Abstract
AIM
To investigate the associations of the genetic polymorphisms of vascular endothelial growth factor A (VEGF-A) -1498C>T and -634G>C, with the survival of patients with colorectal cancer (CRC).
METHODS
A prospective cohort consisting of 131 Brazilians patients consecutively operated on with a curative intention as a result of sporadic colorectal carcinoma was studied. DNA was extracted from peripheral blood and its amplification and allelic discrimination for each genetic polymorphism was performed using the technique of polymerase chain reaction (PCR) in real-time. The real-time PCR technique was used to identify the VEGF-A -1498C>T (rs833031) and -634G>C (rs2010963) polymorphisms. Genotyping was validated for VEGF-A -1498C>T polymorphism in 129 patients and for VEGF-A -634G>C polymorphism in 118 patients. The analysis of association between categorical variables was performed using logistic regression, survival by Kaplan-Meier method and multivariate analysis by the Cox regression method.
RESULTS
In the univariate analysis there was a significant association (OR = 0.32; P = 0.048) between genotype CC of the VEGF-A -1498C>T polymorphism and the presence of CRC liver metastasis. There was no association between VEGF-A -1498C>T polymorphism and VEGF-A -634G>C polymorphism with further clinical or anatomopathologic variables. The genotype CC of the VEGF-A -1498C>T polymorphism was significantly correlated with the 5-year survival (P = 0.032), but not significant difference (P = 0.27) was obtained with the VEGF-A -634G>C polymorphism with the 5-year survival in the univariate analysis. The genotype CT (HR = 2.79) and CC (HR = 4.67) of the polymorphism VEGF-A -1498C>T and the genotype CC (HR = 3.76) of the polymorphism VEGF-A -634C>G acted as an independent prognostic factor for the risk of death in CRC patients.
CONCLUSION
The CT and CC genotypes of the VEGF-A -1498C>T and the CC genotype of the VEGF-A -634C>G polymorphisms are prognostic factors of survival in Brazilians patients with sporadic colorectal carcinoma.
Keywords: Colorectal cancer, Genetic polymorphisms, Vascular endothelial growth factor-A, Colorectal surgery, Genetic variation
Core tip: Vascular endothelial growth factor A (VEGF-A) affects the tumor biological behavior and phenotype. An applied research with relevant achievement that will possibly be favored by such information is the pharmacogenetics impact of VEGF-A polymorphisms. VEGF-A is a significant goal in the anticancer therapy and results about VEGF-A polymorphisms may enhance the targeted therapies. This approach will be of great help to the suitability of individual therapies and improve the quality of post operative treatment. Moreover, since polymorphisms often show a discrepancy between ethnic groups, more studies are also warranted to clarify the association between the VEGF-A polymorphisms and the CRC in diverse ethnic populations.
INTRODUCTION
Colorectal cancer (CRC) is one of the most common malignancies in the world. Despite advances in diagnostic and treatment modalities, patients still face a poor prognosis, and a more individualized treatment approach appears necessary[1-4].
Unlike genetic mutations, polymorphisms represent variations of the naturally occurring DNA sequence. Polymorphisms are found in at least 1% of the healthy population[5]. The vast majority (90%) of DNA polymorphisms is single nucleotide (SNPs)[6,7] and functionally neutral. However, certain polymorphisms have effects on the regulation of gene expression or on the function of the encoded protein[8] and thereby influence the susceptibility and severity of the disease[9,10]. Thus, the polymorphisms may modify the route of angiogenesis and the susceptibility and severity of malignancies[11].
Angiogenesis is a sequence of processes starting with vessel dilatation and pericyte recruitment in the pre-existing vessels, followed by endothelial cell proliferation, formation of new vessels and recruitment of perivascular cells[9]. In the progress toward malignancy, the normal cells must switch to an angiogenic phenotype to attract the nourishing vasculature that they depend on for their growth[9,11].
Malignant tumors depend on angiogenesis for their growth and metastasis[9,11]. It is generally assumed that microvessel formation around the tumor is stimulated by various angiogenic growth factors secreted by the tumor cells[12]. Among them, the vascular endothelial growth factor (VEGF), one of the most potent endothelial cell mitogens, is considered one of the strongest promoters of angiogenesis in CRC[11,12]. The VEGF is vital for the invasion and metastasis of neoplasms through the formation of new blood vessels from mature endothelial cells[13,14]. Moreover, studies have shown that the genetic polymorphisms can be used to predict the clinical outcomes of gastrointestinal[14,15], breast[16], ovary[17] and pancreatic cancer[18].
The cancer targeted therapy with the use of antiangiogenic agents is aimed at inhibition of angiogenic function and tumor dissemination since neoangiogenesis stimulates the growth and invasion of adjacent tissues by tumor cells[9,12]. On the other hand, the inhibition of VEGF limits the tumor growth[9,13]. In colorectal carcinoma, VEGF levels are increased and they are related to further spread and poor prognosis[12,14]. The antiangiogenic agent bevacizumab is a humanized monoclonal immunoglobulin G antibody against recombinant VEGF activity and it is used in the treatment of metastatic CRC[13,14].
Clinical studies have shown that an association between the level of VEGF expression and increased microvessel density in tumors is correlated with an advanced stage of CRC, and with shorter survival. Therefore, it is important to determine the presence of metastasis, and patients with CRC and VEGF overexpression have higher tumor progression and poor prognoses[19-21]. Increased VEGF expression in CRC may predict the risk of multiple liver metastasis and play a role in the spread of CRC cells to the lymph nodes[22]. Due to these properties, VEGF has been used as a therapeutic target for the creation of anticancer drugs and is considered a potential prognostic marker of CRC[13,23].
The VEGF-A gene is located on chromosome 6p21.3 and consists of 8 exons separated by 7 introns that exhibit alternative splicing to form a family of proteins[24]. This gene is a member of the platelet-derived growth factor/VEGF family and encodes a protein that is often found as a disulfide-linked homodimer.
The human VEGF-A gene is highly polymorphic, with more than 15 SNPs described[24,25], thus enabling wide variation in its expression between individuals from different ethnic groups, and there are few studies involving Latinos, Hispanics[20], and particularly Brazilians.
VEGF-A is a dimeric glycoprotein and is considered to be the main, dominant inducer of the growth of blood vessels. VEGF-A is essential for adults during organ remodeling and diseases that involve blood vessels in wound healing, tumor angiogenesis, diabetic retinopathy and age-related macular degeneration.
The -634G>C genetic polymorphism in the promoter region and the -1498C>T genetic polymorphism in the 3’-untranslated region were found to be associated with variations in VEGF-A protein synthesis[24,26,27]. Actually, the VEGF-A -634G/C polymorphism appears to be associated with a higher VEGF-A expression[28,29].
The participation of common genetic polymorphisms of VEGF-A in the prognoses of patients with CRC is not yet clearly established[19,20,23,25,29]. Furthermore, studies investigating the association between VEGF-A genetic polymorphisms and CRC risk report conflicting results[30] and the specific associations still remain controversial[31].
Since VEGF-A is known to be a potent pro-angiogenic factor, we evaluated the potential association of two VEGF-A genetic polymorphisms (-634G>C and -1438C>T) with the clinicopathologic variables and its possible implication for prognosis in a population of Brazilian patients operated on CRC.
MATERIALS AND METHODS
Study design and sample
The present study was conducted according to the ethical principles of the Declaration of Helsinki from the World Medical Association and has been approved by the Institutional Research Ethics Committees.
The study was conducted as a prospective cohort study (observational study) of 131 adult patients of both genders with CRC, without a distinction of ethnicity and operated on consecutively with a curative intention in the period from 2008 to 2011.
The patients were 80 (61.1%) males and 51 (38.9%) females, with a mean age of 58.3 ± 12.5 years (20 to 85 years) and median age of 58 years. Regarding the ethnicity, 121 (92.4%) patients were white and 10 (7.6%) afrodescendants.
The following patients were excluded from the study: Those with familial adenomatous polyposis, colorectal neoplasia other than carcinoma, inflammatory bowel disease, other hereditary CRC syndromes, submitted to neoadjuvant treatment or with synchronous/metachronous tumors elsewhere, except for basal cell carcinoma of skin, and those who were impossible to contact, whether the patients or the patient’s relatives, to obtain the necessary information.
Collection and processing of biological material
Peripheral blood samples were collected by venipuncture into tubes containing 0.1% EDTA and kept in the refrigerator at 4 °C for up to 48 h. If DNA extraction did not take place within this period, the samples were frozen for a maximum of seven days at a temperature of -20 °C. DNA extraction was performed using the method of salting out and storing the material in a freezer at -80 °C.
Surgery data
Regarding the anatomical distribution of the tumors, 98 (74.8%) was found in the colon: 41 (31.3 %) in the right colon, 7 (5.3%) in transverse and 50 (38.2%) in the left colon. In the remaining 33 (25.2%) patients, the tumors were located in the rectum. A right colectomy was performed in 42 (32.1%) cases, a left colectomy in 45 (34.3%), rectosigmoidectomy in 41 (31.3%) and an abdominoperineal resection in 3 (2.3%).
Genotyping
The real-time PCR technique was used to identify genetic polymorphisms in VEGF-A (rs833061 and rs2010963) genes. DNA was amplified using Taqman assays C_10 (rs833061) and C_10 (rs2010963 (Applied Biosystems, Life Technologies Corporation, Foster City, CA, United States) and VIC/FAM dyes (FAM™/ROX™ and VIC®/ROX™ Dye Normalization Plates, Applied Biosystems, Life Technologies Corporation, Foster City, CA, United States). The dye was used as ROX™ passive reference. In each PCR reaction, the following were used: 10 μL of TaqMan Universal Master Mix II (Applied Biosystems, Life Technologies Corporation, Foster City, CA, United States), 1.0 μL of TaqMan Pre-Designed SNP Genotyping Assays 20 × solution (Applied Biosystems, Life Technologies Corporation, Foster City, CA, United States), 30-50 ng of genomic DNA and a final volume of 20 μL of Nuclease-Free Water (Promega, Madison, WI, United States). The equipment used for amplification and allelic discrimination was the Fast ABI-7500 (Applied Biosystems, Life Technologies Corporation, Foster City, CA, United States). Protocols for genotyping were performed according to the manufacturer involving the cycling amplification for 10 min at 95 °C and then 40 cycles for 15 s at 92 °C and for 1 min at 60 °C. At that time, there was the post-run for 1 min allele determination. Negative controls (no template control –NTC) were used, results were analyzed and 10% of all samples were genotyped more than once to ensure there was no contamination. Genotyping was validated for VEGF-A -1498C>T polymorphism in 129 patients and for VEGF-A -634G>C polymorphism in 118 patients.
Statistical analysis
The estimation of the sample power, calculated as the ability to detect a hazard ratio (HR) ≥ 1.7 with an alpha value of 0.05 and a statistical power of 80% provided a minimum sample size of 112 individuals. The estimation was performed using the Stata 11.0 (Stata Statistical Software, StataCorp LP, College Station, TX, United States).
The variable survival was well-defined as the time elapsed between the date of the surgery and the event of interest represented by the patient’s death by CRC. Operationally, this variable is composed of the duration of the follow-up after the operation that was previously established with a maximum duration of 60 mo or the occurrence of death. The clinical variables analyzed were gender, age and presence of metastasis and/or relapse after the operation.
The analyzed histopathological variables were the anatomical site of the CRC in the large intestine, adjacent invasion, degree of cellular differentiation, venous/lymphatic/perineural invasion, and staging of the neoplasia according to the 2010 TNM staging system[32].
Patients who survived until the end of the study follow-up (60 mo) were considered “censored”. In cases in which the event of interest has not occurred after exceeding the maximum observation period of the study (60 mo), loss of observation or occurrence of death were also considered “censored”.
The univariate analysis was made by means of a log-rank test. The significant statistical level was considered as 5% (P < 0.05).
The Cox regression analysis was used to identify the independent effect of the prognostic factors (independent variables). For the selection of independent variables to be included in the multivariate models of Cox regression, each individual variable in the Cox model was tested and the following criteria for inclusion in the multivariate model were verified: Variables with a descriptive level of significance lower than 20% (P < 0.20) in the univariate model and the genetic variations (polymorphisms) used in this study. For the selection of a final model, the automatic selection backwards in Stata 11.0 (Stata Statistical Software, StataCorp LP, College Station, TX, United States) was used.
RESULTS
The follow-up period of 131 patients ranged from 1.8 to 60 mo, with a mean of 33.8 (± 21.9) mo and a median of 34.0 mo. Liver metastasis was found in 26 (19.1%) patients. The involvement of regional lymph nodes occurred in 63 (48.1%) patients and the invasion of adjacent organs was found in 26 (19.8%) cases. At the end of this follow-up period, 70 (53.4%) patients were alive without the disease, 14 (10.7%) were alive with the disease, 42 (32.1%) died with CRC, 3 (2.3%) had died of causes unrelated to CRC and 2 (1.5%) were lost in the follow-up. Thus, censures occurred in 84 (67.9%) patients with a complete observation after 60 mo. The estimate of overall survival at 5 years was 60.7%, with an average of 45.2% (95%CI: 41.5 to 49.0) and a median of 33.0%.
The results of the genotypes’ frequency of the VEGF-A -1498C>T and VEGF-A -634G>C genetic polymorphisms are shown in Table 1. The stages of the CRC and their respective polymorphisms of VEGF -634G>C and the VEGF -1498C>T genes are described in Tables 2 and 3.
Table 1.
Frequency of vascular endothelial growth factor A -1498C>T and vascular endothelial growth factor A -634G>C genetic polymorphisms in blood samples of patients with resected colorectal carcinoma
| Polimorphism | Genotype | n (%) |
| VEGF-A -1498C>T (n = 129) | CC | 27 (20.9) |
| CT | 58 (44.9) | |
| TT | 44 (34.1) | |
| VEGF-A -634G>C (n = 118) | CC | 26 (22.0) |
| CG | 42 (35.6) | |
| GG | 50 (42.4) |
VEGF-A: Vascular endothelial growth factor A.
Table 2.
TNM staging of colorectal tumors and their respective polymorphisms of vascular endothelial growth factor -634G>C gene (n = 118)
| Stage (n) | C/C n (%) | G/C n (%) | G/G n (%) |
| I + II (55) | 10 (18.2) | 22 (40.0) | 23 (4I.8) |
| III + IV (63) | 16 (25.4) | 20 (31.7) | 27 (42.9) |
| Total | 26 (22.0) | 42 (35.6%) | 50 (42.4) |
| P | - | 0.27 | 0.53 |
| OR (95%CI) | 1 | 0.56 (0.21-1.53) | 0.73 (0.27-1.92) |
Cox multiple regression test. OR: Odds ratio.
Table 3.
TNM staging of colorectal tumors and their respective polymorphisms of vascular endothelial growth factor -1498C>T gene (n = 129)
| Stage (n) | C/C n (%) | G/C n (%) | G/G n (%) |
| I + II (61) | 20 (32.8) | 9 (14.8) | 32 (52.5) |
| III + IV (68) | 24 (35.3) | 18 (26.5) | 26 (38.2) |
| Total | 26 (22.0) | 42 (35.6) | 50 (42.4) |
| P | - | 0.31 | 0.33 |
| OR (95%CI) | 1 | 1.66 (0.61-4.51) | 0.67 (0.30-1.48) |
Cox multiple regression test. OR: Odds ratio.
In the univariate analysis there was a significant association (OR = 0.32; P = 0.048) between genotype CC of the VEGF-A -1498C>T polymorphism and the presence of CRC liver metastasis. There was no association between VEGF-A -1498C>T polymorphism with further clinical or anatomopathologic variables (Table 4). The VEGF-A -634G>C polymorphism showed no significant association between clinical or anatomopathologic variables in the univariate analysis.
Table 4.
Univariate analysis of vascular endothelial growth factor A -1498C>T genetic polymorphism and the presence of liver metastasis in patients with resected colorectal carcinoma (n = 129)
| Liver metastasis | Genotype TT n (%) | Genotype CC n (%) | Genotype CT n (%) | |
| Yes (25) | 7 (28.0) | 10 (40.0) | 8 (32.0) | |
| No (104) | 37 (35.6) | 17 (16.3) | 50 (48.1) | |
| P | 0.34 | 0.048 | 0.765 | |
| OR (95%CI) | 1 | 0.32 (0.10-0.98)1 | 1.18 (0.39-3.55) |
Significant. OR: Odds ratio.
Following the previously established criteria, the genotype CC of the VEGF-A -1498C>T polymorphism was significantly correlated with the 5-year survival (P = 0.032) (Table 5), but not significant difference (P = 0.27) was obtained in relation to the VEGF-A -634G>C polymorphism with the 5-year survival in the univariate analysis.
Table 5.
Univariate analysis of vascular endothelial growth factor A -1498C>T genetic polymorphism and 5-year survival in patients with resected colorectal carcinoma (n = 129)
| Polymorphism | Genotype | n (%) | 5-yr survival (%) | P |
| VEGF-A -1498C>T (n = 129) | CC | 27 (20.9) | 46.4 | 0.0321 |
| CT | 58 (44.9) | 61.7 | ||
| TT | 44 (34.1) | 67.1 |
Significant Mantel-Cox log-rank test.
In the multivariate analysis, the genotypes CT (HR = 2.79; 95%CI: 1.01-7.66) and CC (HR = 4.67; 95%CI: 1.51-14.43) of VEGF-A -1498C/T polymorphism and the genotype CC (HR = 3.76; 95%CI: 1.29-10.93) of VEGF-A -634C>G polymorphism were associated with a reduced 5-year survival (Table 6; Figures 1 and 2). The VEGF-A -1498C/T and the VEGF-A -634G>C polymorphisms showed no significant association between clinical or anatomopathologic variables in the multivariate analysis.
Table 6.
Multivariate analysis of vascular endothelial growth factor A -1498C>T genetic polymorphism (n = 129) and vascular endothelial growth factor A -634C>G (n = 118) genotypes and 5-year survival in patients with resected colorectal carcinoma
| Variables | Category | HR (95%CI) | P |
| VEGF-A -1498C>T | TT | 1 (reference) | - |
| CT | 2.79 (1.01- 7.66) | 0.0471 | |
| CC | 4.67 (1.51-14.43) | 0.0071 | |
| VEGF-A -634C>G | GG | 1 (reference) | - |
| CG | 1.44 (0.58-3.55) | 0.433 | |
| CC | 3.76 (1.29-10.93) | 0.0151 |
Significant Cox multiple regression test.
Figure 1.

The Kaplan-Meier survival curves as a function of vascular endothelial growth factor A-1498C>T genetic polymorphism genotypes in colorectal cancer patients. VEGF: Vascular endothelial growth factor.
Figure 2.
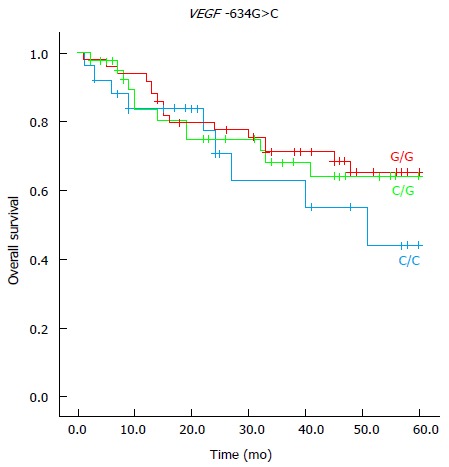
The Kaplan-Meier survival curves as a function of vascular endothelial growth factor A -634G>C genetic polymorphism genotypes in colorectal cancer patients. VEGF: Vascular endothelial growth factor.
DISCUSSION
The selection of studies was carried out by publications on the participation of these SNPs in CRC[25-30]. Due to involvement of angiogenesis in neoplasms, VEGF-A may influence the biology and the phenotype of the CRC[33-36]. The results regarding the association of polymorphisms of VEGF-A with CRC prognosis are controversial[13,29,36-38].
In the present analysis, we examined whether two common VEGF-A -1498C>T and VEGF-A -634G>C polymorphisms were related to prognoses and clinicopathologic features of CRC patients whose tumors had been surgically resected with curative intent.
We used blood samples of patients with colorectal carcinoma because the majority of polymorphism analyses have been carried out on germline DNA extracted from peripheral blood as it is easily obtained and generates large amounts of high quality DNA. Fixation in paraffin-embedded tissue can cause cross-linking and damage of DNA isolated damaging the amplification reaction and primer pattern recognition, besides mutation artifacts, e.g., artificial C-T or G-A transitions[39].
The results of the present study did not support an association of the VEGF-A -1498C>T and VEGF-A -634G>C polymorphisms with tumor size, histological grading, tumor stage, lymph node metastasis and age at diagnosis in CRC cases. In the same way, Hofmann et al[28] and Dassoulas et al[36] found no correlation between the VEGF-A -634G>C and VEGF-A -1498C>T polymorphisms and the tumor characteristics. On the other hand, Chae et al[40] showed that the T allele was related to the advanced stage of CRC.
Kim et al[29] reported that -634GC and -634CC genotypes were associated with a favorable prognosis compared to -634GG genotype VEGF-A polymorphism in CRC patients. Particularly, these authors reported that -634G>C VEGF-A polymorphism is an independent prognostic factor for CRC. Watson et al[24] observed that there is a significant correlation between production of VEGF-A protein from peripheral blood mononuclear cells and VEGF -634G-A>C polymorphism. These authors also reported the decreased production of VEGF-A protein in patients with homozygotes CC VEGF-A gene and increased production of VEGF-A protein in homozygotes GG VEGF-A genes. In the present series, the CC genotype of VEGF-A -634C>G polymorphism was significantly related to survival in patients with resected colorectal carcinoma. In accordance with Kim et al[29], our results pointed out that 634C>C polymorphism was an independent prognostic factor and it was associated with a worse 5-year survival rate compared to the VEGF-A GC genotype. Dassoulas et al[36] analyzed DNA extracted from paraffin-embedded tissue from 312 Greek patients with CRC in all stages and evaluated the prognostic value of five VEGF-A polymorphisms, including the - 634G>C and - 1498C>T polymorphisms. They reported that -634 CC genotype was associated with a poor prognosis in the Greek population, which agree with the results we found with Brazilian patients in the present series. Dassoulas et al[36] concluded that, in Greek patients with CRC, VEGF-A - 634G>C and - 1498C>T, polymorphisms were independent markers of prognosis. Hansen et al[41] demonstrated obvious relationships between genetic variations in the VEGF-A gene and response to first-line capecitabine in patients with metastatic colorectal cancer, which translated to a significant difference in progression-free survival.
Chae et al[40] analyzed the associations of VEGF-A -634G>C polymorphism in patients with CRC. These authors observed that there was no significant correlation between the genotype GC with TNM stage III/IV, lymph node involvement and distant metastasis in CRC. On the other hand, Jang et al[42] genotyped the VEGF-A -634G>C polymorphism in 350 CRC cases from the Korean population. The results suggest that this genetic polymorphism variant is not a potential genetic marker for CRC prognosis.
However, Hansen et al[41] reported opposite results. The authors studied CRC in Danish patients with stages II and III and found that -634GC heterozygote genotype exhibited lower free disease and survival rates compared to the corresponding wild-type homozygote genotypes. This result was the opposite of what we found in the present study.
Kjaer-Frifeldt et al[43] found in a multivariate analysis that VEGF-A -1498C>T and VEGF-A -634G>C polymorphisms were independent prognostic factors for the risk of death of patients by CCR, as we observed in our cases.
The difference between the results of these studies is not sufficiently clear. The discrepancy between the studies of VEGF-A polymorphism and CRC prognosis can be attributed to the differences in disease status, race and size of the sample studied[29,36]. Another possible explanation for these results is the DNA sequence variations in the VEGF-A, variation in the gene locus, action by several other genes and environmental characteristics. All these variables may alter VEGF-A production and/or activity, thereby causing inter-individual differences in the lymphangiogenesis and lymphatic tumor spread and, thus, in the development and progression of the tumors[29,36,39]. The differential role that individual polymorphisms of VEGF-A may play in the biological activity of VEGF-A protein secreted by intratumoral variability of VEGF-A genetic expression is enhanced by these findings. In a large meta-analysis, involving 27 studies Des Guetz et al[19] demonstrated that VEGF-A overexpression is significantly correlated with poor overall survival and with an increased risk of relapse in CRC patients.
The differences between the results of published studies can be attributed to the different sources of DNA, the different ethnic backgrounds of the patients studied, the number of patients tested, the different designs of the studies, a lack of prospective randomized trials, laboratory tests, numerous genetic polymorphisms and errors in the interpretation of results[44-46].
In summary, in the univariate analysis we found an association between genotype CC of the VEGF-A -1498C>T polymorphism and the occurrence of hepatic metastasis of CRC. In the multivariate analysis, genotypes CT and CC of VEGF-A -1498C>T polymorphism and genotype CC of VEGF-A -634C>G genetic polymorphism are independent prognostic factors for the risk of death in Brazilian patients with sporadic CRC.
The study of VEGF-A polymorphisms can bring new impacts on pharmacogenetics as VEGF-A is an important target in antineoplastic therapy, and the results of the VEGF-A polymorphisms may enhance the targeted therapies. This approach will be of great help to physicians in terms of tailoring individual therapies and enhancing the quality of patients’ postoperative treatment. Moreover, since genetic polymorphisms often show a discrepancy between ethnic groups, more studies are also warranted to clarify the association between the VEGF-A polymorphisms and the CRC in diverse ethnic populations.
In conclusion, our data suggested that the CT and CC genotypes of the VEGF-A -1498C>T polymorphisms and the CC genotype of the VEGF-A -634C>G polymorphisms are independent prognostic factors for the risk of death in Brazilian patients with sporadic colorectal carcinoma.
COMMENTS
Background
Polymorphisms are naturally occurring DNA sequence variations, which differ from the gene mutations. The functional polymorphisms could contribute to the difference between individuals according to the susceptibility and severity of diseases. Polymorphisms alone or in combination with environmental factors may affect the angiogenic pathway and, thereby, the susceptibility and severity of cancer. The vascular endothelial growth factor (VEGF), one of the most potent endothelial cell mitogens, is considered one of the strongest promoters of angiogenesis in colorectal cancer (CRC). Given these characteristics, VEGF is a potential marker for determining the prognosis of CRC and it has also been used as a therapeutic target for the new molecular anticancer drugs such as bevacizumab. VEGF-A is considered to be the main, dominant inducer of the growth of blood vessels. The VEGF-A -634G/C polymorphism appears to be associated with a higher VEGF-A expression.
Research frontiers
Studies have shown that the genetic polymorphisms can be used to predict the clinical outcomes of gastrointestinal, breast, ovary and pancreatic cancers. The human VEGF-A gene is highly polymorphic, with more than 15 single nucleotide polymorphisms (SNPs) described, thus enabling wide variation in its expression between individuals from different ethnic groups, and there are few studies involving Latinos, Hispanics, and particularly Brazilians.
Innovations and breakthroughs
The contribution of common VEGF-A genetic polymorphisms to the CRC prognosis remains unclear. Furthermore, studies investigating the association between VEGF-A genetic polymorphisms and CRC risk reporting conflicting results and the specific associations still remain controversial. Because VEGF-A is known to be a potent proangiogenic factor, the authors evaluated the potential association of two VEGF-A genetic polymorphisms (-634G>C and -1438C>T) with the clinicopathologic variables and its possible implication for prognoses in a population of Brazilian patients who had surgical procedures to remove CRC. The authors found an association between genotype CC of the VEGF-A -1498C>T genetic polymorphism and the occurrence of hepatic metastasis of CRC. Moreover, genotypes CT and CC of VEGF-A -1498C>T genetic polymorphism and genotype CC of VEGF-A -634C>G genetic polymorphism were independent prognostic factors for the risk of death in Brazilian patients with sporadic CRC.
Applications
An important translational research field that will benefit from this knowledge is the pharmacogenetics field, in which researches study the impact of VEGF-A polymorphisms. VEGF-A protein is an important target in anticancer therapy and findings about VEGF-A polymorphisms may enhance the targeted therapies. This approach will be of great help to physicians in terms of tailoring individual therapies and enhancing the quality of patients’ postoperative treatments.
Terminology
Polymorphism is the occurrence of two or more clearly different forms or alternative phenotypes in the population of a species. Genetic polymorphism is the occurrence together in the same population of two or more genetically determined phenotypes in such proportions that the rarest of them cannot be maintained merely by recurrent mutation. Most genetic polymorphisms are functionally neutral, but some have effects on the regulation of the gene expression or on the function of the coded protein. SNPs are a genetic polymorphism between two genomes that is based on substitution, deletion, insertion, or exchange of a single nucleotide. Angiogenesis is a sequence of processes starting with vessel dilatation and pericyte recruitment in the pre-existing vessels, followed by endothelial cell proliferation, formation of new vessels, and recruitment of perivascular cells.
Peer-review
This is an excellent article and the findings will definitely add to the existing knowledge.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Research Ethic Committee of UNIC (University of Cuiabá) and in the Research Ethic Committee of Federal University of São Paulo.
Informed consent statement: All study participants, or their legal guardian, provided written consent prior to study enrollment.
Conflict-of-interest statement: The authors of this manuscript having no conflicts of interest to disclose.
Data sharing statement: There is no additional data available.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Brazil
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Peer-review started: June 24, 2016
First decision: August 18, 2016
Article in press: December 14, 2016
P- Reviewer: Kobayashi T, Rastogi A S- Editor: Song XX L- Editor: A E- Editor: Wu HL
References
- 1.Mejia A, Schulz S, Hyslop T, Weinberg DS, Waldman SA. Molecular staging individualizing cancer management. J Surg Oncol. 2012;105:468–474. doi: 10.1002/jso.21858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaton C, Twine CP, Williams GL, Radcliffe AG. Systematic review and meta-analysis of histopathological factors influencing the risk of lymph node metastasis in early colorectal cancer. Colorectal Dis. 2013;15:788–797. doi: 10.1111/codi.12129. [DOI] [PubMed] [Google Scholar]
- 3.Zhang BB, Chen TT, Wei QZ, Wang GC, Lu M. Risk factors for survival after colorectal cancer resection. Hepatogastroenterology. 2013;60:528–532. doi: 10.5754/hge12759. [DOI] [PubMed] [Google Scholar]
- 4.Yuan Y, Li MD, Hu HG, Dong CX, Chen JQ, Li XF, Li JJ, Shen H. Prognostic and survival analysis of 837 Chinese colorectal cancer patients. World J Gastroenterol. 2013;19:2650–2659. doi: 10.3748/wjg.v19.i17.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SC, Lau JY, Chan FK, Suen BY, Leung WK, Tse YK, Ng SS, Lee JF, To KF, Wu JC, et al. Increased risk of advanced neoplasms among asymptomatic siblings of patients with colorectal cancer. Gastroenterology. 2013;144:544–550. doi: 10.1053/j.gastro.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Naylor K, Ward J, Polite BN. Interventions to improve care related to colorectal cancer among racial and ethnic minorities: a systematic review. J Gen Intern Med. 2012;27:1033–1046. doi: 10.1007/s11606-012-2044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins MA, Makalic E, Dowty JG, Schmidt DF, Dite GS, MacInnis RJ, Ait Ouakrim D, Clendenning M, Flander LB, Stanesby OK, et al. Quantifying the utility of single nucleotide polymorphisms to guide colorectal cancer screening. Future Oncol. 2016;12:503–513. doi: 10.2217/fon.15.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider-Stock R, Boltze C, Peters B, Szibor R, Landt O, Meyer F, Roessner A. Selective loss of codon 72 proline p53 and frequent mutational inactivation of the retained arginine allele in colorectal cancer. Neoplasia. 2004;6:529–535. doi: 10.1593/neo.04178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jannuzzi AT, Özhan G, Yanar HT, Alpertunga B. VEGF gene polymorphisms and susceptibility to colorectal cancer. Genet Test Mol Biomarkers. 2015;19:133–137. doi: 10.1089/gtmb.2014.0259. [DOI] [PubMed] [Google Scholar]
- 10.Morris EJ, Penegar S, Whiffin N, Broderick P, Bishop DT, Northwood E, Quirke P, Finan P, Houlston RS. A retrospective observational study of the relationship between single nucleotide polymorphisms associated with the risk of developing colorectal cancer and survival. PLoS One. 2015;10:e0117816. doi: 10.1371/journal.pone.0117816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langsenlehner U, Wolf G, Langsenlehner T, Gerger A, Hofmann G, Clar H, Wascher TC, Paulweber B, Samonigg H, Krippl P, et al. Genetic polymorphisms in the vascular endothelial growth factor gene and breast cancer risk. The Austrian “tumor of breast tissue: incidence, genetics, and environmental risk factors” study. Breast Cancer Res Treat. 2008;109:297–304. doi: 10.1007/s10549-007-9655-z. [DOI] [PubMed] [Google Scholar]
- 12.Roy H, Bhardwaj S, Ylä-Herttuala S. Biology of vascular endothelial growth factors. FEBS Lett. 2006;580:2879–2887. doi: 10.1016/j.febslet.2006.03.087. [DOI] [PubMed] [Google Scholar]
- 13.Hansen TF, Jakobsen A. Clinical implications of genetic variations in the VEGF system in relation to colorectal cancer. Pharmacogenomics. 2011;12:1681–1693. doi: 10.2217/pgs.11.118. [DOI] [PubMed] [Google Scholar]
- 14.Lau TP, Roslani AC, Lian LH, Lee PC, Hilmi I, Goh KL, Chua KH. Association between EGF and VEGF functional polymorphisms and sporadic colorectal cancer in the Malaysian population. Genet Mol Res. 2014;13:5555–5561. doi: 10.4238/2014.July.25.9. [DOI] [PubMed] [Google Scholar]
- 15.Ruzzo A, Graziano F, Kawakami K, Watanabe G, Santini D, Catalano V, Bisonni R, Canestrari E, Ficarelli R, Menichetti ET, et al. Pharmacogenetic profiling and clinical outcome of patients with advanced gastric cancer treated with palliative chemotherapy. J Clin Oncol. 2006;24:1883–1891. doi: 10.1200/JCO.2005.04.8322. [DOI] [PubMed] [Google Scholar]
- 16.Jin Q, Hemminki K, Enquist K, Lenner P, Grzybowska E, Klaes R, Henriksson R, Chen B, Pamula J, Pekala W, et al. Vascular endothelial growth factor polymorphisms in relation to breast cancer development and prognosis. Clin Cancer Res. 2005;11:3647–3653. doi: 10.1158/1078-0432.CCR-04-1803. [DOI] [PubMed] [Google Scholar]
- 17.Hefler LA, Mustea A, Könsgen D, Concin N, Tanner B, Strick R, Heinze G, Grimm C, Schuster E, Tempfer C, et al. Vascular endothelial growth factor gene polymorphisms are associated with prognosis in ovarian cancer. Clin Cancer Res. 2007;13:898–901. doi: 10.1158/1078-0432.CCR-06-1008. [DOI] [PubMed] [Google Scholar]
- 18.Li D, Frazier M, Evans DB, Hess KR, Crane CH, Jiao L, Abbruzzese JL. Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J Clin Oncol. 2006;24:1720–1728. doi: 10.1200/JCO.2005.04.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Des Guetz G, Uzzan B, Nicolas P, Cucherat M, Morere JF, Benamouzig R, Breau JL, Perret GY. Microvessel density and VEGF expression are prognostic factors in colorectal cancer. Meta-analysis of the literature. Br J Cancer. 2006;94:1823–1832. doi: 10.1038/sj.bjc.6603176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen TF, Spindler KL, Andersen RF, Lindebjerg J, Kølvraa S, Brandslund I, Jakobsen A. The prognostic value of haplotypes in the vascular endothelial growth factor a gene in colorectal cancer. Cancers (Basel) 2010;2:1405–1418. doi: 10.3390/cancers2031405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao YJ, Han HZ, Liang Y, Shi CZ, Zhu QC, Yang J. Alternative splicing of VEGFA, APP and NUMB genes in colorectal cancer. World J Gastroenterol. 2015;21:6550–6560. doi: 10.3748/wjg.v21.i21.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saad RS, Kordunsky L, Liu YL, Denning KL, Kandil HA, Silverman JF. Lymphatic microvessel density as prognostic marker in colorectal cancer. Mod Pathol. 2006;19:1317–1323. doi: 10.1038/modpathol.3800651. [DOI] [PubMed] [Google Scholar]
- 23.Giampieri R, Salvatore L, Del Prete M, Prochilo T, D’Anzeo M, Loretelli C, Loupakis F, Aprile G, Maccaroni E, Andrikou K, et al. Angiogenesis genotyping and clinical outcome during regorafenib treatment in metastatic colorectal cancer patients. Sci Rep. 2016;6:25195. doi: 10.1038/srep25195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000;12:1232–1235. doi: 10.1006/cyto.2000.0692. [DOI] [PubMed] [Google Scholar]
- 25.Credidio L, Lima CS, Leal R, de Ayrizono ML, Fagundes JJ, Magna LA, Coy CS. C936T polymorphism of the VEGF gene in relation to the risk and the clinical and biological characteristics of sporadic colorectal adenocarcinoma. BMC Res Notes. 2014;7:768. doi: 10.1186/1756-0500-7-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamori M, Taniguchi M, Maeda S, Nakamura T, Okamura N, Kuwahara A, Iwaki K, Tamura T, Aoyama N, Markova S, et al. VEGF T-1498C polymorphism, a predictive marker of differentiation of colorectal adenocarcinomas in Japanese. Int J Med Sci. 2008;5:80–86. doi: 10.7150/ijms.5.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Li D, Liu Z, Wan X, Wu Y, Jiang C, Qian Q. Vascular endothelial growth factor 1498C/T, 936C/T polymorphisms associated with increased risk of colorectal adenoma: a Chinese case-control study. Mol Biol Rep. 2011;38:1949–1955. doi: 10.1007/s11033-010-0316-7. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann G, Langsenlehner U, Renner W, Langsenlehner T, Yazdani-Biuki B, Clar H, Gerger A, Wehrschuetz M, Samonigg H, Krippl P. Common single nucleotide polymorphisms in the vascular endothelial growth factor gene and colorectal cancer risk. J Cancer Res Clin Oncol. 2008;134:591–595. doi: 10.1007/s00432-007-0322-x. [DOI] [PubMed] [Google Scholar]
- 29.Kim JG, Chae YS, Sohn SK, Cho YY, Moon JH, Park JY, Jeon SW, Lee IT, Choi GS, Jun SH. Vascular endothelial growth factor gene polymorphisms associated with prognosis for patients with colorectal cancer. Clin Cancer Res. 2008;14:62–66. doi: 10.1158/1078-0432.CCR-07-1537. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Z, Ba C, Wang W, Wang X, Xue R, Wu X. Vascular endothelial growth factor (VEGF) gene polymorphisms and colorectal cancer: a meta-analysis of epidemiologic studies. Genet Test Mol Biomarkers. 2012;16:1390–1394. doi: 10.1089/gtmb.2012.0266. [DOI] [PubMed] [Google Scholar]
- 31.Zhou LP, Luan H, Dong XH, Jin GJ, Man DL, Shang H. Vascular endothelial growth factor gene polymorphisms and colorectal cancer risk: a meta-analysis. Genet Mol Res. 2011;10:3674–3688. doi: 10.4238/2011.October.31.13. [DOI] [PubMed] [Google Scholar]
- 32.Asare EA, Washington MK, Gress DM, Gershenwald JE, Greene FL. Improving the quality of cancer staging. CA Cancer J Clin. 2015;65:261–263. doi: 10.3322/caac.21284. [DOI] [PubMed] [Google Scholar]
- 33.Howell WM, Bateman AC, Turner SJ, Collins A, Theaker JM. Influence of vascular endothelial growth factor single nucleotide polymorphisms on tumour development in cutaneous malignant melanoma. Genes Immun. 2002;3:229–232. doi: 10.1038/sj.gene.6363851. [DOI] [PubMed] [Google Scholar]
- 34.Koukourakis MI, Papazoglou D, Giatromanolaki A, Bougioukas G, Maltezos E, Sivridis E. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer. 2004;46:293–298. doi: 10.1016/j.lungcan.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 35.Tzanakis N, Gazouli M, Rallis G, Giannopoulos G, Papaconstantinou I, Theodoropoulos G, Pikoulis E, Tsigris C, Karakitsos P, Peros G, et al. Vascular endothelial growth factor polymorphisms in gastric cancer development, prognosis, and survival. J Surg Oncol. 2006;94:624–630. doi: 10.1002/jso.20619. [DOI] [PubMed] [Google Scholar]
- 36.Dassoulas K, Gazouli M, Rizos S, Theodoropoulos G, Christoni Z, Nikiteas N, Karakitsos P. Common polymorphisms in the vascular endothelial growth factor gene and colorectal cancer development, prognosis, and survival. Mol Carcinog. 2009;48:563–569. doi: 10.1002/mc.20495. [DOI] [PubMed] [Google Scholar]
- 37.Lurje G, Zhang W, Schultheis AM, Yang D, Groshen S, Hendifar AE, Husain H, Gordon MA, Nagashima F, Chang HM, et al. Polymorphisms in VEGF and IL-8 predict tumor recurrence in stage III colon cancer. Ann Oncol. 2008;19:1734–1741. doi: 10.1093/annonc/mdn368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidaurreta M, Sánchez-Muñoz R, Veganzones S, Rafael S, Gutiérrez M, de-la-Orden V, Fernández C, Arroyo M, Cerdán FJ, Maestro de las Casas ML. Vascular endothelial growth factor gene polymorphisms in patients with colorectal cancer. Rev Esp Enferm Dig. 2010;102:20–31. doi: 10.4321/s1130-01082010000100004. [DOI] [PubMed] [Google Scholar]
- 39.Marisi G, Passardi A, Calistri D, Zoli W, Amadori D, Ulivi P. Discrepancies between VEGF -1154 G& gt; A polymorphism analysis performed in peripheral blood samples and FFPE tissue. Int J Mol Sci. 2014;15:13333–13343. doi: 10.3390/ijms150813333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chae YS, Kim JG, Sohn SK, Cho YY, Ahn BM, Moon JH, Jeon SW, Park JY, Lee IT, Choi GS, et al. Association of vascular endothelial growth factor gene polymorphisms with susceptibility and clinicopathologic characteristics of colorectal cancer. J Korean Med Sci. 2008;23:421–427. doi: 10.3346/jkms.2008.23.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen TF, Garm Spindler KL, Andersen RF, Lindebjerg J, Brandslund I, Jakobsen A. The predictive value of genetic variations in the vascular endothelial growth factor A gene in metastatic colorectal cancer. Pharmacogenomics J. 2011;11:53–60. doi: 10.1038/tpj.2010.4. [DOI] [PubMed] [Google Scholar]
- 42.Jang MJ, Kim JW, Jeon YJ, Chong SY, Oh D, Kim NK. Prognostic significance of vascular endothelial growth factor gene polymorphisms in patients with colorectal cancer. Int J Clin Oncol. 2013;18:1032–1041. doi: 10.1007/s10147-012-0493-6. [DOI] [PubMed] [Google Scholar]
- 43.Kjaer-Frifeldt S, Fredslund R, Lindebjerg J, Hansen TF, Spindler KL, Jakobsen A. Prognostic importance of VEGF-A haplotype combinations in a stage II colon cancer population. Pharmacogenomics. 2012;13:763–770. doi: 10.2217/pgs.12.38. [DOI] [PubMed] [Google Scholar]
- 44.Jain L, Vargo CA, Danesi R, Sissung TM, Price DK, Venzon D, Venitz J, Figg WD. The role of vascular endothelial growth factor SNPs as predictive and prognostic markers for major solid tumors. Mol Cancer Ther. 2009;8:2496–2508. doi: 10.1158/1535-7163.MCT-09-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rollin J, Payancé A, Gouilleux-Gruart V, Boisdron-Celle M, Azzopardi N, Morel A, Gruel Y, Paintaud G, Gamelin E, Watier H, et al. Significant effect of VEGFA polymorphisms on the clinical outcome of metastatic colorectal cancer patients treated with FOLFIRI-cetuximab. Pharmacogenomics. 2015;16:2035–2043. doi: 10.2217/pgs.15.139. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, Ji S, Cheng Z. Association between Polymorphisms in Vascular Endothelial Growth Factor Gene and Response to Chemotherapies in Colorectal Cancer: A Meta-Analysis. PLoS One. 2015;10:e0126619. doi: 10.1371/journal.pone.0126619. [DOI] [PMC free article] [PubMed] [Google Scholar]


