Abstract
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the world and the fourth principal cause of cancer deaths worldwide. Currently, there is a lack of low cost and noninvasive screening tests for CRC, becoming a serious health problem. In this context, a potential biomarker for the early detection of CRC has recently gained attention. Circular RNAs (circRNA), a re-discovered, abundant RNA specie, is a type of noncoding covalent closed RNAs formed from both exonic and intronic sequences. These circular molecules are widely expressed in cells, exceeding the abundance of the traditional linear mRNA transcript. They can regulate gene expression, acting as real sponges for miRNAs and also regulate alternative splicing or act as transcriptional factors and inclusive encoding for proteins. However, little is known about circRNA and its relationship with CRC. In this review, we focus on the biogenesis, function and role of these circRNAs in relation to CRC, including their potential as a new biomarker.
Keywords: Circular RNA, Colorectal cancer, Gene regulation, CircRNA, Non-coding RNAs, Long non-coding RNA, Circularization
Core tip: Circular RNAs (circRNAs) are noncoding RNAs, characterized for its circularized shape. These circRNAs are abundant and might play important roles in cancer. In particular, they exhibit altered expression in colorectal cancer, and its activity as miRNA sponge might be involved in the control of cancer progression. Moreover, owing to their stability, could serve as diagnostic or predictive biomarkers for colorectal cancer.
INTRODUCTION
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the world (1.4 million in 2012), and the fourth principal cause of cancer deaths worldwide (694000 deaths for both sexes)[1,2]. In terms of incidence, it represents almost 10% of global cancer diagnoses, and is the third most common cancer in males and second in females (746000 and 614000 respectively), with differences between developed and developing countries[2,3]. More diagnoses occur in developed regions, but less developed regions have higher death rates, reflecting a poorer rate of survival in these countries[1,3]. Sixty-five percent of the new cases occur in developed countries including the United States, Canada, Australia and Europe. In the United States in 2016, the American Cancer Society estimated over 134000 new diagnoses of CRC (95270 for colon, and 39220 for rectal cancer)[2,4]. Also in 2016, CRC is expected to cause over 49190 deaths in the United States[4]. Worldwide the mortality rates are lower in women than in men, except in the Caribbean. But this tendency seems less typical in countries with higher levels of human development[2]. In contrast, CRC incidence and mortality rates in Asia are continually increasing[5]. Besides Asia, in many regions of Europe and North America, the rates of CRC incidence and mortality have decreased for both men and women, due to new screening methods allowing for early diagnosis and treatment[4-6]. These screening tests can prevent the development of CRC, as some have the potential to detect polyps before they can progress into cancer. Not all polyps will progress into cancer tumors, but their removal can prevent the disease[6].
There are several factors that contribute to the development of CRC, some of these are malleable, while others, such as genetic factors, personal medical history, age, racial and ethnic background, or preexisting conditions (inflammatory bowel disease, Lynch syndrome, MUTYH associated polyposis and others) are not[6]. CRC is primarily considered a “lifestyle” disease because its incidence is high in countries with a sedentary population and high-fat diets from animal sources[2]. An association between diet, obesity and carcinogenesis is a very important issue as several studies have shown that obesity promotes inflammatory processes, triggering a cascade of critical mechanisms mediated by proinflammatory cytokines and tumor necrosis factor-α (TNF-α), resulting in CRC development[7-9].
Three fundamental categories of genes have been identified in the carcinogenesis of CRC as follows: (1) APC, DCC, TP53, SMAD2, SMAD4 and p16INK4a (tumor suppressor genes); (2) K-ras and N-ras (proto-oncogenes); and (3) MMR and MUTYH (DNA repair genes). However, little is known about the role of circular RNAs (circRNAs) in the development of CRC. CircRNAs are a class of small group of competing endogenous RNAs (ceRNAs), in the family of the non-coding RNAs (ncRNAs) that function as a class of long non-coding RNA (lncRNA)[10-12]. These molecules have captured the interest of many in the scientific and medical communities, because hundreds of human genes are expressed in circRNA form[13-15]. CircRNA has several functionalities, including the ability to rearrange the order of genomic information, provide protection from exonucleases, and establish constraints on RNA folding[16]. CircRNAs can function as templates for viroid and viral replication, as intermediates in RNA processing reactions, and regulators of transcription in cis. Additionally, it is speculated that circRNAs serve as epigenetic microRNA (miRNA) sponges, negatively regulating miRNAs and thus contributing substantially to the ceRNAs network through RNA-binding protein (RBP) sequestering agents, or nuclear transcriptional regulators, which are frequent in cancer[16-19]. There exists evidence that most circRNAs in the eukaryotic cell are stable, cytoplasmic, lacking the 3’ poly(A) tail and 5’ end cap, arising from pre-mRNA back-splicing exons (downstream 3’ splice donors are covalently linked to upstream 5’ splice acceptors) and host noncoding transcripts generated from protein coding genes[10,12,20-22]. Another study has found evidence that circular isoforms of several human transcripts, exhibiting the non-orderly fashion in the splice junctions, are expressed in similar concentrations to the normal linear isoform[23]. They hypothesized that scrambled exons are a sign of local genomic rearrangements in cancer, and realized this hypothesis by polymerase chain reaction (PCR), where the exon scrambling detected in leukemia patients was also found in normal primary human cell and Hela cells, suggesting that most exon scrambling found in tumor samples are consequences of an active splicing process both in normal and malignant human cells types[23]. This new type of ncRNA can be secreted by cells to the extracellular environment, and can be identified and measured through non-invasive methods, such as stool, blood and other body fluid sampling, suggesting that circRNAs could serve as an effective and cost effective screening test for CRC[15].
In this review, we discuss circRNAs, their biogenesis, possible functions, and implications in CRC.
LITERATURE SEARCH
A systematic literature search was performed of the PubMed database, and the keywords “circRNA”, “circRNA” and “cancer”, “circRNA” and “colorectal cancer” as well other synonyms for circRNAs, with no limitations. The inclusion criteria were as follows: (1) reviews about CRC incidence, statistics, and features of this disease before October 2016; (2) initial studies about circRNAs, its biogenesis, biological functions and techniques used for its identification; (3) studies showing the presence of circRNA in human and other species, detected using different approaches either mathematical, biochemical, biological and technological or a combination; (4) investigations showing the expression of circRNA in different types of biological and cell samples; (5) studies associating circRNA with biological functions and try to explain its biochemical pathway; (6) investigations relating circRNA and CRC; including past, present and future of the disease; and (7) studies associating circRNA with colorectal human samples or cells and discussing its potential use as biomarker for early diagnosis of the disease.
PROPERTIES AND FUNCTIONS OF CIRCRNAS
In the large family of non-protein-coding RNAs, circRNAs are a novel small group of ceRNAs, that could function as a new class of lncRNAs[10-12]. CircRNA is most often composed of exonic sequences containing one or more exons, in contrast to linear RNA which is generally formed by a covalently closed continuous loop containing a 5’-3’ phosphodiester bond and without polyadenylation[10,12,24,25]. This new type of RNA is now known to be expressed in all tissues, at generally low levels but highly represented in the eukaryotic transcriptome[19,26]. Several studies have shown that the circular form of circRNAs gives this molecule a special role in gene expression, but their biological functions have yet to be determined[17,26-29]. In 2013 it was reported that circRNAs are a common, abundant and potentially developmentally regulated component of the gene expression programs in diverse animal species[25]. Salzman et al[23] also demonstrated that circRNAs with scrambled exons compose a significant fraction of cellular RNA for many genes in cancer and normal human cells, suggesting also that the majority of scrambled exons detected in tumor samples were consequential of active splicing processes in both normal and malignant human cell types. Moreover, they subsequently reported that circRNA’s relative abundance is regulated, in a gene and cell type specific manner, at levels comparable to those of canonical linear mRNA, and some of these circRNA species exhibited resistance to a 3’-5’ exoribonuclease - RNase R[25]. Jeck et al[12] demonstrated in the same year, that there exist up to 25000 different circRNAs in human cells, and in some cases are even considerably more abundant (> 10-fold) than associated linear transcripts. Although most circRNAs are expressed at low levels, some have proved to be more abundant than their linear counterparts[25].
A distinctive property of circRNA is they differ structurally from other lncRNAs[19]. In order to create a linear RNA transcript with polarity 5’ to 3’, the eukaryotic cell uses spliceosomal machinery in conjunction with other biochemical process (5’ capping and 3’ polyadenylation) to create a canonical pre-mRNA, removing introns and joint exons[10]. This splicing process is an ubiquitous feature of eukaryotic gene expression, and it is known that the Last Common Eukaryotic Ancestor was relatively intron-rich and had complex spliceosome machinery and splicing signals[21,22]. Conversely, most circRNAs in the eukaryotic cell are stable, cytoplasmic, lacking the 3’ poly(A) tail and 5’ end cap, come from pre-mRNA back-splicing exons and host noncoding transcripts generated from protein coding genes[10,12,20-22]. Guo et al[11] examined the subcellular localization of circRNA, focusing on the 514 circRNAs detected in the K562 whole-cell samples, and found these circRNAs predominantly in the poly(A)-depleted cytoplasmic samples. Jeck et al[12] demonstrated by FISH on Hs68 cells treated with Actinomycin D, against HIPK3; that circular forms of HIPK3 were preferentially localized in the cytoplasm as is consistent with prior studies of RNA circles. Apparently, endogenous circRNAs that are efficiently transported to the cytosol either undergo nuclear export or are released to the cytoplasm during mitosis, where they are extraordinarily stable[12,16,23,30]. This stability, or resistance, to debranching enzymes and RNA exonucleases[12] was also reported by Guo et al[11]; they found that most circRNAs that span < 5 exons are RNase R-resistant. Jeck et al[12] used a method described by Suzuki et al[31] in 2006, using E. coli RNase R, which degrades linear RNAs with short 3′ tails regardless of secondary structure, leaving circRNAs unaffected[12]. Salzman et al[25] found that all the RNA species predicted to be circular are resistant to RNase R whereas all predicted linear sequences are highly sensitive to RNase R.
BIOGENESIS AND REGULATION OF CIRCRNAS
CircRNAs are the product of a back-splicing process also catalyzed by canonical spliceosomal machinery, and it has been reported that this process is modulated by cis-regulatory elements and trans-acting factors, but it is still unclear under which conditions the spliceosomal machinery achieve discrimination between canonical splicing or back-splicing to give rise to a circRNA[10,16,19,20,26,32]. Ashwal-Fluss et al[32] has provided evidence that circRNA are generated co-transcriptionally and their production rate is principally mediated by their flanking intronic sequences. They also demonstrated that canonical pre-mRNA splicing compete with circularization of exons, suggesting that a single pre-mRNA transcript can produce either a linear mRNA or a circular isoform (Figure 1A). Zhang et al[33] indicated that the back-splicing process is generally coupled with canonical splicing, but process happens first depends on the specific spliceosomal machinery and still is under investigation. It is important to know that different types of circRNA molecules are generated by distinct mechanisms, independent of if they arise from intronic or exonic sequences[16,26].
Figure 1.
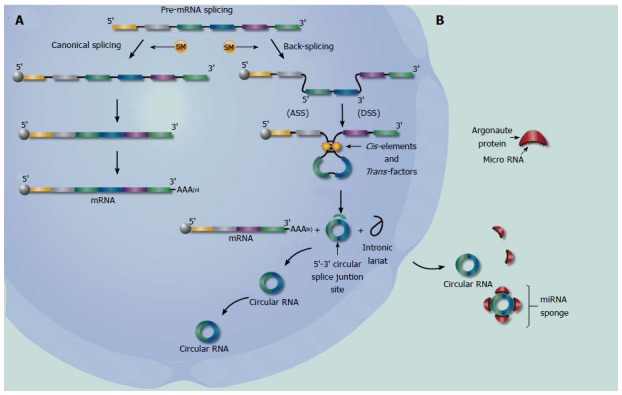
Biogenesis and function of circular RNAs. A: In the nucleus, a single pre-mRNA transcript can produce a linear mRNA and a circular isoform[32]. These two steps, canonical splicing and back-splicing are catalyzed by canonical spliceosomal machinery (Yellow circle, “SM”). In order to create a linear RNA transcript with polarity 5’ to 3’, the eukaryotic cell uses spliceosomal machinery SM together with other biochemical process (5’ capping and 3’ polyadenylation)[10]. Following, circular RNA is produced from a downstream 5’-DSS joined reversely to an upstream 3’-ASS, resulting in a covalently closed circRNA transcript with a 3’, 5’-phosphodiester bond at the junction site (Figure 1)[16,22,26,33,34] and it seems to be modulated by cis-regulatory elements and trans-acting factors[10,16,19,20,26,32]. The final products of these processes are a linear mRNA, a circRNA, and an intron lariat; B: Endogenous circRNAs that are efficiently transported to the cytosol either undergo nuclear export or are released to the cytoplasm during mitosis, where are they are extraordinarily stable[12,16,23,30]. In the cytoplasm circRNAs form a class of post-transcriptional regulators, acting as highly stable epigenetic miRNA sponges competing with the endogenous RNA network (sequestering miRNAs from binding mRNA targets), directly affecting the expression of any related gene[17,18,46].
The process by which a pre-mRNA becomes a circRNA isoform begins with a start codon, and sometimes includes the canonical AUG of the associated linear transcript[12]. When the RNA Pol II recognizes the start codon, it transcribes the exonic circRNA; which is produced from a downstream 5’-DSS inversely joined to an upstream 3’-ASS, resulting in a covalently closed circRNA transcript with a 3’, 5’-phosphodiester bond at the junction site[16,22,26,33,34], or through 2′-5′ linkages formed by a branch-point nucleophilic attack during splicing[35]. The specificity of this process still being studied, but is interesting to note that introns flanking mammalian circRNAs are longer than average[23].
Two other mechanisms, direct back-splicing and lariat intermediate, have also been proposed as associated with circRNA formation from back-spliced exons[12,23]. This suggests that circRNA synthesis is evolutionarily dynamic, providing an alternative generation of circular transcripts[33]. Furthermore, some exons that generally are flanked by longer introns (upstream or downstream), result in a less efficient splicing process. Inverted repeat elements, such as ALU, can facilitate RNA circularization in humans[12,33,34]. Zhang et al[33] identified the phenomenon “Alternative Circularization” (AC), in which a number of multiple exon circularization events can be produced from a single gene loci. This novel process of splicing leads to formation of multiple circRNAs with different expression levels, and it has been associated with diverse biologic processes[12,36,37]. In some events, AC leads to circRNAs retaining introns, adding an additional layer of complexity to the circRNA transcriptome[22,25,26]. It has been suggested that the formation of these isoforms is derived from exon-skipping events. In these events, an exon-containing lariat is formed, which can then be internally spliced to create an exon circle[12,38].
The back-splicing process appears to be regulated by the ratio of circular to linear transcripts, and the relative abundance of differentially spliced circular isoforms is cell-type specific[22,25]. In general, the efficiency of the back-splicing process seems to be much lower than canonical splicing, in steady-state levels of circRNAs; but this efficiency is dependent on the presence of canonical splice sites flanking the exons[20,26,32,33]. Besides, flanking intronic sequences are the main factor determining circularization efficiency of a given exon, and this efficiency is related to the size of the intronic sequence. For example; circRNA flanked by longer introns are less efficiently spliced than other circRNA flanked by smaller introns[32]. Moreover, this circularization appears to have a negative effect on the splicing efficiency in the linear isoform and therefore on gene expression[32]. To overcome this inconvenience, most circRNAs have regulatory elements which reside in the flanking introns of circularized exons to continue with the back-splicing process and facilitate circRNA formation[10,26]. It has been shown that RBPs can regulate and promote circularization, after being recognized by regulatory elements such as short cis elements[26,32].
Another way to facilitate the processing of circRNAs is by RNA base pairing. Complementary short repeat elements, such as inverted ALU repeats, in the flanking introns are used to circularize some intervening exons. This mechanism suggests that the promotion of back-splicing requires for exons to collaborate with intronic repeats and join the back splice sites, thereby facilitating catalysis through base pairing[12,39]. However, in the case of base pairing, the simple presence of these inverted repeats does not necessarily mean that a circRNA will be produced with similar efficiency in all cell types[39]. It is suggested that RBPs, base pairing, and any splicing factor (cis regulatory elements and trans factors), might determine the balance between canonical RNA splicing and back-splicing processes, but whether circRNA is produced co-transcriptionally or post-transcriptionally is still to be determined.
FUNCTIONS OF CIRCULAR RNAS
Little is known about circRNA biology, and a large number of possible functions have been attributed to this circular isoform. In human cells[40], another class of nuclear circRNAs has been identified by Gardner et al[41]: Circular intronic RNAs (ciRNAs) or stable intronic sequences RNA (sisRNA). In human cells (HeLa and human embryonic stem cells hESCs-H9), exon-intron circRNAs were reported, finding that these circRNAs, associated with the elongation Pol II machinery, act as positive regulators of Pol II transcription, suggesting a cis-regulatory role of noncoding intronic products in the efficient transcription of their parent coding genes[42]. CiRNAs do not function only as microRNA sponges, but, some abundant ciRNAs may be able to regulate gene expression in trans under certain circumstances.
A wide variety of studies have demonstrated that abnormal expressions of circRNAs are closely associated with various diseases including CRC[43-45]. It has been speculated that circRNAs form a class of post-transcriptional regulators, acting as epigenetic, highly stable miRNA sponges to compete with the endogenous RNA network, directly affecting the expression of any related gene (Figure 1B)[17,18,46]. CircRNA could be sequestering specific miRNA complexes and releasing them after cleavage[17]. The miRNA sponges ciRS-7 and miR-7, highly expressed in brain, interact to form a miR-7 inhibitor/sponge complex. It is possible that the competition between ciRS-7 and miR-7 effects oncogenesis[18,47]. Similarly, the testis-specific circRNA (Sry) regulates activity of miR-138, a tumor suppressor[17,48]. Therefore, ciRS-7 and Sry complex with miR-7 and miR-138, respectively, to form microRNA sponges blocking functionality, and reducing the invasiveness, metastasis and proliferation of several cancers[47].
CRC shows a negative correlation between global reduction of circRNA and cell proliferation, suggesting that the back-splice machinery responsible for RNA circularization, is dysfunctional in tumor cells[46]. Recently, Xie et al[49] found an association between miR-145 and hsa_circ_001569 and the regulation of CRC progression.
Another circRNA, cir-ITCH acts as a miRNA sponge with an inhibitory effect on esophageal squamous cell carcinoma. Cir-ITCH stimulates ITCH levels, provoking an ubiquitin-mediated Dvl2 degradation, and inhibition of the canonical Wnt/β-catenin pathway, finally, contributing to cancer tumor advancement[50]. The expression of hsa_circ_001988 is downregulated in CRC, and contributes to differentiation and perineural invasion[51]. A study in breast carcinoma cells (MDA-MB-231)[52] shows the possible functions of circ-Foxo3 (hsa_circRNA_104170), which binds to eight miRNAs (miR-22, miR-136*, miR-138, miR-149*, miR-433, miR-762, miR-3614-5p and miR-3622b-5p), as well as Foxo 3 Protein (Foxo3P) and Foxo3 mRNA. Circ-Foxo3 has a sponging effect on these miRNAs, promoting Foxo3 mRNA translation, suppressing both tumor growth and cancer cell proliferation[52].
CIRCRNAS IN CRC
Recent reviews have reported that in CRC cell lines and CRC tissues, a global reduction of circRNA abundance is observed, in comparison to healthy tissue, therefore allowing for the proliferation of CRC cells[53,54].
CircRNAs were associated for the first time with CRC in transcripts of DCC (Deleted in CRC)[30]. In 2015, Bachmayr-Heyda et al[46] reported a global reduction of circRNA abundance in CRC cell lines and tumor samples, as compared to normal mucosa in patients with CRC. Using RNA-seq and the algorithm described by Memczak et al[21] and found by RT-qPCR, 39 circRNAs differentially expressed in the normal colon mucosa and CRC samples, in which 11 of the circRNAs were upregulated and 28 were downregulated. Interestingly, the Bachmayr’s group found that the expression of circRNAs was reduced in tumor samples (circRNA expression 27.8%) compared to normal colon mucosa samples (circRNA expression 78.1%). Similar evidence was found in 11 CRC cell lines, with even wider gaps in expression ratios[46]. In order to validate reduced circRNA expression in CRC compared to normal mucosa samples, they enriched circRNAs through RNase R digestion and subsequent deep sequencing, reporting 21653 distinct back spliced junctions. This method revealed that it is impossible to ensure the detection of all circRNA, and that the actual number of circRNA species is much higher. This was also corroborated in other cell lines in a non-cancerous neo-proliferative disease, showing a similar negative correlation as the circRNA index and proliferation observed in colon tissues and cell lines[46]. Finally, the authors hypothesized that the back-splice machinery responsible for RNA circularization is dysfunctional in tumor cells due to an increased degradation by oncomiRNAs[15,46]. Recently, Wang et al[51] reported similar findings concerning the correlation of hsa_circ_001988 abundance and CRC in tumor tissue and adjacent normal mucosa from 62 CRC patients. By RT-qPCR they verified the presence of hsa_circ_001988 in these tissues, and it was found to be significantly downregulated in tumor tissue compared to healthy samples. The expression level of hsa_circ_001988 was significantly related to differentiation (P < 0.05) and perineural invasion (P < 0.05). Perineural invasion is a predictor of outcome in CRC and negatively associated with survival time and local recurrence in CRC patients. They concluded that hsa_circ_001988 may play a profound role in differentiation and perineural invasion and could be a potential target to regulate cytological behaviors[51]. Recently Xie et al[49] provided information about an up-regulated circRNA (circ_001569) complexing with the tumor suppressor miR-145. MiR-145 has been related with patient survival after CRC diagnosis[55-57]. They explored the expression pattern of circ_001569 by real-time PCR in 30-paired samples of CRC patients, and found expression of circ_001569 was significantly higher in the CRC tissues and found correlation with aggressive characteristics of CRC, including distant metastasis and poor differentiation. Then to detect the function of circ_001569 in the progression and invasion of CRC, they proceeded to over-express and silence the circular isoform in four CRC cell lines (over-expressed in SW480 and HCT116 and silenced in SW620 and LOVO). Accordingly, over-expression increased proliferative and invasive ability in circ_001569 expressing cells, while a sharp reduction in proliferation and invasion rates was shown in circ_001569 silenced cells. In addition, they found the level of miR-145 was significantly lower in CRC tissues[49]. Furthermore, using the bioinformatic algorithms (TargetScan, Pictar and miRANDA), along with Luciferase action, RT-qPCR and Western Blotting assays, they found that circ_001569 increased the protein levels of E2F5, BAG4 and FMNL2 in SW480 and HCT116 cells, and knockdown of the circRNA in SW620 and LOVO cells had the opposite effect. Here, they provided evidence showing tumor promoting functions (proliferation and invasion) of circ_001569 in CRC cells, directly inhibiting the regulatory activity of miR-145, and subsequently up-regulating its protein targets E2F5, BAG4 and FMNL2. Finally, they concluded that this interaction between miR-145 and hsa_circ_001569 in regulating CRC progression may provide new insights and therapeutic strategies for CRC prevention and treatment[49].
CircRNAs that have been related with CRC are shown in Table 1.
Table 1.
Circular RNA found to date in colorectal cancer
| Name | Location | Length [nt] | Tissue/Cell line | Dysregulation | Gene | miRNA associated | Ref. |
| DCC | chr18q21.2 | 253, 948 | HCT116 | None | DCC | None | [30] |
| circ0817 | chr11 | 653 | CRC tissue and CRC cell lines | Downregulated | CUL5 | None | [46] |
| circ3204 | chr15 | 706 | “ | Downregulated | USP3 | None | [46] |
| circ6229 | chr14 | 629 | “ | Downregulated | METTL3 | None | [46] |
| circ7374 | chr17 | 288 | “ | Upregulated | TNS4 | None | [46] |
| circ7780 | chr7 | 317 | “ | Downregulated | -- | None | [46] |
| circ_001988 | chr4q3 | - | CRC tissue | Downregulated | CDR1as | miRNA-7 | [21,51] |
| circ_001569 | chr16q13.1 | - | CRC tissue | Upregulated | ABCC1 | miRNA-145 | [49] |
CRC: Colorectal cancer.
CONCLUSION
The information reviewed is the beginning of a better understanding of mechanisms and functions of this recently re-discovered circular isoform. CircRNAs can act as competing endogenous RNAs or miRNA sponges, regulating alternative splicing or transcription, modulating the expression of parental gene, managing local concentration of RBPs and RNAs through RNA transport. In cancer these molecules complex with miRNAs as inhibitor/sponge, with profound effects on oncogenesis and regulation of cancer pathways. It is possible that more important functions will be associated to this circular transcript in the near future. Still, the molecular mechanisms concerning the positive or negative relationship between circRNAs and miRNAs in CRC are poorly understood.
The search for a suitable circRNA biomarker for CRC is an important topic requiring further study. It is probable that the principal role of circRNAs in the progression of cancer is the modulation of the gene expression of oncogenes and or tumor suppressor genes, through the control of miRNA targets.
Summarizing, we conclude that microRNAs expression levels are regulated by circRNAs in a cell-type specific way, playing an important role in oncogenesis and the malignant behavior of cancer. Nevertheless, the function between circRNA and cancer, as well as the regulatory mechanisms is still elusive and needs to be further explored. Someday this circular isoform might become an important biomarker with a potential use in the diagnosis, prognosis and therapy of CRC.
Footnotes
Conflict-of-interest statement: The authors declare no conflict of interest.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Chile
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Peer-review started: May 4, 2016
First decision: July 20, 2016
Article in press: December 14, 2016
P- Reviewer: Huang ZH, Lakatos PL, Sameer AS S- Editor: Gong ZM L- Editor: A E- Editor: Wu HL
References
- 1.World Health Organization. Estimated Incidence, Mortality and Prevalence Worldwide in 2012. Available from: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx.
- 2.Bosman FT, Hamilton SR, Lambert R. World Cancer Report 2014. 5.5 Colorectal Cancer. In: Stewart BW, Wild CP. World Cancer Report. 2014;IARC. 2014:560–576. [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.American Cancer Society. What is Colorectal cancer (Am Cancer Soc 2015; 74)? Available from: http://www.cancer.org/cancer/colonandrectumcancer/detailedguide/colorectal-cancer-risk-factors.
- 5.Sung JJ, Ng SC, Chan FK, Chiu HM, Kim HS, Matsuda T, Ng SS, Lau JY, Zheng S, Adler S, et al. An updated Asia Pacific Consensus Recommendations on colorectal cancer screening. Gut. 2015;64:121–132. doi: 10.1136/gutjnl-2013-306503. [DOI] [PubMed] [Google Scholar]
- 6.American Cancer Society. Colorectal Cancer Facts & Figures 2014-2016. Available from: http://www.cancer.org/research/cancerfactsstatistics/colorectal-cancer-facts-figures.
- 7.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pietrzyk L, Torres A, Maciejewski R, Torres K. Obesity and Obese-related Chronic Low-grade Inflammation in Promotion of Colorectal Cancer Development. Asian Pac J Cancer Prev. 2015;16:4161–4168. doi: 10.7314/apjcp.2015.16.10.4161. [DOI] [PubMed] [Google Scholar]
- 9.Roberts DL, Dive C, Renehan AG. Biological mechanisms linking obesity and cancer risk: new perspectives. Annu Rev Med. 2010;61:301–316. doi: 10.1146/annurev.med.080708.082713. [DOI] [PubMed] [Google Scholar]
- 10.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Song YX, Ma B, Wang JJ, Sun JX, Chen XW, Zhao JH, Yang YC, Wang ZN. Regulatory Roles of Non-Coding RNAs in Colorectal Cancer. Int J Mol Sci. 2015;16:19886–19919. doi: 10.3390/ijms160819886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ragusa M, Barbagallo C, Statello L, Condorelli AG, Battaglia R, Tamburello L, Barbagallo D, Di Pietro C, Purrello M. Non-coding landscapes of colorectal cancer. World J Gastroenterol. 2015;21:11709–11739. doi: 10.3748/wjg.v21.i41.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 18.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 19.Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 22.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 27.Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin Z, Ma Q, Ren S, Wang G, Li F. The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct Genomics. 2016 doi: 10.1093/bfgp/elw001. Feb 13; Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 29.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 33.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA. 1996;2:603–610. [PMC free article] [PubMed] [Google Scholar]
- 35.Flores R, Grubb D, Elleuch A, Nohales MÁ, Delgado S, Gago S. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol. 2011;8:200–206. doi: 10.4161/rna.8.2.14238. [DOI] [PubMed] [Google Scholar]
- 36.Evsyukova I, Somarelli JA, Gregory SG, Garcia-Blanco MA. Alternative splicing in multiple sclerosis and other autoimmune diseases. RNA Biol. 2010;7:462–473. doi: 10.4161/rna.7.4.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 38.Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci USA. 1996;93:6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 41.Gardner EJ, Nizami ZF, Talbot CC, Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012;26:2550–2559. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 43.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hancock JM. Circles within circles: commentary on Ghosal et al. (2013) “Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits”. Front Genet. 2014;5:459. doi: 10.3389/fgene.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (Review) Oncol Rep. 2015;33:2669–2674. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 48.Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2015 doi: 10.1080/15476286.2015.1122162. Dec 9; Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, Yan Y, Jia B, Liu H, Li S, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 52.Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–3931. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- 53.Yang Y, Du Y, Liu X. Non-coding RNAs in Colorectal Cancer (2016) Available from: http: //link.springer.com/10.1007/978-3-319-42059-2.
- 54.Arvelo F, Sojo F, Cotte C. Biology of colorectal cancer. Ecancermedicalscience. 2015;9:520. doi: 10.3332/ecancer.2015.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li JM, Zhao RH, Li ST, Xie CX, Jiang HH, Ding WJ, Du P, Chen W, Yang M, Cui L. Down-regulation of fecal miR-143 and miR-145 as potential markers for colorectal cancer. Saudi Med J. 2012;33:24–29. [PubMed] [Google Scholar]
- 56.Slattery ML, Wolff E, Hoffman MD, Pellatt DF, Milash B, Wolff RK. MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer. 2011;50:196–206. doi: 10.1002/gcc.20844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]


