrations of this transcription factor had a mild inhibiting effect (Fig. 3D, lanes 7–12), even if the inhibition was less dramatic than what had been observed previously in yeast (22).
From our analysis in Fig. 3A, it was clear that bubble templates with mismatches farther upstream than −3 showed no transcription in the absence of TFB2M (Fig. 3A, lanes 4 and 5). These templates also displayed substantially lower levels of transcription as compared with other bubble constructs when all three transcription proteins were present (Fig. 3E, lanes 3 and 4), indicating that mismatches upstream of −3 may disturb the POLRMT recognition site.
All templates with bubbles extending downstream to +3 and +4 were active, but failed to produce the promoter-specific 19- and 20-nt products (Fig. 3A, LP1, LP5, LP6, and LP9, lanes 3, 7, 8, and 11, respectively). These templates also showed similar transcription patterns when both TFAM and TFB2M were omitted (Fig. 3F, lanes 3, 7, 8, and 11). As with template LP1, it is possible that these four constructs present a sufficiently long single-stranded stretch of thymidines for non-promoter-specific transcription initiation to occur.
The precise position of the mismatch was important for transcription activity and specificity. A 4-bp mismatch covering positions −3 to +1 (LP7) produced a strong, TFAM-dependent, but TFB2M-independent transcription reaction. When the mismatch region was moved one step downstream (−2 to +2, LP8), transcription initiation was reduced, and a further move to −1 to +3 (LP9) completely abolished the reaction (Fig. 3A, compare lanes 9–11). Even if the LP8 construct (mismatch at −2 to +2) still supported promoter-specific transcription, initiation was mainly seen from the most downstream start site (+2, 19-nt run-off), indicating that melting at −3 is important for transcription initiation at position +1. Interestingly, when TFB2M was present, the LP8 transcription from +1 and +2 was restored to similar levels, indicating that TFB2M still affects transcription initiation on this template (Fig. 3E, lane 9).
Decreasing the size of the bubble to 3 bp abolished initiations in two out of three templates (Fig. 3A, LP10, LP11, and LP12, lanes 12, 13, and 14, respectively). Template LP10 (mismatch at −3 to −1) supported initiation, but at a lower level as compared with template LP7 (mismatch −3 to +1), (Fig. 3A, compare lanes 9 and 12); hence, melting at +1 is important for TFB2M-independent transcription.
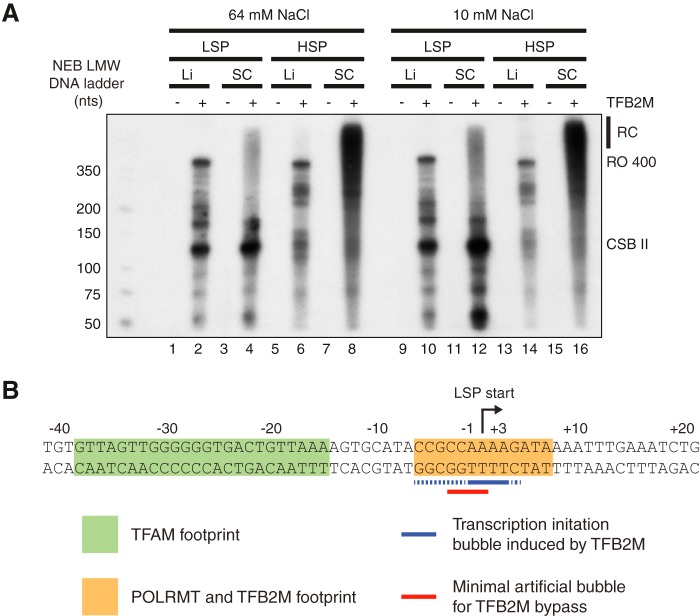
No TFB2M-independent Transcription from Supercoiled Templates
In yeast, the RNA polymerase Rpo41 can initiate transcription in the absence of the TFB2M homologue Mtf1 on a supercoiled template (22). To investigate whether POLRMT behaves in a similar manner, we performed in vitro transcription from the LSP and HSP promoters, using linear and supercoiled templates. As expected, we observed transcription from all four templates in the presence of POLRMT, TFAM, and TFB2M (Fig. 4A, lanes 2, 4, 6, and 8). Omission of TFB2M abolished all transcription from the linear templates (Fig. 4A, lanes 1 and 5). Interestingly, the effects were the same on supercoiled templates, where no transcripts could be observed in the absence of TFB2M (Fig. 4A, lanes 3 and 7). In an attempt to further facilitate promoter melting, we also performed the reactions at low salt concentrations (10 mm NaCl), which have previously been shown to stimulate promoter melting and promote TFAM-independent transcription (28). Lowering the salt did not change the outcome of the experiment, because TFB2M was still needed for promoter-specific initiation of transcription on the supercoiled templates (Fig. 4A, lanes 9–16).
FIGURE 4.
POLRMT is dependent on TFB2M for promoter melting on both linear and supercoiled templates. A, in vitro transcription from linear and supercoiled LSP and HSP templates in the presence or absence of TFB2M. The reactions were performed at 64 mm NaCl (lanes 1–8) or 10 mm NaCl (lanes 9–16). Template topology (linear (Li) or supercoiled (Sc)) and promoter (LSP or HSP) are indicated. Rolling circle transcription products (RC) from supercoiled templates, run-off products (RO 400, ∼400 nt) from linear templates, and conserved sequence block II (CSB II) pre-terminated products from LSP templates are indicated. B, promoter melting at a human mitochondrial promoter. The LSP sequence with the indicated TFAM DNase I footprint (green shading) and POLRMT/TFB2M DNase I footprint (in the presence of TFAM, orange shading) are shown. The blue solid line indicates definite TFB2M-induced melting area observed by potassium permanganate footprinting, and the blue dotted line indicates possible TFB2M-induced melting. The red line indicates the minimal pre-melted region for TFB2M bypass.
Discussion
Here we demonstrate that TFB2M is required for transcription start site unwinding at the human LSP promoter. Potassium permanganate footprinting suggests that the melting induced by TFB2M at least stretches from positions −1 to +3, but not farther than to positions −6 and +4 (Fig. 4B). The requirement of TFB2M can be bypassed when a pre-melted bubble is introduced at −3 to +1 (Fig. 4B), but TFAM is still needed for full strength initiation on this template.
We show that human POLRMT and TFAM produce a footprint covering the transcription start site, which is not notably altered upon the addition of TFB2M (Fig. 1). This finding is in agreement with previous studies, demonstrating that POLRMT interacts sequence-specifically with DNA sequences surrounding the transcription start site, whereas TFB2M interacts with the priming nucleotide, without forming specific interactions with promoter DNA (17, 19). Hence, the addition of TFB2M does not extend the footprint, because the transcription start site is already completely covered by POLRMT. It should be noted that our observations with human transcription factors differ in one important aspect from those obtained with the mouse transcription system as mouse TFB2M stimulates POLRMT binding to the transcription start site (6). It seems unlikely that the mechanism of promoter recognition differs between these two closely related systems. Instead, mouse POLRMT may be less stable in solution and require the TFB2M factor to adopt a conformation that allows it to properly interact with the region surrounding the transcription start site.
In a previous study, we used a FRET-based assay to examine structural changes at the transcription start site of the HSP promoter (6). In the presence of TFAM and POLRMT, TFB2M was required to induce structural changes at the transcription start site. In the light of the data presented in the current study, we believe that these previous observations suggest that TFB2M is also required for promoter melting at HSP. This conclusion is also supported by a recent study, which demonstrates that the TFAM-POLRMT pre-initiation complexes at LSP and HSP are identical (29). Taken together it therefore seems likely that the results from LSP obtained in this study are also of relevance for our understanding of HSP activation.
A previous study suggested that neither TFAM nor TFB2M is required for transcription initiation on a 7-bp bubble template (17). We verify this observation, but also find that when we exclude the short RNA primers used in the previously published experiments, the 7-bp bubble template is a poor template for promoter-specific initiations. Instead, this long bubble behaves like a single-stranded region, which can be used for non-sequence-specific transcription initiation by POLRMT. This observation explains the lack of TFAM dependence in these previous experiments. Here, using templates with shorter bubbles, we can clearly distinguish the function of TFAM from TFB2M in transcription initiation. We also noticed that transcription from bubble templates is stronger than from a duplex template. This observation could indicate that POLRMT binds tighter to a bubble template, and that TFB2M binding and/or promoter melting could be rate-limiting steps in the human mitochondrial transcription initiation process. Our screen for TFB2M-independent pre-melted bubble templates demonstrated that the −4 and −5 bases have to be in duplex form for promoter-specific transcription initiation, an effect that could be explained if this region constitutes part of the POLRMT recognition site.
In contrast to the yeast system, TFB2M is needed for POLRMT to initiate transcription from mitochondrial promoters on both linear and supercoiled templates. This observation, in addition to the presence of a second transcription factor not present in the yeast system, TFAM, suggests that human mitochondrial transcription is under more complex control.
In conclusion, our results show that TFB2M is the promoter melting-inducing factor of human mitochondria and indicate that this process may be a rate-limiting step in transcription initiation. These findings demonstrate that TFAM is needed for POLRMT recruitment but not sufficient for promoter melting, and that TFB2M is required for promoter melting but dispensable for POLRMT recruitment. In addition, TFB2M is not required for promoter-dependent transcription, once melting has been achieved. Our observations are in excellent agreement with the sequential model for initiation of mitochondrial transcription (6, 14) (Fig. 1C). Our new bubble template LP7 may also prove valuable for continued structural and functional studies of mitochondrial transcription initiation.
Experimental Procedures
Recombinant Proteins
TFAM, TFB2M, and POLRMT were purified as described previously (6).
Potassium Permanganate Footprinting
The footprinting template was produced by PCR with the pUC18 human LSP vector as template (30) using the primer pair 5′-GCA CTT AAA CAC ATC TCT GCC AAA CCC C (forward) and 5′-GTA AAA CGA CGG CCA GTG CCA AGC (reverse). The forward primer (template strand) or the reverse primer (non-template strand) was labeled with polynucleotide kinase (PNK) enzyme (New England Biolabs) and [γ-32P]ATP (3000 Ci/mmol). The footprinting reactions were performed in 20 μl containing 25 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 1 mm ATP, 100 μg/ml BSA, 1 mm DTT, 50 mm NaCl, labeled template (22,500 cpm), and proteins as indicated (1 pmol of TFAM, 2 pmol of POLRMT, 2 pmol of TFB2M). The mixture was incubated for 20 min at room temperature followed by 2 min on ice, 2 min at 32 °C, the addition of 2 μl of 50 mm KMnO4, and 2 min at 32 °C. Reactions were stopped by the addition of 2.4 μl of β-mercaptoethanol followed by 17.6 μl of stop buffer (5.3 μl of EDTA (from 0.5 m stock), 2 μl of NaOAc (0.3 m, pH 5.5), 0.5 μl of glycogen (20 mg/ml) and 9.8 μl of water). The mixture was ethanol-precipitated, and the resulting wet pellet was dissolved in 70 μl of 10% piperidine followed by incubation at 90 °C for 30 min. The reaction was dried, dissolved in 30 μl of water, repeatedly dried, and dissolved in 50 μl of water with the addition of 4 μl of NaOAc (0.3 m, pH 5.5), followed by ethanol precipitation. The dried pellets were dissolved in 10 μl of gel loading buffer (98% formamide, 10 mm EDTA, 0.025% xylene cyanol FF, and 0.025% bromphenol blue) and heated at 95 °C for 3 min. The resulting products were analyzed on 8% denaturing polyacrylamide sequencing gels (1× TBE and 7 m urea). The gels were dried followed by exposure on photo film. Densitometric scanning profiles of the autoradiographs were performed in the program Multi Gauge (FUJI FILM), and background signals for template and non-template strands in lanes 2 and 7, respectively, were subtracted from the other lanes to detect differences only upon protein addition. All experiments were performed at least three times and showed similar results.
DNase I Footprinting
The template (forward primer labeled) and reaction conditions were the same as those used for potassium permanganate footprinting. After 20 min at room temperature, 2 μl of 50 milliunits/μl of DNase I diluted in 2.5 × DNase I buffer with MgCl2 (Thermo Scientific) was added. The DNase I reaction was stopped after 2 min by the addition of 20 μl of stop buffer (200 mm NaCl, 20 mm EDTA, 1% SDS, and 100 μg/ml yeast tRNA (Ambion)) directly followed by incubation on ice. The DNA fragments were recovered with phenol extraction and ethanol precipitation. The resulting products were analyzed as the potassium permanganate footprinting products. All experiments were performed at least three times and showed similar results.
GA Ladder
One μl of footprinting template was mixed with 4 μl of NaOH loading solution (10 mm EDTA, 1 mm NaOH, 0.1% bromphenol blue, and 98% formamide) and 20 μl of water. The mixture was boiled for 30 min with an open cap followed by incubation on ice before loading on the sequencing gel.
Templates for in Vitro Transcription
The oligonucleotides used were ordered from Eurofins MWG Operon. Annealing of strands was performed in 10 mm Tris-HCl, pH 8.0, 10 mm NaCl, 5 min at 95 °C with slow cooling to room temperature. All templates were formed by annealing a wild type template strand with a complementary or partly complementary non-template strand containing the indicated mismatches. The following sequences were used: template strand, WT (GGA TTT CAA ATT TTA TCT TTT GGC GGT ATG CAC TTT TAA CAG TCA CCC CCC AAC TAA CAC AT); non-template strands, WT (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG CCA AAA GAT AAA ATT TGA AAT CC); LP1 mismatch, −3 to +4 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG AAG GGG AAT AAA ATT TGA AAT CC); LP2 mismatch, −5 to −1 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CAA AAC AAA GAT AAA ATT TGA AAT CC); LP3 mismatch, −4 to +1 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCA AAG GAA GAT AAA ATT TGA AAT CC); LP4 mismatch, −3 to +2 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG AAG GGA GAT AAA ATT TGA AAT CC); LP5 mismatch, −2 to +3 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG CAG GGG GAT AAA ATT TGA AAT CC); LP6 mismatch, −1 to +4 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG CCG GGG AAT AAA ATT TGA AAT CC); LP7 mismatch, −3 to +1 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG AAG GAA GAT AAA ATT TGA AAT CC); LP8 mismatch, −2 to +2 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG CAG GGA GAT AAA ATT TGA AAT CC); LP9 mismatch, −1 to +3 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG CCG GGG GAT AAA ATT TGA AAT CC); LP10 mismatch, −3 to −1 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG AAG AAA GAT AAA ATT TGA AAT CC); LP11 mismatch, −2 to +1 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG CAG GAA GAT AAA ATT TGA AAT CC); LP12 mismatch, −1 to +2 (ATG TGT TAG TTG GGG GGT GAC TGT TAA AAG TGC ATA CCG CCG GGA GAT AAA ATT TGA AAT CC). The plasmid templates used were pUC18 vectors with human mitochondrial promoter region inserts (human mtDNA 1–477 for LSP and 499–742 for HSP, cloned between the BamHI and HindIII restriction sites). Restriction enzymes BamHI (for LSP) and NdeI (for HSP) were used to linearize templates for run-off transcription experiments.
In Vitro Transcription
Transcription reactions (25 μl) contained 25 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 64 mm NaCl (or 10 mm where indicated in Fig. 4A), 100 μg/ml BSA, 1 mm DTT, 400 μm ATP, 150 μm GTP, 150 μm CTP 10 μm UTP, 0,02 μm [α-32P]UTP (3000 Ci/mmol), 4 units of RNase Inhibitor, Murine (New England Biolabs). For transcription with oligonucleotide templates, 500 fmol of the indicated template was added together with 500 fmol of POLRMT, 500 fmol of TFAM, and 500 fmol of TFB2M, where indicated. For plasmid templates, 100 fmol of the indicated template was added together with 500 fmol of POLRMT, 5 pmol of TFAM, and 1.5 pmol of TFB2M, where indicated. The reactions were stopped after 30 min at 32 °C by the addition of 200 μl of stop buffer (10 mm Tris-HCl, pH 8.0, 0.2 m NaCl, 1 mm EDTA, and 100 μg/ml proteinase K) followed by incubation at 42 °C for 45 min. The transcripts were ethanol-precipitated, and the pellets were dissolved in 20 μl of gel loading buffer (98% formamide, 10 mm EDTA, 0.025% xylene cyanol FF, and 0.025% bromphenol blue) and then heated at 95 °C for 3 min. The samples were analyzed on 10% denaturing polyacrylamide sequencing gels (1× TBE and 7 m urea) for oligonucleotide templates and 4% denaturing polyacrylamide gels (1× TBE and 7 m urea) for plasmid templates, followed by exposure on photo film. Quantification of transcript levels was performed with the program Multi Gauge (FUJI FILM) with images generated from a FUJI FILM FLA-7000 instrument. All experiments were performed at least three times and showed similar results. The quantifications of transcript levels indicated are based on the displayed autoradiograms (Fig. 3, A and C).
Author Contributions
V. P. planned and conducted the experiments. V. P. and C. M. G. analyzed the data and wrote the paper. Both authors reviewed the results and approved the final version of the manuscript.
This work was supported by Swedish Research Council Grant 2012-2583; Swedish Cancer Foundation Grant CAN 2013/855; European Research Council Grant 268897; the IngaBritt and Arne Lundberg Foundation; and the Knut and Alice Wallenbergs Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
- HSP
- heavy-strand promoter
- LSP
- light-strand promoter
- NTE
- N-terminal extension
- nt
- nucleotide(s)
- TBE
- Tris borate-EDTA.
References
- 1. Gustafsson C. M., Falkenberg M., and Larsson N. G. (2016) Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 85, 133–160 [DOI] [PubMed] [Google Scholar]
- 2. Masters B. S., Stohl L. L., and Clayton D. A. (1987) Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell 51, 89–99 [DOI] [PubMed] [Google Scholar]
- 3. Ringel R., Sologub M., Morozov Y. I., Litonin D., Cramer P., and Temiakov D. (2011) Structure of human mitochondrial RNA polymerase. Nature 478, 269–273 [DOI] [PubMed] [Google Scholar]
- 4. Litonin D., Sologub M., Shi Y., Savkina M., Anikin M., Falkenberg M., Gustafsson C. M., and Temiakov D. (2010) Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 285, 18129–18133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N. G., and Gustafsson C. M. (2002) Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31, 289–294 [DOI] [PubMed] [Google Scholar]
- 6. Posse V., Hoberg E., Dierckx A., Shahzad S., Koolmeister C., Larsson N. G., Wilhelmsson L. M., Hällberg B. M., and Gustafsson C. M. (2014) The amino terminal extension of mammalian mitochondrial RNA polymerase ensures promoter specific transcription initiation. Nucleic Acids Res. 42, 3638–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fisher R. P., and Clayton D. A. (1988) Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 8, 3496–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fisher R. P., Lisowsky T., Parisi M. A., and Clayton D. A. (1992) DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 267, 3358–3367 [PubMed] [Google Scholar]
- 9. Fisher R. P., Topper J. N., and Clayton D. A. (1987) Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell 50, 247–258 [DOI] [PubMed] [Google Scholar]
- 10. Parisi M. A., and Clayton D. A. (1991) Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 252, 965–969 [DOI] [PubMed] [Google Scholar]
- 11. Ngo H. B., Kaiser J. T., and Chan D. C. (2011) The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 18, 1290–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rubio-Cosials A., Sidow J. F., Jiménez-Menéndez N., Fernández-Millán P., Montoya J., Jacobs H. T., Coll M., Bernadó P., and Solà M. (2011) Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 18, 1281–1289 [DOI] [PubMed] [Google Scholar]
- 13. Dairaghi D. J., Shadel G. S., and Clayton D. A. (1995) Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim. Biophys. Acta 1271, 127–134 [DOI] [PubMed] [Google Scholar]
- 14. Morozov Y. I., Agaronyan K., Cheung A. C., Anikin M., Cramer P., and Temiakov D. (2014) A novel intermediate in transcription initiation by human mitochondrial RNA polymerase. Nucleic Acids Res. 42, 3884–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaufman B. A., Durisic N., Mativetsky J. M., Costantino S., Hancock M. A., Grutter P., and Shoubridge E. A. (2007) The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell 18, 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kukat C., Davies K. M., Wurm C. A., Spåhr H., Bonekamp N. A., Kühl I., Joos F., Polosa P. L., Park C. B., Posse V., Falkenberg M., Jakobs S., Kühlbrandt W., and Larsson N. G. (2015) Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 112, 11288–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sologub M., Litonin D., Anikin M., Mustaev A., and Temiakov D. (2009) TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell 139, 934–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mangus D. A., Jang S. H., and Jaehning J. A. (1994) Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J. Biol. Chem. 269, 26568–26574 [PubMed] [Google Scholar]
- 19. Gaspari M., Falkenberg M., Larsson N. G., and Gustafsson C. M. (2004) The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 23, 4606–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morozov Y. I., Parshin A. V., Agaronyan K., Cheung A. C., Anikin M., Cramer P., and Temiakov D. (2015) A model for transcription initiation in human mitochondria. Nucleic Acids Res. 43, 3726–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu B., and Clayton D. A. (1992) Assignment of a yeast protein necessary for mitochondrial transcription initiation. Nucleic Acids Res. 20, 1053–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsunaga M., and Jaehning J. A. (2004) Intrinsic promoter recognition by a “core” RNA polymerase. J. Biol. Chem. 279, 44239–44242 [DOI] [PubMed] [Google Scholar]
- 23. Borowiec J. A., Zhang L., Sasse-Dwight S., and Gralla J. D. (1987) DNA supercoiling promotes formation of a bent repression loop in lac DNA. J. Mol. Biol. 196, 101–111 [DOI] [PubMed] [Google Scholar]
- 24. Chan B., Minchin S., and Busby S. (1990) Unwinding of duplex DNA during transcription initiation at the Escherichia coli galactose operon overlapping promoters. FEBS Lett. 267, 46–50 [DOI] [PubMed] [Google Scholar]
- 25. Rubin C. M., and Schmid C. W. (1980) Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 8, 4613–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wanrooij S., Fusté J. M., Farge G., Shi Y., Gustafsson C. M., and Falkenberg M. (2008) Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl. Acad. Sci. U.S.A. 105, 11122–11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paratkar S., Deshpande A. P., Tang G. Q., and Patel S. S. (2011) The N-terminal domain of the yeast mitochondrial RNA polymerase regulates multiple steps of transcription. J. Biol. Chem. 286, 16109–16120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi Y., Dierckx A., Wanrooij P. H., Wanrooij S., Larsson N. G., Wilhelmsson L. M., Falkenberg M., and Gustafsson C. M. (2012) Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc. Natl. Acad. Sci. U.S.A. 109, 16510–16515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morozov Y. I., and Temiakov D. (2016) Human mitochondrial transcription initiation complexes have similar topology on the light and heavy strand promoters. J. Biol. Chem. 291, 13432–13435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Posse V., Shahzad S., Falkenberg M., Hällberg B. M., and Gustafsson C. M. (2015) TEFM is a potent stimulator of mitochondrial transcription elongation in vitro. Nucleic Acids Res. 43, 2615–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]