Abstract
The serine/threonine kinase Unc-51-like kinase-1 (Ulk1) is thought to be essential for induction of autophagy, an intracellular bulk degradation process that is activated by various stresses. Although several proteins have been suggested as Ulk1 substrates during autophagic process, it still remains largely unknown about Ulk1's physiological substrates. Here, by performing in vitro and in vivo phosphorylation assay, we report that the co-chaperone cell division cycle protein 37 (Cdc37) is a Ulk1 substrate. Ulk1-mediated phosphorylation of Ser-339 in Cdc37 compromised the recruitment of client kinases to a complex comprising Cdc37 and heat shock protein 90 (Hsp90) but only modestly affected Cdc37 binding to Hsp90. Because the recruitment of protein kinase clients to the Hsp90 complex is essential for their stability and functions, Ser-339 phosphorylation of Cdc37 disrupts its ability as a co-chaperone to coordinate Hsp90. Hsp90 inhibitors are cancer chemotherapeutic agents by inducing depletion of clients, many of which are oncogenes. Upon treatment with an Hsp90 inhibitor in cancer cells, Ulk1 promoted the degradation of Hsp90-Cdc37 client kinases, resulting in increased cellular sensitivity to Hsp90 inhibitors. Thus, our study provides evidence for an anti-proliferative role of Ulk1 in response to Hsp90 inhibition in cancer cells.
Keywords: AMP-activated kinase (AMPK), autophagy, cell death, heat shock protein 90 (Hsp90), protein kinase, Hsp90 inhibitor, Ulk1, cdc37
Introduction
Autophagy is a tightly regulated pathway that can be stimulated by multiple forms of cellular stress (1, 2). During autophagic process, cells form double-membraned vesicles, autophagosomes, that sequester organelles, proteins, or portions of the cytoplasm for delivery to the lysosome (3, 4). The molecular mechanism of autophagosome formation is conserved in evolution and depends on several autophagy-related proteins (ATGs)2 (5). Atg1 is a serine/threonine protein kinase that plays important roles in autophagy initiation (6, 7). In mammalians cells, Atg1's functional homologues are Unc-51-like kinases 1 (Ulk1) and 2 (8–13). During autophagy process, the activated Ulk1/2 phosphorylates Atg13, Fip200, and Beclin-1, leading to autophagy induction (14–16).
Besides autophagy-related protein substrates, several autophagy-unrelated substrates of Ulk1 were also found. For example, during deprivation of amino acid and growth factors, Ulk1/2 directly phosphorylates key glycolytic enzymes to sustain glycolysis (17). Stimulator of interferon genes (STING) is also phosphorylated by Ulk1 to prevent the persistent transcription of innate immune genes (18). Ulk1/2 phosphorylates SEC16A and regulates endoplasmic reticulum export, which is essential for cellular homeostasis (19).
Because Ulk1 has a variety of substrates, it plays important roles in many biological processes, especially in cell death and proliferation. Autophagy facilitates cell survival by maintaining cellular metabolism and eliminating harmful damaged proteins and organelles; therefore, Ulk1-autophagy axis is generally considered as a cyto-protective response. However, several reports showed that Atg1-induced autophagy inhibits cell growth and induces apoptotic cell death in Drosophila (20). Overexpression of wild-type Ulk but not kinase-dead Ulk changed cell morphology and caused cytotoxicity in NIH3T3 cells (12). Recent studies also showed that Ulk1 contributes to cell death in an autophagy-dependent or -independent manner (21–23). For example, upon DNA damage, p53 up-regulated Ulk1 is necessary for the sustained autophagy, which results in subsequent cell death (22). In addition, nuclear Ulk1 can also promote cell death by regulating the activity of the DNA damage repair protein poly(ADP-ribose) polymerase 1 (PARP1) (23). Therefore, the mechanism of Ulk1-induced cell death is very complex and needs further exploration.
In this study we identified cochaperone Cell Division Cycle Protein 37 (Cdc37) as a new phosphorylation target of Ulk1. Phosphorylation of Cdc37 at Ser-339 by Ulk1 decreases its interaction with client kinases, resulting in the instability of the clients. In addition, we also found that Ulk1 kinase affected loss of client stability and activity upon Hsp90 inhibition. Finally, silencing Ulk1 decreased cancer cell sensitivity to Hsp90 inhibitors, showing that Ulk1 plays an important role in cellular response to Hsp90 inhibition.
Results
Ulk1 Phosphorylates Cdc37
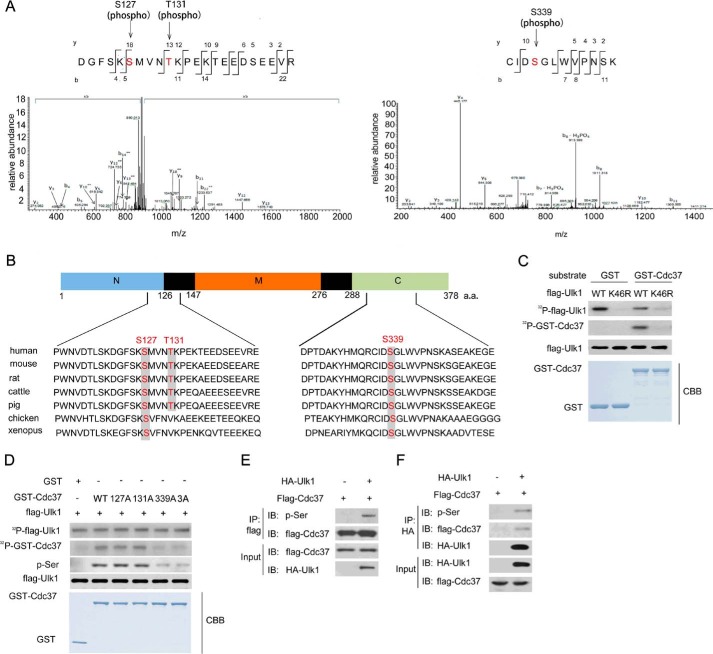
It has been reported that the interaction between Ulk1 and Hsp90-Cdc37 stabilizes and activates Ulk1 (24) (supplemental Fig. 1). Because Ulk1 is a well known serine/threonine kinase, we tested whether Ulk1 was able to phosphorylate Cdc37. As shown in Fig. 1, A and B, mass spectrometry analysis indicated that highly conserved sites Ser-127, Thr-131, and Ser-339 of Cdc37 were phosphorylated by Ulk1. In addition, by performing in vitro phosphorylation assay, we found that GST-Cdc37-WT was phosphorylated by wild-type Ulk1 but not the kinase-impaired K64R Ulk1 mutant (Fig. 1C). To further confirm these phosphorylation sites, Cdc37(WT), Cdc37(S127A), Cdc37(T131A), Cdc37(S339A), or Cdc37(S127A/T131A/S339A)-GST fusion proteins were purified. As shown in Fig. 1D, after incubation with Ulk1(WT), the radiolabeling of Cdc37(WT), Cdc37(S127A), and Cdc37(T131A) was much higher than that of Cdc37(S339A) and Cdc37(S127A/T131A/S339A), indicating that Ser-339 of Cdc37 is a phosphorylation target of ULK1. Moreover, we also tested whether Cdc37 is the target of Ulk2, an Ulk1 close homolog. As shown in supplemental Fig. 2, Cdc37 was phosphorylated by Ulk2; however, the difference of phosphorylation signal intensity between Cdc37-WT and Cdc37–3A was not obvious, indicating that Ser-339 of Cdc37 is a unique substrate site of Ulk1 in vitro.
FIGURE 1.
Ulk1 phosphorylated Cdc37. A, identification of S127, T131 and S338 phosphorylation on GST-Cdc37 incubated in vitro with Ulk1 by using mass spectrometry analysis. B, schematic of the phosphorylation sites Ser-127, Thr-131, and Ser-338 in human and other species Cdc37. a.a., amino acids. C, FLAG-Ulk1(WT) or FLAG-Ulk1(K46R) (a mutant Ulk1 lacking kinase activity) proteins were immunopurified from transfected HEK293 cells. Then in vitro kinase assays were performed in the presence of GST-Cdc37. Phosphorylated proteins were visualized with autoradiography. CBB, Coomassie Brilliant Blue. D, FLAG-Ulk1(WT) protein was immunopurified from transfected HEK293 cells. In vitro kinase assays were then performed in the presence of GST-Cdc37 proteins. Phosphorylated proteins were visualized with autoradiography. E, an empty plasmid and an expression plasmid for FLAG-Ulk1 were cotransfected with or without GFP-Cdc37 into HCT116 cells for 24 h. Proteins were immunoprecipitated (IP) with an antibody to GFP followed by immunoblot (IB) with an antibody to phosphorylated-serine. F, an empty plasmid and an expression plasmid for FLAG-Ulk1 were cotransfected with or without GFP-Cdc37 into HCT116 cells for 24 h. Proteins were immunoprecipitated with an antibody to HA followed by immunoblot with an antibody to FLAG or phosphorylated serine.
Next, we tried to determine whether Cdc37 is phosphorylated by Ulk1 in vivo. Protein extraction of HCT116 cells was immunoprecipitated with an anti-Cdc37 and then probed with anti-phosphorylated serine antibody. A significant increase of phosphorylated Cdc37 was detected in Ulk1-overexpressed HCT116 cells (Fig. 1, E and F), suggesting that Ulk1 plays a critical role in regulating Cdc37 phosphorylation.
Phosphorylation of Cdc37 Decreases Its Interaction with Clients
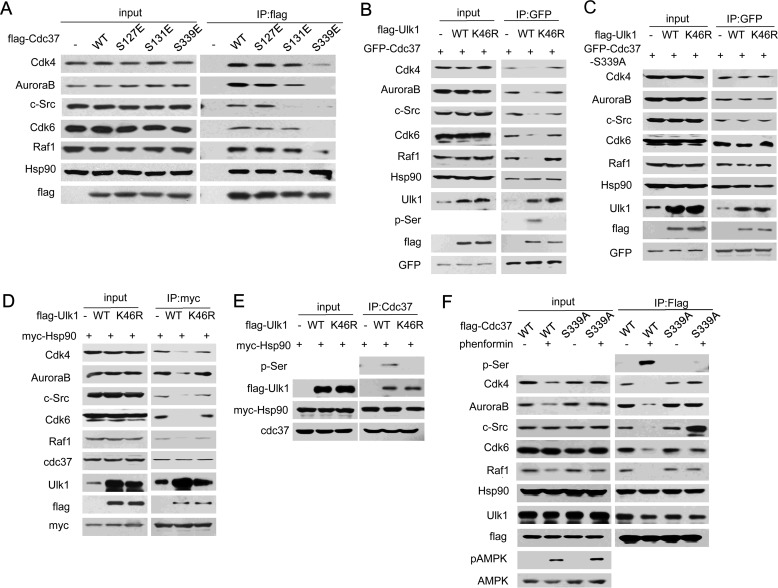
We next tested whether the Ulk1-mediated phosphorylation site on Cdc37 is necessary for its function. It has been reported that Cdc37 is a general cochaperone for protein kinases by recruiting client kinases into the Hsp90 system (25). As shown in Fig. 2A, although the interaction between Hsp90 and Cdc37 was not dramatically changed, several protein kinases, for example, Cdk4, AuroraB, Src, Cdk6, and Raf1, cannot be recruited to Cdc37(S339E), which mimics Cdc37 phosphorylation, indicating that Ser-339 dephosphorylation is essential for Cdc37 binding to these client kinases. We then tried to further confirm the role of Ulk1 in regulation of Cdc37-client binding. As shown in Fig. 2, B and C, expression of wild-type Ulk1 decreased the interaction between Cdc37 and its client kinases Cdk4, AuroraB, Src, Cdk6, and Raf1. Consistently, we also found that Ulk1 suppressed the interaction of clients with chaperone Hsp90 (Fig. 2, D and E). It has been reported that AMPK directly phosphorylates and activates Ulk1 (26–28). Therefore, we used AMPK activator phenformin to treat HCT116 cells. We found that phenformin also blocked Cdc37(WT) (but not S339A) binding to its client kinases (Fig. 2F). These data suggested that Ulk1-induced Cdc37 phosphorylation at Ser-339 negatively regulates the recruitment of client kinases to Hsp90-Cdc37 complex but only modestly affects Cdc37's binding to Hsp90.
FIGURE 2.
Phosphorylation of Cdc37 decreased its interaction with clients. A, HCT116 cells transfected with FLAG-Cdc37(WT), FLAG-Cdc37(S127E), FLAG-Cdc37(T131E), or FLAG-Cdc37(S339E) were subjected to FLAG immunoprecipitation (IP). B and C, HCT116 cells were cotransfected with FLAG-Ulk1 plus GFP-Cdc37 (B) or GFP-Cdc37-S339A (C). 24 h after transfection cells were subjected to GFP immunoprecipitation. D, HCT116 cells transfected with FLAG-Ulk1 and myc-Hsp90 were subjected to myc immunoprecipitation. E, HCT116 cells transfected with FLAG-Ulk1 and myc-Hsp90 were subjected to immunoprecipitated with an antibody to Cdc37. F, a set of RNAi-resistant rescue forms of Cdc37 plasmids were transfected into stable Cdc37-RNAi HCT116 cells. 24 h after transfection cells were treated with phenformin and then subjected to FLAG immunoprecipitation.
Ulk1 Suppresses the Stability of Cdc37 Clients
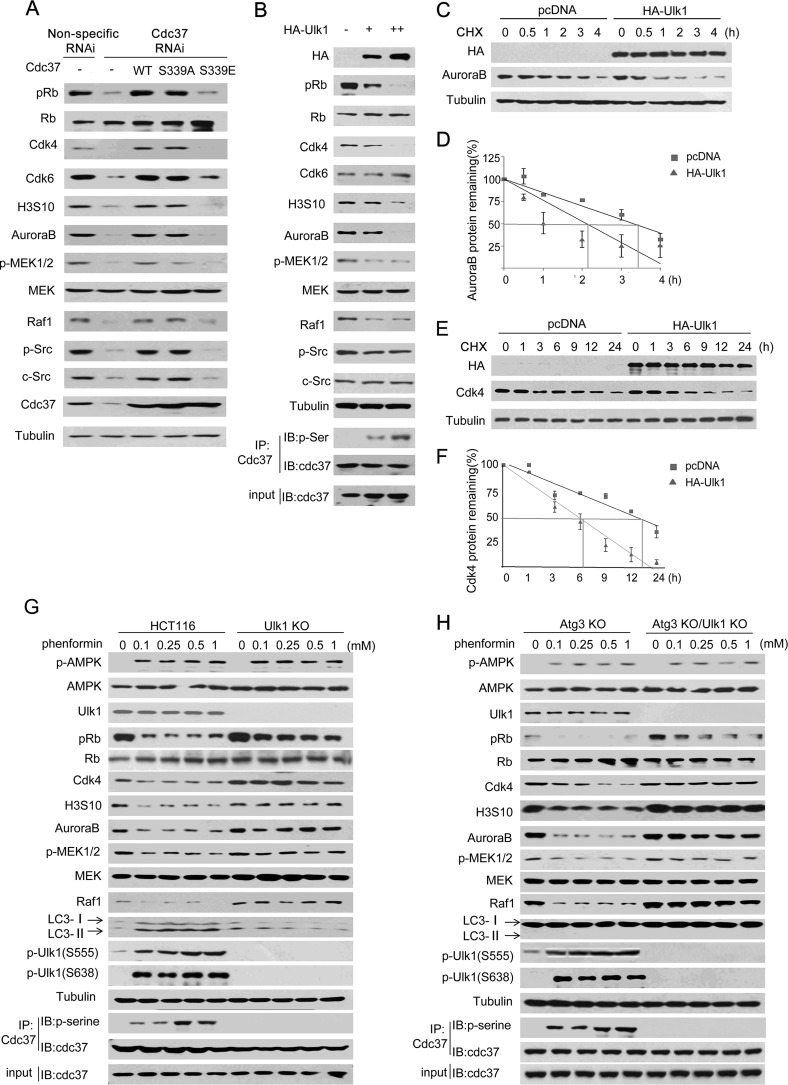
Because Cdc37 is important for maintaining the stability and functions of its client protein kinases (29, 30), we next tested the effect of Cdc37 Ser-339 phosphorylation on client kinase stability. Wild-type Cdc37, Cdc37-S339E(phosphorylation-mimic), or Cdc37-S339A(phosphorylation-incompetent) was stably transfected into Cdc37 knockdown HCT116 cells. As shown in Fig. 3A, compared with that of wild-type Cdc37 or Cdc37-S339A, expression of Cdc37-S339E was not able to rescue the protein level of client kinases. Consistently, we found that p-RB (Thr-826), H3S10, p-MEK1/2 (Ser-217/Ser-211) and p-Src (Tyr-416), which are direct downstream phosphorylated products of CDK4/6, AuroraB, RAF-1, and Src, respectively, were rescued by Cdc37-S339A but not Cdc37-S339E (Fig. 3A), indicating that Cdc37 Ser-339 phosphorylation compromised the stability and activity of these clients in cancer cells.
FIGURE 3.
Ulk1 suppressed the stability of Cdc37's clients. A, a set of RNAi-resistant rescue forms of Cdc37 plasmids were transfected into stable Cdc37-RNAi HCT116 cells. Western blotting was then performed to detect Cdc37 clients and their targets. B, different amounts HA-Ulk1 plasmid were transfected into Ulk1-KO HCT116 cells. Western blotting (IB) was then performed to detect Cdc37's clients and their targets. IP, immunoprecipitation. C–F, HA-Ulk1 plasmid or an empty plasmid was transfected into Ulk1 KO HCT116 cell. 24 h after transfection cycloheximide (CHX) was added at 10 μg/ml for different times as indicated. Western blotting was then performed to detect AuroraB (C) or CDK4 (E). The line graph shows the change in AuroraB (D) or CDK4 (F) levels at different times. G, wild-type and Ulk1-KO HCT116 cells were treated at different concentrations of phenformin for 24 h. H, Atg3-KO and Atg3-KO/Ulk1-KO HCT116 cells were treated at different concentrations of phenformin for 24 h.
We next examined whether Ulk1 reduces the amount of protein kinases. As shown in Fig. 3B, in the Ulk1 knock-out cell line, which was generated by using CRISPR/Cas9 technology, Cdk4, AuroraB, and Raf-1 were diminished after Ulk1 overexpression. In addition, we also tested the half-life of Cdc37 client kinases in the presence of cycloheximide, a protein synthesis inhibitor. We found that Cdk4 and AuroraB half-lives were decreased dramatically after Ulk1 was expressed (Fig. 3, C–F). Consistently, phenformin decreased Cdc37 clients' expression in wild-type HCT116 cells but not in Ulk1 KO HCT116 cells (Fig. 3G). Because AMPK activator phenformin is known to phosphorylate Ulk1 to induce autophagy (26–28), we then tested whether Ulk1 mediated autophagy is related with Cdc37 clients' regulation. By using the autophagy-deficient Atg3 knock-out HCT116 cell line, we found that Ulk1 was still required for phenformin-induced Cdc37's client degradation (Fig. 3H), supporting the autophagy-independent role of Ulk1 in regulating the stability of Cdc37's client kinases.
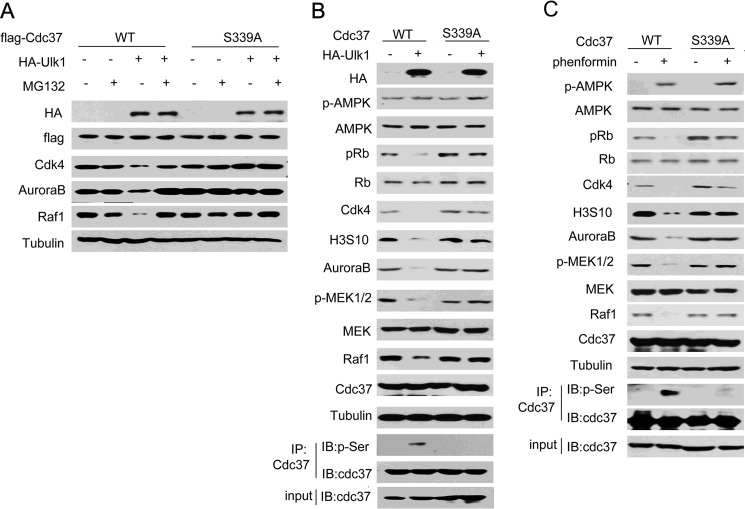
Next, to determine whether Cdc37 Ser-339 phosphorylation is a necessary modification for Ulk1-suppressed kinases stability, Cdc37(WT) or Cdc37(S339A) were transfected into Cdc37-KD HCT116 cells. As shown in Fig. 4A, Ulk1 overexpression reduced the protein level of clients in a proteasome-dependent manner. Furthermore, the reduction was almost abolished when Cdc37(S339A) was overexpressed (Fig. 4, B and C), indicating that Ulk1 suppressed the stability of Cdc37's clients by inducing its phosphorylation.
FIGURE 4.
Ulk1 suppressed the stability of Cdc37's clients by inducing its phosphorylation. A, Ulk1-KO HCT116 cells were transfected with the indicated plasmids in the presence or absence of MG132. 24 h after transfection proteins were extracted for Western blotting. B, HCT116 cells were transfected with the indicated plasmids. 24 h after transfection proteins were extracted for Western blotting (IB). IP, immunoprecipitation. C, HCT116 cells were transfected with the indicated plasmids. 24 h after transfection cells were treated with phenformin for another 24 h. Proteins were then extracted for Western blotting.
Ulk1 Is Involved in the 17-AAG-induced Degradation of Several Kinase Clients of Cdc37
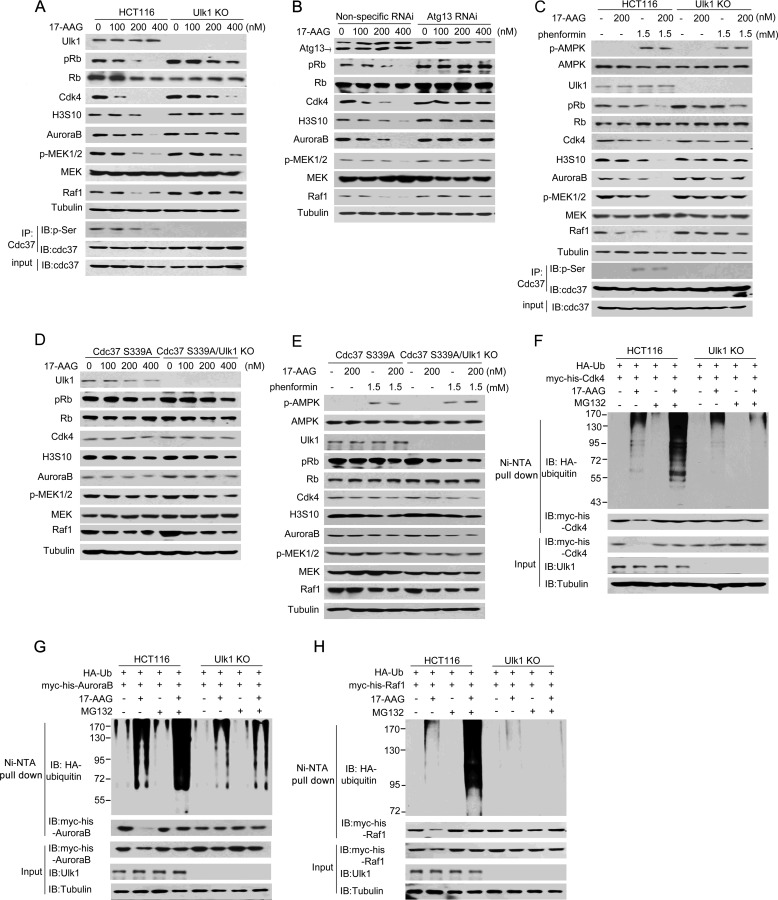
As Cdc37 is required for Hsp90-mediated chaperoning of many clients, we next investigated whether Ulk1 affects Hsp90 inhibitor-mediated abrogation of kinase clients. For this purpose, Ulk1-knock-out HCT116 (Ulk1 KO) cells were stably transfected with wild-type Ulk1 plasmid or an empty plasmid. As shown in Fig. 5A, compared with Ulk1(WT)-expressing HCT116 cells, 17-AAG-induced client degradation was alleviated in Ulk1 KO cells. In the autophagy induction process, the kinase activity of Ulk1 is highly dependent on its binding partner, for example Atg13 (15, 31). We, therefore, knocked down Atg13 in HCT116 cells and then observed a similar reduction of the 17-AAG effect that caused by depletion of Ulk1 (Fig. 5B), pointing to a crucial role of Ulk1 complex in sustaining the 17-AAG effect. Additionally, we also used phenformin to activate AMPK-Ulk1 in HCT116 cells. Consistently, upon 17-AAG treatment, the presence of phenformin induced more obvious degradation of Cdc37's clients in wild-type HCT116 cells but not in Ulk1 KO HCT116 cells (Fig. 5C). Next, to determine whether Cdc37 Ser-339 phosphorylation is a necessary modification for these events, Cdc37(S339A) was stable transfected into Cdc37-KD HCT116 cells. As shown in Fig. 5, D–E, Ulk1-dependent client degradation was almost abolished in Cdc37(S339A)-expressing HCT116 cells, indicating that Ulk1 sustained the 17-AAG effect by inducing Cdc37 phosphorylation.
FIGURE 5.
Ulk1 is involved in the 17-AAG-induced degradation of several kinase clients of Cdc37. A, wild-type Ulk1 or an empty plasmid was stably transfected into Ulk1-knock-out HCT116 (Ulk1 KO) cells. Cells were treated at different concentrations of 17-AAG for 12 h, and proteins were then extracted for Western blot (IB). IP, immunoprecipitation. B, Atg13 RNAi was performed in Ulk1(WT)-expressing HCT116 cells. Cells were treated at different concentrations of 17-AAG for 12 h, and proteins were then extracted for Western blotting. C, wild-type and Ulk1-KO HCT116 cells were treated with the combination of 17-AAG and phenformin for 12 h. Proteins were then extracted for Western blotting. D, RNAi-resistant rescue form of Cdc37(S339) plasmid was transfected into Cdc37-KD or Cdc37-KD/Ulk1-KO HCT116 cells. Cells were treated at different concentrations of 17-AAG for 12 h, and proteins were then extracted for Western blotting. E, RNAi-resistant rescue form of Cdc37(S339) plasmid was transfected into Cdc37-KD or Cdc37-KD/Ulk1-KO HCT116 cells. Cells were treated with the combination of 17-AAG and phenformin for 12 h. Proteins were then extracted for Western blotting. F–H, Ulk1-expressing and Ulk1-KO HCT116 cells were transfected with the indicated plasmids. 24 h after transfection cells were treated for 12 h with MG132 and/or 17-AAG. Then CDK4 (F), AuroraB (G), and Raf-1 (H) conjugated to ubiquitin was purified by using Ni-NTA beads under denaturing conditions and then detected by Western blotting.
Furthermore, to determine whether Ulk1 regulates 17-AAG-mediated kinase degradation via the ubiquitin-proteasome system, HCT116 cells were co-treated with 17-AAG and proteasome inhibitor MG132. As shown in Fig. 5, F–H, less ubiquitin was associated with Cdk4, AuraroB, and Raf-1 in Ulk1 KO cells compared with Ulk1(WT)-expressing HCT116 cells. Taken together, our data indicated that Ulk1 plays a role in the degradation of several kinase clients after Hsp90 inhibition.
Ulk1 Sensitizes Cancer Cells to Hsp90 Inhibitor
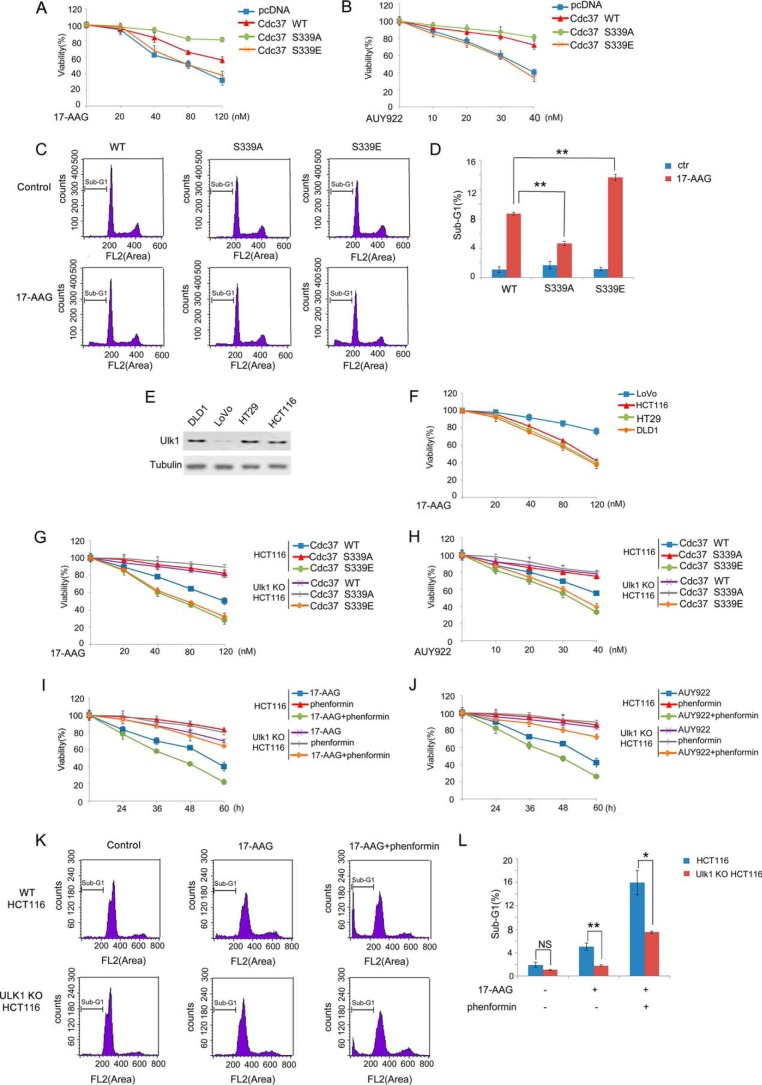
It has been reported that silencing the Cdc37 destabilizes kinase clients and sensitizes cancer cells to Hsp90 inhibitors (32). Therefore, we next detected whether Cdc37 phosphorylation is involved in cellular sensitivity to Hsp90 inhibitors. Wild-type Cdc37, Cdc37-S339E, or Cdc37-S339A were stably transfected into Cdc37-KD HCT116 cells. As shown in Fig. 6A, the cellular sensitivity to Hsp90 inhibitor 17-AAG is much higher in Cdc37-S339E-expressing cells compared with the wild-type Cdc37 or Cdc37-S339A-expressing cells. Similarly, the increased sensitivity was reproduced with another clinical Hsp90 inhibitors AUY922 (33), indicating that the effect was likely general to Hsp90 inhibitors (Fig. 6B). In addition, the cell death population (sub-G1 population) was then measured by using FACS analysis. As shown in Fig. 6, C and D, 17-AAG treatment induced higher sub-G1 cell population in Cdc37-S339E-expressing cells. Collectively, these data suggested that cellular sensitivity to Hsp90 inhibitor is regulated by Cdc37 Ser-339 phosphorylation.
FIGURE 6.
Ulk1 sensitized cancer cells to Hsp90 inhibitor. A and B, a set of RNAi-resistant rescue forms of Cdc37 plasmids was transfected into stable Cdc37-RNAi HCT116 cells. Cells were then treated with 17-AAG (A) or AUY922 (B) at different concentration for 36 h, and cell proliferation was tested with the MTT assay. C and D, a set of RNAi-resistant rescue forms of Cdc37 plasmids were transfected into stable Cdc37-RNAi HCT116 cells. Cells were treated with 100 nm 17-AAG for 72 h, and the cells were then harvested for flow cytometry analysis. The percentage of sub-G1 was shown in D. E, Western blotting was performed to determine Ulk1 expression inhuman colon cancer cell lines DLD1, LoVo, HT29, and HCT116. F, DLD1, LoVo, HT29, and HCT116 cells were treated with 17-AAG at different concentration for 36 h, and cell proliferation was tested with the MTT assay. G and H, a set of the RNAi-resistant rescue form of Cdc37 plasmids was transfected into Cdc37-KD or Cdc37-KD/Ulk1-KO HCT116 cells. Cells were then treated with 17-AAG (G) or AUY922 (H) at different concentration for 36 h, and cell proliferation was tested with the MTT assay. I and J, Wild-type and Ulk1-KO HCT116 cells were treated with 40 nm 17-AAG (I) or 20 nm AUY922 (J) combined with phenformin, and cell proliferation was tested with the MTT assay. K and L, wild-type and Ulk1-KO HCT116 cells were treated with 100 nm 17-AAG combined with phenformin for 72 h, and the cells were then harvested for flow cytometry analysis. The percentage of sub-G1 was shown in L. NS, not significant. *, p < 0.05; **, p < 0.01.
Because Cdc37's cochaperone activity was dampened by Ulk1 induced phosphorylation, we next tested whether Ulk1 is involved in the cancer cell growth inhibitory effects of pharmacological Hsp90 inhibitor. First, we compared the levels of Ulk1 expression in four human colon cancer cell lines including HCT116, DLD1, HT29, and LoVo. Ulk1 was highly expressed in HCT116, DLD1, and HT29 cells, but virtually no expression was detected in LoVo cells (Fig. 6E). As shown in Fig. 6F, Ulk1 levels seemed to be related to the Hsp90 inhibitor sensitivity in that Hsp90 inhibitor decreased cell viability in HCT116, DLD1, and HT29 cells but not in LoVo cells. To further clarify the role of Ulk1 in cancer cell's sensitivity to Hsp90 inhibitor, DLD1 and HCT116 cells were treated with Hsp90 inhibitors. We found that knocking-out Ulk1 decreased the cellular sensitivity to 17-AAG or AUY922 in DLD1 and HCT116 cells (Fig. 6, G and H, and supplemental Fig. 3, A and B). To further determine the effect of Ulk1-induced Cdc37 Ser-339 phosphorylation, Cdc37-WT-expressing and Cdc37-S339A-expressing cells were used. As shown in Fig. 6, G and H, Ulk1-reduced cellular sensitivity to Hsp90 inhibitors is dependent on Cdc37 Ser-339 phosphorylation, indicating the importance of the Ulk1-Cdc37 axis in Hsp90 inhibitor-decreased cell viability.
Because Ulk1 plays important roles in autophagy, we next tried to determine autophagy's role in regulation of 17-AAG sensitivity. As shown in supplemental Fig. 3, C and D, silencing Atg3, another essential gene in autophagy, cannot change the cellular sensitivity to Hsp90 inhibitor, indicating that Ulk1 sensitized cancer cells to 17-AAG independent of autophagy. On the other hand, we found that phenformin, which induces Ulk1 phosphorylation and activation (27), increased the 17-AAG or AUY922 sensitivity in DLD1 and HCT116 cells but not in Ulk1 KO cells (Fig. 6, I–J, and supplemental Fig. 3, E and F). In addition, the combination of phenformin with 17-AAG increased the sub-G1 cell population, indicative of increased cell death (Fig. 6, K–L). Taken together, these data suggested that Ulk1 promotes the anti-proliferative response caused by Hsp90 inhibitor in cancer cells.
Discussion
Although Ulk1 is mainly understood to mediate autophagy, we found here that Ulk1 also has a function to negatively regulate Hsp90-Cdc37 system. We demonstrated that Ulk1 induces Cdc37 Ser-339 phosphorylation and inhibits the cochaperone activity of Cdc37. As a result, Hsp90-Cdc37's client kinases' stability decreased, leading to the hypersensitivity of cancer cells to Hsp90 inhibitors.
Cdc37 is a cochaperone that promotes the association of Hsp90 with the protein kinases to maintain their stability and functions (29, 30). Many of these kinases are oncogenes; therefore, the molecular mechanism of their activation/inactivation is important for cancer therapy. Our results illustrated an Ulk1-regulated-pathway in Hsp90-Cdc37 complex-dependent kinase activation. The results presented here identified Ulk1-mediated Cdc37 phosphorylation as a requirement for release of clients from the Hsp90 chaperone (Fig. 2, B–D). It has been reported that phosphorylation of N-terminal Ser-13 is required for the proper function of Cdc37 (34). This site is phosphorylated by the constitutively active kinase CK2 (35) and dephosphorylated by PP5 (36, 37). Although Ser-13 is responsible for binding client kinases, Cdc37 also interacts with several client kinases (for example B-Raf) dependent on its C terminus (38). A previous study also showed that phosphorylation on Tyr-4 and Tyr-298 of Cdc37 disrupts client-Cdc37 association (39). Here in our study we showed that Ulk1-mediated phosphorylation of Cdc37 is also one of PTMs (post-translational modification) that controls Hsp90-Cdc37 chaperone function by affecting cochaperone and client association. We identified Ser-339 as a new phosphorylation site of Cdc37, which was also important for its activity. Although it is not essential for the interactions between Cdc37 and each kinase client (supplemental Fig. 4), Ser-339 dephosphorylation status is obviously a requirement for several clients' recognition, at least for the kinases CDK4, AuroraB, Src, Cdk6, and Raf1 (Fig. 2A). However, among these kinase only Cdk4, AuroraB, and Raf-1 were diminished by Ulk1 overexpression (Fig. 3B), raising the possibility that Ulk1 may regulate Src and Cdk6 via other pathways besides Cdc37 phosphorylation.
Recently, a growing number of studies have shown that ATG proteins also play a role in autophagy-independent functions (40–43). Besides initiation of autophagy, Ulk1's autophagy-independent functions were also expanded by several reports (17–19). Here in our study, we found that Ulk1 affects many kinases' stability and activity. Moreover, knocking-down of Atg13, an essential gene for the kinase activity of endogenous Ulk1, also reduced the 17-AAG effect on Hsp90-cdc37's client kinase (Fig. 5B). However, no significant change of 17-AAG effect was detected in the Atg3 KD cells (supplemental Fig. 5, A and B). In addition, Ulk1-medicated Cdc37 phosphorylation on Ser-339 was not required for phenformin induced autophagy (supplemental Fig. 5C), indicating that Ulk1 regulates Hsp90-cdc37's clients independent of the downstream autophagic events. Moreover, compared with wild-type cells, cellular sensitivity to Hsp90 inhibitor was not changed in autophagy-deficient cell line, further supporting Ulk1's autophagy-independent role (supplemental Fig. 3, C and D).
Inhibition of mitochondrial Hsp90 activates AMPK and Ulk1/FIP200, which in turn inhibits FAK (focal adhesion kinase)-directed tumor cell motility and metastasis (44). Here we showed that Hsp90 inhibition-induced cytotoxicity is also related with Ulk1. It has been reported that Ulk1 is a client of the Hsp90-cdc37 complex chaperone (24). We also observed that the Ulk1 protein level gradually decreased upon Hsp90 inhibitor treatment, although it was less prominent than other clients such as CDK4 (supplemental Fig. 6). Therefore, we treated colon cancer cells with the combination of Hsp90 inhibitor and Ulk1 activator. The data showed that AMPK-Ulk1 activation promotes the anti-proliferative response caused by Hsp90 inhibitor (Fig. 6, I–L). In contrast to our findings, the Akt-mTOR-Ulk1 signaling pathway was reported to be involved in Hsp90 inhibitor BIIB021-triggered autophagy, which played a cytoprotective role in BIIB021-treated CML cells (45). However, our results showed that silencing of Ulk1 increased the cell viability upon Hsp90 inhibitor treatment in colon cancer cell lines HCT116 and DLD1 cells (Fig. 6, G and H, supplemental Fig. 3, A and B). Moreover, autophagy deficiency did not change the cellular sensitivity to Hsp90 inhibitor (supplemental Fig. 3, C and D). We believed the conflicting results uncovered by our group and others could be explained by the different cell lines used in the respective studies; solid tumor cell lines were used in our study, and leukemia cells were used in the other study.
It has been reported that Hsp90-Cdc37-Akt form a complex and the degradation of Akt is controlled by Hsp90 (46–48). In our study we also tested whether Ulk1 can regulate the Akt level by phosphorylating Cdc37. As shown in supplemental Fig. 7A, we did not observe that Ulk1 had any noticeable effect on Akt or p-Akt in HCT116 cells. Moreover, the human colon cancer cell lines HCT116 and DLD1, which we used to harbor activating PI3KCA mutations, results in a constitutively active form of Akt (49). Therefore, in our system the Akt pathway is unlikely to play an important role in 17-AAG's effect on regulation. Although the mechanism of how Hsp90 inhibitors affect cell viability is very complex, Ulk1-Cdc37 axis apparently acts as an important component in regulating Hsp90 inhibitors' effect.
Experimental Procedures
Cell Culture and Plasmid Transfection
Cells were grown in DMEM or McCoy's 5A with 10% (v/v) fetal bovine serum and the appropriate amount of penicillin/streptomycin in a 37 °C incubator with a humidified 5% CO2 atmosphere. Transient and stable transfections were performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol.
Plasmids and siRNA
cDNA of Cdc37 was amplified and cloned into p3xFLAG-CMV-10, pEGFP-C1, and pGEX-6p1. Ulk1/2, Raf1, Cdk4, and AuroraB DNA were inserted into p3xFLAG-CMV-10. Cdc37 and Ulk1/2 mutation constructs were generated with a Fast mutagenesis kit (Vazyme). Ulk1 Cas9 cell lines were generated by using Ulk1 sgRNA (5′-CGAAGGCGCCGTGGCCGATC-3′): Cdc37 siRNA sequence 5′-CGGCAGUUCUUCACUAAGA-3′; Atg13 siRNA sequence 5′-CCAUGUGUGUGGAGAUUUCACUUAA-3′; Atg3 siRNA sequence 5′-CAUUGAGACUGUUGCAGAA-3′.
Antibodies and Reagents
The antibodies used were anti-HA tag, anti-GFP tag, anti-Myc tag, anti-α-tubulin (MBL); anti-FLAG tag (Sigma); anti-CRAF (Cell Signaling, #9422S), anti-phospho-MEK1/2(Ser-217/221) (Cell Signaling, #9154P), anti-Src (Cell Signaling, #2123P), anti-phospho-Src(Tyr-416) (Cell Signaling, #6943P), anti-Atg13 (Cell Signaling, #13468S), anti-Atg3 (Cell Signaling, #3415S), anti-phospho-AMPK (T172) (Cell Signaling, #2535S), anti-Ulk1 (Cell Signaling, #8054S); anti-phosphoserine (Abcam, #ab9332), anti-AuroraB (Abcam, #ab45145), anti-phospho-H3S10 (Abcam, #ab32107), anti-MRK (Abcam, #ab166895); anti-MOK(Proteintech, #23926-1-AP), anti-MAK (Proteintech, #13638-1-AP), anti-AKT1 (Proteintech, #10176-2-AP), anti-Hsp90(Proteintech, #13171-1-AP); anti-phospho-pRb(Thr-826) (Ruiying, #RLP0556). Phenformin(#S2542), MG132(#S2619), 17-AAG(#S1141), and AUY922(#S1069) were purchased from Selleck.
Co-immunoprecipitation
Cells were harvested and then lysed in lysis buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris at pH 7.5, 5 mm EDTA, 0.05% SDS, and 1% EDTA-free protease and phosphatase inhibitor cocktails (Roche Applied Science)) on ice for 30 min. After centrifugation at 4 °C at 12,000 rpm for 15 min, 2 μg of the indicated antibody was added into the supernatant and incubated at 4 °C overnight. Then, 30 μl of protein G- or A-Sepharose slurry (GE healthcare) was added and incubated for 2 h at 4 °C. The beads were washed by Nonidet P-40 buffer three times. The precipitated components were analyzed by Western blotting.
HA-Ub Immunoprecipitation
The cells were harvested and washed with PBS. After centrifugation at 1000 rpm for 5 min, the cell pellet was resuspended in lysis buffer (50 mm Tris, pH 8.0, 137 mm NaCl, 1 mm EDTA, 1% Triton X-100, 10% glycerol, and 1% EDTA-free protease and phosphatase inhibitor cocktails (Roche Applied Science)) and incubated on ice for 30 min. After centrifugation at 12,000 rpm for 10 min, the supernatant was harvested as a whole cell extract. The pellet was resuspended in buffer S (20 mm Tris, pH 7.4, 150 mm NaCl, and 1% SDS), sonicated for 10 s, and then left on a rocking platform at room temperature for 30 min. The lysate was passed through a 22-gauge needle 30 times to break the DNA. After centrifugation at 12,000 rpm for 10 min, the supernatant was harvested as SDS extract. The SDS extract was diluted 10 times with lysis buffer and then mixed with whole cell extract for immunoprecipitation with anti-FLAG (M2) affinity gel (Sigma). Immunoprecipitations were carried out for 2 h at 4 °C. Immunoprecipitates were washed 5 times with lysis buffer. The beads were then eluted with 3×FLAG peptide (Sigma) for 8 h at 4 °C.
Western Blotting
Equal amounts of proteins (20–150 μg) were size-fractionated by 6–15% SDS-polyacrylamide gel electrophoresis.
Protein Purification
GST fusion proteins were expressed in Escherichia coli induced with isopropyl-d-thio-galactoside and purified by glutathione-Sepharose 4B beads (GE Healthcare) and then washed with TEN buffer (20 mm Tris-HCl, pH 7.4), 0.1 mm EDTA, and 100 mm NaCl). Recombinant His-tagged proteins were expressed in and purified from E. coli by Ni2+-Sepharose affinity (GE Healthcare), and the bound protein was eluted with 250 mm imidazole in PBS and desalted by buffer exchange with PBS. For GST pulldown assays, GST fusion proteins were incubated with His-tagged proteins in TEN buffer. The proteins were incubated at 4 °C overnight. The beads were washed 3 times with TEN buffer and boiled with 2×SDS loading buffer, and the proteins were analyzed by Western blot with an anti-His or anti-GST antibody.
In Vitro Kinase Assay
HCT116 cells were grown, and each dish was transfected with 8 μg of FLAG-Ulk1/2. After 24 h post-transfection, cells were lysed in MLB (10 mm Tris at pH 7.5, 2 mm EDTA, 100 mm NaCl, 1% Nonidet P-40, 50 mm NaF, 1 mm Na3VO4, and 1% EDTA-free protease and phosphatase inhibitor cocktails (Roche Applied Science)). Ulk1/2 proteins were immunoprecipitated with anti-FLAG-tag (Sigma) antibodies and then washed with MLB once and radioimmune precipitation assay buffer (50 mm Tris at pH 7.5, 150 mm NaCl, 50 mm NaF, 1 mm EDTA, 1 mm EGTA, 0.05% SDS, 1% Triton X-100, and 0.5% deoxycholate) twice followed by washing with kinase assay buffer containing 20 mm HEPES at pH 7.4, 1 mm EGTA, 0.4 mm EDTA, 5 mm MgCl2, and 0.05 mm dithiothreitol. For Ulk1/2 autophosphorylation assay, the immunoprecipitated Ulk1 bead was incubated in kinase assay buffer containing 10 μm cold ATP and 2 μCi of [γ-32P]ATP per reaction. For kinase assays with GST-Cdc37 and GST-Cdc37-S339A, GST-Cdc37, and GST-Cdc37-S339A were bacterially purified. The kinase reaction was performed at 37 °C for 30 min, and the reaction was terminated by adding SDS sample buffer and subjected to SDS-PAGE and autoradiography.
Cell Proliferation Assay
Cellular sensitivity to compounds was measured by MTT cell counting kit (Vazyme).
Ni-NTA-agarose Purification
Cells were lysed in buffer A (6 m guanidinium chloride, 0.1 m Na2HPO4, NaH2PO4, pH 8.0, 10 mm imidazole). The cell lysates were incubated with 50 μl of Ni-NTA beads for 3 h at room temperature; this was followed by the intense washing of beads with buffer A and 25 mm Tris-HCl, pH 6.8, 20 mm imidazole. The bound proteins were eluted by 5× SDS sample buffer supplemented with 200 mm imidazole at 100 °C for 5 min, resolved on SDS-PAGE, and detected by the appropriate antibodies.
Author Contributions
R. L. performed most of the experimental work. F. Y. and W. F. performed the quantifications and Western blots. L. Z. and Y. W. assisted in protein purification. K. M., X. L., and L. W. assisted in autophagy-deficient cell line generation. W.-G. Z. supervised the project. Y. Z. designed and supervised experiments and wrote the manuscript. All authors critically read the paper.
Supplementary Material
This work was supported by the National Natural Science Foundation of China (81222028, 81621063, 81472581, 81672712, 91319302, and 31261140372). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. 1–7.
- ATG
- autophagy-related proteins
- Ulk1
- Unc-51-like kinase-1
- Cdc37
- cell division cycle protein 37
- Hsp90
- heat shock protein 90
- AMPK
- adenosine monophosphate activated protein kinase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
References
- 1. Kroemer G., Mariño G., and Levine B. (2010) Autophagy and the integrated stress response. Mol. Cell 40, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaur J., and Debnath J. (2015) Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 16, 461–472 [DOI] [PubMed] [Google Scholar]
- 3. He C., and Klionsky D. J. (2009) Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Navone F., Genevini P., and Borgese N. (2015) Autophagy and neurodegeneration: insights from a cultured cell model of ALS. Cells 4, 354–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mizushima N., Yoshimori T., and Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 [DOI] [PubMed] [Google Scholar]
- 6. Mizushima N. (2010) The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–139 [DOI] [PubMed] [Google Scholar]
- 7. Suzuki K., Kubota Y., Sekito T., and Ohsumi Y. (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12, 209–218 [DOI] [PubMed] [Google Scholar]
- 8. Yan J., Kuroyanagi H., Kuroiwa A., Matsuda Y., Tokumitsu H., Tomoda T., Shirasawa T., and Muramatsu M. (1998) Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem. Biophys. Res. Commun. 246, 222–227 [DOI] [PubMed] [Google Scholar]
- 9. Yan J., Kuroyanagi H., Tomemori T., Okazaki N., Asato K., Matsuda Y., Suzuki Y., Ohshima Y., Mitani S., Masuho Y., Shirasawa T., and Muramatsu M. (1999) Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene 18, 5850–5859 [DOI] [PubMed] [Google Scholar]
- 10. Chan E. Y., Longatti A., McKnight N. C., and Tooze S. A. (2009) Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell Biol. 29, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan E. Y., Kir S., and Tooze S. A. (2007) siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282, 25464–25474 [DOI] [PubMed] [Google Scholar]
- 12. Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., and Mizushima N. (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Young A. R., Chan E. Y., Hu X. W., Köchl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., and Tooze S. A. (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900 [DOI] [PubMed] [Google Scholar]
- 14. Russell R. C., Tian Y., Yuan H., Park H. W., Chang Y. Y., Kim J., Kim H., Neufeld T. P., Dillin A., and Guan K. L. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J. L., Oshiro N., and Mizushima N. (2009) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., and Kim D. H. (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li T. Y., Sun Y., Liang Y., Liu Q., Shi Y., Zhang C. S., Zhang C., Song L., Zhang P., Zhang X., Li X., Chen T., Huang H. Y., He X., et al. (2016) ULK1/2 constitute a bifurcate node controlling glucose metabolic fluxes in addition to autophagy. Mol. Cell 62, 359–370 [DOI] [PubMed] [Google Scholar]
- 18. Konno H., Konno K., and Barber G. N. (2013) Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155, 688–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Joo J. H., Wang B., Frankel E., Ge L., Xu L., Iyengar R., Li-Harms X., Wright C., Shaw T. I., Lindsten T., Green D. R., Peng J., Hendershot L. M., Kilic F., Sze J. Y., Audhya A., and Kundu M. (2016) The noncanonical role of ULK/ATG1 in ER-to-Golgi trafficking is essential for cellular homeostasis. Mol. Cell 62, 491–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scott R. C., Juhász G., and Neufeld T. P. (2007) Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ci Y., Shi K., An J., Yang Y., Hui K., Wu P., Shi L., and Xu C. (2014) ROS inhibit autophagy by down-regulating ULK1 mediated by the phosphorylation of p53 in selenite-treated NB4 cells. Cell Death Dis. 5, e1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao W., Shen Z., Shang L., and Wang X. (2011) Up-regulation of human autophagy-initiation kinase ULK1 by tumor suppressor p53 contributes to DNA-damage-induced cell death. Cell Death Differ. 18, 1598–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Joshi A., Iyengar R., Joo J. H., Li-Harms X. J., Wright C., Marino R., Winborn B. J., Phillips A., Temirov J., Sciarretta S., Kriwacki R., Peng J., Shelat A., and Kundu M. (2016) Nuclear ULK1 promotes cell death in response to oxidative stress through PARP1. Cell Death Differ. 23, 216–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joo J. H., Dorsey F. C., Joshi A., Hennessy-Walters K. M., Rose K. L., McCastlain K., Zhang J., Iyengar R., Jung C. H., Suen D. F., Steeves M. A., Yang C. Y., Prater S. M., Kim D. H., et al. (2011) Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol. Cell 43, 572–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hunter T., and Poon R. Y. (1997) Cdc37: a protein kinase chaperone? Trends Cell Biol. 7, 157–161 [DOI] [PubMed] [Google Scholar]
- 26. Kim J., Kundu M., Viollet B., and Guan K. L. (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Egan D. F., Shackelford D. B., Mihaylova M. M., Gelino S., Kohnz R. A., Mair W., Vasquez D. S., Joshi A., Gwinn D. M., Taylor R., Asara J. M., Fitzpatrick J., Dillin A., Viollet B., Kundu M., Hansen M., and Shaw R. J. (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao M., and Klionsky D. J. (2011) AMPK-dependent phosphorylation of ULK1 induces autophagy. Cell Metab. 13, 119–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caplan A. J., Mandal A. K., and Theodoraki M. A. (2007) Molecular chaperones and protein kinase quality control. Trends Cell Biol. 17, 87–92 [DOI] [PubMed] [Google Scholar]
- 30. Karnitz L. M., and Felts S. J. (2007) Cdc37 regulation of the kinome: when to hold 'em and when to fold 'em. Sci. STKE 2007, pe22. [DOI] [PubMed] [Google Scholar]
- 31. Ganley I. G., Lam du H., Wang J., Ding X., Chen S., and Jiang X. (2009) FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith J. R., Clarke P. A., de Billy E., and Workman P. (2009) Silencing the cochaperone CDC37 destabilizes kinase clients and sensitizes cancer cells to HSP90 inhibitors. Oncogene 28, 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eccles S. A., Massey A., Raynaud F. I., Sharp S. Y., Box G., Valenti M., Patterson L., de Haven Brandon A., Gowan S., Boxall F., Aherne W., Rowlands M., Hayes A., Martins V., Urban F., et al. (2008) NVP-AUY922: a novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 68, 2850–2860 [DOI] [PubMed] [Google Scholar]
- 34. Shao J., Prince T., Hartson S. D., and Matts R. L. (2003) Phosphorylation of serine 13 is required for the proper function of the Hsp90 co-chaperone, Cdc37. J. Biol. Chem. 278, 38117–38120 [DOI] [PubMed] [Google Scholar]
- 35. Miyata Y., and Nishida E. (2004) CK2 controls multiple protein kinases by phosphorylating a kinase-targeting molecular chaperone, Cdc37. Mol. Cell Biol. 24, 4065–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vaughan C. K., Mollapour M., Smith J. R., Truman A., Hu B., Good V. M., Panaretou B., Neckers L., Clarke P. A., Workman P., Piper P. W., Prodromou C., and Pearl L. H. (2008) Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Mol. Cell 31, 886–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oberoi J., Dunn D. M., Woodford M. R., Mariotti L., Schulman J., Bourboulia D., Mollapour M., and Vaughan C. K. (2016) Structural and functional basis of protein phosphatase 5 substrate specificity. Proc. Natl. Acad. Sci. U.S.A. 113, 9009–9014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keramisanou D., Aboalroub A., Zhang Z., Liu W., Marshall D., Diviney A., Larsen R. W., Landgraf R., and Gelis I. (2016) Molecular mechanism of protein kinase recognition and sorting by the Hsp90 kinome-specific cochaperone Cdc37. Mol. Cell 62, 260–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu W., Mollapour M., Prodromou C., Wang S., Scroggins B. T., Palchick Z., Beebe K., Siderius M., Lee M. J., Couvillon A., Trepel J. B., Miyata Y., Matts R., and Neckers L. (2012) Dynamic tyrosine phosphorylation modulates cycling of the HSP90-P50(CDC37)-AHA1 chaperone machine. Mol. Cell 47, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bestebroer J., V'kovski P., Mauthe M., and Reggiori F. (2013) Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic 14, 1029–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J., Kim H. R., Quinley C., Kim J., Gonzalez-Navajas J., Xavier R., and Raz E. (2012) Autophagy suppresses interleukin-1β (IL-1β) signaling by activation of p62 degradation via lysosomal and proteasomal pathways. J. Biol. Chem. 287, 4033–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frémont S., Gérard A., Galloux M., Janvier K., Karess R. E., and Berlioz-Torrent C. (2013) Beclin-1 is required for chromosome congression and proper outer kinetochore assembly. EMBO Rep. 14, 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hwang S., Maloney N. S., Bruinsma M. W., Goel G., Duan E., Zhang L., Shrestha B., Diamond M. S., Dani A., Sosnovtsev S. V., Green K. Y., Lopez-Otin C., Xavier R. J., Thackray L. B., and Virgin H. W. (2012) Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 11, 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Caino M. C., Chae Y. C., Vaira V., Ferrero S., Nosotti M., Martin N. M., Weeraratna A., O'Connell M., Jernigan D., Fatatis A., Languino L. R., Bosari S., and Altieri D. C. (2013) Metabolic stress regulates cytoskeletal dynamics and metastasis of cancer cells. J. Clin. Invest. 123, 2907–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. He W., Ye X., Huang X., Lel W., You L., Wang L., Chen X., and Qian W. (2016) Hsp90 inhibitor, BIIB021, induces apoptosis and autophagy by regulating mTOR-Ulk1 pathway in imatinib-sensitive and -resistant chronic myeloid leukemia cells. Int. J. Oncol. 48, 1710–1720 [DOI] [PubMed] [Google Scholar]
- 46. Basso A. D., Solit D. B., Chiosis G., Giri B., Tsichlis P., and Rosen N. (2002) Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J. Biol. Chem. 277, 39858–39866 [DOI] [PubMed] [Google Scholar]
- 47. Dickey C. A., Koren J., Zhang Y. J., Xu Y. F., Jinwal U. K., Birnbaum M. J., Monks B., Sun M., Cheng J. Q., Patterson C., Bailey R. M., Dunmore J., Soresh S., Leon C., Morgan D., and Petrucelli L. (2008) Akt and CHIP coregulate tau degradation through coordinated interactions. Proc. Natl. Acad. Sci. U.S.A. 105, 3622–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramsey A. J., Russell L. C., Whitt S. R., and Chinkers M. (2000) Overlapping sites of tetratricopeptide repeat protein binding and chaperone activity in heat shock protein 90. J. Biol. Chem. 275, 17857–17862 [DOI] [PubMed] [Google Scholar]
- 49. Samuels Y., Diaz L. A. Jr, Schmidt-Kittler O., Cummins J. M., Delong L., Cheong I., Rago C., Huso D. L., Lengauer C., Kinzler K. W., Vogelstein B., and Velculescu V. E. (2005) Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 7, 561–573 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.