Abstract
Insulin-dependent translocation of glucose transporter 4 (Glut4) to the plasma membrane plays a key role in the dynamic regulation of glucose homeostasis. We recently showed that this process is critically dependent on palmitoylation of Glut4 at Cys-223. To gain further insights into the regulation of Glut4 palmitoylation, we set out to identify the palmitoyl acyltransferase (PAT) involved. Here we report that among 23 mammalian DHHC proteins, DHHC7 is the major Glut4 PAT, based on evidence that ectopic expression of DHHC7 increased Glut4 palmitoylation, whereas DHHC7 knockdown in 3T3-L1 adipocytes and DHHC7 KO in adipose tissue and muscle decreased Glut4 palmitoylation. Moreover, inactivation of DHHC7 suppressed insulin-dependent Glut4 membrane translocation in both 3T3-L1 adipocytes and primary adipocytes. Finally, DHHC7 KO mice developed hyperglycemia and glucose intolerance, thereby confirming that DHHC7 represents the principal PAT for Glut4 and that this mechanism is essential for insulin-regulated glucose homeostasis.
Keywords: adipose tissue, gene knockout, glucose transport, glucose transporter type 4 (GLUT4), membrane trafficking, protein acylation, DHHC protein, palmitoylation.
Introduction
DHHC3 proteins are Asp-His-His-Cys domain-containing palmitoyl acyltransferases (PATs) that catalyze thioacylation of substrate proteins with the 16-carbon fatty acid palmitate (1). Protein palmitoylation equips substrate proteins with a bulky hydrophobic moiety that serves as a lipid anchor (2). Thus, an important function of DHHC proteins is to regulate the interaction between proteins and membranes, a process that is critical for vesicle trafficking, intracellular signaling, and protein membrane compartmentalization (3, 4). Emerging evidence indicates that DHHC proteins also play important roles in the regulation of cell function, as mutations in DHHC proteins have been linked to diverse pathological conditions (5).
There are 23 DHHC proteins in the mammalian genome (6, 7). They were initially recognized as a family of proteins that contain a conserved DHHC motif within a cysteine-rich domain (CRD); therefore, this domain is also referred to as DHHC-CRD. DHHC proteins contain four to six transmembrane domains and are variably associated with intracellular, membrane-enclosed compartments or the plasma membrane. These proteins were named DHHC1 to DHHC 24 (without 10) according to the approximate order by which they were deposited into GenBankTM. Thus, the numbers associated with the name of DHHC proteins do not suggest any structural and functional relationship. The functions of mammalian DHHC proteins had remained a mystery until 2002 when the first yeast DHHC genes were demonstrated to encode cysteine palmitoyl acyltransferase with the DHHC-CRD as the catalytic domain (8, 9). Based on this notion, mammalian DHHC proteins were thereafter soon shown to also represent palmitoyl acyltransferases (10, 11). Since then, a number of newly developed chemical in vitro assays were developed to facilitate the identification of putative DHHC enzyme-substrate pairs (7, 12).
DHHC7 was first cloned as a zinc finger domain protein from Sertoli cells and given the name SERZ-β (for Sertoli cell gene with a zinc finger protein β) (13). Among the 23 mammalian DHHC proteins, DHHC7 is most closely related to DHHC3 (also known as GODZ for Golgi-specific DHHC zinc finger protein), as they exhibit the highest peptide sequence similarity (77%), the same membrane topology (10, 14), and largely the same tissue and cell type-specific expression patterns (14). It has, therefore, been suggested that DHHC3 and DHHC7 constitute a subfamily of DHHC palmitoyl acyltransferases (7). Supporting this view, ectopically expressed DHHC3 and DHHC7 have been shown to palmitoylate many of the same substrates (7, 14). However, emerging evidence suggests that the substrate specificity of DHHC3 and DHHC7 in vivo is distinct (15).
Glut4 (glucose transporter 4) belongs to the 12-member facilitative glucose transporter family and is predominantly expressed in adipose and muscle tissues (16). Unlike other glucose transporters that constitutively reside in the plasma membrane, Glut4 under basal conditions is stored in intracellular compartments that include the trans-Golgi network (TGN) and tubular-vesicular structures in the form of small membrane vesicles. It is distributed to the plasma membrane only after insulin stimulation (17, 18). As an insulin-regulated glucose transporter expressed dominantly in adipose tissue and muscle tissue, Glut4 plays a critical role in the regulation of whole body glucose homeostasis and peripheral insulin sensitivity. In mice and humans, impaired Glut4 function results in hyperglycemia and insulin resistance, a hallmark of type II diabetes (19–21).
Insulin-dependent Glut4 membrane translocation is a multistep process. After insulin stimulation, Glut4 vesicles first leave their intracellular storage compartment (22). Upon stimulation of cells with glucose, they are then sequestered to the plasma membrane and docked on the plasma membrane through interaction with a membrane targeting-SNARE complex that consists of Syntaxin4, SNAP23, and VAMP2 (23). Once docked onto the plasma membrane, Glut4 vesicles are fused with the plasma membrane to enable glucose influx. Among these different steps of Glut4 membrane translocation, Glut4 vesicle-sorting, membrane-docking, and fusion depends on the interaction between membrane and proteins including (24, 25). In addition, DHHC proteins have been implicated in the regulation of vesicular protein trafficking in neurons (26). Thus, it is conceivable that DHHC proteins play a role in translocation of intracellular Glut4 vesicles to the plasma membrane.
We have been interested in the role of protein palmitoylation in adipose tissue as it is a vital organ in the regulation of body glucose homeostasis and peripheral insulin sensitivity (27, 28). Toward this, we analyzed the palmitoylated proteome of adipose tissues through thiopropyl captivation (TPC)/mass spectrum studies and identified >800 putative palmitoylated proteins (29). Among them were enzymes for lipid and energy metabolism as well as proteins that are important for insulin-dependent Glut4 plasma membrane translocation such as Glut4, insulin-responsive amino peptidase (IRAP), the major cargo of Glut4 vesicles and SNAP23, the key component of targeting-SNARE for Glut4 vesicle membrane docking and fusion (29). In addition, we recently found that Glut4 is palmitoylated at Cys-223 and that Glut4 palmitoylation is important for insulin-dependent Glut4 membrane translocation, as substitution of Cys-223 with a serine residue abolishes Glut4 palmitoylation and insulin-dependent Glut4 membrane translocation (30). To further investigate this mechanism we have screened all mammalian DHHC proteins for their ability to palmitoylate Glut4. Here we report that DHHC7 is the major Glut4 palmitoyl transferase that regulates insulin-dependent Glut4 membrane translocation and glucose transport in vivo.
Results
DHHC7 Is the Major PAT for Glut4
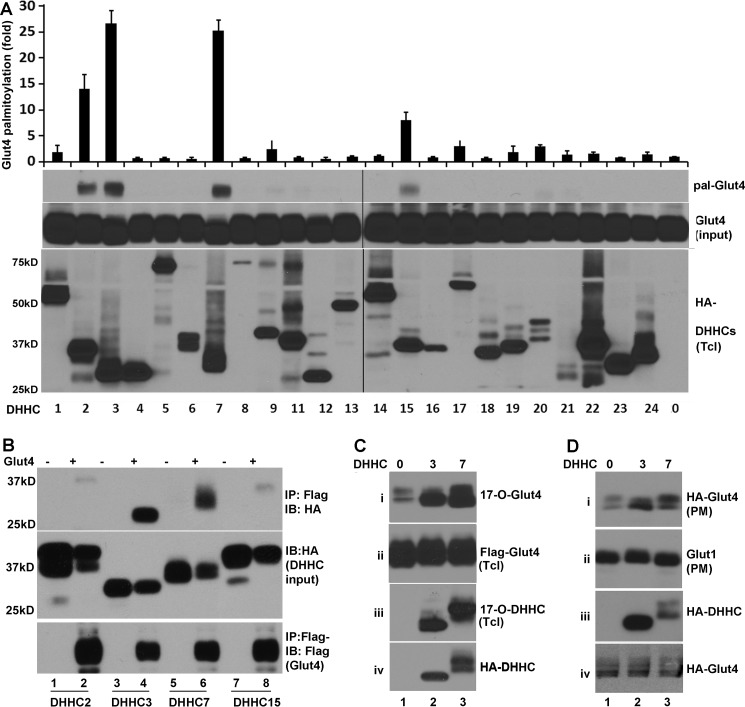
Toward identification of Glut4 PAT(s), we screened all 23 mammalian DHHC proteins for their ability to palmitoylate Glut4. In co-transient transfection and TPC assays (“Experimental Procedures”), the level of Glut4 palmitoylation was increased to different degrees by ectopic expression of different DHHC proteins. It was increased the most by DHHC3 and DHHC7 followed by DHHC2 and DHHC15 (Fig. 1A), suggesting that DHHC3 and DHHC7 served as candidate PATs for Glut4.
FIGURE 1.
Identification of DHHC PATs for Glut4. A, TPC assays of HEK 293T cells co-transfected with FLAG-Glut4; HA-DHHC proteins are indicated. Shown are from the top. 1) Summary quantitation of palmitoylated Glut4 catalyzed by DHHC proteins #1–9 and 11–24, normalized to the amount of palmitoylated Glut4 observed with empty vector (#0). 2) Western blot of palmitoylated Glut4 (pal-Glut4) of a representative TPC assay testing all 23 DHHC proteins. 3) A corresponding immunoblot comparing total amounts of Glut4 in the same reactions (Glut4 input). 4) A corresponding immunoblot probed for the different DHHC proteins (HA-DHHCs). Note that significant palmitoylation of Glut4 was observed only with DHHC2, -3, -7, and -15. Also note that the apparent molecular mass of DHHC proteins typically is lower than that predicted based on amino acid sequences, presumably because of the highly hydrophobic nature of these proteins. Bar graphs and error bars indicate means ± S.D. (n = 3). B, DHHC3 and DHHC7 form stable complexes with Glut4. HEK 293T cells were co-transfected with FLAG-Glut4 (+) or the corresponding empty vector (−) and either HA-DHHC2, -3, -7, or -15 and subjected to immunoprecipitation (IP) with anti-FLAG antibody, and the precipitates were analyzed by immunoblotting (IB) for co-immunoprecipitated DHHC proteins using anti-HA antibody (top panel). Aliquots of total extracts were probed with anti-HA and anti-FLAG to verify faithful expression of HA-DHHC (middle panel) and FLAG-Glut4 (bottom). Note the efficient co-immunoprecipitation of Glut4 with both DHHC3 and DHHC7. C, COS-7 cells transfected with FLAG-Glut4 and HA-DHHC3 or HA-DHHC7 were metabolically labeled with 17-ODYA followed by the Click chemistry assay. The labeled proteins were purified over streptavidin-agarose and analyzed by immunoblotting with anti-FLAG (Glut4) (i) and anti-HA (DHHC3 and DHHC7) antibodies (iii). Immunoblots of total lysates were probed in parallel for FLAG-Glut4 (ii) and HA-DHHC (iv). Note the efficient palmitoylation of Glut4 by both DHHC3 and DHHC7. D, palmitoylation of Glut4 by DHHC3 and DHHC7 facilitates membrane association. CHO-IR cells were co-transfected with HA-Glut4-GFP and HA-DHHC3 or HA-DHHC7 and treated with insulin (15 min) followed by comparison by immunoblot of crude membranes (i, ii) and total cell lysates (iii, iv) probed for HA-Glut4-GFP (i), endogenous Glut1 (ii), HA-DHHC3 and -7 (iii), and HA-Glut4 (iv). Note that both DHHC3 and DHHC7 facilitate Glut4 membrane association, whereas Glut2 accumulates in membranes independently of DHHC protein transfection.
Given our previous findings that overexpression PATs can form stable complexes with cognate substrates on overexpression in HEK cells (14), we next examined whether DHHC proteins form similar substrates with Glut4. Accordingly, HEK293T cells were co-transfected with empty vector or vector encoding FLAG-Glut4 and plasmids coding for the indicated HA-tagged DHHC proteins. Immunoprecipitation of FLAG-Glut4 from total cell lysates with FLAG antibody resulted in efficient co-immunoprecipitation of both HA-DHHC3 and HA-DHHC7 without corresponding bands in FLAG-Glut4-lacking, negative controls (Fig. 1A, top panel). Similar assays that probed for association of DHHC2 and DHHC15 with Glut4 detected only trace amounts of DHHC2 and DHHC15 in anti-FLAG-Glut4 immunoprecipitates (top panel). These differences in HA-DHHC proteins detected in anti-FLAG-Glutl4 immunoprecipitates were not due to sample variations, as comparable levels of FLAG-Glut4 and HA-DHHC proteins were present in all test samples (middle and bottom panels).
To further examine whether DHHC3 and DHHC7 indeed promoted Glut4 palmitoylation, we next carried out 17-octadecynoic acid (17-ODYA) metabolic labeling and Click chemistry to examine how ectopic expression of DHHC3 or DHHC7 affects Glut4 palmitoylation. 17-ODYA metabolic labeling and Click chemistry allows examining protein palmitoylation, as 17-ODYA specifically reacts with palmitoylated cysteine residue(s) (31). Specifically, COS-7 cells were transiently co-transfected with vectors coding for FLAG-tagged Glut4 and the indicated HA-tagged DHHC proteins followed by metabolic labeling with 17-ODYA and Click chemistry purification. In agreement with the notion that Glut4 is palmitoylated, FLAG-tagged Glut4 (panel i) was detected in 17-ODCA-labeled cells (Fig. 1C). Overexpression of either DHHC3 or DHHC7 increased the level of 17-ODYA labeled Glut4. In these experiments we also examined the palmitoylation of DHHC3 and DHHC7. In agreement with the view that DHHC proteins are also subject to palmitoylation themselves (32), both DHHC3 and DHHC7 were labeled by 17-ODYA. Taken together, these data demonstrate that DHHC3 and DHHC7 promote Glut4 palmitoylation upon overexpression of enzymes and substrate in heterologous cells.
To determine whether the increase in Glut4 palmitoylation contributes to Glut4 plasma membrane (PM) translocation, we next examined the impact of ectopic expression of DHHC3 or DHHC7 on the level of Glut4 in the PM of CHO-IR cells. CHO-IR cells are fibroblast-like Chinese hamster ovary cells stably transfected with a human insulin receptor (IR) (33). They lack endogenous Glut4 but contain insulin responsive vesicles. Upon transfection of these cells with Glut4, insulin induces translocation of Glut4 to the PM, analogous to insulin-induced Glut4 membrane translocation in adipocytes and muscle cells (34–36). Specifically, CHO-IR cells were co-transfected with FLAG-tagged Glut4 and either DHHC3 or DHHC7. The transfected cells were then treated with 10 nm insulin for 15 min, and the amount of Glut4 in crude PM fractions was analyzed by immunoblotting. As shown in Fig. 1D, ectopic expression of either DHHC3 or DHHC7 increased PM levels of Glut4 (panel i), but not that of Glut1 (panel ii), consistent with Glut4 palmitoylation contributing to insulin-dependent Glut4 membrane translocation, whereas Glut1 is known not to be regulated by insulin (37). Total FLAG-Glut4 and HA-DHHC amounts expressed were comparable in the different samples (Fig. 1D, panels iii and iv).
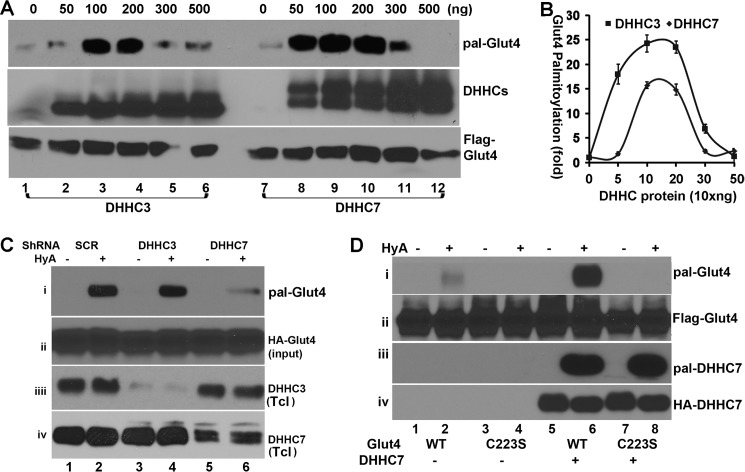
To further compare the efficacy by which DHHC3 and DHHC7 palmitoylate Glut4, we next evaluated how Glut4 palmitoylation responded to different doses of co-transfected DHHC3 and DHHC7. A constant amount of expression vector coding for FLAG-Glut4 was co-transfected into HEK293T cells with different amounts of expression vectors coding for either HA-DHHC3 or HA-DHHC7. At 36 h post transfection, total cell lysates were prepared and subjected to TPC assay to assess the levels of Glut4 palmitoylation. As shown in Fig. 2A, DHHC7 was much more effective than DHHC3 in palmitoylating Glut4, especially at the lower dosages. For instance, at 50 ng of expression vector, DHHC7 increased Glut4 palmitoylation by >20-fold above basal levels, whereas DHHC3 did so only 2-fold even though comparable amounts of Glut4 and DHHC proteins were present in these samples (Fig. 2, A and B). To our surprise, both DHHC3 and DHHC7 were less effective in palmitoylating Glut4 at higher enzyme-to-substrate ratios. At present, the reason for this is not known. These results nevertheless indicated that DHHC7 is more effective than DHHC3 as a PAT of Glut4.
FIGURE 2.
DHHC7 is the major palmitoyl acyltransferase for Glut4. A, enzyme dose dependence of DHHC3- and DHHC7-mediated palmitoylation of Glu4. HEK 293T cells were transfected with Glut4 and the different amounts of DHHC3 or DHHC7 plasmid indicated. Images are representative of three experiments. B, summary quantitation of three experiments analogous to the data in A. Note the much more efficient palmitoylation of vGlut4 in the presence of 10 versus 30 ng of transfected DHHC vector and the fact that DHHC7 is more effective than DHHC3 as a PAT of Glut4. C, impact of DHHC3 and DHHC7 shRNA on Glut4 palmitoylation in CHO-IR cells. CHO-IR cells transduced with adenoviral vectors that express HA-Glut4 and the shRNA indicated were subjected to TPC assay to examine the role of DHHC3 and DHHC7 for Glut4 palmitoylation. shRNA treatment effectively reduced the expression of DHHC3 and DHHC7 in total cell lysates (Tcl). Note that DHHC7 shRNA, but not DHHC3 or scrambled (SCR) shRNA, reduced palmitoylated Glut4 (pal-Glut4). D, DHHC7-mediated palmitoylation of GluT4 is dependent on Cys-223 of Glut4. TPC assays of HEK 293T cells transfected with expression vectors that encode Glut4 (lanes 1, 2, 5, and 6) or Glut4C223S (lanes 3, 4, 7, and 8) and DHHC7 (lanes 5–8) demonstrate that GluT4 palmitoylation requires Cys-223.
The preceding data suggested that DHHC7 is likely a principal PAT for Glut4. To evaluate this further, we examined how knockdown of DHHC3 or DHHC7 affects Glut4 palmitoylation. Adenoviral vectors that simultaneously express HA-Glut4-GFP and either scrambled shRNA (control) or DHHC3 or DHHC7 shRNA were generated (see “Experimental Procedures”) and used to transduce CHO-IR cells. At 48 h post transduction, the palmitoylation status of Glut4 in total cell lysates was evaluated by TPC assays. As shown in Fig. 2C, Glut4 palmitoylation was reduced by >70% when HA-Glut4 was co-expressed with DHHC7 shRNA versus the scrambled control shRNA (panel i). Unexpectedly, the DHHC3 shRNA had no effect on Glut4 palmitoylation (Fig. 2C, panel i) even though overexpression of DHHC3 strongly augmented Glut4 palmitoylation (Fig. 1A). The differences in Glut4 palmitoylation observed here were not due to variable sample loading, as comparable levels of total HA-Glut4 were seen in each sample (panel ii). Both DHHC3 and DHHC7 shRNA were effective, as the levels of DHHC3 and DHHC7 were reduced by >75% in shRNA versus scrambled shRNA-expressing cells (Fig. 2C, panels iii and vi). Taken together, the data indicate that DHHC7 is the principal PAT for Glut4.
Glut4 is palmitoylated at Cys-223 (30). We, therefore, examined the impact of DHHC7 on palmitoylation of WT Glut4 and Glut4C223S, respectively. As shown in Fig. 2D, ectopically expressed DHHC7 strongly increased the palmitoylation of WT Glut4 but had no effect on Glut4C223S, confirming that DHHC7 palmitoylates Glut4 at Cys-223. These differences in Glut4 palmitoylation were not due to variance in sample loading or TPC assay as indicated by comparable levels of total Glut4 (panel ii) and unaltered DHHC7 autopalmitoylation (panel iii). Taken together, our data demonstrate that DHHC7 is the major PAT for Glut4 in heterologous cells.
DHHC7 Knockdown Suppresses Insulin-dependent Glut4 Membrane Translocation
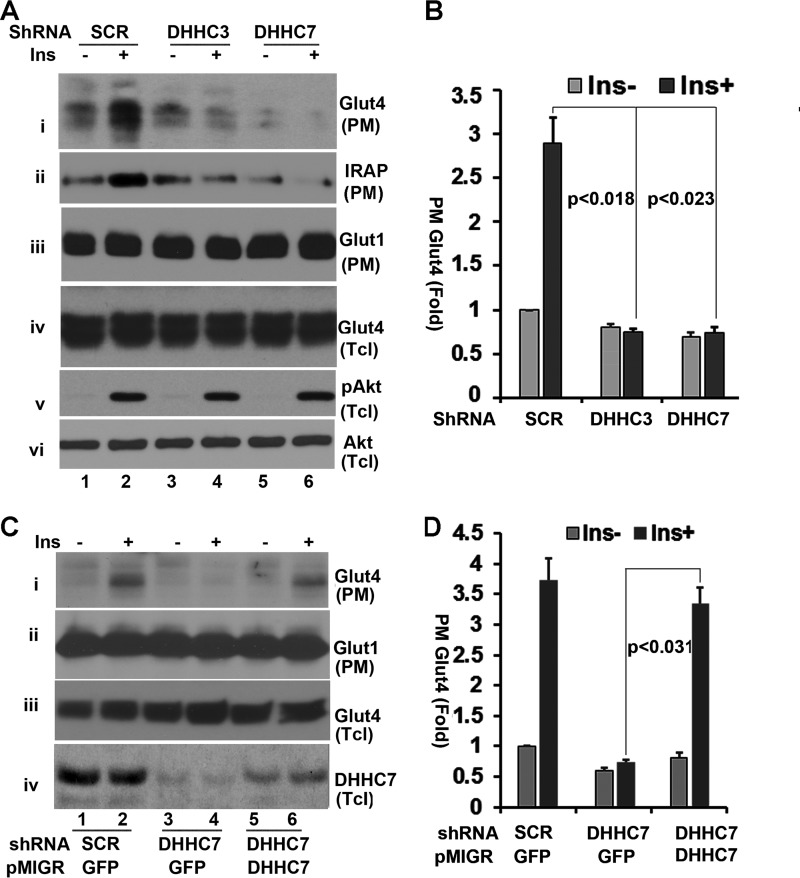
In the preceding experiment we found that ectopic expression of DHHC7 enhanced Glut4 palmitoylation and Glut4 PM association, thereby indicating that DHHC7 is involved in Glut4 translocation to the PM (Fig. 1). We next examined how silencing of DHHC7 would affect insulin-dependent Glut4 PM translocation. CHO-IR cells were transduced with adenoviral vectors that simultaneously express HA-Glut4 and either scrambled shRNA or DHHC7 shRNA followed by treatment with or without 10 nm insulin for 15 min. The levels of Glut4, Glut1, and IRAP (the major cargo of Glut4 vesicles) in the PM were determined by Western blotting. As shown in Fig. 3, insulin treatment of scrambled shRNA-expressing cells increased the levels of Glut4 as well as IRAP in the PM ∼3.5-fold (Fig. 3A, panels i and ii, and B). By contrast, insulin had no such effect on PM-associated Glut4 in DHHC7 shRNA-expressing cells (Fig. 3A, panels i and ii). We also examined the impact of DHHC3 shRNA on Glut4 membrane distribution. Unexpectedly, although DHHC3 shRNA expression had little influence on Glut4 palmitoylation (Fig. 2B), it also suppressed insulin-induced Glut4 membrane translocation (Fig. 3A, i and ii). The differences in PM Glut4 observed here were not due to sample variations, as comparable levels of PM Glut1 and total lysate Glut4 were seen in each sample (Fig. 3A, panels ii and iv). Moreover, the membrane localization of Glut1 was not affected by insulin, and comparable levels of Akt Ser473 phosphorylation were seen in all samples (Fig. 3A, panels v and vi), indicating that insulin signaling through Akt did not contribute to the effects of DHHC3 and DHHC7 knockdown on Glut4 PM association.
FIGURE 3.
Regulation of insulin (Ins)-dependent Glut4 membrane translocation by DHHC7. A, CHO cells co-transduced with AAV HA-Glut4 and either scrambled (SCR) shRNA or DHHC3 shRNA or DHHC7 shRNA were serum-deprived and treated with or without insulin (10 nm, 15 min). PM fractions and total lysates (Tcl) were analyzed by Western blotting with anti-Glut4 (i, iii, and iv). In addition, immunoblots of total cell lysates were probed for phospho-Akt (S373) (v) and total Akt (vi), whereas PM fractions were probed for Glut1 as loading control (iii). B, summary statistics of PM Glut4 data illustrated in A. The data were normalized to PM Glut4 of insulin-untreated scrambled RNA-transfected cells and to PM Glut1 levels of each sample. C and D, analyses analogous to A and B except that the cells were, in addition, infected with pMIGR1 retrovirus vector expressing GFP or shRNA-resistant FLAG-DHHC7. PM Glut4 levels were normalized to PM Glut4 in scrambled shRNA-expressing cells without insulin treatment and total levels of Glut4. Note that shRNA-resistant FLAG-DHHC7 restores PM association of Glut4 in DHHC7 shRNA-treated cells. Bar graphs and error bars represent means ± S.D. (n = 3).
The preceding studies have demonstrated that silencing of DHHC7 suppresses insulin-induced Glut4 membrane translocation. To rule out possible shRNA off-target effects, we next reintroduced an shRNA-resistant DHHC7 cDNA into DHHC7 shRNA-expressing cells and examined whether re-expression of DHHC7 would restore insulin-dependent Glut4 PM translocation in DHHC7 shRNA-expressing cells. Specifically, CHO-IR cells were transduced with adenoviral vectors that simultaneously express HA-Glut4 and either scrambled or DHHC7 shRNA as described in Fig. 3A. At 24 h post transduction, the cells were infected with pMGR1 retrovirus encoding either shRNA-resistant FLAG-DHHC7 or GFP (as a control). One day later the cells were serum-deprived overnight followed by treatment with insulin for 15 min. The PM was subjected to Western blotting with an anti-Glut4 antibody. As shown in Fig. 3, C and D, reintroducing DHHC7 into DHHC7 shRNA-expressing cells rescued insulin-induced Glut4 PM association (panel i). By contrast, comparable levels of Glut1 were seen in each sample (panel ii). The expression of HA-Glut4 and DHHC7 was verified by Western blotting of total cell lysates (panels iii and vi).
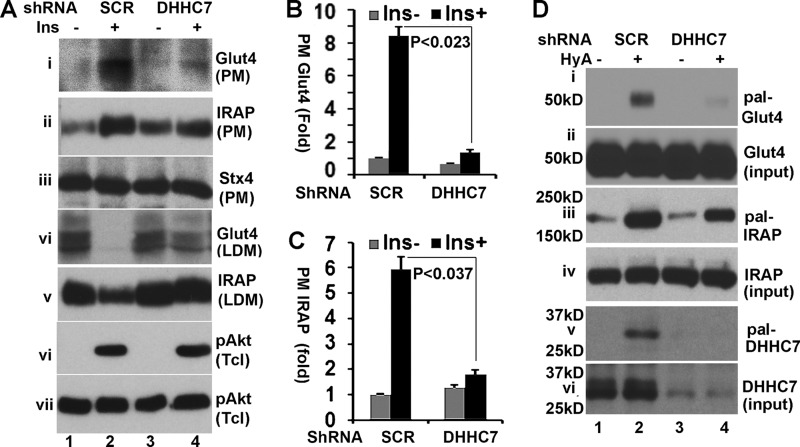
We next examined how DHHC7 affected insulin-dependent Glut4 membrane translocation in fully differentiated 3T3-L1 adipocytes, a cell line that maintains most properties of primary adipocytes (38). Specifically, 3T3-L1 adipocytes were transduced with adenoviral vectors expressing scrambled or DHHC7 shRNA. PM and low density microsome (LDM) fractions were prepared from adipocytes after 30 min of treatment with or without 10 nm insulin. Western blotting indicated that DHHC7 shRNA reduced the amount of insulin-induced PM-associated Glut4 by >80% (Fig. 4, A, panel i, and B), with a corresponding increase of Glut4 in the LDM fraction (panel iv). We further examined the membrane translocation of IRAP, a major cargo of insulin-responsive Glut4 vesicles that is translocated to the PM after insulin stimulation in a Glut4-dependent manner (39). As expected, IRAP PM translocation was impaired in DHHC7 shRNA-expressing cells (Fig. 4, A, panels ii and v, and C). As a control, we examined the level of syntaxin 4 in the PM (panel iii in Fig. 4A); comparable levels of syntaxin 4 were seen in each sample, ruling out PM sample variation. Finally, we examined insulin-induced Akt phosphorylation, the key kinase in insulin-regulated Glut4 membrane translocation. No appreciable changes in insulin-induced Akt phosphorylation were observed in DHHC7 shRNA versus scramble shRNA-expressing cells (panels vi and vii in Fig. 4A).
FIGURE 4.
DHHC7 regulates insulin-dependent Glut4 membrane translocation in adipocytes. A, 3T3-L1 adipocytes were transduced with AAV-scrambled (SCR) shRNA or DHHC7 shRNA, serum-deprived, and insulin-treated (10 nm, 30 min). PM, LDM fractions, and total lysates were subjected to quantitation of Glut4 and IRAP by immunoblotting. Syntaxin 4 (Stx4) was quantitated as a loading control of PM fractions. B and C, quantitation of Glut4 and IRAP in PM fractions. PM Glut4 and PM IRAP were normalized to the corresponding values of insulin (Ins)-untreated cells expressing scrambled RNA and to total Glut4 and IRAP as well as PM syntaxin 4 (Stx4). Bar graphs and error bars represent the means ± S.D. (n = 3). D, TPC assay of 3T3-L1 adipocytes transduced with AAV scrambled (SCR) shRNA or DHHC7 shRNA. Eluates from thiopropyl beads were analyzed by Western blotting with anti-Glut4 (i), anti-IRAP (iii), and anti-DHHC7 (v) antibodies. Panels ii, iv, and v show input levels of Glut4, IRAP, and DHHC7.
DHHC7 modulates Glut4 membrane distribution in adipocytes. Therefore, we examined the Glut4 palmitoylation status in DHHC7 shRNA-expressing 3T3-L1 adipocytes. As shown in Fig. 4D, in agreement with the notion that DHHC7 mediates Glut4 palmitoylation, the levels of Glut4 palmitoylation were reduced by >80% (panel i). In these experiments we also examined IRAP palmitoylation. IRAP palmitoylation was reduced by ∼40% (panel iii), implying that DHHC7 also contributes to IRAP palmitoylation. Finally, we examined DHHC7 expression and its palmitoylation. As expected, the levels of DHHC7 and its palmitoylation in DHHC7 shRNA-expressing cells were reduced (Fig. 4D, panels v and vi). Taken together, these results indicate that DHHC7 regulates Glut4 palmitoylation and insulin-dependent Glut4 membrane translocation.
DHHC7 Knock-out Mice Show Impaired Glut4 Palmitoylation and Glut4 Membrane Translocation in Adipocytes
In the preceding experiments we demonstrated that DHHC7 palmitoylates Glut4 and thereby regulates insulin-dependent Glut4 membrane translocation. To further examine the role of DHHC7 in insulin-dependent Glut4 membrane trafficking, we next took advantage of DHHC3 and DHHC7 knock-out (KO) mice (see Ref. 15 under Experimental Procedures). Notably, we found that like DHHC3 KO mice (15), DHHC7 KO mice showed a modest reduction in body weight compared with their WT littermate controls (DHHC7 WT, 24.71 ± 0.21 g; KO, 23.36 ± 0.27 at 12 weeks of age; DHHC7 WT, 30.11 ± 0.48 g; KO, 28.79 ± 0.06 at 18 weeks of age; p < 0.05, n = 6–8 male mice, t tests, for both comparisons).
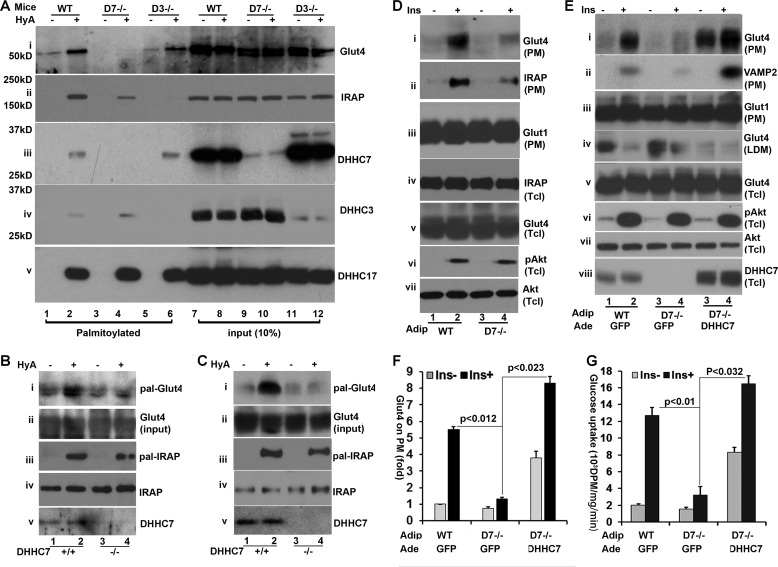
We first examined the status of Glut4 palmitoylation in white adipose tissue isolated from epididymal fat pads of mice. As shown in Fig. 5A, palmitoylation of Glut4 (panel i) was completely abolished in DHHC7 KO adipose tissue, whereas it was unaffected in DHHC3 KO mice (Fig. 5A, panel i, left). By contrast, IRAP palmitoylation was decreased in adipose tissue of both DHHC7 KO and DHHC3 KO versus their respective littermate control mice (panel i, left), an indication that both DHHC3 and DHHC7 contributed to IRPA phosphorylation. The differences in Glut4 and IRAP palmitoylation were not due to sample variations, as comparable levels of these proteins were present in input samples (Fig. 5A, panel i, right). Finally, we verified the loss of DHHC3 and DHHC7 palmitoylation and expression. As expected, the palmitoylation and expression were diminished in DHHC3 KO and DHHC7 KO adipose tissues (panels iii and vi and Fig. 5B). However, faint residual protein bands with molecular weights similar to those of DHHC3 and DHHC7 were observed in the respective DHHC3−/− and DHHC7−/− adipose tissues. These bands likely are due to cross-reactivity of anti-DHHC3 and DHHC7 antibodies with DHHC7 and DHHC3, respectively, as these two proteins exhibited 77% amino acid sequence identity. Finally, as a control, we examined DHHC17 autopalmitoylation. Comparable levels of DHHC17 autopalmitoylation were observed in each sample (Fig. 5A, panel v). Next, we compared the palmitoylation status of Glut4 in skeletal muscle and brown fat of DHHC7 KO versus WT mice, as Glut4 also plays important role in these tissues. As expected, palmitoylation of Glut4 and IRAP was impaired in DHH7 KO muscle (Fig. 5B) and brown fat tissue (Fig. 5C).
FIGURE 5.
Glut4 palmitoylation in adipose tissues of DHHC7 knock-out mice. A, TPC assay of primary adipocytes from epididymal tissue of wild type, DHHC3 KO, and DHHC7 KO mice. The palmitoylation levels of Glut4 (i), IRAP (ii), DHHC7 (iii), DHHC3 (iv), and DHHC7 as well as 10% input of each protein were analyzed by Western blotting. Data shown are representative of three to four experiments. DHHC17 was analyzed as a positive autopalmitoylation and loading control (v). Note that, compared with wild type cells, Glut4 palmitoylation (HyA+ samples) is decreased selectively in DHHC7 KO but not DHHC3 KO adipocytes, whereas IRAP palmitoylation is abolished in DHHC3 KO but not DHHC7 KO adipocytes. B and C, TPC assays showing that Glut4 palmitoylation is impaired in skeletal muscle (B) and brown fat (C) of DHHC7 KO versus wild type mice. D, insulin (Ins)-mediated Glut4 PM translocation in DHHC7 KO adipocytes. Primary adipocytes from epididymal fat pads of DHHC7 KO and wild type littermate mice were treated with insulin (10 nm, 30 min) followed by quantitation by immunoblot of Glut4 (i), IRAP (ii), and Glut1 (iii) in PM fractions and Glut4 (iv), IRAP (v), phospho-Akt (vi), and Akt (vii) in total cell lysates (Tcl). E, re-expression of DHHC7 in DHHC7 KO adipocytes rescues insulin-mediated Glut4 PM translocation. DHHC7 KO adipocytes were transduced with adenoviral vectors expressing GFP (control) or FLAG-DHHC7. At 36 h post transduction, the adipocytes were serum-deprived for 6 h and treated with insulin (10 nm, 30 min) followed by subcellular fractionation assay. The PM levels of Glut4 (i), VAMP (ii), and Glut1 (iii), the LDM levels of Glut4 (iv), and the total levels of Glut4 (v), phospho-Akt (vi), Akt (vii), and DHHC7 (viii) in total cell lysates (Tcl) were determined by Western blotting with antibodies to each protein. F, summary statistics of the data in D. Insulin-induced PM Glut4 levels were normalized to PM Glut4 levels of WT adipocytes without insulin treatment and to PM Glut1 and total Glut4. Note the reduced insulin induced PM association of Glut4 in DHHC7 KO versus wild type adipocytes (n = 3). G, glucose uptake measurements of primary adipocytes from DHHC7 KO versus wild type mice, performed as described under “Experiment Procedures.” Uptake measurements in counts per min/mg of protein/min of KO adipocytes were normalized to wild type. Note the significantly reduced glucose uptake in DHHC KO versus WT adipocytes (n = 3). Bar graphs and error bars indicate means ± S.D.
Next, we examined insulin-dependent Glut4 membrane translocation in adipocytes isolated from DHHC7 KO epididymal fat. In agreement with the notion that DHHC7 is involved in insulin-dependent Glut4 membrane translocation, the PM-associated Glut4 levels induced by insulin in DHHC7 KO adipocytes were reduced by >70% (Fig. 5D, panel i). As expected, IRAP PM translocation was also impaired (panel ii).
To further examine the role of DHHC7 in Glut4 membrane translocation in primary adipocytes, we next examined whether DHHC7 re-expression in DHHC7 KO adipocytes would rescue the defect in basal and insulin-dependent Glut4 membrane translocation. DHHC7 KO adipocytes were transduced with adenoviral vectors that expressed FLAG-tagged DHHC7 and compared with wild type and DHHC7 KO adipocytes that had been transduced with adenoviral vector expressing GFP. PM and LDM fractions were examined separately. Reintroduction of FLAG-DHHC7 into DHHC7 KO adipocytes increased both the basal and insulin-induced PM levels of Glut4 (Fig. 5E), and these effects were accompanied with corresponding reductions of Glut4 in the LDM fractions (Fig. 5E, panel iv).
As a positive control, we examined the level of VAMP2, a SNARE protein that is associated with Glut4 vesicles. As expected, DHHC7 KO impaired VAMP2 membrane translocation and re-expression of DHHC7 restored VAMP2 membrane translocation (Fig. 5E, panel ii). The increased expression of Glut4 and VAMP2 in the PM was not due to sample variation, as comparable levels of PM Glut1 (panel iii) and that of total cellular Glut4 (v) were seen in each sample.
In these experiments we also examined the total cellular levels of DHHC7 in DHHC7 KO adipocytes transduced with DHHC7 expressing adenoviral vectors and observed that the level of DHHC7 was 2–3 times higher than in wild type adipocytes (Fig. 5E, panel viii). This observation is consistent with the previous notion (Fig. 1D) that overexpression of DHHC7 increases the expression of Glut4 in the PM. In further agreement with the notion that DHHC7 does not affect Akt activation, comparable levels of phospho-Akt and total Akt were seen in corresponding samples (Fig. 5E, panels vi and vii). The defect of insulin-dependent Glut4 PM translocation in DHHC7 KO adipocytes resulted in impaired insulin-induced glucose uptake (Fig. 5G), and this defect was normalized when DHHC7 was reintroduced, indicating complete restoration of GluT4 function.
DHHC7 Knock-out Mice Are Hyperglycemic and Glucose-intolerant
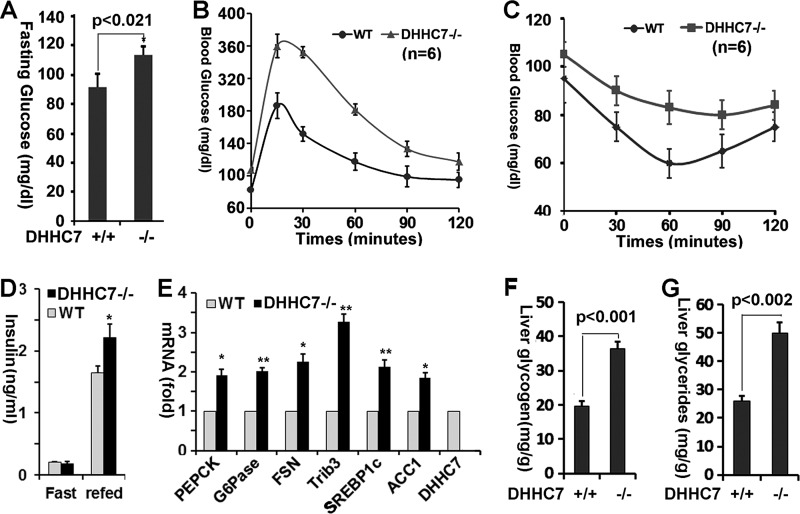
Insulin-dependent Glut4 membrane translocation plays an essential role in the maintenance of systemic glucose homeostasis, as impaired Glut4 function in mice results in hyperglycemia and glucose intolerance (19–21). To assess the impact of DHHC7 inactivation on whole body glucose homeostasis and peripheral insulin sensitivity, we measured serum glucose levels of DHHC7 KO mice. We observed that the fasting serum glucose level of DHHC7 KO mice was significantly increased compared with that of WT littermate controls (Fig. 6A).
FIGURE 6.
DHHC7 knock-out mice show hyperglycemia and glucose intolerance. A, blood glucose levels of male DHHC7 KO mice and wild type littermates. B, glucose tolerance test of DHHC7 KO mice and WT littermates (n = 6). The mice were challenged with d-glucose (2 g/kg, ip) followed by glucose measurements of tail vein blood at the times indicated. C, insulin tolerance test of DHHC7 KO versus WT mice. The mice were challenged with human insulin (0.75 units/kg, ip) and then subjected to glucose measurements in tail vein blood at the times indicated. Note the elevated glucose concentrations (areas under the curve) in both B and C, indicative of impaired glucose tolerance (B) and increased insulin resistance (C) of DHH7 KO versus WT mice. D, blood insulin levels of KO versus wild type littermates (n = 6). All experiments were performed with 6-month-old male mice that were fasted for 6 h. E, quantitative RT-PCR analysis of PEPCK, G6Pase, ACC1, fatty acid synthetase (FSN), SERBP1C, Trib3, and DHHC7 mRNA in liver from KO versus wild type mice (n = 3. *, p < 0.02; **, p < 0.03). F and G, glycogen (F) and triglyceride (G) levels in liver of DHHC7 KO mice versus wild type littermates (n = 6). Graphs with error bars indicate the means ± S.D.
Next, we used glucose and insulin tolerance tests to determine how DHHC7 KO mice respond to glucose and dispose of blood glucose in response to insulin. In the glucose tolerance test, DHHC7 KO mice showed significantly higher glucose blood concentrations than WT controls, indicating glucose intolerance (Fig. 6B). In the insulin tolerance test, DHHC7 KO mice showed increased blood glucose, indicating that DHHC7 KO mice have a lower glucose disposal rate than WT controls (Fig. 6C) and hence are insulin-resistant. The observation that DHHC7 KO mice showed glucose intolerance and insulin resistance confirms that Glut4 palmitoylation and membrane translocation is impaired in these mice.
The preceding results suggested that DHHC7 KO mice developed insulin resistance. We, therefore, measured fasting and refed serum levels of insulin in DHHC7 KO mice and observed that the levels of circulating insulin in DHHC7 KO mice were significant higher than in wild type mice (Fig. 6D).
To further assess whether DHHC7 KO mice develop peripheral insulin resistance, we next examined the expression of glucose and lipid metabolic genes in the liver of DHHC7 KO mice and found that the expression levels of the key gluconeogenesis genes PEPCK and G6Pase and the lipogenesis genes ACC1, SREBP1c and C/EBPα were significantly increased in DHHC7 KO compared with wild type mice (Fig. 6E).
The increase in expression of gluconeogenesis and lipogenesis genes in the liver implied that glucose and lipid metabolisms were altered in DHHC7 KO liver. To directly examine this, we quantitated triglycerides and glycogen in liver of DHHC7 KO liver. As shown in Fig. 6, F and G, the levels of triglycerides and glycogen in DHHC7 KO liver were significantly elevated compared with wild type. Collectively, the results indicate that KO mice developed a type II diabetic syndrome.
Insulin Regulates DHHC7 Autopalmitoylation
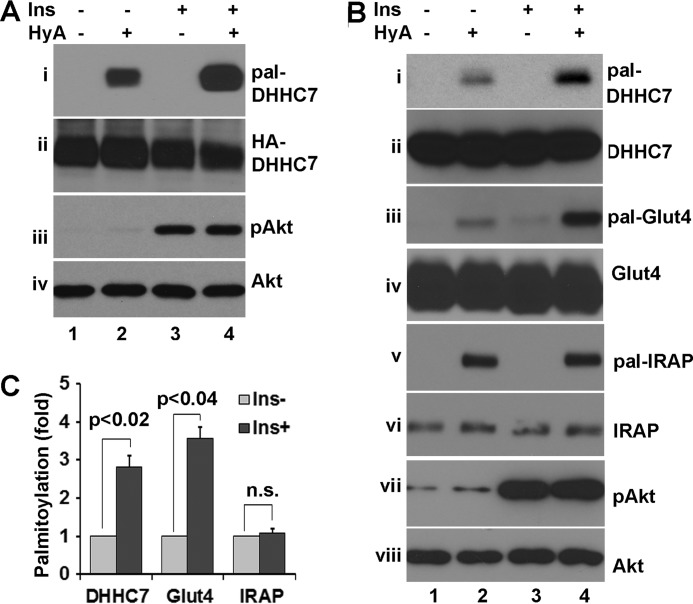
On several occasions (Figs. 2D, 4D, and 5A) we noticed that autopalmitoylation of ectopically expressed DHHC7 was significantly increased compared with that of endogenous DHHC7, as judged by the ratio of autopalmitoyated DHHC7 and its input. DHHC protein PAT activity is critically dependent on autopalmitoylation at the DHHC motif (32, 40, 41). Substitution of the DHHC cysteine residue abolishes autopalmitoylation and results in failure of DHHC enzymes to interact with and palmitoylate their substrates (11, 14). Thus, DHHC protein autopalmitoylation serves as an indicator of DHHC PAT activity. Increased autopalmitoylation of overexpressed DHHC proteins versus autopalmitoylation of endogenous DHHC protein could indicate that autopalmitoylation is normally inhibited by an unknown inhibitory protein. Alternatively, overexpression of DHHC proteins is known to lead to their aggregation, which favors autopalmitoylation over substrate palmitoylation (42). A previous study revealed reduced PAT activity in diabetic and obese mice (43). More recently, insulin was found to promote protein palmitoylation (44). Based on these results, we investigated whether insulin modulates DHHC7 protein autopalmitoylation. HEK293T cells transfected with HA-DHHC7 were treated with 10 nm insulin for 15 min followed by TPC assays to examine HA-DHHC7 autopalmitoylation. As shown in Fig. 7A, we observed that the level of DHHC7 autopalmitoylation was increased ∼3-fold (panel i) by insulin stimulation. Next, we examined the endogenous DHHC7 autopalmitoylation in adipocytes. Similar to assays in heterologous cells, we observed that DHHC7 autopalmitoylation was increased ∼2.5-fold after insulin stimulation (Fig. 7, B, panel i, and C). These changes in DHHC7 autopalmitoylation were not due to altered expression of DHHC7, as comparable levels of input DHHC7 were seen in each sample (panel ii in Fig. 7, A and B). Insulin treatment was effective, as phospho-Akt was induced in insulin-treated cells (Fig. 7A, panels iii and vi, and B, panels vii and viii).
FIGURE 7.
Insulin induced DHHC7 autopalmitoylation. A, HEK293 transfected with HA-DHHC7 were serum-deprived, treated with insulin (In; 10 nm, 10 min), and subjected to TPC assay to detect palmitoylated HA-DHHC (i). The input levels of HA-DHHC7, phospho-Akt, and total Akt are presented in panels ii, iii, and iv. The experiments were repeated three times with similar results. Note the insulin-induced increase in HA-DHHC7 autopalmitoylation. B, TPC assay of 3T3-L1 adipocytes showing insulin-induced palmitoylation of DHHC7 and Glut4. 3T3-L1 adipocytes were treated with or without insulin (10 nm, 10 min). Palmitoylated DHHC7 (i), Glut4 (iii), and IRAP (v) as well as phospho-Akt are shown next to the corresponding total protein levels (panels ii, iv, vi, and viii). C, summary statistics of DHHC7, Glut4, and IRAP in B (n = 3). Insulin-induced palmitoylated levels of each protein were normalized to the corresponding levels without insulin treatment and total amounts of each protein. Bar graphs and error bars indicate means ± S.D.
The notion that insulin potentially induces DHHC7 activity suggests that insulin promotes Glut4 palmitoylation. As expected, Glut4 palmitoylation was increased ∼3-fold after insulin stimulation (Fig. 7, B, panel iii, and C). We also examined IRAP palmitoylation; no appreciable changes of IRAP palmitoylation was seen after insulin treatment (Fig. 7B, panel v). Thus, these data suggest that insulin regulates DHHC7 PAT activity and, thereby, palmitoylation of DHHC7 substrates.
Discussion
In the present report we set out to identify the PAT for Glut4. In our initial screening, we found that 4 of 23 ectopically expressed DHHC proteins were able to palmitoylate Glut4, including DHHC2, DHHC3, DHHC7, and DHHC15 (Fig. 1A). Because DHHC3 and -7, but not DHHC2 and -15, showed strong interaction with Glut4 (Fig. 1B), our first conclusion was that DHHC3 and DHHC7 might represent PATs for Glut4. However, because silencing of DHHC7, but not DHHC3, abolished Glut4 palmitoylation (Fig. 2) and because there was a defect in Glut4 palmitoylation in DHHC7, but not in DHHC3 KO mouse tissues (Fig. 6A), our final conclusion was that DHHC7 is the major PAT for Glut4.
To determine the importance of DHHC7 in insulin-induced Glut4 translocation to the plasma membrane, we examined how DHHC7 inactivation impacts insulin-dependent Glut4 membrane translocation. Our studies revealed that knockdown of DHHC7 in CHO-IR cells (Fig. 3A), 3T3-L1 adipocytes (Fig. 3C), and in primary adipocytes (Fig. 5, D and F) impaired insulin-dependent Glut4 membrane translocation. Because of the importance of Glut4 in the regulation of body glucose homeostasis, we have examined the blood glucose levels and performed glucose tolerance and insulin tolerance tests on DHHC7 KO mice (Fig. 6). Our results indicated that DHHC7 KO mice developed hyperglycemia and showed glucose intolerance, similar to Glut4 adipose and muscle-specific KO mice (19–21), confirming that DHHC7 regulates Glut4 palmitoylation and plasma membrane insertion. At present, we have not yet examined DHHC7 KO mice in terms of glucose and energy metabolism. Regardless, our studies for the first time show that DHHC7 is a regulator of body glucose homeostasis.
An interesting observation was that higher levels of overexpression of DHHC proteins did not simply result in more efficient substrate palmitoylation (Fig. 2B). Oligomerization of DHHC proteins are known to suppress PAT activity (42). Consistent with this idea, DHHC3 and -7 are known to oligomerize on overexpression in HEK293 cells (14). Because of this, our studies of DHHC7 have largely relied on DHHC7 loss of function experiments rather than overexpression.
PAT activity is critically dependent on autopalmitoylation of DHHC proteins (32, 40, 41). Based on the finding that ectopically expressed DHHC7 autopalmitoylation was markedly higher than that of endogenous DHHC7 (Figs. 1D, 2D, and 5A), we postulated that DHHC7 autopalmitoylation is regulated by extracellular signaling. Accordingly, we found that insulin induced DHHC7 autopalmitoylation (Fig. 7). At present, the mechanism by which insulin promotes DHHC7 autopalmitoylation remains to be determined. One explanation is that DHHC7 exists as a multisubunit complex that includes regulatory and catalytic subunits, with DHHC7 as the catalytic subunit. When overexpressed, other subunits might then be titrated out. As a result, the ectopically expressed DHHC7 becomes super-active, which will compromise its enzyme specificity. This probably explains why DHHC2, DHHC3, and DHHC7 have been found to palmitoylate many proteins in in vitro screening (7). Recent studies of DHHC3 revealed that DHHC3 is subject to tyrosine phosphorylation at multiple tyrosine residues, which influences DHHC3 autopalmitoylation (45). Thus, it is possible that insulin promotes DHHC7 tyrosine phosphorylation at specific tyrosine residues that regulates DHHC7 PAT activity. Further studies are required to address this issue.
It is noteworthy that our studies of DHHC protein subcellular localization revealed that DHHC7 is normally localized in trans-Golgi and DHHC3 in cis-Golgi. However, on overexpression, DHHC3 also accumulates in trans-Golgi, and Glut4 resides in the trans-Golgi. Therefore, DHHC3 encounters Glut4 only when it is overexpressed, whereas DHHC7 is naturally expressed in the same Golgi compartment as Glut4, providing a logical explanation of why DHHC7 is a default PAT for Glut4.
There are 100s of palmitoylated proteins in each type of cell (29, 46, 47) and only 23 DHHC PATs in the mammalian genome identified to date (48). At present we cannot rule out that there are other DHHC7 substrates that are involved in insulin-dependent Glut4 membrane translocation and thereby contribute to the glucose intolerance observed in DHHC7 KO mice. Further studies of DHHC7 substrates will address this issue.
Finally, we point out that the stoichiometry of Glut4 palmitoylation is markedly lower than that of IRAP (Figs. 4D and 5A). This might indicate that only a portion of Glut4 is palmitoylated or that Glut4 palmitoylation is highly dynamic and rapidly reversible. Both possibilities may exist, as Glut4 vesicles are highly heterogeneous (22), and only a portion of Glut4 vesicles are insulin-responsive, and Glut4 vesicles constantly cycle from the plasma membrane to intracellular storage compartments, which may affect the Glut4 palmitoylation status. It is noteworthy that DHHC3 also regulated Glut4 membrane translocation, although its deletion had little effect on Glut4 palmitoylation in DHHC3 KO mice. Thus, it is likely that DHHC3 regulates insulin-induced Glut4 membrane translocation through palmitoylation of proteins other than Glut4.
In summary, we have identified DHHC7 as the major palmitoyl acyltransferase for Glut4 and have demonstrated that DHHC7 plays an important role in the regulation of insulin-dependent Glut4 PM translocation and the maintenance of body glucose homeostasis. Our studies for first time implicate DHHC proteins in glucose metabolism. We are only at the beginning of understanding the function of DHHC7 and of its regulation in glucose metabolism. Questions remaining to be addressed include how DHHC7 autopalmitoylation is regulated by insulin and how inactivation of DHHC7 in mice affects body glucose and energy metabolism.
Experimental Procedures
Reagents
All commonly used chemicals and streptavidin-agarose were from Fisher. Dexamethasone, insulin, 3-isobutyl-1-methylxanthine, anti-FLAG- and HRP conjugated anti-HA, anti-DHHC17, and anti-DHHC7 (SAB1101901) antibodies were from Sigma. Anti-IRAP, anti-Akt, and anti-phospho-Akt antibodies were from Cell Signaling. Anti-Glut4 (1F8), anti-β-tubulin, anti-Glut1, anti-syntaxin 4, anti-VAMP2, and mouse monoclonal anti-DHHC3 (sc-377378) antibodies were from Santa Cruz. Methyl methanethiosulfonate, hydroxylamine hydrochloride, and thiopropyl-Sepharose 6B were from GE Healthcare. 17-ODYA was from Caymen. Biotin azide was from Invitrogen. Anti-HA antibody was from Covance.
Plasmid Constructions
HA-tagged DHHC plasmids and FLAG-Glut4 and its mutant expression vectors have been described previously (49). HA-Glut4 expression vectors were gifts from Dr. Joshua J. Zimmerberg (NICHD, National Institutes of Health) (50). DHHC3 shRNA and DHHC7 shRNA were gifts of Dr. Orly Rainer of The Weizmann Institute of Science (51). Adenoviral vectors that express DHHC3 or DHHC7 shRNA or scramble shRNA were generated as previously described procedures (52). Adenoviral vectors that simultaneously express HA-Glut4 and shRNA were used to ensure co-expression of shRNA and HA-Glut4 in the same cells. Specifically, the H1 promoter-driven shRNA was inserted between NruI and MfeI sites upstream of the CMV promoter of pCDNA3 such that the H1 promoter was in the direction opposite that of the CMV promoter. HA-Glut4 was then inserted into the resulting vector between HindIII and XbaI sites to produce pcDNAshRNA-H1-CMV-HA-Glut4. To generate an adenoviral vector, the shRNA-H1-CMV-HA-Glut4 fragment was released from shRNA-pcDNA3-HA-Glut4 with BglII and XbaI and cloned into pShuttle. The HA-tagged DHHC proteins have been described (53). To generate a DHHC7 shRNA-resistant DHHC3 construct, FLAG-tagged DHHC7 was subjected to site-directed mutagenesis with primers forward, 5′-CATGACGTGGCTCCTTGTTGTCTATGAGACTTC-3 and reverse 5′-GAAGTCTGCATAGACAACAAGGGCCACGTCATG-3′ (the bolded sequences correspond to DHHC7 shRNA, and the underlined nucleotides are mutated ones). Then DHHC3 shRNA-resistant FLAG-tagged DHHC7 cDNA was cloned into pMIGR1 retroviral vector (addgene) between BglII and EcoR1 sites. To generate DHHC7-expressing adenoviral vector, FLAG-DHHC7 cDNA was cloned into BglII and EcoRV site of pAdtrack (addgene).
DHHC3 and DHHC7 Knock-out Mice
DHHC3 and DHHC7 knock-out mice were generated as conditional knock-out mice in the 129SvJ genetic background. DHHC3 and DHHC7 global knock-out mice were generated through crossing DHHC3 and DHHC7 conditional knock-out mice with B6.FVB-Tg(EIIa-cre) transgenic mice. In the course of breeding DHHC3 and DHHC7 global knock-out mice, both knock-out mice have been crossing C57BL/6 mice at least >4 generations (15). To ensure that DHHC3 and DHHC7 knock-out mice are in at least >95% C57BL/6 genetic background, these mice were backcrossed with C57BL/6 mice for three more generations. Homozygous DHHC KO mice were then generated through inbreeding of heterozygous mice. The resultant DHHC KO mice and wild type littermates were used for the experiments. The mice were genotyped by PCR of tail biopsies as described (15). The mice were bred and housed at the Tufts University animal facility. Only male mice were used for experimentation. All animal experiments were approved by the Institutional Animal Care and Use Committees (IACUC) of Tufts University and performed in accordance with all relevant guidelines and regulations of the National Institute of Health.
Cell Culture and Transient Transfection
Culture and differentiation of 3T3-L1 pre-adipocytes into adipocytes and subcellular fractionation have been described previously (49). CHO-IR cells were cultured in RPMI 1640 supplemented with 10% (v/v) FBS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). HEK293T cells and COS-7 cells were cultured in DMEM supplemented with 15% FBS, 100 units/ml penicillin, and 100 units/ml streptomycin.
Primary Adipocyte Isolation and Glucose Uptake Assay
The isolation of primary adipocytes from fat pads and subcellular fractionation assays have been described (54). Briefly, the epididymal fat pads were taken from mice and minced. Then the minced adipose tissue was digested with collagenase type IIa in TESCA buffer (50 mm TES, 0.36 mm calcium chloride, pH 7.4) at 37 °C for 2 h. After digestion, the digested tissues were filtered and cultured in DMEM. Transient transfection was carried out with Lipofectamine 2000 according to the manufacturer's instructions (BioTechniques). For subcellular fractionation assay, the primary adipocytes were serum-deprived for 6 h and then treated with or without 10 nm insulin for 30 min. The glucose uptake assay has been described (54). Briefly, primary adipocytes were serum-deprived overnight and then treated with or without insulin for 30 min. The cells were then washed twice with Krebs buffer (50 mm HEPES, pH 7.4, 136 mm NaCl, 4.7 mm KCl, 1.25 mm MgSO4, 1.25 mm CaCl2) and further incubated with Krebs buffer plus 100 mm 2-deoxyglucose, 1 μm [2-3H]deoxyglucose (PerkinElmer Life Sciences). After 5 min of incubation, the cells were washed 3 times with PBS and lysed with 0.05 m NaOH. [2-3H]Deoxyglucose was quantitated using a scintillation counter and normalized to protein concentrations in the lysates determined separately. In addition, the final results were corrected by subtracting background glucose uptake, which was determined by the amount of [2-3H]deoxyglucose in the lysates when the cells were pretreated with 10 μm cytochalasin B (Sigma).
Adenovirus and pMIGR1 Retrovirus Production and Cell Transduction
The adenoviruses were produced and purified by a double CsCl gradient as previously described (52). pMIGR retrovirus was prepared by co-transfection of pMIGR1 vectors with packaging plasmid Ecopack. The viruses were concentrated by centrifuging the medium at 28,000 rpm for 3 h with a SW28 rotor.
To transduce CHO cells with retrovirus, the cells were incubated with viral particles (10 million) in 6 μg/ml Polybrene overnight. Under this condition, infection efficiency was >80% based on GFP expression in the cells as pMIGR virus expresses GFP peptides.
To transduce the cells with adenovirus, 10 μl of CsCl double-banded adenoviruses (∼10 million viral particles) were incubated with the cells in a 10-cm dish containing 5 ml of culture medium. 8 h post transfection, 5 ml of culture medium was added to the cells and incubated overnight. Under this condition, the infection efficiency of adipocytes was ∼75%, and that of CHO cells was >95% as judged by expression of GPF, which was expressed by adenoviral constructs as a separate open reading frame.
Western Blotting
Cells were washed twice with PBS and lysed with cell lysis buffer (20 mm Tris, pH 7.6, 150 mm NaCl, 0.5 mm EDTA, 0.5 mm DTT, 10 mm, 1% Triton X-100 or 1% Nonidet P-40, 10% glycerol, and protease and phosphatase inhibitors). Equal amounts of protein (20–30 μg) from total cell lysates or different fractions in the subcellular fractionation assay were subjected to SDS-PAGE electrophoresis and transferred to polyvinylidene fluoride membrane (Bio-Rad). The membranes were incubated with each primary antibody (1:1000 dilution) followed by incubation with a horseradish peroxidase-conjugated secondary antibody (Bio-Rad). The protein bands were visualized using the ECL detection system (Life Technology, BioTechniques). For quantification, the densities of bands were measured with Image J software.
Crude Plasma Membrane Isolation and Subcellular Fractionation Assay
To isolate crude membranes, the cells were homogenized in HES I buffer (0.25 m sucrose, 20 mm Tris-HCl, pH 7.6, 1 mm EDTA plus protease inhibitors). The nuclei were removed by spinning the homogenates for 10 min at 1000 × g. Then the supernatants were centrifuged for 20 min at 14,000 × g. The pellets represent the crude membrane fraction. For subcellular fractionation assays, CHO cells (in 15-cm dishes) or 3T3-L1 adipocytes (in 10-cm dishes) with or without insulin treatment were suspended into HES I buffer. The cells were homogenized by passing them through a 23-gauge needle 10 times, and then the homogenates were centrifuged at 19,000 × g for 20 min. To isolate membrane fractions, the resultant pellets were resuspended into HES I buffer and layered on HES II buffer (1.12 m sucrose, 20 mm Tris-HCl, pH 7.6, 1 mm EDTA) and centrifuged at 100,000 × g for 60 min. The resultant pellets were designated as nuclear and mitochondria fraction. The plasma membrane layers were removed from the sucrose cushion and suspended into HES I buffer and centrifuged at 41,000 × g for 20 min. The resultant pellets are referred to as the plasma membrane fraction. To isolate microsomes, the supernatant from the 19,000 × g centrifugation was centrifuged at 175,000 × g for 75 min, and the pellets were collected as LDMs. The supernatant from the 175,000 × g centrifugation was saved and designated as cytosol.
Protein Palmitoylation Assay
TPC of S-palmitoylated proteins has been described (29, 31). Briefly, total cytoplasmic extracts were prepared from cultured cells or mouse tissues. Then the extracts were incubated with blocking buffer (100 mm HEPES, pH 7.6, 2.5% SDS, 1 mm EDTA 1 μl/ml methyl methanethiosulfonate) for 20 min at 42 °C, and the proteins were precipitated with 3 volumes of 100% acetone. After centrifugation the protein pellets were dissolved into binding buffer (100 mm HEPES, pH 7.6, 1 mm EDTA) plus thiopropyl-Sepharose beads and incubated for 4 h. The beads were washed three times with binding buffer. Then the proteins on the Sepharose beads were eluted with 100 mm DTT and analyzed by Western blotting with appropriate antibodies.
The 17-ODCA metabolic labeling and Click chemistry was carried out as described (31). Briefly, the cells were cultured in culture medium supplemented with 50 μm 17-ODCA overnight to label palmitoylated proteins. After overnight labeling, the total cell lysates were prepared for Click chemistry to biotinylate labeled proteins with biotin azide. Then the biotinylated labeled proteins were purified with streptavidin-agarose and analyzed by Western blotting with appropriate antibodies.
Glucose and Insulin Measurement, Glucose Tolerance Assay, and Insulin Tolerance Assay
Blood glucose was measured with One Touch ultra2 glucose meter. Blood insulin was measured with EZRMI-13K rat/mouse insulin ELISA kit (EMD Millipore) according to the manufacturer's instruction. For the glucose tolerance test, age-matched WT and KO mice (6 per group) were subjected to fasting for 6 h. The mice were then weighed, fasting blood glucose was measured, and the mice were injected intraperitoneally with a 20% d-glucose (2 g of d-glucose/kg body weight) in saline buffer. After 15, 30, 60, and 120 min, the blood was sampled from the tail vein, and glucose levels were measured as previously described (56). For insulin tolerance assays, baseline glucose levels were determined as described above. Subsequently, the mice were injected intraperitoneally with human insulin (Lilly) (0.75 units/kg body weight). After 15, 30, 60, and 120 min, blood was sampled from the tail, and glucose levels were measured as described (52).
RNA Analysis
Total RNA was isolated from mouse tissues using RNeasy kit (Qiagen). First Strand cDNA was synthesized using the SuperScript® III First-Strand Synthesis System with random decamers as primers (ThermoFisher). cDNAs were amplified using specific primers for the indicated genes. The primers for DHHC7 were 5′-GCGAGGTGATCTACAAGTG-3 (forward) and 5′-CAGAGCTATGTACATGTG-3′ (reverse), which were within the deleted exons. The primers for G6Pase, PEPCK, Trib3, ACC1, SREBP1c, and fatty acid synthetase and the procedure for quantitative PCR have been described (55). Real time PCR was carried out as 94 °C for 45 min, 56 °C for 45 min, and 72 °C for 30 min for 29 cycles.
The Liver Triglycerides and Glycogen Assays
The liver triglycerides and glycogen assay were carried out using the Cayman Chemical kit according to the manufacturer's protocol. Briefly, the liver (200 mg) was homogenized in 1 ml of 1× standard diluent buffer. After homogenizing, the specimen was centrifuged at 10,000 × g for 10 min, the supernatant was diluted 2 times, and 10 μl of the liver extracts were used for the assay. The triglyceride assay was carried out with triglycerides colorimetric assay kit (Cayman Chemical catalog #10010303), and the glycogen assay was carried out with the glycogen assay kit (Cayman Chemical catalog #700480).
Statistical Analyses
Means ± S.D. were calculated, and statistically significant differences among groups were determined by one-way analysis of variance followed by post hoc comparisons or by two-tailed unpaired Student's t tests as appropriate, with significance set at p < 0.05.
Author Contributions
K. D. participated in the study design and performed the experiments, analyzed the data, supervised the study, and wrote the manuscript. S. M. participated in generating KO mice. Y. S participated in the study design, performed the experiments, and analyzed the data. C. L. K. participated in breeding mice and writing the manuscript. B. L. participated in data analyses and wrote the manuscript.
Acknowledgments
We are grateful to Dr. Wenying Ren for technical assistance. We thank Dr. Masaki Fukata (National Institute for Physiological Sciences, Okazaki, Japan) for the HA-tagged DHHC expression vectors and Dr. Samuel W. Cushman (NIDDK, National Institutes of Health, Bethesda, MD) for the HA-tagged-Glut4 expression vector.
This work was supported, in whole or in part, by National Institutes of Health Grants DK084319 (NIDDK; to K. D.) and MH062391 and MH083911 (to B. L.). This work was also supported by a grant from the Pennsylvania Department of Health using Tobacco Settlement Funds. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- DHHC
- Asp-His-His-Cys
- PAT
- palmitoyl acyltransferase
- CRD
- cysteine-rich domain
- Glut4
- glucose transporter 4
- IRAP
- insulin-responsive amino peptidase
- SNAP23
- soluble NSF attachment protein 23 kDa
- PM
- plasma membrane
- LDM
- low density microsomes
- IR
- insulin receptor
- TPC
- thiopropyl captivation
- 17-ODYA
- 17-octadecynoic acid
- G6Pase
- glucose 6-phosphatase
- PEPCK
- phosphoenolpyruvate carboxykinase
- TES buffer
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid
- ACC1
- aceto-CcA caboxylase 1.
References
- 1. Mitchell D. A., Vasudevan A., Linder M. E., and Deschenes R. J. (2006) Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 47, 1118–1127 [DOI] [PubMed] [Google Scholar]
- 2. Nadolski M. J., and Linder M. E. (2007) Protein lipidation. FEBS J. 274, 5202–5210 [DOI] [PubMed] [Google Scholar]
- 3. Sorek N., Bloch D., and Yalovsky S. (2009) Protein lipid modifications in signaling and subcellular targeting. Curr. Opin. Plant Biol. 12, 714–720 [DOI] [PubMed] [Google Scholar]
- 4. Aicart-Ramos C., Valero R. A., and Rodriguez-Crespo I. (2011) Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta. 1808, 2981–2994 [DOI] [PubMed] [Google Scholar]
- 5. Hornemann T. (2015) Palmitoylation and depalmitoylation defects. J. Inherited Metab. Dis. 38, 179–186 [DOI] [PubMed] [Google Scholar]
- 6. Korycka J., Łach A., Heger E., Bogusławska D. M., Wolny M., Toporkiewicz M., Augoff K., Korzeniewski J., and Sikorski A. F. (2012) Human DHHC proteins: a spotlight on the hidden player of palmitoylation. Eur. J. Cell Biol. 91, 107–117 [DOI] [PubMed] [Google Scholar]
- 7. Greaves J., and Chamberlain L. H. (2011) DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem. Sci. 36, 245–253 [DOI] [PubMed] [Google Scholar]
- 8. Roth A. F., Feng Y., Chen L., and Davis N. G. (2002) The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lobo S., Greentree W. K., Linder M. E., and Deschenes R. J. (2002) Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277, 41268–41273 [DOI] [PubMed] [Google Scholar]
- 10. Keller C. A., Yuan X., Panzanelli P., Martin M. L., Alldred M., Sassoè-Pognetto M., and Lüscher B. (2004) The γ2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ. J. Neurosci. 24, 5881–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fukata M., Fukata Y., Adesnik H., Nicoll R. A., and Bredt D. S. (2004) Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 12. Martin B. R. (2013) Chemical approaches for profiling dynamic palmitoylation. Biochem. Soc. Trans. 41, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chaudhary J., and Skinner M. K. (2002) Identification of a novel gene product, Sertoli cell gene with a zinc finger domain, that is important for FSH activation of testicular Sertoli cells. Endocrinology 143, 426–435 [DOI] [PubMed] [Google Scholar]
- 14. Fang C., Deng L., Keller C. A., Fukata M., Fukata Y., Chen G., and Lüscher B. (2006) GODZ-mediated palmitoylation of GABA(A) receptors is required for normal assembly and function of GABAergic inhibitory synapses. J. Neurosci. 26, 12758–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kilpatrick C. L., Murakami S., Feng M., Wu X., Lal R., Chen G., Du K., and Luscher B. (2016) Dissociation of Golgi-associated DHHC-type zinc finger protein (GODZ) and Sertoli cell gene with a zinc finger domain-beta (SERZ-β)-mediated palmitoylation by loss of function analyses in knockout mice. J. Biol. Chem. 291, 27371–27386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thorens B., and Mueckler M. (2010) Glucose transporters in the 21st Century. Am. J. Physiol. Endocrinol. Metab. 298, E141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adekola K., Rosen S. T., and Shanmugam M. (2012) Glucose transporters in cancer metabolism. Curr. Opin. Oncol. 24, 650–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leto D., and Saltiel A. R. (2012) Regulation of glucose transport by insulin: traffic control of GLUT4. Nat. Rev. Mol. Cell Biol. 13, 383–396 [DOI] [PubMed] [Google Scholar]
- 19. Graham T. E., and Kahn B. B. (2007) Tissue-specific alterations of glucose transport and molecular mechanisms of intertissue communication in obesity and type 2 diabetes. Horm. Metab. Res. 39, 717–721 [DOI] [PubMed] [Google Scholar]
- 20. Shulman G. I. (2000) Cellular mechanisms of insulin resistance. J. Clin. Invest. 106, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nandi A., Kitamura Y., Kahn C. R., and Accili D. (2004) Mouse models of insulin resistance. Physiol. Rev. 84, 623–647 [DOI] [PubMed] [Google Scholar]
- 22. Bryant N. J., Govers R., and James D. E. (2002) Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell Biol. 3, 267–277 [DOI] [PubMed] [Google Scholar]
- 23. Cheatham B. (2000) GLUT4 and company: SNAREing roles in insulin-regulated glucose uptake. Trends Endocrinol. Metab. 11, 356–361 [DOI] [PubMed] [Google Scholar]
- 24. Hoffman N. J., and Elmendorf J. S. (2011) Signaling, cytoskeletal, and membrane mechanisms regulating GLUT4 exocytosis. Trends Endocrinol. Metab. 22, 110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Foley K., Boguslavsky S., and Klip A. (2011) Endocytosis, recycling, and regulated exocytosis of glucose transporter 4. Biochemistry 50, 3048–3061 [DOI] [PubMed] [Google Scholar]
- 26. Fukata Y., and Fukata M. (2010) Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 11, 161–175 [DOI] [PubMed] [Google Scholar]
- 27. Galic S., Oakhill J. S., and Steinberg G. R. (2010) Adipose tissue as an endocrine organ. Mol. Cell. Endocrinol. 316, 129–139 [DOI] [PubMed] [Google Scholar]
- 28. Kajimura S., and Saito M. (2014) A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu. Rev. Physiol. 76, 225–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ren W., Jhala U. S., and Du K. (2013) Proteomic analysis of protein palmitoylation in adipocytes. Adipocyte 2, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ren W., Sun Y., and Du K. (2015) Glut4 palmitoylation at Cys-223 plays a critical role in Glut4 membrane trafficking. Biochem. Biophys. Res. Commun. 460, 709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin B. R., and Cravatt B. F. (2009) Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 6, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitchell D. A., Hamel L. D., Ishizuka K., Mitchell G., Schaefer L. M., and Deschenes R. J. (2012) The Erf4 subunit of the yeast Ras palmitoyl acyltransferase is required for stability of the Acyl-Erf2 intermediate and palmitoyl transfer to a Ras2 substrate. J. Biol. Chem. 287, 34337–34348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ebina Y., Edery M., Ellis L., Standring D., Beaudoin J., Roth R. A., and Rutter W. J. (1985) Expression of a functional human insulin receptor from a cloned cDNA in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. U.S.A. 82, 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bogan J. S., Hendon N., McKee A. E., Tsao T. S., and Lodish H. F. (2003) Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature 425, 727–733 [DOI] [PubMed] [Google Scholar]
- 35. Omata W., Shibata H., Suzuki Y., Tanaka S., Suzuki T., Takata K., and Kojima I. (1997) Subcellular distribution of GLUT4 in Chinese hamster ovary cells overexpressing mutant dynamin: evidence that dynamin is a regulatory GTPase in GLUT4 endocytosis. Biochem. Biophys. Res. Commun. 241, 401–406 [DOI] [PubMed] [Google Scholar]
- 36. Lampson M. A., Racz A., Cushman S. W., and McGraw T. E. (2000) Demonstration of insulin-responsive trafficking of GLUT4 and vpTR in fibroblasts. J. Cell Sci. 113, 4065–4076 [DOI] [PubMed] [Google Scholar]
- 37. Bell G. I., Kayano T., Buse J. B., Burant C. F., Takeda J., Lin D., Fukumoto H., and Seino S. (1990) Molecular biology of mammalian glucose transporters. Diabetes Care 13, 198–208 [DOI] [PubMed] [Google Scholar]
- 38. Green H., and Meuth M. (1974) An established pre-adipose cell line and its differentiation in culture. Cell 3, 127–133 [DOI] [PubMed] [Google Scholar]
- 39. Ross S. A., Herbst J. J., Keller S. R., and Lienhard G. E. (1997) Trafficking kinetics of the insulin-regulated membrane aminopeptidase in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 239, 247–251 [DOI] [PubMed] [Google Scholar]
- 40. González Montoro A., Quiroga R., and Valdez Taubas J. (2013) Zinc co-ordination by the DHHC cysteine-rich domain of the palmitoyltransferase Swf1. Biochem. J. 454, 427–435 [DOI] [PubMed] [Google Scholar]
- 41. Ohno Y., Kashio A., Ogata R., Ishitomi A., Yamazaki Y., and Kihara A. (2012) Analysis of substrate specificity of human DHHC protein acyltransferases using a yeast expression system. Mol. Biol. Cell 23, 4543–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lai J., and Linder M. E. (2013) Oligomerization of DHHC protein S-acyltransferases. J. Biol. Chem. 288, 22862–22870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pouliot J. F., and Béliveau R. (1995) Palmitoylation of the glucose transporter in blood-brain barrier capillaries. Biochim. Biophys. Acta 1234, 191–196 [DOI] [PubMed] [Google Scholar]
- 44. Wei X., Song H., and Semenkovich C. F. (2014) Insulin-regulated protein palmitoylation impacts endothelial cell function. Arterioscler. Thromb. Vasc. Biol. 34, 346–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lievens P. M., Kuznetsova T., Kochlamazashvili G., Cesca F., Gorinski N., Galil D. A., Cherkas V., Ronkina N., Lafera J., Gaestel M., Ponimaskin E., and Dityatev A. (2016) ZDHHC3 tyrosine phosphorylation regulates NCAM palmitoylation. Mol. Cell. Biol. 36, 2208–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang W., Di Vizio D., Kirchner M., Steen H., and Freeman M. R. (2010) Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell. Proteomics 9, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R. 3rd, Davis N. G., and El-Husseini A. (2008) Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Greaves J., and Chamberlain L. H. (2010) S-Acylation by the DHHC protein family. Biochem. Soc. Trans. 38, 522–524 [DOI] [PubMed] [Google Scholar]
- 49. Ren W., Cheema S., and Du K. (2012) The association of ClipR-59 protein with AS160 modulates AS160 protein phosphorylation and adipocyte Glut4 protein membrane translocation. J. Biol. Chem. 287, 26890–26900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stenkula K. G., Lizunov V. A., Cushman S. W., and Zimmerberg J. (2010) Insulin controls the spatial distribution of GLUT4 on the cell surface through regulation of its postfusion dispersal. Cell Metab. 12, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shmueli A., Segal M., Sapir T., Tsutsumi R., Noritake J., Bar A., Sapoznik S., Fukata Y., Orr I., Fukata M., and Reiner O. (2010) Ndel1 palmitoylation: a new mean to regulate cytoplasmic dynein activity. EMBO J. 29, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Du K., Herzig S., Kulkarni R. N., and Montminy M. (2003) TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 53. Ren W., Sun Y., and Du K. (2013) DHHC17 palmitoylates ClipR-59 and modulates ClipR-59 association with the plasma membrane. Mol. Cell. Biol. 33, 4255–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ding J., and Du K. (2009) ClipR-59 interacts with Akt and regulates Akt cellular compartmentalization. Mol. Cell Biol. 29, 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ren W., Sun Y., Cheema S., and Du K. (2013) Interaction of constitutive photomorphogenesis 1 protein with protein-tyrosine phosphatase 1B suppresses protein-tyrosine phosphatase 1B activity and enhances insulin signaling. J. Biol. Chem. 288, 10902–10913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Du K., and Yingmin S. (2015) ClipR-59 plays a critical role in the regulation of body glucose homeostasis. Adipocyte 4, 286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]