Abstract
Golgin45 is required for normal Golgi structure and the transportation of protein from the ER. It forms a specific complex with GRASP55 in vivo. Little is known regarding the molecular details of this interaction and its structural role in stacking of the Golgi complex. Here, we present the crystal structure of the GRASP domains of GRASP55 in complex with the Golgin45 C-terminal peptide, determined at 1.33 Å resolution. Similar to the structure of GRASP65 bound to GM130 reported recently, this structure reveals more than one interacting site and involves both PDZ1 and PDZ2 domains of the GRASP simultaneously. The C-terminal peptides of Golgin45 and GM130 present a conserved PDZ domain binding motif sequence and recognize the canonical PDZ-peptide binding groove of the PDZ1 domains of GRASP55 and GRASP65. A main difference in this recognition process resides in a structural rearrangement of GRASP65-GM130 that does not occur for the GRASP55-Golgin45 complex. The binding site at the cleft between the PDZ1 and PDZ2 domains of GRASP65 is dominated by hydrophobic interactions with GM130 that are not observed in the GRASP55-Golgin45 complex. In addition, a unique zinc finger structure is revealed in the GRASP55-Golgin45 complex crystal structure. Mutagenesis experiments support these structural observations and demonstrate that two of these sites are required to form a stable complex. Finally, a novel Golgi stacking model is proposed according to these structural findings.
Keywords: Golgi, metal ion-protein interaction, PDZ domain, protein trafficking (Golgi), protein-protein interaction, zinc finger, BLZF1, GRASP55, Golgi stacking, Golgin45
Introduction
In mammalian cells, the Golgi apparatus forms closely aligned stacked flattened cisternae laterally linked into a ribbon-like structure, and this organelle is required for accurate sequential glycosylation in coordination with protein trafficking and sorting. Golgi peripheral membrane proteins called GRASPs (Golgi reassembly stacking proteins) mediate the stacking of such cisternae structure and ribbon formation (1–3). They have a well conserved N-terminal domain comprised of two PDZ domains in tandem (PDZ1 and PDZ2) and a non-conserved serine- and proline-Rich C-terminal domain with several phosphorylation sites. Two GRASP isoforms are found in vertebrates, namely, GRASP55 and GRASP65. Both GRASPs are anchored to the cisternae membranes of Golgi via a myristoylated glycine at their N terminus (2–4).
GRASP55 and GRASP65 form specific complexes with Golgin45 and GM130 in vivo, respectively. GRASP55 interacts with Golgin45 and is mainly present in the mid-cisternae part of the Golgi, whereas GRASP65 binds cis-Golgin GM130 and is located in the cis-cisternae (5, 6). Both Golgin45 and GM130 are coiled-coil proteins. Although GM130 is involved in vesicle tethering, cell cycle control, centrosome organization, cell signaling, growth control, cell polarization, and directed cell migration, less is known regarding Golgin45. This latter protein was first reported as a leucine zipper nuclear transcription factor named BLZF1 or Jem-1 (7). Golgin45 was then demonstrated to co-localize with GRASP55 on the medial Golgi and to interact specifically with GRASP55 and the GTP-bound form of rab2. Golgin45 is a Golgi matrix protein that is required not only for the normal Golgi structure but also for protein transport from the ER. Knockdown of Golgin45 by RNAi disrupts the Golgi apparatus and blocks the transport of secretory proteins (6). In other reports, Golgin45 was identified as a substrate of tankyrase and RNF146, a RING domain E3 ubiquitin ligase that is important for the activation of Wnt/β-catenin signaling (8). Golgin45 was also demonstrated to interact with DNA (cytosine-5)-methyltransferase 3-like protein DNMT3L and AP-1 (9, 10). Gene profile analysis found that Golgin45 expression was up-regulated in liver cells that responded to dengue virus infection (11), in transcription factor Gfi-1B (growth factor independence 1B) overexpressing leukemia cells (12), and in EBLN1 (Endogenous bornavirus-like nucleoprotein elements) silencing oligodendroglia cells (13), and it was reduced in hepatocellular carcinoma tissues (14). The exact role of Golgin45 still needs further confirmation and investigation.
It is generally accepted that mammalian GRASPs form transoligomers mediated by the transinteraction of the protruding surface of PDZ2 in one GRASP molecule with the PDZ1 peptide binding pocket of another GRASP molecule (15–18). Surprisingly, we recently found, from the structure of GRASP65 bound to a GM130 C terminus peptide, that GM130 interacts with both PDZ1 and PDZ2 domains concurrently (19). In this crystal structure, GM130 forms a canonical PDZ-peptide interaction with the PDZ1 peptide binding pocket rather than PDZ2. Therefore, GRASP molecules cannot cluster through PDZ1 domains when GM130 is present. We thus wondered whether Golgin45 could interact with GRASP55 in a similar manner. Although several crystal structures of the GRASP domains have been recently reported (18, 20), little or no information regarding the molecular recognition of Golgin45 by GRASP55 is available thus far.
Here, we present the crystal structure of GRASP55 GRASP domains in complex with the Golgin45 C-terminal tail. This structure reveals that Golgin45 interacts with GRASP55 in a similar but different manner when compared with the GRASP65-GM130 crystal structure. This interaction also involves both PDZ1 and PDZ2 domains, but no conformational changes are revealed in the GRASP domains of GRASP55. Moreover, an unexpected zinc finger structure is also shown in this crystal structure, indicating an additional role for this interaction. Mutagenesis experiments support these structural observations and demonstrate that two of these sites are crucial for this GRASP55-Golgin45 interaction. These new findings allow us to hypothesize that GRASP55 clustering occurs through its PDZ2 domain rather than the PDZ1-PDZ2 interaction, implying a novel Golgi stacking model.
Results
Overall Structure of GRASP55 in Complex with Golgin45 C Peptide
Crystals of GRASP55 in complex with Golgin45 C terminus peptide were obtained and diffracted to 1.33 Å resolution at the Shanghai Synchrotron Radiation Facility. The space group is C2 (a = 97.68 Å, b = 35.85 Å, c = 64.03 Å, α = 90.0°, β = 93.54°, and γ = 90.0°) (Table 1). The structure was determined by molecular replacement with the program PHASER (21, 22) using a previously reported free form of GRASP55 structure (PDB code 3RLE)2 as a search model. The final model contains one GRASP55 molecule that binds with a Golgin45 C terminus peptide per asymmetric unit. Residues 8–204 of GRASP55 molecule were clearly visible in the electron density map, whereas the last 20 residues (383–403) of the Golgin45 C-terminal peptide were clearly assigned in the structure (Fig. 1A). The bound Golgin45 peptide exhibits a mirrored S-like shape in the complex, and its highly conserved residues among vertebrates (Fig. 1B) exhibit clear electron density. The side chains of several residues not conserved in the peptide are pointing outward in the structure of the complex.
TABLE 1.
Data collection and refinement statistics
| GRASP55-Golgin45 complex | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 97.68, 35.85, 64.03 |
| α, β, γ (°) | 90.0, 93.54, 90.0 |
| Resolution (Å) | 33.65–1.33 (1.377–1.33)a |
| Rmerge | 0.107 (0.587) |
| I/σI | 11.64 (3.08) |
| Completeness (%) | 99.85 (99.35) |
| Redundancy | 7.3 (7.2) |
| CCa | 0.998 (0.974) |
| Refinement | |
| Resolution (Å) | 31.95–1.33 |
| No. unique reflections | 51129 (5043) |
| Rwork/Rfree | 0.137/0.162 |
| No. atoms | 2102 |
| Macromolecules | 1715 |
| Ligand/ion | 1 |
| Water | 386 |
| B-factors | 22.50 |
| Macromolecules | 20.10 |
| Ligand/ion | 15.70 |
| Water | 33.30 |
| RMSDs | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.24 |
| Ramachandran plot | |
| Favored regions (%) | 98 |
| Allowed regions (%) | 2 |
| Outliers (%) | 0 |
a The values in parentheses are statistics for highest resolution shell.
One crystal was used for this data set.
FIGURE 1.
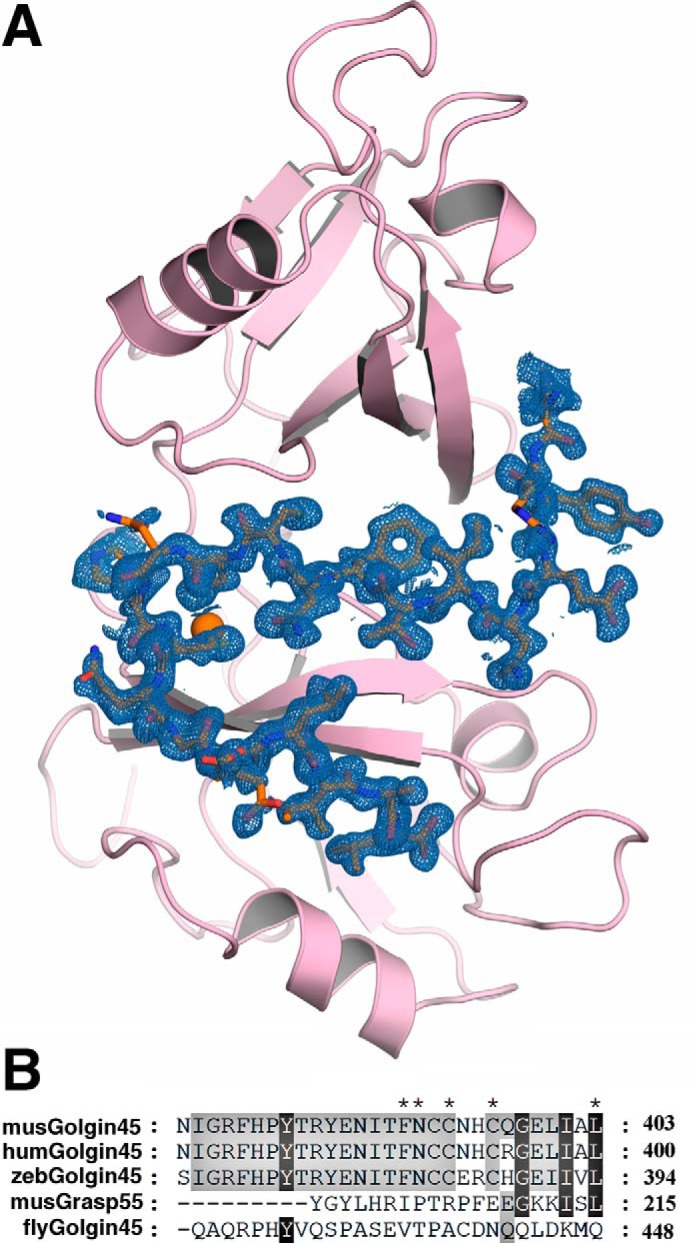
Overall crystal structures of GRASP55 in complex with Golgin45 C-terminal peptide and sequence alignment of Golgin45 C-terminal peptide. A, cartoon representation of the overall structure of GRASP55 (pink) bound to Golgin45 C-terminal peptide (orange). The electron density map of the bound Golgin45 C-terminal peptide is also shown (sky blue, 1.5 σ). B, sequence alignment of Golgin45 C terminus from mouse (NP_001153680), human (NP_003657.1), zebrafish (NP_001138275), Drosophila (NP_649889.1), and GRASP55 (196–215) from mouse.
The 3D conformation of the two PDZ domains are nearly identical to the previously reported free form of the GRASP55 structure. Each of the two PDZ domains exhibited high structural similarity to the canonical prokaryotic PDZ fold with a partially opened β-sandwich capped on each end by two α-helices. Among these domains, the PDZ1 domain forms a well folded, recognizable peptide-binding groove that was formed between the α2-helix and β2-strand. The Golgin45 C terminus peptide binds at this peptide-binding groove and at the cleft between the PDZ1 and PDZ2 domains (Fig. 1A).
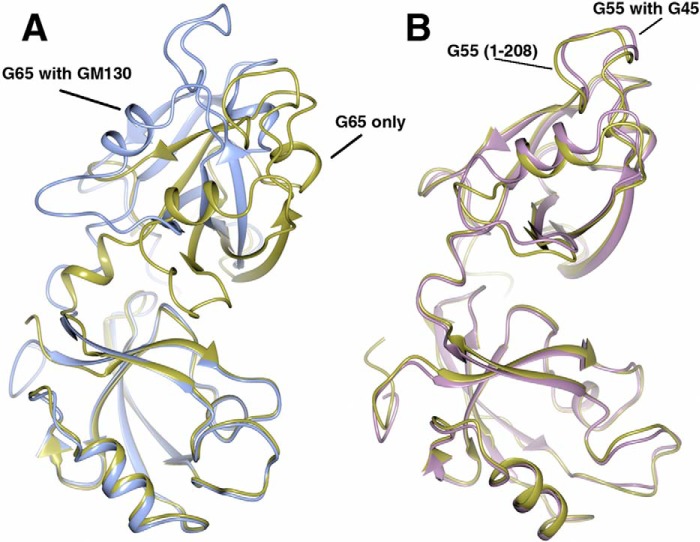
Remarkably, we observed no conformational changes upon Golgin45 C-terminal peptide binding, unlike the ones observed for GRASP65 bound to GM130 (the global RMSD between GRASP65 with and without GM130 binding was over 2.7 Å; Fig. 2A). The structures of GRASP55 (1–208) with and without Golgin45 are virtually superimposable. The global RMSD between the two structures is only 0.8 Å (Fig. 2B). This distance is a major difference in the mode of action of GRASP55 versus GRASP65.
FIGURE 2.
The superimposed structures of GRASP55 before and after Golgin45 C peptide binding. The software Chimera was used to superimpose the structures and generate figures. The RMSD between two structures is calculated using the CCP4 program. A, superimposed structures of GRASP65 with and without GM130 C-terminal peptide using PDZ1 domain as template. Unbound GRASP65 structure (PDB code 4KFV) is colored in gold; bound GRASP65 structure (PDB code 4REY) is colored in blue. B, superimposed structures of GRASP55 from the different PDBs using PDZ1 domain as template. The unbound GRASP55 (1–208, PDB code 3RLE) is colored in gold; GRASP55 from the complex bound with Golgin45 C-terminal peptide (PDB code 5H3J) is colored in lilac.
Golgin45 Mainly Recognizes the Canonical PDZ-Peptide Binding Groove of GRASP55 PDZ1 Domain
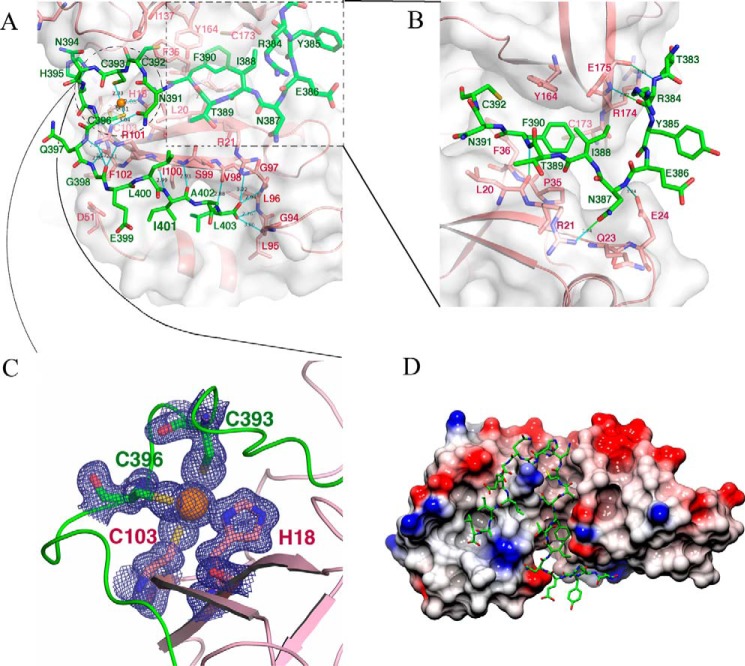
Previous biochemical studies showed that GM130 interacts exclusively with the PDZ2 domain of GRASP65 (5); no similar reports regarding the molecular interaction between GRASP55 and Golgin45 are available in the literature. Thus, it follows that Golgin45 should interact with the second PDZ domain of GRASP55 (15, 17). However, our structure unambiguously shows that Golgin45 interacts with both PDZ domains (PDZ1 and PDZ2) of GRASP55. Indeed, in the structure of the complex, the last C-terminal residues of Golgin45, QGELIAL, insert into the canonical PDZ-peptide binding pocket in the PDZ1 domain and form multiple hydrogen bonds with the β2-strand of GRASP55. The C-terminal residue Leu403 is located at the bottom of the pocket (Fig. 3, A and D), and the carboxylate oxygen atoms of Leu403 form a 4-hydrogen bond network with the amide nitrogen atoms Leu95, Leu96, Gly97, and Val98 of the carboxylate-binding loop, which is well conserved in the canonical PDZ domain. In the carboxylate-binding loop, Gly97 adopts a left-handed conformation and thus cannot be replaced by other amino acids at this position. The side chain of Leu403 is stabilized by Leu55 and Lys56 from the α2 helix of GRASP55 via hydrophobic interactions. The last C-terminal residue of Golgin45 forms a supplementary β-strand with the β2-strand of GRASP55. The main chain of residues from the peptide, Gln397, Gly398, and Ile401, forms 4 hydrogen bonds with the main chain and Ile100 and Phe102 from the β2-strand of GRASP55.
FIGURE 3.
the molecular interactions between GRASP55 and Golgin45 C-terminal peptide. A, detailed interactions between GRASP55 and Golgin45 C-terminal residues at the conventional peptide-binding groove of the PDZ1 domain. The residues involved in the PDZ1-Golgin45 peptide interaction are shown with a stick model in which GRASP55 and Golgin45 peptide backbones are colored pink and green, respectively, and nitrogen and oxygen atoms are colored blue and red, respectively. The surface of GRASP55 PDZ1 domain is also illustrated. A zoom of the area in the dashed square is shown in B, whereas a zoom of the area in the dashed circle is shown in C. B, detailed interactions between GRASP55 and Golgin45 C-terminal peptide at the cleft between PDZ1 and PDZ2 domains. The residues involved in the interaction are shown with a stick model. GRASP55 and Golgin45 peptide backbones are colored pink and green, respectively, and nitrogen and oxygen atoms are colored blue and red, respectively. The GRASP55 surface is also illustrated. C, detailed interactions of the zinc finger formed between GRASP55 and Golgin45. Residues involved in the zinc finger are shown with a stick model. Electron density map of the zinc finger and ion are also illustrated (1.5 σ). Zinc ion, GRASP55 and Golgin45 peptide backbones are colored orange, pink, and green, respectively. D, surface topology of the GRASP domain of GRASP55 highlighting the canonical PDZ-peptide binding pocket of PDZ1 and the cleft between PDZ1 and PDZ2. The C-terminal sequence of the Golgin45 peptide is shown with a stick model and colored green. Electrostatic surface potential is shown too.
Additionally, the crystal structure shows that the Arg101 residue from the β2-strand of the PDZ2 domain of GRASP55 is a critical residue because it interacts simultaneously with two different parts of the peptide. The Arg101 side chain forms three hydrogen bonds with the side chain of the Asn391 residue and the hydroxyl oxygen atom of Gly398 from the Golgin45 peptide. The detailed interactions between Golgin45 and PDZ1 at the binding groove are shown in Fig. 3A.
Golgin45 Also Interacts with the Cleft in GRASP55 between PDZ1 and PDZ2
The upstream residues of the C-terminal sequence of Golgin45, TRYENITFNCCNHC, insert into the cleft between PDZ1 and PDZ2 in GRASP55, interacting with both PDZ domains. Specifically, the hydrophobic side chain of Phe390 is found deeply buried within a hydrophobic cavity that is comprised of residues Pro35/Phe36 from PDZ1 and residues Tyr164/Cys173 from the second PDZ domain of GRASP55 (Fig. 3B). Concomitantly, the Phe390 (amide nitrogen atom) from Golgin45 establishes hydrogen bonds with Leu20 (hydroxyl oxygen) from the first PDZ domain of GRASP55, whereas Arg384 (hydroxyl oxygen and amide nitrogen atoms) establishes hydrogen bonds with Glu175 (amide nitrogen atom and hydroxyl oxygen) from the second PDZ domain of GRASP55. Finally, Asn387 (amide nitrogen atom and oxygen atom of the side chain) forms hydrogen bonds with the side chain of Glu24 (oxygen atom) and the side chain of Arg21 (amide nitrogen atom) from the first PDZ domain of GRASP55. The detailed molecular interaction between Golgin45 and the cleft between PDZ1 and PDZ2 is highlighted in Fig. 3B.
A Zinc Finger-like Structure Is Formed between Golgin45 and the GRASP55 PDZ2 Domain
Notably, there are three conserved cysteines in the C-terminal sequence of Golgin45. Our crystal structure highlights that a zinc finger-like structure is formed between Golgin45 and GRASP55. Cys393/Cys396 of Golgin45 and His18 (β1)/Cys103 (β2) of GRASP55 are involved in coordinating a zinc ion absent in the free form (Fig. 3C). This is the third (and a unique) interaction site between Golgin45 and GRASP55. The presence of a zinc atom was confirmed by an anomalous signal in the structure. The zinc binding is crucial for the interaction of Golgin45 and GRASP55, because alanine residue substitutions in Cys393 and Cys396 disrupt the interaction between the two partners. A previous study reported a zinc finger-like structure in the free form of GRASP65, shaped by residues His17, His19, and Cys102 and a chloride ion (substituted in cysteine) (17). The structure of GRASP65 with the C-terminal sequence of GM130 demonstrated that His18 (His17 in rat), His20 (His19 in rat), and Cys103 (Cys102 in rat) interact directly with GM130 (19); thus, no zinc finger-like structures were formed in this complex. Zinc binding properties of GRASP55 and GRASP65 might thus work by an opposite mode of action. The zinc ion might be needed for Golgin45-GRASP55 complex formation (not for the free GRASP55 protein), whereas the zinc ion would be crucial for the free GRASP65 but not for its complex with GM130. This would imply an opposite role in physiological events related to zinc ion concentration. Nonetheless, this unique binding site identified in Golgin45-GRASP55 complex formation is a major difference between GRASP55 and GRASP65.
Each of the Three Binding Sites Plays a Different Role in GRASP55-Golgin45 Molecular Recognition
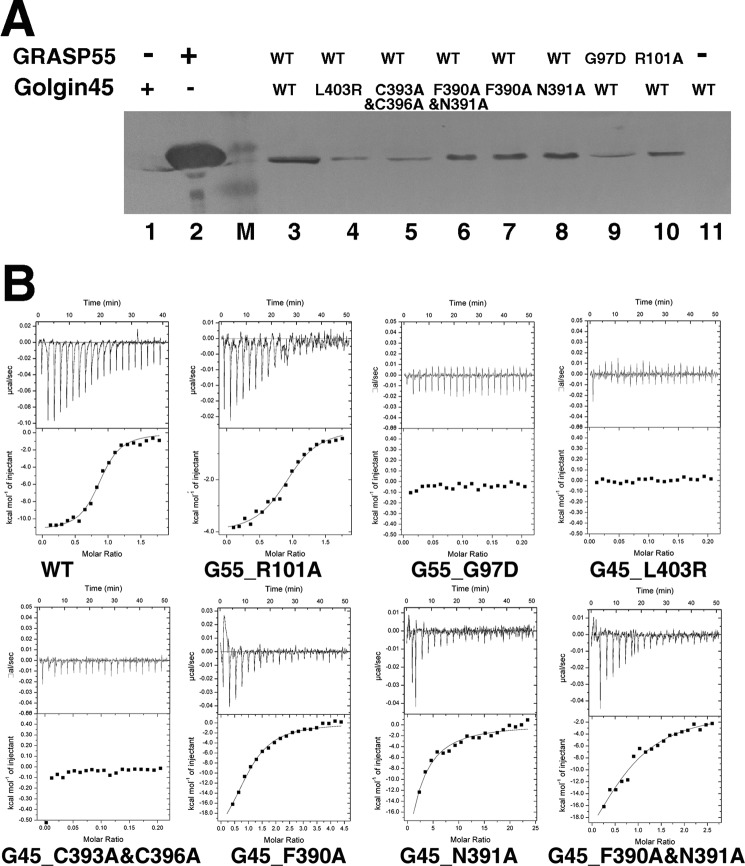
We used isothermal titration calorimetry (ITC) and pulldown assays to assess the relevance of the binding sites observed in our crystal structure. Because the molecular weights of our Golgin45 construct (C-terminal part of Golgin45 fused to a GST tag) and GRASP55 (2–208) with a His6 tag are too similar for SDS-PAGE experiments, we used an anti-His antibody to detect the His tag of GRASP55 with Western blotting and evaluated its binding affinity for different sequences of Golgin45. Pulldown results involving mutations at the C terminus (L403R) or in the zinc binding site (C393A/C396A) of the Golgin45 peptide revealed faint bands with dramatically decreased binding affinity. A similar observation was made when the Golgin45 peptide was kept unchanged, whereas the highly conserved Gly97 of the carboxylate binding loop on PDZ1 was mutated to an aspartic acid. Conversely, mutations in the cleft binding site of Golgin45, either single point mutations (F390A or N391A) or the double mutation (F390A/N391A), exhibited a less dramatic effect. Considering the importance of residue Arg101, a single point mutation R101A was introduced in GRASP55. The pulldown result also revealed a lesser effect of this single point mutation (Fig. 4A).
FIGURE 4.
ITC experiments and pull down analysis of GRASP55-Golgin45 C-terminal peptide interaction. A, Western blotting results obtained from pulldown analysis of GRASP55-Golgin45 C-terminal peptide interaction. Lanes 1 and 2 represent the purified GST-Golgin45 and His6-GRASP55 proteins; lanes 3–8 represent wild type His6-GRASP55 co-purified with the different Golgin45 mutants; lanes 9 and 10 display wild type GST-Golgin45 co-purified with the different GRASP55 mutants; in lane 11 only GST-Golgin45 protein was added. B, ITC binding curves obtained for the binding of the Golgin45 C-terminal peptide and its mutants (C393A & C396A, L403R, F390A, N391A, and F390A & N391A) to GRASP55 and its mutants (G97D and R101A). The top graphs illustrate the raw data, and the y axes indicate the heat released per second during Golgin45 and GRASP55 binding. The bottom graphs illustrate the integrated heat for each injection of GRASP55 together with a fit, whereas the y axes represent the heat released per mole in each injection. The association constants were fitted from the curves obtained by the titration of Golgin45 with GRASP55.
Results from ITC experiments are also consistent with pulldown results. We measured a dissociation constant (Kd) of 0.27 ± 0.05 μm between the wild type GRASP55 (2–208) and the wild type Golgin45 C peptide, whereas mutations L403R or C393A/C396A in the Golgin45 peptide or G97D in GRASP55 dramatically decreased their binding affinities (Table 2). The single point mutation R101A in GRASP55 or the mutations at the cleft binding site of Golgin45, either the single point mutation (N391A or F390A) or double mutation (F390A/N391A) exhibited a smaller impact, 2–3-fold, on their binding affinities (Fig. 4B and Table 2).
TABLE 2.
The dissociation constants of GRASP55-Golgin45 and their mutants measured and calculated from ITC experiments
| GRASP55 | Golgin45 | Kd |
|---|---|---|
| μm | ||
| WT | WT | 0.27 ± 0.05 |
| G97D | WT | None |
| R101A | WT | 0.61 ± 0.10 |
| WT | L403R | None |
| WT | C393A and C396A | None |
| WT | N391A | 0.65 ± 0.28 |
| WT | F390A | 0.34 ± 0.06 |
| WT | F390A and N391A | 0.66 ± 0.20 |
These results validate our structural model and indicate that the interaction between GRASP55 and Golgin45 requires not only the conventional PDZ-peptide interaction but also the additional zinc finger interaction, as indicated by the crystal structure. However, the interaction observed at the cleft between PDZ1 and PDZ2 is less important for this molecular recognition.
Golgin45 Acts as Molecular Glue Promoting Oligomerization of GRASP55 through Its PDZ2 Domain
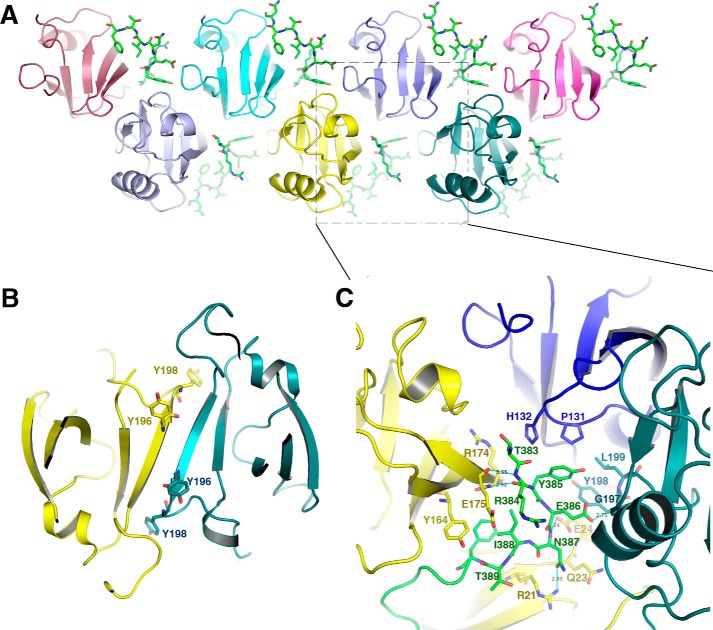
The C-terminal sequence of Golgin45 interacts not only with both PDZ1 and PDZ2 at the cleft between them but also with the PDZ2 domains of two neighboring molecules in the crystal (Fig. 5A). Previous studies presented a PDZ2-PDZ2 dimer interface in both GRASP65 (1–210) and GRASP55 (1–215) “free” structures (Fig. 5B). We could not demonstrate such a PDZ2-PDZ2 dimer interface in the structure of GRASP55 bound to Golgin45. In contrast, we found that the key residue Tyr198 involved in the dimer interaction of the free form does not interact with another PDZ2 but instead interacts with the C-terminal part of Golgin45. Indeed, its amide nitrogen atom forms hydrogen bonds with the side chain (oxygen atom) of Glu386 from the peptide. Residues Glu386 and Tyr385 from Golgin45 are also involved in additional hydrophobic interactions with Gly197, Tyr198, and Leu199 of the neighboring PDZ2 domain. Concomitantly, Thr383, Arg384, and Tyr385 of Golgin45 are also involved in hydrophobic interactions with Pro131 and His132 of the second PDZ domain of GRASP55 (the third molecule) (Fig. 5C). Thus, the binding of Golgin45 to GRASP55 not only overrides the canonical GRASP55 C terminus PDZ1 interaction but also diminishes the PDZ2 dimer formation observed in the free form of GRASP55. Golgin45 acts as molecular glue promoting the oligomerization of GRASP55 through the PDZ2 domain by creating a new interaction with two neighboring PDZ2 molecules, certainly playing an important role in Golgi stacking.
FIGURE 5.
Golgin45 acts as molecular glue promoting the oligomerization of GRASP55 through PDZ2 domain. A, oligomerization of PDZ2 domain of GRASP55 upon Golgin45 binding in the crystal of the complex. The Golgin45 peptides are shown with a stick model colored green. The PDZ2 domains of GRASP55 are shown with different colors. B, the PDZ2-PDZ2 dimer interface of GRASP55 (1–215). The two neighboring molecules are colored yellow and teal, respectively, and relevant tyrosine 196 and 198 are shown with a stick model. C, detailed interactions between Golgin45 C-terminal peptide with neighboring PDZ2 domains of GRASP55. The residues involved in the interaction are shown with a stick model. Golgin45 peptide is colored green, whereas the three different PDZ2 domains of GRASP55 are colored yellow, blue, and teal, respectively.
Discussion
The crystal structure of GRASP55 bound to the Golgin45 C-terminal peptide solved in this study indicates that Golgin45 unexpectedly recognizes the classical PDZ-peptide binding groove formed between the α2-helix and the β2-strand of PDZ1 and the cleft between PDZ1 and PDZ2. In addition, we discovered that a zinc binding site (zinc finger) forms between GRASP55 and Golgin45. The recognition mode of GRASP55-Golgin45 is very similar to GRASP65-GM130, which was reported recently; both C termini of Golgins present conserved PDZ-peptide binding motif sequences and recognize the canonical PDZ-peptide binding groove by anchoring to the carboxylate binding loop of the first PDZ domain. However, additional contacts are revealed, forming the basis for explaining the biological role for each complex.
As a first example, the related position of the tandem PDZ domains of GRASP65 is rotated by 32.6° upon GM130 binding. A deep and hydrophobic pocket that accommodates three conserved consecutive aromatic residues (FFY) of the GM130 is formed between the PDZ domains as a consequence of this conformational change. This binding site is dominated by hydrophobic interactions. Any single aromatic residue mutation abolishes this interaction. Golgin45 does not present this hydrophobic motif and thus does not undergo conformational changes upon Golgin45 binding. It still presents the hydrophobic residue phenylalanine, which inserts into the pocket formed by residues Pro35, Phe36, Tyr164, and Cys173 of GRASP55 (Fig. 3B). However, this recognition mode is not dominated by hydrophobic interactions. The mutagenesis experiments reveal that the hydrophobic interactions and the hydrogen bonds of Asn391 at the cleft binding site between PDZ1 and PDZ2 are less important for the Golgin45-GRASP55 interaction.
The GRASP65-GM130 structure forms a superhelical structure upon GM130 binding, and the N terminus of the GM130 C-terminal peptide points to the helical axis in the center and thus does not interact with neighboring GRASP65 molecules. However, the relative position of the Golgin45 C-terminal peptide is very different. It interacts not only at the cleft between both PDZ1 and PDZ2 but also with the PDZ2 domains of two neighboring molecules in the crystal (Fig. 5A). Golgin45 thus acts as a molecular glue promoting the oligomerization of GRASP55 through its PDZ2 domain.
It is generally accepted that oligomers are formed by the first PDZ domain of GRASP molecules, whereas the second PDZ domain interacts with golgins during Golgi stacking (17, 18, 23). All the current Golgi stacking models are based on these two assumptions, but in our crystal structure, both C termini of Golgin45 and GM130 peptides bind to the PDZ1 peptide binding pocket through the canonical PDZ-peptide binding pocket rather than PDZ2. Thus, GRASP molecules cannot form oligomers through PDZ1 domains when Golgin45 or GM130 are present. Previous studies indicated that GRASP55 and GRASP65 always co-localize with Golgin45 and GM130, respectively, in the Golgi apparatus (3, 6), implying that a new model is needed for Golgi cisternae stacking.
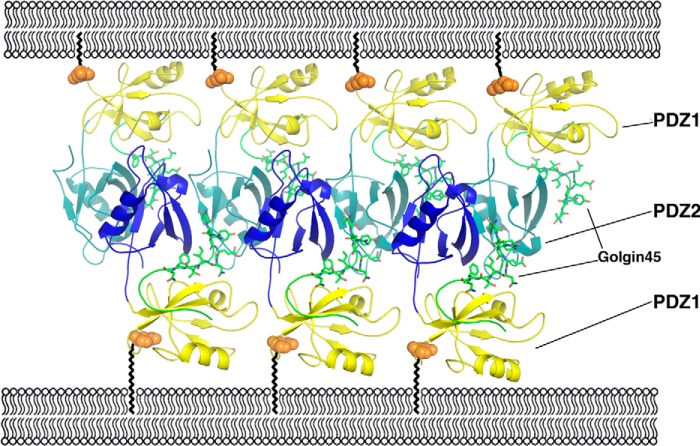
Our crystal structure shows that Golgin45 can interact with 3 PDZ2 domains at the same time and cluster the PDZ2 domain of different GRASP55 together, acting as a molecular glue. We thus propose that oligomers can be formed by the second PDZ domain with Golgin45 playing a crucial role rather than the PDZ1 of GRASP55. In this model, the first PDZ domain of GRASP55 is anchored to the Golgi cisternae membrane through myristoylation of its N terminus (glycine at position 2), whereas its second PDZ domain is clustered by Golgin45. Golgin45 not only acts as a molecular glue, but it also keeps two tandem PDZ domains (PDZ1 and PDZ2) rigid, binding to them tightly at the same time. The mid-cisternae stacking is thus mediated by a series of GRASP55 and Golgin45 molecules. This forms ordered membrane-associated protein arrays between two apposing membranes. The second PDZ domains of the GRASP55 molecules are clustered in the middle with high density, whereas the first PDZ domains of GRASP55 molecules lie at the two sides anchored to the membrane (Fig. 6).
FIGURE 6.
Molecular models of GRASP55-Golgin45-mediated mid-cisternae membrane stacking. PDZ1 domains are colored yellow, whereas PDZ2 domains of GRASP55 are colored blue. Golgin45 C-terminal peptides are shown with a stick model and colored green. The ball and zigzag lines represent GRASP55 N-terminal myristoylation. A series of GRASP55 and Golgin45 molecules form ordered membrane-associated protein arrays between two apposing membranes.
Surprisingly, our crystal structure reveals that a zinc ion may play an important role in the oligomerization process of GRASPs, its effect on GRASP55 being the opposite of that observed for GRASP65. The binding of a zinc ion is required for complex formation between GRASP55 and Golgin45, whereas it is not observed nor necessary for the GRASP65-GM130 recognition process. This difference may relate to some other bioprocesses that are yet unclear. We propose herein, from our structural study, a new model of Golgi mid-cisternae stacking. Further studies and direct in cellulo evidence are still required to validate this new model.
Although both GRASPs have a clear role in Golgi organization, some studies revealed that they seemed dispensable for Golgi stacking. However, depletion of GRASP55 or GRASP65 has no significant effect on cisternal stacking or general protein secretion (24–28). In addition, plant cells present perfectly stacked Golgi cisternae without GRASP molecules. Moreover, in addition to mediating the Golgi stacking and ribbon formation, GRASPs are also involved in other cellular processes, such as the Golgi mitotic checkpoint and membrane trafficking. Their roles in unconventional secretion may be even more important because deletion of GRASPs blocks the unconventional protein secretion pathway. Previous studies confirmed that the C-terminal valine motifs of membrane proteins, such as CD83, CD8a, TGF-α, CFTR, and p24 cargo receptor, bind directly to the PDZ domains of GRASPs (29–34). It is thus elusive whether other proteins secreted through the GRASP-related unconventional pathway bind to GRASPs with a similar recognition mode. From our crystal structures, it is predicted that these C-terminal valine motif sequences should bind to the conventional PDZ-peptide binding groove of PDZ1 and compete with GM130 and Golgin45 because their motif sequences are very similar to the C termini of GM130 and Golgin45. Thus, their binding to the first PDZ domain of GRASPs will change the oligomerization state and the localization of GRASPs. How the transition of these processes is regulated remains unknown. The different affinity of GRASPs for their biological partners may control the balance between the structural role of the Golgi and the unconventional secretion role. It is thus important to elucidate the interaction between GRASP and its different biological partners, a key to understanding molecular mechanisms of its regulation. The structure of GRASP55 bound to Golgin45 C-terminal peptide presented here reveals one of these molecular interactions. This model opens new perspectives for understanding this biological switch and the regulation of these processes.
Experimental Procedures
Construct Generation
The GRASP domain (residues 2-208) of mouse GRASP55 was amplified and cloned into the modified pRSFDuet-1 vector (Novagen) with an N-terminal His6 tag and a PreScission Protease cleavage site. Mouse Golgin45 C-terminal peptide (residues 380–403) was amplified from the full-length gene, cloned into the pGEX-6P-1 (Novagen) vector, and expressed as a GST fusion protein. Point mutations were introduced using the QuikChange protocol (Stratagene). All constructs were confirmed by DNA sequencing.
Protein Expression and Purification
The protein complex was obtained by co-expression of GRASP55 and Golgin45 C-terminal peptide in BL21 (DE3) Escherichia coli (Novagen) induced with 0.4 mm isopropyl-β-d-thiogalactopyranoside for 12 h at 16 °C. The cell lysate was clarified after lysis (50 mm Tris-HCl, 150 mm NaCl (pH 8.0) containing 1 mm β-mercaptoethanol, 1 mm PMSF; Sigma), sonication, and centrifugation and was incubated with glutathione-Sepharose beads (GE Healthcare) and washed with lysis buffer. The protein complex without a tag was obtained by incubating the beads with PreScission Protease at 5 °C for 3 h and further separated by ion exchange chromatography on a RESOURCE Q column (GE Healthcare) and size exclusion chromatography on a Superdex 75 column (GE Healthcare) pre-equilibrated in a buffer with 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 1 mm DTT. Peak fractions corresponding to pure protein were concentrated to 20 mg/ml for crystallization.
All the proteins for pulldown and ITC experiments were expressed and purified individually. The Golgin45 C-terminal peptide with GST tag and its mutants were obtained by following a procedure similar to the one described for the complex (without adding PreScission Protease). GRASP55 (2–208) with a His6 tag and its mutants were purified using affinity beads (nickel-nitrilotriacetic acid agarose). All samples were collected at each step and analyzed by SDS-PAGE.
Crystallization and Data Collection
The purified complex proteins were crystallized by the sitting drop vapor diffusion method mixed 1:1 with reservoir solution. Crystals appeared in reservoir buffer containing 0.1 m Tris (pH 8.5), 0.2 m potassium sodium tartrate, and 30% PEG4K in 3 days at 18 °C and were frozen in a cryoprotectant consisting of the reservoir solution supplemented with 20% PEG400. The data were collected on the BL17U1 station of the Shanghai Synchrotron Radiation Facility and then were processed using the HKL2000 and XDS software.
Structure Determination and Refinement
The structures were determined by the molecular replacement program PHASER (21, 22) using the GRASP domain structures of GRASP55 as an initial search model. Model building and iterative refinement were performed with the COOT and PHENIX refinement programs (35, 36). The orientations of the amino acid side chains and bound water molecules were modeled on the basis of 2Fobs - Fcalc and Fobs - Fcalc difference Fourier maps. Detailed data collection and refinement statistics are listed in Table 1. The model figures were generated with PyMOL and CCP4mg (37). The interactions were analyzed with PyMOL and LigPlus (38).
Pulldown Assay and Western Blotting
The Golgin45 C-terminal peptide with a GST tag, GRASP55 (2–208) with a His6 tag, and their mutants were expressed and purified. Pulldown experiments were performed by using Golgin45 C-terminal peptide or its mutants with GST tag as bait on glutathione-Sepharose 4B beads. After washing with equilibration buffer containing 50 mm Tris and 150 mm NaCl, His-tagged GRASP55 or its mutants were added into the column and incubated for ∼30 min. The beads were washed with equilibration buffer before elution with the buffer containing 10 mm reduced glutathione. The eluted samples (proteins in complex) were separated by 15% SDS-PAGE and then transferred to a PVDF membrane. The Western blotting results were analyzed using mouse anti-His tag antibody as the primary antibody and HRP-conjugated sheep anti-mouse antibody as the secondary antibody. The bound conjugate on the blot was detected by reaction with 3,3′,5,5′-tetramethylbenzidine substrate in the presence of H2O2 and potassium nitroprusside.
Isothermal Titration Calorimetry
The measurements were conducted using an ITC-200 microcalorimeter (MicroCal) at 23 °C. All the samples were dialyzed into 20 mm Tris, 100 mm NaCl buffer prior to the ITC experiments. The sample cell (300 μl in volume) was filled with the Golgin45 C-terminal peptide or its mutants in a 20 mm Tris, 100 mm NaCl buffer (pH 8.0). The injection syringe (40 μl) was filled with GRASP55 (residues 2–208) or its mutants. The concentration of GRASP55 was three times higher compared than the Golgin45 C-terminal peptide. The experimental parameters were as follows: 20 injections, 2 μl, and 1 s per injection, with an interval of 150 s and stirring speed of 1000 rpm. The data were analyzed and fitted using the Microcal Origin software suite.
ACCESSION NUMBER
Atomic coordinates and structure factors for the reported crystal structures have been deposited in the Protein Data Bank with accession code 5H3J.
Author Contributions
J. Z. and X. H. performed the protein purification, crystallization, and biochemical experiments; N. S. supervised the project, performed the structural determination and structure refinement, and wrote the paper; B. L. generated the constructs; and J. Z., B. L., X. M., and N. S. analyzed the data, discussed the results, and commented on the manuscript.
Acknowledgments
The X-ray data were collected at Beamline BL17U1 of the Shanghai Synchrotron Radiation Facility. We are grateful to Prof. Jiahuai Han for the plasmids.
This work was supported by National Natural Science Foundation of China Grant 31370738 and Hundred Talents Program of the Chinese Academy of Sciences and Campus France (Partenariats Hubert Curien CAI YUANPEI 2013 International program between France and China) Project 26203WD. This work was also supported by the Agence Nationale de la Recherche Grant NR-15-CE18-0023. The authors declare that they have no conflicts of interest with the contents of this article.
- PDB
- Protein Data Bank
- RMSD
- root mean square deviation
- ITC
- isothermal titration calorimetry.
References
- 1. Rabouille C., Misteli T., Watson R., and Warren G. (1995) Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system. J. Cell Biol. 129, 605–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shorter J., Watson R., Giannakou M. E., Clarke M., Warren G., and Barr F. A. (1999) GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18, 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barr F. A., Puype M., Vandekerckhove J., and Warren G. (1997) GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91, 253–262 [DOI] [PubMed] [Google Scholar]
- 4. Bachert C., and Linstedt A. D. (2010) Dual anchoring of the GRASP membrane tether promotes trans pairing. J. Biol. Chem. 285, 16294–16301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barr F. A., Nakamura N., and Warren G. (1998) Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 17, 3258–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Short B., Preisinger C., Körner R., Kopajtich R., Byron O., and Barr F. A. (2001) A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155, 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duprez E., Tong J. H., Dérré J., Chen S. J., Berger R., Chen Z., and Lanotte M. (1997) JEM-1, a novel gene encoding a leucine-zipper nuclear factor upregulated during retinoid-induced maturation of NB4 promyelocytic leukaemia. Oncogene 14, 1563–1570 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y., Liu S., Mickanin C., Feng Y., Charlat O., Michaud G. A., Schirle M., Shi X., Hild M., Bauer A., Myer V. E., Finan P. M., Porter J. A., Huang S. M., and Cong F. (2011) RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 13, 623–629 [DOI] [PubMed] [Google Scholar]
- 9. Tong J. H., Duprez E., and Lanotte M. (1999) JEM-1, a novel nuclear co-factor: localisation and functional interaction with AP-1. Leukemia 13, 1982–1992 [DOI] [PubMed] [Google Scholar]
- 10. Pacaud R., Sery Q., Oliver L., Vallette F. M., Tost J., and Cartron P. F. (2014) DNMT3L interacts with transcription factors to target DNMT3L/DNMT3B to specific DNA sequences: role of the DNMT3L/DNMT3B/p65-NFκB complex in the (de-)methylation of TRAF1. Biochimie 104, 36–49 [DOI] [PubMed] [Google Scholar]
- 11. Ekkapongpisit M., Wannatung T., Susantad T., Triwitayakorn K., and Smith D. R. (2007) cDNA-AFLP analysis of differential gene expression in human hepatoma cells (HepG2) upon dengue virus infection. J. Med. Virol. 79, 552–561 [DOI] [PubMed] [Google Scholar]
- 12. Koldehoff M., Zakrzewski J. L., Klein-Hitpass L., Beelen D. W., and Elmaagacli A. H. (2008) Gene profiling of growth factor independence 1B gene (Gfi-1B) in leukemic cells. Int. J. Hematol. 87, 39–47 [DOI] [PubMed] [Google Scholar]
- 13. He P., Sun L., Zhu D., Zhang H., Zhang L., Guo Y., Liu S., Zhou J., Xu X., and Xie P. (2016) Knock-down of endogenous bornavirus-like nucleoprotein 1 inhibits cell growth and induces apoptosis in human oligodendroglia cells. Int. J. Mol. Sci. 17, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang R. Y., Su S. G., Wu D. C., Fu J., and Zeng X. (2015) BLZF1 expression is of prognostic significance in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 467, 602–609 [DOI] [PubMed] [Google Scholar]
- 15. Rabouille C., and Linstedt A. D. (2016) GRASP: A multitasking tether. Front. Cell Dev. Biol. 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X., and Wang Y. (2015) GRASPs in Golgi structure and function. Front. Cell Dev. Biol. 3, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Truschel S. T., Sengupta D., Foote A., Heroux A., Macbeth M. R., and Linstedt A. D. (2011) Structure of the membrane-tethering GRASP domain reveals a unique PDZ ligand interaction that mediates Golgi biogenesis. J. Biol. Chem. 286, 20125–20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng Y., Yu W., Li X., Lin S., Zhou Y., Hu J., and Liu X. (2013) Structural insight into Golgi membrane stacking by GRASP65 and GRASP55 proteins. J. Biol. Chem. 288, 28418–28427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu F., Shi X., Li B., Huang X., Morelli X., and Shi N. (2015) Structural basis for the interaction between the Golgi reassembly-stacking protein GRASP65 and the Golgi matrix protein GM130. J. Biol. Chem. 290, 26373–26382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Truschel S. T., Zhang M., Bachert C., Macbeth M. R., and Linstedt A. D. (2012) Allosteric regulation of GRASP protein-dependent Golgi membrane tethering by mitotic phosphorylation. J. Biol. Chem. 287, 19870–19875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Read R. J. (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57, 1373–1382 [DOI] [PubMed] [Google Scholar]
- 23. Tang D., Yuan H., Vielemeyer O., Perez F., and Wang Y. (2012) Sequential phosphorylation of GRASP65 during mitotic Golgi disassembly. Biol. Open 1, 1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee I., Tiwari N., Dunlop M. H., Graham M., Liu X., and Rothman J. E. (2014) Membrane adhesion dictates Golgi stacking and cisternal morphology. Proc. Natl. Acad. Sci. U.S.A. 111, 1849–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Behnia R., Barr F. A., Flanagan J. J., Barlowe C., and Munro S. (2007) The yeast orthologue of GRASP65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic. J. Cell Biol. 176, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Puthenveedu M. A., Bachert C., Puri S., Lanni F., and Linstedt A. D. (2006) GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 8, 238–248 [DOI] [PubMed] [Google Scholar]
- 27. Sütterlin C., Polishchuk R., Pecot M., and Malhotra V. (2005) The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol. Biol. Cell 16, 3211–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kondylis V., Spoorendonk K. M., and Rabouille C. (2005) dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol. Biol. Cell 16, 4061–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stein M. F., Blume K., Heilingloh C. S., Kummer M., Biesinger B., Sticht H., and Steinkasserer A. (2015) CD83 and GRASP55 interact in human dendritic cells. Biochem. Biophys. Res. Commun. 459, 42–48 [DOI] [PubMed] [Google Scholar]
- 30. Kim J., Noh S. H., Piao H., Kim D. H., Kim K., Cha J. S., Chung W. Y., Cho H. S., Kim J. Y., and Lee M. G. (2016) Monomerization and ER relocalization of GRASP is a requisite for unconventional secretion of CFTR. Traffic 17, 733–753 [DOI] [PubMed] [Google Scholar]
- 31. Gee H. Y., Noh S. H., Tang B. L., Kim K. H., and Lee M. G. (2011) Rescue of DeltaF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell 146, 746–760 [DOI] [PubMed] [Google Scholar]
- 32. Kuo A., Zhong C., Lane W. S., and Derynck R. (2000) Transmembrane transforming growth factor-α tethers to the PDZ domain-containing, Golgi membrane-associated protein p59/GRASP55. EMBO J. 19, 6427–6439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barr F. A., Preisinger C., Kopajtich R., and Körner R. (2001) Golgi matrix proteins interact with p24 cargo receptors and aid their efficient retention in the Golgi apparatus. J. Cell Biol. 155, 885–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. D'Angelo G., Prencipe L., Iodice L., Beznoussenko G., Savarese M., Marra P., Di Tullio G., Martire G., De Matteis M. A., and Bonatti S. (2009) GRASP65 and GRASP55 sequentially promote the transport of C-terminal valine-bearing cargos to and through the Golgi complex. J. Biol. Chem. 284, 34849–34860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McNicholas S., Potterton E., Wilson K. S., and Noble M. E. (2011) Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wallace A. C., Laskowski R. A., and Thornton J. M. (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]