Abstract
Upon T cell receptor stimulation, CD4+ T helper (Th) lymphocytes release extracellular vesicles (EVs) containing microRNAs. However, no data are available on whether human CD4+ T cell subsets release EVs containing different pattern of microRNAs. The present work aimed at filling this gap by assessing the microRNA content in EVs released upon in vitro T cell receptor stimulation of Th1, Th17, and T regulatory (Treg) cells. Our results indicate that EVs released by Treg cells are significantly different compared with those released by the other subsets. In particular, miR-146a-5p, miR-150-5p, and miR-21-5p are enriched, whereas miR-106a-5p, miR-155-5p, and miR-19a-3p are depleted in Treg-derived EVs. The in vitro identified EV-associated microRNA signature was increased in serum of autoimmune patients with psoriasis and returned to healthy levels upon effective treatment with etanercept, a biological drug targeting the TNF pathway and suppressing inflammation. Moreover, Gene Set Enrichment Analysis showed an over-representation of genes relevant for T cell activation, such as CD40L, IRAK1, IRAK2, STAT1, and c-Myb in the list of validated targets of Treg-derived EV miRNAs. At functional level, Treg-derived (but not Th1/Th17-derived) EVs inhibited CD4+ T cell proliferation and suppressed two relevant targets of miR-146a-5p: STAT1 and IRAK2. In conclusion, our work identified the miRNAs specifically released by different human CD4+ T cell subsets and started to unveil the potential use of their quantity in human serum to mark the pathological elicitation of these cells in vivo and their biological effect in cell to cell communication during the adaptive immune response.
Keywords: autoimmunity, cell-cell interaction, extracellular vesicles, lymphocyte, microRNA (miRNA)
Introduction
Similarly to most cell types, cells of the immune system release vesicles of nanometric size (20–200 nm, possibly referred to as nanovesicles). Nanovesicles that are formed by the inward budding and subsequent fusion to the plasma membrane of multivesicular endosomes are more specifically called exosomes. Vesicles of larger size (0.2–1 micron, often referred to as “microvesicles”) can bud directly from the plasma membrane, but their biogenesis is still unclear (for an exhaustive description of vesicle classification and properties, see Ref. 1). Extracellular vesicles (EVs)5 are known to mediate cell to cell communication in the immune system, playing a role in antigen presentation in vivo (2, 3), T lymphocyte stimulation (4), cell killing (5), shuttling of cytokines (6–10), T regulatory cell differentiation (11), induction of antigen specific tolerance (12), and modulation of allograft rejection (13).
In addition to proteins and lipids, a component invariably found in association with EVs is microRNA (miRNA). Although the role of miRNAs as key regulators of gene expression in eukaryotic cells and in cells of the immune system specifically is well recognized (14–16), the biological function of extracellular EV-associated miRNAs has started to be fully explored only in recent time. From the first description in mast cell-derived vesicles (17), miRNAs have been demonstrated to regulate several different immune physiological and pathological processes, such as monocyte to macrophage maturation (18), viral infection (19, 20), formation of the T-B lymphocyte immune synapse (21, 22), cross-talk of dendritic cells for the fine-tuning of antigen presentation (23), and cellular response to endotoxin (24).
CD4+CD25+FoxP3+ regulatory T (Treg) cells are critical for sustaining immunological homeostasis, representing a distinct cell lineage that is committed to suppressive functions (25). A recent study in mouse demonstrated that Treg cells release EVs containing miRNAs that differ from those released by effector T cells. More importantly, Treg cells deficient for Dicer (necessary for miRNA maturation) but also for Rab27 (necessary for vesicle release) show impaired ability to suppress Th1, suggesting that non-cell-autonomous gene silencing, mediated by EV-associated miRNAs, is a required mechanism employed by Treg cells to suppress T cell-mediated disease (26). Nonetheless, this mechanism has not been yet described in humans.
EV-associated miRNAs are believed to mostly function in a paracrine manner, with vesicles traveling very short distances or even passing from one cell to the other upon cell to cell direct interaction, as may be the case for Treg-mediated immune regulation. Nonetheless, EV-associated miRNAs can also travel in blood and thus mirror the activity of releasing cells at a distance. In the past years, blood circulating miRNAs have become the most promising biomarkers for the diagnosis, prognosis, and therapeutic options of a variety of pathological conditions such as cancer (27–29), cardiovascular diseases (30, 31), diabetes (32), liver pathologies (33, 34), and sepsis (35, 36), among others (reviewed in Refs. 37 and 38).
Upon the identification of EV-associated miRNA signature of human CD4+ T lymphocyte subsets (the pro-inflammatory Th1 and Th17 and the immune suppressive Treg cells), our work indicates that Treg-derived EVs have a distinct miRNA profile, are highly enriched with the anti-inflammatory miR-146a-5p, and have the ability to down-modulate miR-146a-5p mRNAs target IRAK2 and STAT1 and inhibit proliferation in EV-target cells. We then decided to evaluate the clinical relevance of the in vitro identified CD4+ T cell-derived miRNA signature by quantifying it in serum of patients with psoriasis, a common chronic relapsing inflammatory cutaneous disease characterized by thickened, scaly skin patches (39). Recent studies have pointed out that psoriasis inflammation is primarily caused by skin-resident pathogenic lymphocytes, in particular lymphocytes Th1, Th17, and their cytokines (TNF-α, IFN-γ, IL-6, IL-8, IL-12, IL-17, IL-22, and IL-23). The activity of these cells may be inhibited by Treg cells, which are pivotal in preventing the autoimmune response against self-antigens and maintaining the cutaneous immunological homeostasis (40). Importantly, miRNAs belonging to the T cell-derived miRNA signature were found significantly increased in sera of patients with acute psoriasis and then returned to healthy donor levels upon effective treatment with etanercept, a biological drug targeting TNF pathway and inflammation. These results suggest that in vitro CD4+ T cell-derived miRNA signature may be quantified to monitor disease activity in the blood of autoimmune patients.
Results
EV-associated miRNAs Contribute to Classify Treg and Th1/Th17 Cell Subsets
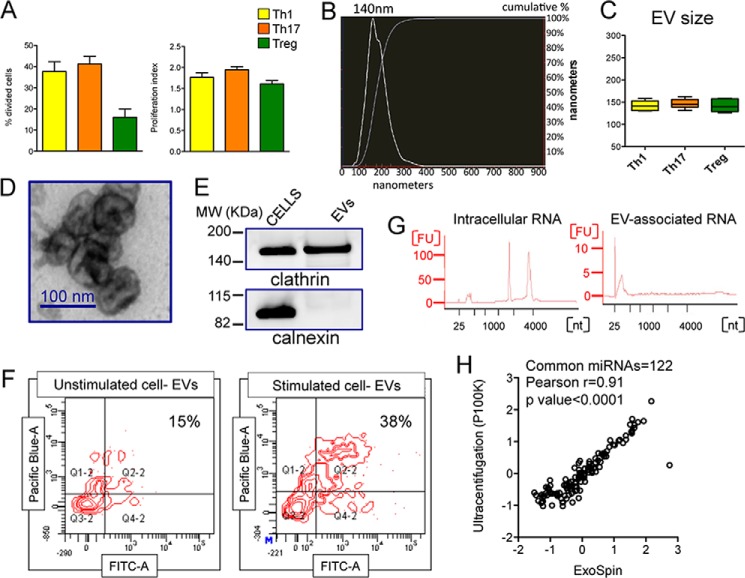
To characterize the EVs released by human CD4+ T helper cell subsets, we purified Th1, Th17, and Treg cells from buffy coats of healthy donors, cultured, and stimulated them with anti-CD3 and anti-CD28. Upon 72 h of stimulation, the different subsets were proliferating at a comparable rate, although a lower percentage of Treg cells was going through division compared with Th1 and Th17, as expected because of their lower responsiveness to in vitro stimulation (Fig. 1A). EVs were then isolated and characterized for size, morphology, and protein markers (Fig. 1, B–E). EV-producing cells were also stained with SYTO RNASelect green fluorescent dye (that stains all RNA molecules in the cell (41)), and purified EVs were directly incubated with Pacific Blue cell Trace, which stains only intact vesicles. CD4+ T cell-derived EVs did prove to have an intact membrane and contain RNA by FACS analysis (Fig. 1F). As already demonstrated for total CD4+ T cells (42), EV-associated RNA from sorted CD4+ T cell subsets was enriched in small molecules compared with intracellular RNA (Fig. 1G). Importantly, the miRNA content of EVs isolated by our elected method of differential precipitation (ExoSpin, Cell Guidance System) was very similar to that of vesicles purified by ultracentrifugation (Fig. 1H).
FIGURE 1.
Characterization of human CD4+ T helper cell-derived extracellular vesicles. A, FlowJo proliferation tool was used to calculate the percentage of divided cells (left panel) and the proliferation index (right panel; n = 6) of CD4+ T cell subsets (as indicated) analyzed for Cell Trace Violet fluorescence by flow cytometry upon 72 h of aCD3-aCD28 stimulation. B, a representative NTA-size distribution of CD4+ T cell-derived EVs. C, whisker plots showing EV size (mode, n = 6) determined by NTA for the indicated subset derived EVs. D, representative image by transmission electron microscopy of CD4+ T cell-derived EVs. E, Western blot from protein lysates from either CD4+ T cells or derived EVs for the indicated proteins. F, FACS analysis of purified EVs released by either unstimulated or stimulated CD4+ T cells previously stained with SYTO RNASelect green fluorescent. Isolated EVs were subsequently stained with Pacific Blue Cell Trace for assessing membrane integrity. G, bioanalyzer qualitative analysis of CD4+ T cell intracellular (left panel) and EV-associated (right panel) RNA. One representative sample is reported. H, correlation of Log10 relative (to miRNA global mean) Quantity values (evaluated by RT-qPCR) for 122 miRNAs (each circle is a single miRNA) coexpressed (Ct < 35) in extracellular vesicles isolated from an expanded Treg cell line conditioned medium by ultracentrifugation (final speed, 100000 × g) versus ExoSpin protocol (mean of 4 samples/group). The p value and Pearson r value are reported.
To analyze miRNA differential expression in EVs released by stimulated T cell subsets, we divided the study into two phases: a discovery phase (number of biological replicates = 5/group), in which EVs were purified by ExoSpin, and a validation phase (number of new biological replicates = 4/group), in which EVs were purified by the alternative commercial kit ExoMiR (based on microfiltration and formerly shown to produce comparable results with ultracentrifugation (42)). The use of two different EV isolation procedures aimed at cross-validating our results based on a total number of nine healthy donors.
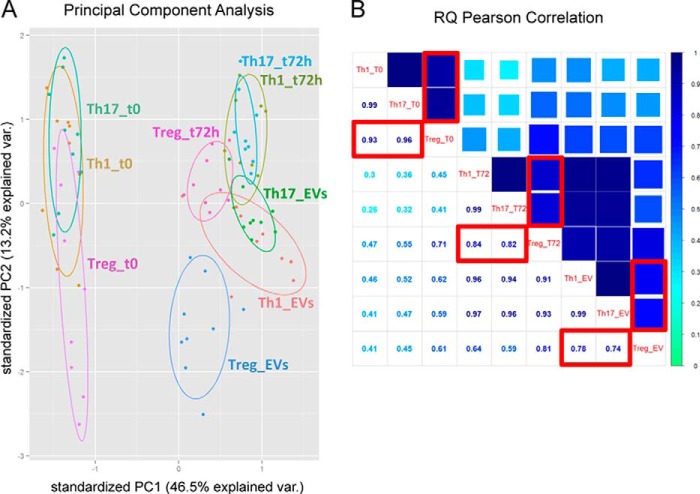
A stringent T cell EV-associated miRNA signature was composed of 26 miRNAs, detectable in all samples independently of the specific isolation method/subset evaluated (Table 1). For each EV sample, we also profiled the miRNA content of resting (immediately after ex vivo isolation) and anti-CD3/anti-CD28 stimulated Th1, Th17, and Treg cells. The principal component analysis (PCA) showed that, in terms of miRNA quantitative levels, samples were divided in three major clusters: (i) resting cells from all subsets, (ii) in vitro stimulated cells from all subsets together with EV samples derived from Th1 and Th17, and (iii) Treg-derived EVs only (Fig. 2A). This result was confirmed by an analysis of correlation with Pearson metric performed on miRNA normalized relative quantities (NRQs): the distance between Treg and Th1/Th17 miRNome was growing when analyzing in vitro stimulated cells compared with ex vivo isolated resting cells and was the highest when looking at EV-associated miRNome (Fig. 2B).
TABLE 1.
miRNAs of CD4+ T cell-derived EVs
Shown is a list of miRNAs (n = 26) found invariably detectable (Ct<35) in all samples (commonly in discovery and validation phase) of CD4+ T cell-derived EVs independently of the subset evaluated/vesicle isolation procedure.
| T cell-derived EV-associated common miRNA signature | ||||
|---|---|---|---|---|
| let-7a-5p | miR-142-5p | miR-19a-3p | miR-25-3p | miR-92a-3p |
| let-7g-5p | miR-146a-5p | miR-20a-5p | miR-29a-3p | miR-93-5p |
| miR-103a-3p | miR-150-5p | miR-21-5p | miR-29b-3p | |
| miR-106a-5p | miR-155-5p | miR-222-3p | miR-29c-3p | |
| miR-1260a | miR-15a-5p | miR-23b-3p | miR-30b-5p | |
| miR-142-3p | miR-16-5p | miR-24-3p | miR-342-3p | |
FIGURE 2.
EV-associated miRNAs contribute to classify Treg and Th1/Th17 cell subsets. A, bi-dimensional score plot resulting by the PCA performed on the miRNA profiling dataset from the entire in vitro CD4+ T cell subset experiment. Each dot represents a sample, and each color represents one experimental group as reported. For each T cell subset (Th1, Th17, and Treg) three conditions are reported: intracellular miRNA profile at time 0 and 72 h after anti-CD3/anti-CD28 stimulation; and vesicle miRNA profile at 72 h (EVs). The percentage of variation explained by each principal component is reported close to the axes. The 95% confidence ellipse is drawn for each experimental group. B, the average correlation between miRNA NRQs values of each in vitro CD4+ T cell subset experimental group is calculated by Pearson Correlation algorithm, and it is shown in the matrix of correlation, where the coefficient of correlation r is reported both numerically and through color code. In the red boxes, the correlation between Treg cells and the other subsets for ex vivo isolated resting cells (upper left), in vitro stimulated cells (middle), and EVs (lower right).
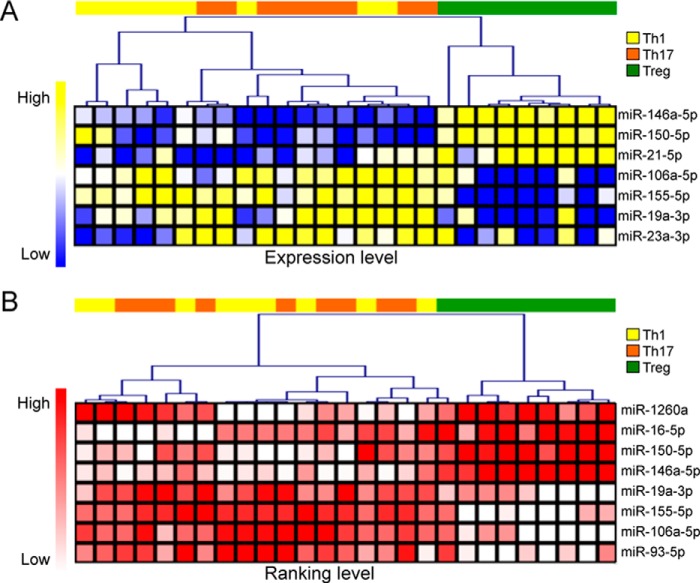
miRNA differential expression in EVs released by the different CD4+ T cell subsets was analyzed by the parametric one-way analysis of variance (ANOVA). We selected miRNAs significant at both discovery and validation phase independently (p < 0.05) and when evaluating the whole dataset (p < 0.01). miR-150-5p, miR-146a-5p, and miR-21-5p were found increased; miR-155-5p, miR-106a-5p, and miR-19a-3p decreased in EVs derived from Treg cells compared with Th1 and Th17 cells (Fig. 3A). ANOVA and post-ANOVA test results are summarized in Table 2. Moreover, the non-parametric Kruskal-Wallis analysis on ranked values (not dependent on any normalization procedure) confirmed the differentially expressed miRNAs, except for miR-21-5p that was ranking as the most expressed miRNA in the majority of samples and thus could not be discriminative in this specific analytical setting (Fig. 3B).
FIGURE 3.
Differential expression analysis of EV-associated miRNAs. Heat maps showing hierarchical clustering of differentially expressed miRNAs (p < 0.05) tested by either ANOVA (A) or Kruskal-Wallis Test (B). For the former analysis, a Pearson metric distance (complete average method) was used on normalized (by miRNA global mean) log-transformed miRNA values; for the latter, a Spearman metric distance was used on ranked Ct data.
TABLE 2.
miRNAs differentially expressed in Treg versus Th1/Th17 cell-derived EVs
Shown are results from ANOVA and Newman-Keuls Multiple comparison post-ANOVA test for miRNAs differentially expressed in Treg versus Th1/Th17 cell-derived EVs. NS, not significant.
| miRNA | T cell-derived EV-associated subset specific miRNA signature |
|||||
|---|---|---|---|---|---|---|
|
p value ANOVA |
Post-ANOVA testing (whole data set) |
|||||
| Discovery | Validation | Whole data set | Th1 vs. Th17 | Th1 vs. Treg | Th17 vs. Treg | |
| miR-106a-5p | 0.0335 | 0.0047 | 4.62E-05 | NS | <0.001 | <0.01 |
| miR-155-5p | 0.0001 | 0.0003 | 1.47E-08 | NS | <0.001 | <0.001 |
| miR-19a-3p | 0.0157 | 0.0011 | 2.03E-05 | NS | <0.01 | <0.001 |
| miR-146a-5p | 0.0013 | 0.00001 | 2.00E-09 | NS | <0.001 | <0.001 |
| miR-150-5p | 0.0068 | 0.0011 | 1.41E-04 | NS | <0.001 | <0.001 |
| miR-21-5p | 0.0074 | 0.00003 | 1.07E-05 | NS | <0.001 | <0.001 |
Treg- and Th1/Th17 Cell-specific EV-associated miRNA Signature Does Not Mirror Differences among Subsets at the Intracellular Level
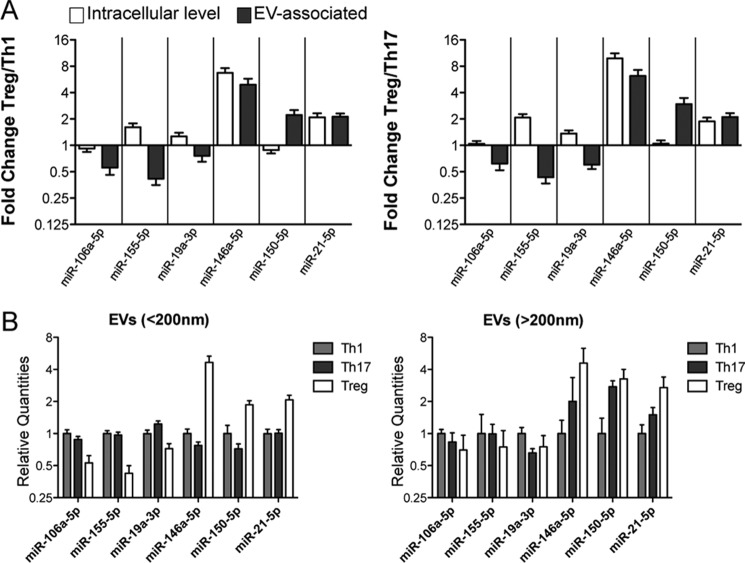
Although miR-146a-5p and miR-21-5p were found to be expressed at higher level not only in Treg-derived EVs but also in Treg compared with Th1 and Th17 at intracellular level (as shown also by other groups (43)), the same was not true for miR-150-5p. Moreover, miR-106a-5p, miR-155-5p, and miR-19a-3p were not expressed at lower levels in Treg compared with Th1 and Th17 cells (Fig. 4A). These results suggest that EV differential expression is actively maintained and does not passively depend on intracellular differences. Furthermore, miRNA differential expression revealed in nano-sized EVs (purified for being smaller than 200 nm) was lost when analyzing miRNA content of larger vesicles (having a diameter >200 nm, purified in parallel with our EVs by ExoMiR through microfiltration, see “Experimental Procedures”), suggesting that Treg versus Th1/Th17 cell-derived extracellular miRNA signature is confined to “exosome-like” vesicles of smaller size (<200 nm) (Fig. 4B).
FIGURE 4.
CD4+ T cell subset-specific EV-associated miRNA signature does not mirror differences among subsets at the intracellular level. A, histogram graphs (means with S.E.) showing the fold change (evaluated by RT-qPCR and normalized relative to global mean) of the indicated extracellular miRNAs when comparing Treg with either Th1 (left panel) or Th17 (right panel) at the intracellular level (in ex vivo purified resting cells, white) compared with EV-associated level (gray). B, column bars (mean with S.E.) showing the relative quantities (evaluated by RT-qPCR and normalized relative to global mean) of the indicated extracellular miRNAs in smaller (<200 nanometers, left panel) versus larger (>200 nanometers, right panel) vesicles. The results were obtained by nine biological replicates.
Significant Modulation of the in Vitro Identified CD4+ T Cell EV-associated miRNA Signature in Sera of Psoriasis Patients
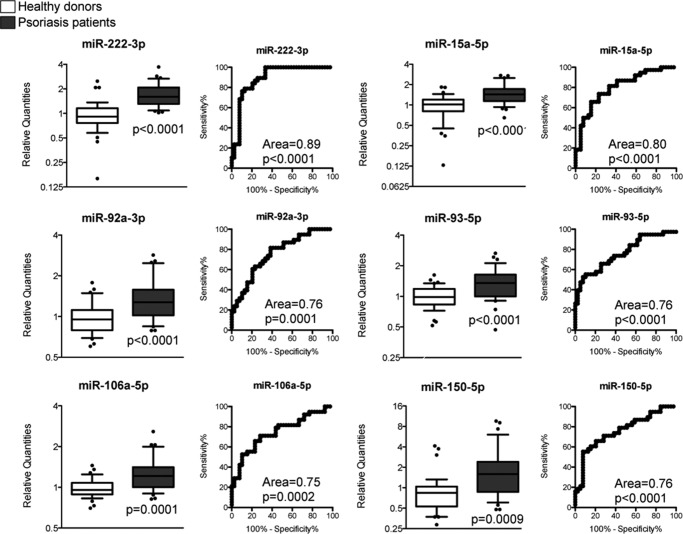
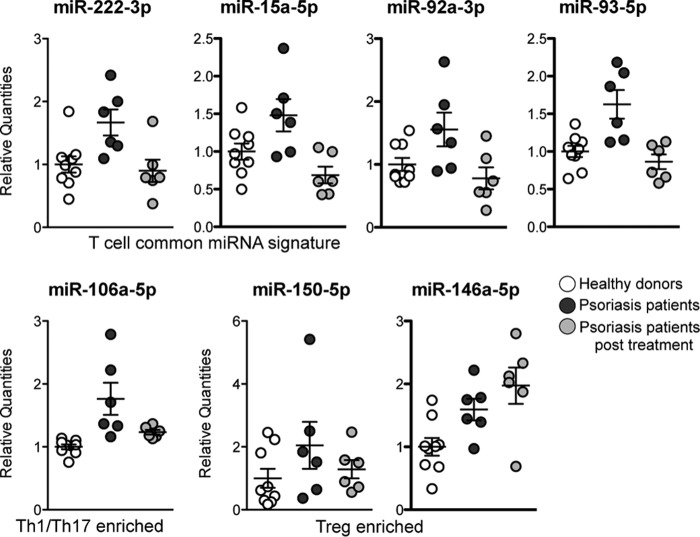
The in vitro identified CD4+ T cell-derived miRNA signature was then quantified in the serum of healthy donors (HDs) and psoriasis patients (PSs, with a psoriasis area severity index (PASI) of >10) as representative of a pathologic condition in which CD4+ T cells are well known to be aberrantly activated (44). To this aim, we constructed an ad hoc designed custom RT-qPCR plate containing the in vitro CD4+ T cell-derived miRNAs (the 26 miRNAs being invariably expressed in CD4+ T cell-derived EVs; Table 1). miRNA quantification in the first set of serum samples (HDs = 4; PSs = 4) (i) showed that the in vitro CD4+ T cell-derived miRNA signature by itself was sufficient to discriminate HDs from PSs as much as whole serum miRNome and (ii) helped us to identify miR-18a-5p as a valuable endogenous normalization factor in our experimental setting (Fig. 5). With our Extracellular CD4+ T Cell-miRNA Signature RT-qPCR plate, we then profiled the rest of the samples, divided in a discovery phase (HD, n = 9; PS, n = 9), in which data were normalized by global miRNA quantity (see “Experimental Procedures” and Ref. 45) followed by a validation phase (HD, n = 29; PS, n = 30) in which data were normalized by the selected endogenous normalization factor miR-18a-5p. We found that six miRNAs, being part of the in vitro CD4+ T cell-derived miRNA signature (but none of the control miRNAs), were consistently up-regulated in serum of PSs compared with HDs (t test < 0.05 in both discovery and validation phase; differential expression was not affected by either gender or age, not shown) and showed an area under the receiving operating characteristic (ROC) curve ≥ 0.75 (Fig. 6). Notably, among these six miRNAs, there were miR-150-5p (that we have previously found increased in serum of flu vaccinated individuals (42)) and miR-106a-5p (that we have here identified as specifically enriched in in vitro Th1/Th17 cell-derived EVs). Furthermore, a subset of PS patients (n = 6) was followed, and serum was analyzed upon effective psoriasis treatment with etanercept (registered as a PASI decrease of ≥ 80%), a biological drug that blocks the TNF-α pathway and suppresses lymphocyte activation and tissue inflammation. miRNAs with a significant ROC curve were found decreased to a quantity comparable with that present in HDs. Differently, in vitro Treg-derived miR-150-5p and miR-146a-5p did not decrease in patients treated with etanercept (or actually showed a trend of increase, as for miR-146a-5p) (Fig. 7, post-test analysis in Table 3)
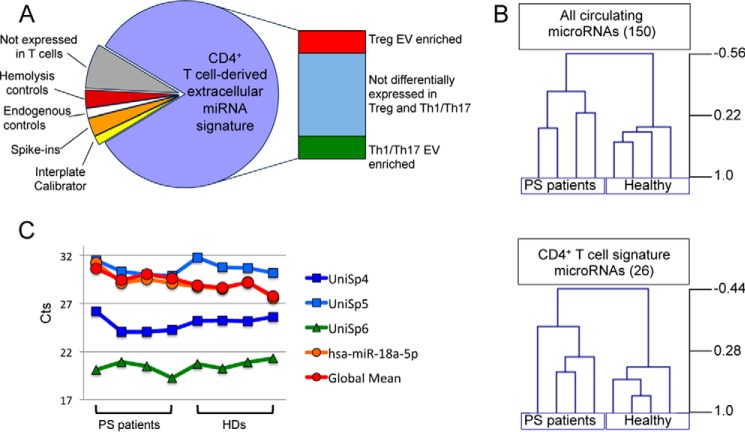
FIGURE 5.
Extracellular T cell-miRNA signature RT-qPCR plate to profile psoriasis patient sera. A, scheme of miRNA representation in the RT-qPCR custom plate used to profile serum circulating miRNome of psoriasis patients. B, unsupervised hierarchical clustering of log-transformed normalized relative quantities of either all detectable serum circulating miRNAs (upper panel) or the 26 miRNAs belonging to the in vitro CD4+ T cell-derived miRNA signature (lower panel). The distance was determined by Pearson correlation with average linkage. C, Ct values variability among 8 samples (four psoriasis patients and four healthy donors) for the exogenous spike-ins added before extraction (in blue: UniSp4 and UniSp5) and before retro-transcription (in green: UniSp6), miRNA global mean (red), and the endogenous normalization factor (in orange: miR-18a-5p).
FIGURE 6.
Significant modulation of miRNAs of the in vitro identified CD4+ T cell subset extracellular signature in serum of psoriasis patients. For each of the six validated serum miRNAs, two panels are reported. Left panels, whisker plots showing the fold change in 39 psoriasis patients (gray) versus 38 healthy donors (white) with p values referring to discovery and validation phase data together. Right panels, ROC curves (showing an area under the curve ≥ 0.75) with the associated p values.
FIGURE 7.
Serum quantity of miRNAs up-represented in psoriasis patients upon effective treatment with etanercept. Shown are serum miRNA quantities in psoriasis patients (dark gray) compared with the same individuals upon effective treatment with etanercept (light gray) and healthy donors (white) n = 9 for HDs and n = 6 for patients. The post-ANOVA test results are reported in Table 3.
TABLE 3.
Newman-Keuls Multiple comparison post-ANOVA test results for sample groups of Fig. 7
miRNAs belonging to T cell subset specific EV-miRNA signature are highlighted in bold. NS, not significant.
| miRNA | HD vs. PS | HD vs. PS (treated) | PS vs. PS (treated) |
|---|---|---|---|
| miR-222-3p | <0.01 | NS | <0.05 |
| miR-15a-5p | <0.05 | NS | <0.01 |
| miR-93-5p | <0.01 | NS | <0.01 |
| miR-92a-3p | <0.05 | NS | <0.05 |
| miR-106a-5p | <0.01 | NS | <0.05 |
| miR-146a-5p | <0.05 | <0.01 | NS |
Specific Molecular Functions Are Defined by Treg-derived miRNA Target Genes
To evaluate the potential regulatory role of Th1/Th17- and Treg-derived EV miRNAs, we selected the mRNAs whose miRNA interaction has been validated in literature as reported by miRSearch V3.0 (Exiqon), obtaining a list of 172 genes as targets of the Treg-derived and 142 of the Th1/Th17-derived EV miRNA signature, and we made sure that publications referenced by miRSearch to select for validated targets did not show any bias toward “immune related processes” when comparing Treg-derived versus Th1/Th17-derived miRNA targets (supplemental Table S1). Using a binomial exact test to determine the over-representation of biological processes reported in Gene Ontology classification categories, we found that for Th1/Th17-derived miRNAs, mRNA targets were enriched in general functions such as “cellular process,” “biological regulation,” and “transcription”; on the other hand, in the list of Treg-derived miRNA targets, “immune system process” and “immune response” were significantly over-represented (data not shown).
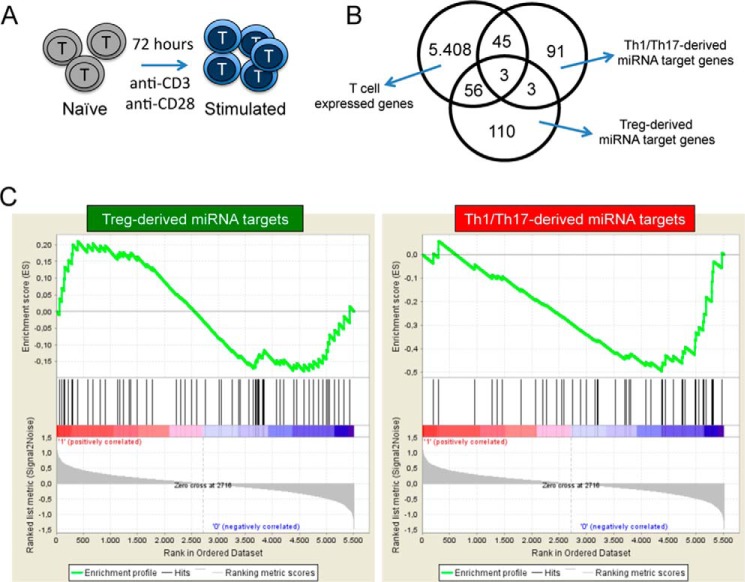
To focus our analysis on the potential effect of Treg-derived miRNAs in suppressing effector CD4+ T cell function, we restricted our attention on genes relevant for the activation of these cells. To this aim, naïve CD4+ T cells purified from healthy donors were in vitro stimulated by plate-bound anti-CD3 and anti-CD28, and gene expression analysis was performed on both resting (ex vivo purified) and stimulated cells (72 h upon stimulation; Fig. 8A). We considered the 5512 genes expressed in all samples. The Venn diagram of Fig. 8B indicates the number of genes belonging to this list that were found in either Treg- (n = 59) or Th1/Th17-derived (n = 48) miRNA target gene set. The Gene Set Enrichment Analysis (GSEA) performed on gene expression modulation upon naïve CD4+ T cell in vitro stimulation showed a significant enrichment of genes up-regulated in the list of validated targets of Treg- (but not Th1/Th17)-derived EV-associated miRNAs, suggesting a suppressive potential of the Treg-derived EVs only (Fig. 8C).
FIGURE 8.
Gene Set Enrichment Analysis. A, schematic model of CD4+ T cell in vitro stimulation. B, Venn diagram showing the overlap between the genes expressed in all samples of CD4+ T naïve and in vitro stimulated cells (n = 5512), the genes that are validated targets of Th1/Th17-derived miRNA signature (n = 142) and Treg-derived miRNA signature (n = 172) (see supplemental Table S1 for gene lists). C, the gene sets obtained by the list of targets of Treg- (left panels) and Th1/Th17-derived (right panels) EV miRNA signature were imposed on the CD4+ T cell in vitro activation gene expression data by GSEA. Genes whose expression levels are modulated the most by CD4+ T cell stimulation get the highest metric scores with positive or negative sign and are located at the left or right edge of the list. At the top, the enrichment score shows a significant enrichment of genes that get up-regulated by CD4+ T cell stimulation in the list of targets of Treg- (but not Th1/Th17)-derived EV miRNA signature.
Biological Effect of Treg-derived EVs
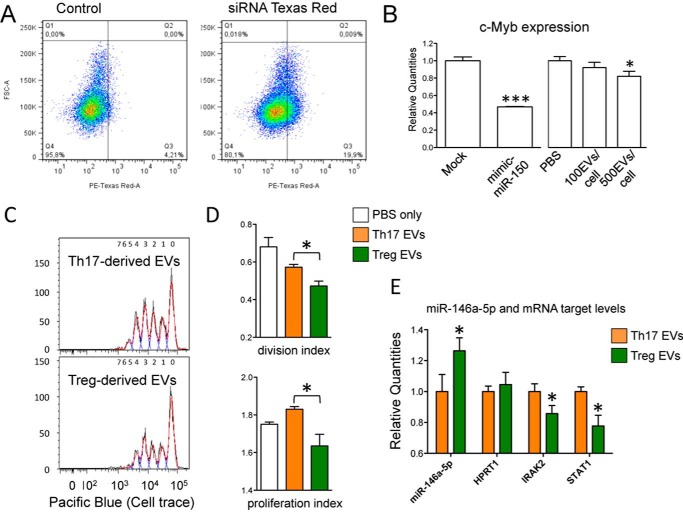
As a proof of concept that EV-associated miRNAs do play a regulatory role in target cells, we started evaluating the effect of Treg-derived EVs (that highly express miR-150-5p) on miR-150-5p validated target c-Myb, in HuH-7 (a hepatocyte derived cellular carcinoma cell line) that do not express this miRNA (average Ct >35, not shown). Treg-derived EVs were detectably picked up by HuH-7 cells and were indeed able to significantly down-modulate c-Myb mRNA quantity at a ratio of 500 EVs/cell (0.82 ± 0.06, p value = 0.03; Fig. 9, A and B). Next, we anti-CD3/anti-CD28-stimulated CD4+ T cells for 96 h in the presence of either Treg- or Th17-derived EVs (and PBS only as a control). Although the percentage of cell division was not significantly different between the different treatments, both division index and proliferation index were found decreased in cells treated with Treg- compared with Th17-derived EVs (0.82 ± 0.03, p value = 0.01 and 0.89 ± 0.03, p value = 0.02, respectively; Fig. 9, C and D). The quantity of miR-146a-5p (high in Treg-derived EVs) relative to a miRNA that is not differentially expressed in T cell subset-derived EVs (miR-15a-5p) was actually higher in cells treated with Treg- compared with Th17-derived EVs (Fig. 9E). We then checked the amount of mRNAs that are (i) validated targets of miR-146a-5p and (ii) known to play a central role in CD4+ T cell activation: CD40L, IRAK1, IRAK2, and STAT1. Notably, two of these targets, IRAK2 and STAT1, were found significantly decreased in cells treated with Treg- compared with Th17-derived EVs (0.86 ± 0.05, p value < 0.05 and 0.82 ± 0.03, p value = 0.02, respectively), whereas an unrelated mRNA (hypoxanthine phosphoribosyltransferase 1) was used as control and found to be unaffected (Fig. 9E). In conclusion, Treg-derived EVs may cooperate with Treg cells themselves in inhibiting T cell proliferation at least in part via the down-modulation of miR-146a-5p targets IRAK2 and STAT1.
FIGURE 9.
Biological effect of Treg-derived EVs. A, FACS analysis showing the uptake of Treg-derived EVs (transfected with positive control siRNA TexRed) by HuH-7 cells upon 20 h of incubation. B, histogram graph (means with S.E.) showing the fold change (evaluated by RT-qPCR and normalized relative to RNA 18S) of c-Myb when comparing Mock and mimic mir-150-5p transfected HuH-7 cells (left panel, n = 4) or PBS and EVs-treated cells (right panel, n = 6). *, p value < 0.05; ***, p value < 0.001. C, Cell Trace Violet fluorescence CD4+ T cells analyzed by flow cytometry upon 96 h of stimulation with CD3/CD28 Dynabeads in presence of either Th17 or Treg-derived EVs. For each group one representative sample is reported. D, FlowJo proliferation tool was used to calculate the percentage of dividing cells, the division index cells and the proliferation index of Cell Trace Violet Fluorescent CD4+ T cells upon 96 h of stimulation with CD3/CD28 Dynabeads in the presence of either Th17- or Treg-derived EVs (n = 4). PBS only was used as negative controls. E, histogram graph (means with S.E.) showing the fold change of miR-146a-5p and miR-146a validated targets STAT1 and IRAK2 in CD4+ T cells stimulated for 96h with CD3/CD28 Dynabeads in the presence of either Th17- (set to 1) or Treg-derived EVs (n = 4). All parameters were evaluated by RT-qPCR and normalized relative to EV commonly expressed miR-15a-5p (miR-146a-5p) and RNA 18S (all mRNAs). *, p value ≤ 0.05.
Discussion
Specialized human CD4+ T helper cell subsets are known to express specific miRNA signatures at the intracellular level (46). In the present work, we have described the miRNA signature associated with EVs released by these subsets. The 26 miRNAs found to be invariably present in CD4+ T cell-derived EVs are also expressed at high level in the intracellular milieu, but the relative enrichment in EVs is different for the different subsets. When comparing Treg and Th1/Th17 subsets, it is at the extracellular (EV-associated) level that Treg miRNome varies the most, suggesting a regulatory mechanism that maintains these significant extracellular differences. In addition to the release of suppressor cytokines, IL-2 consumption, granzyme-mediated cytolysis, and cell-cell interaction through specific surface molecules, Treg cells may make use of EV-associated miRNAs to regulate immune tolerance. Recently, Okoye et al. (26) have shown that non-cell-autonomous gene silencing, mediated by miRNA-containing EVs, is a required mechanism employed by Treg cells to suppress T cell-mediated disease, at least in mouse.
Among the miRNAs that we have found enriched in human Treg-derived EVs, miR-146a-5p stands up as particularly notable, because it is known to target crucial genes that are up-regulated upon stimulation of naïve cells, such as IRAK1, IRAK2, STAT1, and CD40L. The observation that Treg cells keep miR-146a-5p high in the released EVs suggests that miR-146a-5p may participate in Treg-directed suppression also through a cell to cell passage from Treg to Th1/Th17 target cells. This hypothesis finds support in our data shown here that demonstrate how Treg-derived EVs are specifically able to inhibit CD4+ T cell proliferation and down-regulate relevant miR-146a-5p targets, IRAK2 and STAT1. Notably, miR-146a-5p loss in Treg cells has been demonstrated to lead to failure of Treg-directed suppression of pathological Th1 activation (47) and to cause spontaneous autoimmunity in aged mice (48). Although this miRNA is considered to exert these biological effects at the intracellular level, our work now is suggestive of an additional role that miR-146a-5p may be importantly playing in the extracellular milieu by passing from Treg to Treg target cells in association with protective EVs. The fact that Okoye et al. (26) do not find miR-146a either to characterize mouse Treg-derived EVs or to have a role in Treg-triggered suppression does not come as a surprise. Although the global miRNome of T lymphocytes of mice and human is relatively well conserved, a poor concordance between specific T cell subset miRNA signatures of the two species has been described, and significant species divergence may be expected (46). It is also important to point out that even though our study here is focused on the communication between Treg and T effector cells, Treg-derived miRNAs may well be targeting other cells in vivo.
Another interesting miRNA we identified is miR-155-5p that is significantly enriched in EVs released by Th1/Th17 compared with Treg cells. miR-155-deficient mice show less severe asthma, arthritis, experimental autoimmune encephalomyelitis, and reduced response to bacteria, such as Salmonella and Toxoplasma, and this miRNA has been variably associated to inflammatory pathways (reviewed in Ref. 49). The hypothesis originating by our results that Th1/Th17 may augment the level of inflammation also through the release of massive amounts of the pro-inflammatory miR-155-5p surely deserves future further investigation.
Although the EV-mediated cell to cell communication is believed to occur mostly through paracrine mechanisms, EV-associated miRNAs have the ability to travel long distances and become endocrine messages (50). Hence Treg-triggered immune suppression is hypothesized to be primarily happening through the direct cell to cell interaction, but miRNAs released by T cells in pathologic conditions (such as autoimmunity) can be monitored in blood and have the potential to give relevant information about the inflammatory status of the patient.
In our ex vivo approach, it is not possible to demonstrate that miRNAs increased in serum of psoriasis patients have indeed been released by CD4+ T cells in vivo. We do acknowledge that, before it will become feasible to univocally track down the cell origin of blood circulating extracellular miRNAs, we cannot rule out the possibility that there exist a contribution from cells other than CD4+ T lymphocytes as miRNA releasing cells in psoriasis. Our study here is correlating miRNA quantity in serum with the knowledge that CD4+ T effector cells (in particular Th1 and Th17) are dramatically augmented in the skin of psoriasis patients, whereas the proportion of Treg cells is significantly increased upon anti-TNF therapies (51–53), which suggests our results may depend on an augmented number of CD4+ T cells in vivo, an enhanced miRNA release on a single cell basis, or a combination of the two phenomena.
We had previously shown that both B and T lymphocytes release EV-associated miR-150 that increases significantly in the blood of humans after flu vaccination and correlates with a higher immune response (42). Here we have got a step forward and described groups of miRNAs that can be associated to the activation of pro-inflammatory Th1/Th17 versus anti-inflammatory Treg cells. In particular, the relative quantities of serum miR-106a-5p (enriched in in vitro Th1/Th17-derived EVs) and of serum miR-150-5p and miR-146a-5p (enriched in in vitro Treg-derived EVs) may be used as a readout of the inflammatory state in psoriasis patients before and after effective treatment with an anti-TNF-α treatment, such as etanercept.
In conclusion, the identification of miRNAs specifically released by different human CD4+ T helper cell subsets is relevant not only because it uncovers new potential mechanisms of cell to cell communication important for the activation and regulation of the adaptive immune response but also because it renders virtually possible to mark the pathological elicitation of these cells through the ex vivo (blood) assessment of their specific in vitro extracellular miRNA signatures. In the longer run, the possibility of monitoring inflammation through the specific assessment of CD4+ T cell subset specific extracellular miRNA signature is worthy of deeper investigation not only in larger sets of psoriasis patients but also in other autoimmune diseased individuals.
Experimental Procedures
CD4+ T Cell Subset Purification and Stimulation
Human CD4+ T cells have been isolated from peripheral blood mononuclear cells using CD4 MultiSort kit (Miltenyi Biotec), and then specific CD4+ T cell subsets (Th1, Th17, and Treg) were purified by sorting on a FACSAria (BD) by various combinations of surface markers: Th1 (CD127+, CXCR3+, CCR6−), Th17 (CD127+, CCR6+, CXCR3−), and Treg (CD127dim, CD25+). T cell subsets were then cultured separately in AIM V medium (Life Technologies) with plate-bound anti-CD3 and anti-CD28 antibodies (Biolegend). To monitor proliferation upon stimulation, the cells were stained with Cell Trace Violet (Life Technologies) according to the manufacturer's instructions. We collected cells (0 and 72 h after activation) for RNA extracts and conditioned medium (72 h after activation) for EV isolation. For gene expression analysis, resting naïve CD4+ T cells were purified >95% by negative selection with magnetic beads (Miltenyi) and in vitro stimulated with plastic-bound anti-CD3 and anti-CD28 antibodies in presence of IL-2 (n = 4). Gene expression of naive and in vitro activated CD4+ T cells was analyzed by Illumina direct hybridization assays: cRNA was generated according to the Illumina protocol (Ambion); an Illumina iScan System was used for hybridization and scanning; and data were processed with BeadStudio v.3.
In Vitro Expansion of Naive Treg Cells
Lymphocytes were isolated from buffy coats, obtained from the blood of healthy donors by Ficoll-Paque, and purified by sorting on a FACSAria III (BD) by specific markers of naïve Treg cells (CD4+, CD45RO−, CD62L+, CD25+, CD127dim). Then, purified cells were expanded using anti-CD3 (30 ng/ml, OKT3 clone), IL-2 (200 units/ml), irradiated peripheral blood mononuclear cells as feeders, and irradiated EBV cell lines. IL-2 was replenished every 3 days. On day 15, expanded cells were cultured in AIM V medium (Life Technologies) and stimulated with plate-bound anti-CD3 and anti-CD28 antibodies (2 μg/ml; Biolegend). Conditioned media were collected at 72 h, and EVs were isolated using differential centrifugation or ExoSpin.
Extracellular Vesicle Isolation
Conditioned medium was centrifuged to eliminate floating cells (300 × g) and dead cells (2,000 × g). Then three different EV isolation methods were used: differential centrifugation, ExoMir Mini kit (Bioo Scientific), and ExoSpin kit (Cell Guidance systems). For differential centrifugation, larger vesicles were eliminated by a centrifugation at 10,000 × g, and the final supernatant was then ultracentrifuged at 100,000 × g (Beckman Coulter Swinging-Bucket rotor SW 60 Ti, polyallomer tubes) for 70 min to pellet the nanovesicles. The pellet was then resuspended, filtered through a 0.2-micron filter to eliminate residual larger particles, and centrifuged one last time at the same high speed. For microfiltration (ExoMir kit), supernatants were digested by proteinase K to eliminate protein complexes and then passed over two filters connected in series (top filter pore size of ∼200 nm, bottom filter pore size of 20 nm). After washing the filters, they were disconnected. The bottom filter was flushed with 1 ml of BiooPure-MP plus isolation spike-in (Sp2, Sp4, Sp5 Exiqon) to lyse the vesicles of nanometric size, whereas the top filter was flushed to lyse the vesicles of micrometric size (Micro-EV). For differential precipitation (ExoSpin kit), supernatants after a centrifugation at 20,000 × g were used to directly precipitate nanovesicles.
EV Characterization
For Western blotting, isolated EVs were lysed in 10 mm Tris, pH 7, 50 mm NaCl, 30 mm sodium pyrophosphate, 50 mm NaF, 5 μm ZnCl2, 0.1 mm, 1% Triton, protease inhibitor. After protein quantification by BCA assay, 5 μg of protein extracts (cells or EVs) were loaded on 4–12% Bolt Bis-Tris gel (Life technologies) and transferred onto nitrocellulose membranes using iBlot Dry Blotting System (Life Technologies). The membranes were blocked with 5% milk and incubated with specific primary antibodies (rabbit to calnexin ab22595 and rabbit to clathrin ab21679; Abcam) over night at 4 °C. Bound antibodies were detected with anti-rabbit IgG HRP-linked secondary antibody (Cell Signaling) and visualized by Amersham BiosciencesTM ECLTM (GE Healthcare). For electron microscopy, EV visualization by transmission electron microscope was performed according to Thery et al. (54) at Dipartimento di Medicina Sperimentale, Department of Experimental Medicine, University of Genoa. For nanoparticle tracking analysis (nanoparticle tracking analysis (NTA)), EV sizing and counting were performed by NTA (Malvern NanoSight) in collaboration with the Department of Clinical Sciences and Community Health, University of Milan and IRCCS Ca' Granda Policlinico Ospedale Maggiore (Milan, Italy). For flow cytometry, CD4+ T cells were stained with SYTO RNASelect green fluorescent cell stain (Life Technologies) according to the manufacturer's protocol for in vivo labeling of RNA and treated with IL-2 (20 units/ml), phorbol 12-myristate 13-acetate (25 ng/ml), and ionomycin (1 μg/ml). After 72 h, isolated EVs were labeled with Cell Trace Violet and analyzed on a FACS Canto (BD) calibrated with nanometric fluorescent beads (FluoSpheres size kit 2; Life Technologies).
Cell Treatment with EVs
For Huh-7 cells, reverse transfection of Huh-7 cells with 20 nm miscript mir-150 mimic (Qiagen) was performed according to Lipofectamine RNAiMax (Invitrogen) protocol. In other experiment, Huh-7 cells were treated with EVs isolated from CD4+ T cell conditioned medium and transfected with positive control siRNA TexRed, miscript mir-150 mimic, or mirVana mir-150-5p inhibitor (Exo-Fect exosome transfection reagent; System Biosciences). EV to cell ratio, 2000:1. For CD4+ T cells, CD4+ T cells were plated in 8-well ibiTreat microscopy chamber (450,000/well) and incubated with double-labeled (Sytogreen/Cell Trace) EVs (EV to cell ratio, 200:1). After 24 h, the cells were labeled with DAPI to stain nuclei and fixed with cold methanol. The images were obtained using inverted microscope TI-E Eclipse Nikon 100× (NA 1.5).
Suppression Assay
CD4+ T cells were stained with Cell Trace Violet, activated with Dynabeads human T-activator CD3/CD28 (Invitrogen) at a bead to cell ratio of 0.1:1 in presence/absence of EVs derived from Th17 or Treg (125 EVs/CD4+ cell). PBS was used as negative control. Proliferation was assessed by FACS at 96 h.
Human Serum Samples
Blood was collected from 38 healthy donors (maximum age, 60 years) at the IRCCS Ca' Granda Ospedale Maggiore Policlinico (Milan, Italy). After eliminating the clot, the serum was centrifuged at 3000 × g to obtain cell free starting material.
Psoriasis Patients
Moderate to severe psoriasis patients (followed by Professor Emilio Berti's team at the Dermatology Department of Fondazione IRCCS Ca' Granda-Ospedale Maggiore Policlinico, Milan, Italy) were selected for being 18 years old or older (n = 39); presenting a PASI of >10; not responding to conventional therapy and being not treated with systemic drugs for at least a month before enrollment. A group of psoriasis patients (n = 6) was successfully treated with etanercept, a fusion protein acting as a TNF-α inhibitor.
RNA Extraction and miRNA Profiling
Serum RNA was extracted using miRCURY RNA isolation kit Biofluids (Exiqon) according to the manufacturer's instructions. RNA from cells was extracted using either miRVana miRNA isolation kit (Ambion) or using miRCURY RNA isolation kit (Exiqon). EV RNA was extracted as specified in ExomMir kit protocol or using miRCURY RNA isolation kit (Exiqon) and evaluated by Agilent Bioanalyzer. Because the RNA yield from small serum volumes (i.e. 200 μl) or from EV was usually below the limit of accurate quantitation, we used a fixed volume of eluted RNA sample as input for reverse transcription reaction (Universal cDNA synthesis kit II; Exiqon). 752 miRNAs were then profiled by using either the complete human miRNome PCR panels I and II, V3 (Exiqon) or our designed custom plates (Pick & Mix microRNA PCR panel; Exiqon).
miRNA RT-qPCR Data Analysis
The obtained data were preprocessed and quality controlled with GenEx software (Exiqon) and analyzed using MultiExperimentViewer, GraphPad Prism and specific Bioconductor packages in R statistical environment (i.e. ggplot2, H. Wickham; ggplot2, Elegant Graphics for Data Analysis; Springer-Verlag; HTqPCR (55), limma (56), corrplot). We considered only miRNAs whose Ct (threshold cycle) was below 35 in all samples (45). Global mean was used for normalization and NRQ calculation (NRQ = 2 − ΔCT), and sample similarity was described by Pearson correlation, multivariate PCA, and Spearman's rank correlation coefficient on raw Ct data. For differential expression, EV-associated miRNA quantities were analyzed by one-way ANOVA (p value< 0.01) on NRQ data and, in parallel by non-parametric Kruskal-Wallis tests on ranked raw Ct data.
Target Prediction and Functional Analysis
miRNA putative targets were predicted using TargetScan algorithm (57, 58) and further selected for targets whose miRNA interaction is supported by experimental evidence, as reported by miRSearch v3.0 web tool (Exiqon). On the Exiqon website a list of reference for each interaction is available. miRNA targets were then classified by the PANTHER (Protein Analysis th rough Evolutionary Relationships) Gene Ontology System (59, 60), and a binomial distribution test was used to compare them to a reference list representing the experimental T cell genomic profile to statistically determine over- or under-representation of the ontology categories. We used GSEA (61) to analyze the T cell activation gene expression data ranking genes on the base of their expression values and their fold change after activation.
Throughout the ranked list, we evaluated the distribution of gene sets belonging to the 1910 C7 MSigDB immunologic signatures (n = 1910) collection, composed of gene sets representing cell types, states, and perturbations within the immune system (Human Immunology Project Consortium). The distribution of gene sets including interesting miRNA targets was also investigated calculating the enrichment score of each gene. The relationship between the Treg-derived EV miRNA targets with a positive enrichment score in activated T cells was further explored by both STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) and GeneMANIA (62, 63).
Ethics Statement
The use of buffy coat from healthy donors for research purposes has been approved by the Ethics Committee of IRCCS Ca' Granda Policlinico Ospedale Maggiore (Milan, Italy). The use of sera of psoriasis patients has been independently approved by Ethics Committee of IRCCS Ca' Granda Policlinico Ospedale Maggiore (Milan, Italy). Written informed consent regarding study participation was obtained from all involved individuals.
Author Contributions
A. T., E. B., M.-C. C., M. M., P. G., G. R., M. H., C. G., M. Par., J. G., and P. d. C. performed experiments; D. C., R. L. R., and M. Pag. performed bioinformatics analysis; L. C., L. V., and E. B. supervised the clinical study and provided human samples; D. D. V., V. B., and C. T. performed and supervised the EV characterization; and G. M., S. A., and P. d. C. planned the experiments, supervised the study, and wrote the paper.
Supplementary Material
Acknowledgments
We thank Chiara Zingaretti, Veronica De Rosa, and Gaia Spinetti for helpful discussions and Dr. Angelo Cattaneo (chief of the psoriasis center) and Dr. Simona Tavecchio (Policlinico Ospedale Maggiore, Milan, Italy) for collaboration. We also thank Fabio Grassi for careful revision of the manuscript and for kind support.
This work was supported by funds from the European Foundation for the Study of Diabetes/Juvenile Diabetes Research Foundation/Lilly program 2015 (to G. M. and P. d. C.); by Institute Merieux Grant 2009-3603 and European Research Council Advanced Grant 269022 (to S. A.); and by Fondazione Invernizzi. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- EV
- extracellular vesicle
- Th
- T helper
- Treg
- T regulatory
- miRNA
- microRNA
- PCA
- principal component analysis
- NRQ
- normalized relative quantity
- ANOVA
- analysis of variance
- HD
- healthy donor
- PS
- psoriasis patient
- PASI
- psoriasis area severity index
- qPCR
- quantitative PCR
- ROC
- receiving operating characteristic
- GSEA
- Gene Set Enrichment Analysis
- NTA
- nanoparticle tracking analysis.
References
- 1. Raposo G., and Stoorvogel W. (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., and Geuze H. J. (1996) B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blanchard N., Lankar D., Faure F., Regnault A., Dumont C., Raposo G., and Hivroz C. (2002) TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/ζ complex. J. Immunol. 168, 3235–3241 [DOI] [PubMed] [Google Scholar]
- 4. Wahlgren J., Karlson Tde L., Glader P., Telemo E., and Valadi H. (2012) Activated human T cells secrete exosomes that participate in IL-2 mediated immune response signaling. PLoS One 7, e49723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor D. D., Akyol S., and Gercel-Taylor C. (2006) Pregnancy-associated exosomes and their modulation of T cell signaling. J. Immunol. 176, 1534–1542 [DOI] [PubMed] [Google Scholar]
- 6. MacKenzie A., Wilson H. L., Kiss-Toth E., Dower S. K., North R. A., and Surprenant A. (2001) Rapid secretion of interleukin-1β by microvesicle shedding. Immunity 15, 825–835 [DOI] [PubMed] [Google Scholar]
- 7. Qu Y., Franchi L., Nunez G., and Dubyak G. R. (2007) Nonclassical IL-1β secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J. Immunol. 179, 1913–1925 [DOI] [PubMed] [Google Scholar]
- 8. Kandere-Grzybowska K., Letourneau R., Kempuraj D., Donelan J., Poplawski S., Boucher W., Athanassiou A., and Theoharides T. C. (2003) IL-1 induces vesicular secretion of IL-6 without degranulation from human mast cells. J. Immunol. 171, 4830–4836 [DOI] [PubMed] [Google Scholar]
- 9. Gulinelli S., Salaro E., Vuerich M., Bozzato D., Pizzirani C., Bolognesi G., Idzko M., Di Virgilio F., and Ferrari D. (2012) IL-18 associates to microvesicles shed from human macrophages by a LPS/TLR-4 independent mechanism in response to P2X receptor stimulation. Eur. J. Immunol. 42, 3334–3345 [DOI] [PubMed] [Google Scholar]
- 10. Zhang H. G., Liu C., Su K., Yu S., Zhang L., Zhang S., Wang J., Cao X., Grizzle W., and Kimberly R. P. (2006) A membrane form of TNF-α presented by exosomes delays T cell activation-induced cell death. J. Immunol. 176, 7385–7393 [DOI] [PubMed] [Google Scholar]
- 11. Wang G. J., Liu Y., Qin A., Shah S. V., Deng Z. B., Xiang X., Cheng Z., Liu C., Wang J., Zhang L., Grizzle W. E., and Zhang H. G. (2008) Thymus exosomes-like particles induce regulatory T cells. J. Immunol. 181, 5242–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karlsson M., Lundin S., Dahlgren U., Kahu H., Pettersson I., and Telemo E. (2001) “Tolerosomes” are produced by intestinal epithelial cells. Eur. J. Immunol. 31, 2892–2900 [DOI] [PubMed] [Google Scholar]
- 13. Pêche H., Renaudin K., Beriou G., Merieau E., Amigorena S., and Cuturi M. C. (2006) Induction of tolerance by exosomes and short-term immunosuppression in a fully MHC-mismatched rat cardiac allograft model. Am. J. Transplant 6, 1541–1550 [DOI] [PubMed] [Google Scholar]
- 14. He L., and Hannon G. J. (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 15. Xiao C., and Rajewsky K. (2009) MicroRNA control in the immune system: basic principles. Cell 136, 26–36 [DOI] [PubMed] [Google Scholar]
- 16. Bronevetsky Y., and Ansel K. M. (2013) Regulation of miRNA biogenesis and turnover in the immune system. Immunol. Rev. 253, 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., and Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 18. Ismail N., Wang Y., Dakhlallah D., Moldovan L., Agarwal K., Batte K., Shah P., Wisler J., Eubank T. D., Tridandapani S., Paulaitis M. E., Piper M. G., and Marsh C. B. (2013) Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 121, 984–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pegtel D. M., Cosmopoulos K., Thorley-Lawson D. A., van Eijndhoven M. A., Hopmans E. S., Lindenberg J. L., de Gruijl T. D., Würdinger T., and Middeldorp J. M. (2010) Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Narayanan A., Iordanskiy S., Das R., Van Duyne R., Santos S., Jaworski E., Guendel I., Sampey G., Dalby E., Iglesias-Ussel M., Popratiloff A., Hakami R., Kehn-Hall K., Young M., Subra C., et al. (2013) Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J. Biol. Chem. 288, 20014–20033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mittelbrunn M., Gutiérrez-Vázquez C., Villarroya-Beltri C., González S., Sánchez-Cabo F., González M. Á., Bernad A., and Sánchez-Madrid F. (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gutiérrez-Vázquez C., Villarroya-Beltri C., Mittelbrunn M., and Sánchez-Madrid F. (2013) Transfer of extracellular vesicles during immune cell-cell interactions. Immunol. Rev. 251, 125–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Montecalvo A., Larregina A. T., Shufesky W. J., Stolz D. B., Sullivan M. L., Karlsson J. M., Baty C. J., Gibson G. A., Erdos G., Wang Z., Milosevic J., Tkacheva O. A., Divito S. J., Jordan R., Lyons-Weiler J., et al. (2012) Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119, 756–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alexander M., Hu R., Runtsch M. C., Kagele D. A., Mosbruger T. L., Tolmachova T., Seabra M. C., Round J. L., Ward D. M., and O'Connell R. M. (2015) Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 6, 7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohkura N., Kitagawa Y., and Sakaguchi S. (2013) Development and maintenance of regulatory T cells. Immunity 38, 414–423 [DOI] [PubMed] [Google Scholar]
- 26. Okoye I. S., Coomes S. M., Pelly V. S., Czieso S., Papayannopoulos V., Tolmachova T., Seabra M. C., and Wilson M. S. (2014) MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 41, 89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boeri M., Verri C., Conte D., Roz L., Modena P., Facchinetti F., Calabrò E., Croce C. M., Pastorino U., and Sozzi G. (2011) MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 3713–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moussay E., Wang K., Cho J. H., van Moer K., Pierson S., Paggetti J., Nazarov P. V., Palissot V., Hood L. E., Berchem G., and Galas D. J. (2011) MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 108, 6573–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taylor D. D., and Gercel-Taylor C. (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21 [DOI] [PubMed] [Google Scholar]
- 30. D'Alessandra Y., Devanna P., Limana F., Straino S., Di Carlo A., Brambilla P. G., Rubino M., Carena M. C., Spazzafumo L., De Simone M., Micheli B., Biglioli P., Achilli F., Martelli F., Maggiolini S., et al. (2010) Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur. Heart J. 31, 2765–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goretti E., Vausort M., Wagner D. R., and Devaux Y. (2013) Association between circulating microRNAs, cardiovascular risk factors and outcome in patients with acute myocardial infarction. Int. J. Cardiol 168, 4548–4550 [DOI] [PubMed] [Google Scholar]
- 32. Guay C., and Regazzi R. (2013) Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 9, 513–521 [DOI] [PubMed] [Google Scholar]
- 33. Starkey Lewis P. J., Dear J., Platt V., Simpson K. J., Craig D. G., Antoine D. J., French N. S., Dhaun N., Webb D. J., Costello E. M., Neoptolemos J. P., Moggs J., Goldring C. E., and Park B. K. (2011) Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 54, 1767–1776 [DOI] [PubMed] [Google Scholar]
- 34. Chen Y. J., Zhu J. M., Wu H., Fan J., Zhou J., Hu J., Yu Q., Liu T. T., Yang L., Wu C. L., Guo X. L., Huang X. W., and Shen X. Z. (2013) Circulating microRNAs as a fingerprint for liver cirrhosis. PLoS One 8, e66577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J. F., Yu M. L., Yu G., Bian J. J., Deng X. M., Wan X. J., and Zhu K. M. (2010) Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun. 394, 184–188 [DOI] [PubMed] [Google Scholar]
- 36. Roderburg C., Luedde M., Vargas Cardenas D., Vucur M., Scholten D., Frey N., Koch A., Trautwein C., Tacke F., and Luedde T. (2013) Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One 8, e54612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reid G., Kirschner M. B., and van Zandwijk N. (2011) Circulating microRNAs: Association with disease and potential use as biomarkers. Crit. Rev. Oncol. Hematol. 80, 193–208 [DOI] [PubMed] [Google Scholar]
- 38. Wang J., Chen J., and Sen S. (2016) MicroRNA as biomarkers and diagnostics. J. Cell. Physiol. 231, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lowes M. A., Bowcock A. M., and Krueger J. G. (2007) Pathogenesis and therapy of psoriasis. Nature 445, 866–873 [DOI] [PubMed] [Google Scholar]
- 40. Mattozzi C., Salvi M., D'Epiro S., Giancristoforo S., Macaluso L., Luci C., Lal K., Calvieri S., and Richetta A. G. (2013) Importance of regulatory T cells in the pathogenesis of psoriasis: review of the literature. Dermatology 227, 134–145 [DOI] [PubMed] [Google Scholar]
- 41. Hergenreider E., Heydt S., Tréguer K., Boettger T., Horrevoets A. J., Zeiher A. M., Scheffer M. P., Frangakis A. S., Yin X., Mayr M., Braun T., Urbich C., Boon R. A., and Dimmeler S. (2012) Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 14, 249–256 [DOI] [PubMed] [Google Scholar]
- 42. de Candia P., Torri A., Gorletta T., Fedeli M., Bulgheroni E., Cheroni C., Marabita F., Crosti M., Moro M., Pariani E., Romanò L., Esposito S., Mosca F., Rossetti G., Rossi R. L., et al. (2013) Intracellular modulation, extracellular disposal and serum increase of MiR-150 mark lymphocyte activation. PLoS One 8, e75348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhairavabhotla R., Kim Y. C., Glass D. D., Escobar T. M., Patel M. C., Zahr R., Nguyen C. K., Kilaru G. K., Muljo S. A., and Shevach E. M. (2016) Transcriptome profiling of human FoxP3 regulatory T cells. Hum. Immunol. 77, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ghoreschi K., Weigert C., and Röcken M. (2007) Immunopathogenesis and role of T cells in psoriasis. Clin. Dermatol. 25, 574–580 [DOI] [PubMed] [Google Scholar]
- 45. Marabita F., de Candia P., Torri A., Tegnér J., Abrignani S., and Rossi R. L. (2016) Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief. Bioinform. 17, 204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rossi R. L., Rossetti G., Wenandy L., Curti S., Ripamonti A., Bonnal R. J., Birolo R. S., Moro M., Crosti M. C., Gruarin P., Maglie S., Marabita F., Mascheroni D., Parente V., Comelli M., et al. (2011) Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat. Immunol. 12, 796–803 [DOI] [PubMed] [Google Scholar]
- 47. Lu L. F., Boldin M. P., Chaudhry A., Lin L. L., Taganov K. D., Hanada T., Yoshimura A., Baltimore D., and Rudensky A. Y. (2010) Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 142, 914–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boldin M. P., Taganov K. D., Rao D. S., Yang L., Zhao J. L., Kalwani M., Garcia-Flores Y., Luong M., Devrekanli A., Xu J., Sun G., Tay J., Linsley P. S., and Baltimore D. (2011) miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 208, 1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mashima R. (2015) Physiological roles of miR-155. Immunology 145, 323–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cortez M. A., Bueso-Ramos C., Ferdin J., Lopez-Berestein G., Sood A. K., and Calin G. A. (2011) MicroRNAs in body fluids: the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 8, 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Keijsers R. R., Joosten I., Hendriks A. G., Koenen H. J., van Erp P. E., and van de Kerkhof P. C. (2015) Balance of Treg versus T-effector cells during systemic treatment with adalimumab and topical treatment with calcipotriol-betamethasone dipropionate ointment. Exp. Dermatol. 24, 65–67 [DOI] [PubMed] [Google Scholar]
- 52. Cordiali-Fei P., Bianchi L., Bonifati C., Trento E., Ruzzetti M., Francesconi F., Bultrini S., D'Agosto G., Bordignon V., Francavilla V., Tripiciano A., Chiricozzi A., Campione E., Cavallotti C., Orlandi A., et al. (2014) Immunologic biomarkers for clinical and therapeutic management of psoriasis. Mediators Inflamm. 2014, 236060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Quaglino P., Bergallo M., Ponti R., Barberio E., Cicchelli S., Buffa E., Comessatti A., Costa C., Terlizzi M. E., Astegiano S., Novelli M., Cavallo R., and Bernengo M. G. (2011) Th1, Th2, Th17 and regulatory T cell pattern in psoriatic patients: modulation of cytokines and gene targets induced by etanercept treatment and correlation with clinical response. Dermatology 223, 57–67 [DOI] [PubMed] [Google Scholar]
- 54. Thery C., Amigorena S., Raposo G., and Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3, Unit 3.22 [DOI] [PubMed] [Google Scholar]
- 55. Dvinge H., and Bertone P. (2009) HTqPCR: high-throughput analysis and visualization of quantitative real-time PCR data in R. Bioinformatics 25, 3325–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., and Smyth G. K. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lewis B. P., Burge C. B., and Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 58. Garcia D. M., Baek D., Shin C., Bell G. W., Grimson A., and Bartel D. P. (2011) Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 18, 1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mi H., Muruganujan A., and Thomas P. D. (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mi H., Muruganujan A., Casagrande J. T., and Thomas P. D. (2013) Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Warde-Farley D., Donaldson S. L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C. T., Maitland A., Mostafavi S., Montojo J., Shao Q., Wright G., et al. (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.