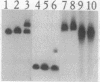
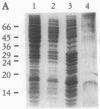
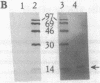
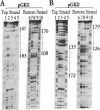
Abstract
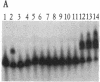
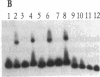
The yeast linear DNA plasmids pGKl1 and pGKl2 contain inverted terminal repeats (ITRs) and terminal proteins covalently bound to the 5' termini of each plasmid. The presence of these features suggests a protein-primed mechanism of DNA replication, similar to that exemplified by mammalian adenovirus and phi 29 phage of Bacillus subtilis. In this paper, we report the identification of an activity in cytoplasmic extracts of yeast harboring the pGKl plasmids that recognizes the termini of both pGKl1 and pGKl2. We call this activity TRF1, for terminal region recognition factor 1. Deletion analyses and DNase I protection experiments demonstrate that the activity recognizes base pairs 107-183 within the ITR of pGKl1, and base pairs 126-179 within the ITR of pGKl2. The presence of T-tracts within these two regions, but otherwise dissimilar nucleotide sequences, suggests that TRF1 recognizes a common structural feature within the ITRs of the two plasmids. TRF1 has been partially purified from yeast cytoplasmic extracts and Southwestern analysis indicates that the apparent molecular mass of the protein is 16 kDa. By expressing three open reading frames from pGKl2 in Escherichia coli, we found that open reading frame 10 (ORF10) of pGKl2 encodes TRF1. The sequence of the ORF10 gene product indicates that TRF1 is a highly basic protein of small molecular mass. Comparison of TRF1 with other DNA-binding proteins known to recognize the terminal regions of linear DNAs, such as NFI and NFIII involved in adenovirus DNA replication, and phi 29 p6, involved in phi 29 DNA replication, indicates that TRF1 has a different mode of binding.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Chen M., Mermod N., Horwitz M. S. Protein-protein interactions between adenovirus DNA polymerase and nuclear factor I mediate formation of the DNA replication preinitiation complex. J Biol Chem. 1990 Oct 25;265(30):18634–18642. [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- Dodson M., Dean F. B., Bullock P., Echols H., Hurwitz J. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science. 1987 Nov 13;238(4829):964–967. doi: 10.1126/science.2823389. [DOI] [PubMed] [Google Scholar]
- Dodson M., Echols H., Wickner S., Alfano C., Mensa-Wilmot K., Gomes B., LeBowitz J., Roberts J. D., McMacken R. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: localized unwinding of duplex DNA by a six-protein reaction. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7638–7642. doi: 10.1073/pnas.83.20.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounari F., De Francesco R., Schmitt J., van der Vliet P., Cortese R., Stunnenberg H. Amino-terminal domain of NF1 binds to DNA as a dimer and activates adenovirus DNA replication. EMBO J. 1990 Feb;9(2):559–566. doi: 10.1002/j.1460-2075.1990.tb08143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunge N., Tamaru A., Ozawa F., Sakaguchi K. Isolation and characterization of linear deoxyribonucleic acid plasmids from Kluyveromyces lactis and the plasmid-associated killer character. J Bacteriol. 1981 Jan;145(1):382–390. doi: 10.1128/jb.145.1.382-390.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Hishinuma F., Hirai K. Genome organization of the linear plasmid, pSKL, isolated from Saccharomyces kluyveri. Mol Gen Genet. 1991 Apr;226(1-2):97–106. doi: 10.1007/BF00273592. [DOI] [PubMed] [Google Scholar]
- Hishinuma F., Nakamura K., Hirai K., Nishizawa R., Gunge N., Maeda T. Cloning and nucleotide sequences of the linear DNA killer plasmids from yeast. Nucleic Acids Res. 1984 Oct 11;12(19):7581–7597. doi: 10.1093/nar/12.19.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen H. M., Admon A., Bell S. P., Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990 Apr 26;344(6269):830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Jung G. H., Leavitt M. C., Ito J. Yeast killer plasmid pGKL1 encodes a DNA polymerase belonging to the family B DNA polymerases. Nucleic Acids Res. 1987 Nov 11;15(21):9088–9088. doi: 10.1093/nar/15.21.9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Jentoft J. E. What is the role of the cys-his motif in retroviral nucleocapsid (NC) proteins? Bioessays. 1989 Dec;11(6):176–181. doi: 10.1002/bies.950110605. [DOI] [PubMed] [Google Scholar]
- Kitada K., Gunge N. Palindrome-hairpin linear plasmids possessing only a part of the ORF1 gene of the yeast killer plasmid pGKL1. Mol Gen Genet. 1988 Dec;215(1):46–52. doi: 10.1007/BF00331301. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. S., Hill W. S., Ng A. S., Vogel U. S., Schaber M. D., Scolnick E. M., Dixon R. A., Sigal I. S., Gibbs J. B. A C-terminal domain of GAP is sufficient to stimulate ras p21 GTPase activity. EMBO J. 1989 Apr;8(4):1105–1110. doi: 10.1002/j.1460-2075.1989.tb03480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskimins W. K., Roberts M. P., McClelland A., Ruddle F. H. Use of a protein-blotting procedure and a specific DNA probe to identify nuclear proteins that recognize the promoter region of the transferrin receptor gene. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6741–6744. doi: 10.1073/pnas.82.20.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Enomoto T., Lichy J. H., Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Hurwitz J. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6177–6181. doi: 10.1073/pnas.80.20.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O., Sakaguchi K., Gunge N. Curing of the killer deoxyribonucleic acid plasmids of Kluyveromyces lactis. J Bacteriol. 1981 Dec;148(3):988–990. doi: 10.1128/jb.148.3.988-990.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E. A., Kelly T. J. Purification and characterization of nuclear factor III (origin recognition protein C), a sequence-specific DNA binding protein required for efficient initiation of adenovirus DNA replication. J Biol Chem. 1988 Jan 15;263(2):931–937. [PubMed] [Google Scholar]
- Prieto I., Serrano M., Lázaro J. M., Salas M., Hermoso J. M. Interaction of the bacteriophage phi 29 protein p6 with double-stranded DNA. Proc Natl Acad Sci U S A. 1988 Jan;85(2):314–318. doi: 10.1073/pnas.85.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K. Invertrons, a class of structurally and functionally related genetic elements that includes linear DNA plasmids, transposable elements, and genomes of adeno-type viruses. Microbiol Rev. 1990 Mar;54(1):66–74. doi: 10.1128/mr.54.1.66-74.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Salas M., Hermoso J. M. A novel nucleoprotein complex at a replication origin. Science. 1990 May 25;248(4958):1012–1016. doi: 10.1126/science.2111580. [DOI] [PubMed] [Google Scholar]
- Sor F., Wésolowski M., Fukuhara H. Inverted terminal repetitions of the two linear DNA associated with the killer character of the yeast Kluyveromyces lactis. Nucleic Acids Res. 1983 Aug 11;11(15):5037–5044. doi: 10.1093/nar/11.15.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam J. C., Kwakman J., Meijer M., Stuitje A. R. Efficient isolation of the linear DNA killer plasmid of Kluyveromyces lactis: evidence for location and expression in the cytoplasm and characterization of their terminally bound proteins. Nucleic Acids Res. 1986 Sep 11;14(17):6871–6884. doi: 10.1093/nar/14.17.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. J., Mileham A. J., Romanos M. A., Boyd A. Nucleotide sequence and transcription analysis of a linear DNA plasmid associated with the killer character of the yeast Kluyveromyces lactis. Nucleic Acids Res. 1984 Aug 10;12(15):6011–6030. doi: 10.1093/nar/12.15.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasino M. Killer system of Kluyveromyces lactis: the open reading frame 10 of the pGK12 plasmid encodes a putative DNA binding protein. Yeast. 1991 Apr;7(3):245–252. doi: 10.1002/yea.320070306. [DOI] [PubMed] [Google Scholar]
- Tommasino M., Ricci S., Galeotti C. L. Genome organization of the killer plasmid pGK12 from Kluyveromyces lactis. Nucleic Acids Res. 1988 Jul 11;16(13):5863–5878. doi: 10.1093/nar/16.13.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer C. P., Kal A. J., Van der Vliet P. C. The DNA binding domain (POU domain) of transcription factor oct-1 suffices for stimulation of DNA replication. EMBO J. 1990 Jun;9(6):1883–1888. doi: 10.1002/j.1460-2075.1990.tb08314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]