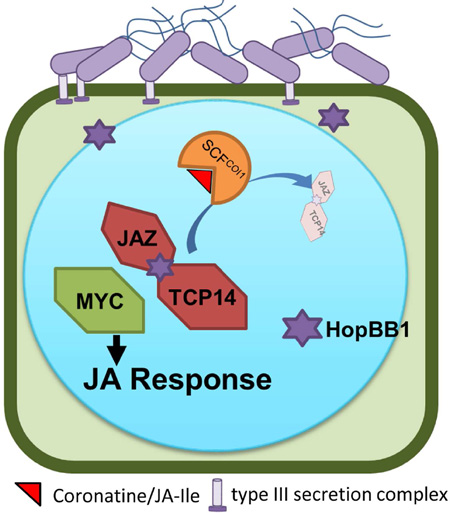
Summary
Independently evolved pathogen effectors from three branches of life (ascomycete, eubacteria and oomycete) converge onto the Arabidopsis TCP14 transcription factor to manipulate host defense. However, the mechanistic basis for defense control via TCP14 regulation is unknown. We demonstrate that TCP14 regulates the plant immune system by transcriptionally repressing a subset of the Jasmonic acid (JA) hormone signaling outputs. A previously unstudied Pseudomonas syringae (Psy) type III effector, HopBB1, interacts with TCP14 and targets it to the SCFCOI1- degradation complex by connecting it to the JA signaling repressor JAZ3. Consequently, HopBB1 de-represses the TCP14-regulated subset of JA response genes and promotes pathogen virulence. Thus, HopBB1 fine-tunes host phytohormone crosstalk by precisely manipulating part of the JA regulon to avoid pleiotropic host responses, while promoting pathogen proliferation.
eTOC blurb
Yang et al. demonstrate that the Pseudomonas syringae type III effector HopBB1 modulates two negative regulators of plant Jasmonic acid (JA) signaling, TCP14 and JAZ3, and ‘glues’ them together for degradation, resulting in precise activation of a subset of JA output responses that promote bacterial virulence.
Graphical Abstract
Introduction
A robust immune system defends plants against most microbes. Plants deploy surface-localized pattern recognition receptors to detect conserved microbe-associated molecular patterns (MAMPs), which leads to the activation of MAMP-triggered immunity (MTI). To counteract MTI, pathogenic microbes deploy virulence factors, often termed effector proteins, into plant cells where they interact with host factors to subvert defense responses or to alter nutrition distribution. To counteract effector protein action, plants evolved a large, polymorphic family of intracellular receptors with a nucleotide-binding domain and leucine-rich repeats, termed NLRs. Plant NLR receptors are analogous to animal NLR innate immune receptors. NLR receptors in both kingdoms are activated by either direct interactions with a ligand, including effector proteins, or by recognition of effector-modified host cellular machines that are the nominal effector target, or decoys of those targets (Bentham et al., 2016;Jones and Dangl, 2006; van der Hoorn and Kamoun, 2008). NLR activation initiates effector-triggered immunity (ETI). Deciphering the mechanisms by which effector repertoires from divergent pathogens act will provide a more comprehensive view of the host cellular machinery responsible for plant immune system function.
Interactome studies revealed that candidate effector repertoires from three evolutionarily diverse pathogens [P. syringae (Psy; eubacteria), H. arabidopsidis (Hpa; oomycete) and Golovinomyces orontii (Go; ascomycete)] converge onto a limited set of interconnected Arabidopsis proteins (Dreze et al., 2011; Mukhtar et al., 2011; Wessling et al., 2014). TCP14, a transcription factor belonging to the conserved TCP (teosinte branched1, CYCLOIDEA, PROLIFERATING CELL FACTORS 1 and 2) family is one of the convergent host targets. The reference Arabidopsis Col-0 genome encodes 24 TCP family members that share a basic helix-loop-helix (bHLH) domain (the TCP domain) and are versatile regulators of plant development and hormone signaling (Lopez et al., 2015). TCP14 physically interacts with SRFR1 and contributes to effector-triggered immunity (Kim et al., 2014). In various tissues, TCP14 promotes cytokinin and gibberellic acid growth hormone responses (Kieffer et al., 2011; Resentini et al., 2015; Steiner et al., 2012). TCP14 is localized to sub-nuclear foci and its co-expression resulted in the re-localization of 22/33 tested nuclear-localized effectors from the three pathogens noted above (Wessling et al., 2014). Additionally, the phytoplasma SAP11 effector associates with other members of the TCP family to repress JA biosynthesis which ultimately enhances the feeding behavior of its insect vector, the leaf hopper (Sugio et al., 2011).
Plant cells integrate growth and division cues with defense cues via phytohormone signaling interactions (Belkhadir et al., 2014; Robert-Seilaniantz et al., 2011). The antagonistic regulatory relationship between the defense hormones Jasmonic acid (JA) and Salicylic acid (SA) endows a plant with the flexibility to prioritize defense responses against pathogens with diverse life styles (Robert-Seilaniantz et al., 2011). In Arabidopsis, activation of SA-dependent responses limits the growth of biotrophic or hemibiotrophic pathogens. On the other hand, JA-dependent responses limit the growth of necrotrophic pathogens and herbivorous insects. Hence, biotrophic or hemibiotrophic pathogens inhibited by SA-mediated immune responses will benefit from activation of JA-dependent responses (Browse, 2009; He et al., 2004; Zheng et al., 2012). Host cellular machines regulating the SA-JA balance are therefore attractive targets for effectors that mimic the action of either hormone, misdirect the defense response, and thus facilitate pathogen or pest proliferation (Kazan and Lyons, 2014).
The transcriptional outputs of JA response are repressed by a group of JASMONATE ZIM DOMAIN (JAZ) proteins through their association with transcription factors (Chini et al., 2007; Zhang et al., 2015). Three MYC transcription factors (MYC2/3/4) repressed by JAZ proteins, are positive regulators of JA-mediated responses in Arabidopsis (Kazan and Manners, 2013). JAZ proteins directly or indirectly recruit a transcription co-repressor complex, containing Topless (TPL) and histone deacetylase, to repress MYC activity (Pauwels et al., 2010). JA biosynthesis is induced during normal development or following either MAMP treatment or Pseudomonas syringae infection (Lewis et al., 2015; Schmelz et al., 2003). JAZ proteins bind isoleucine-conjugated JA, which facilitates their physical interaction with the CORONATINE-INSENSITIVE 1 (COI1) F-box component of an SCF-(Skip-cullin-F-box)-type E3 ubiquitin ligase. This results in proteasome-mediated degradation of JAZ proteins, and allows MYC-dependent activation of JA response genes (Thines et al., 2007; Wasternack and Hause, 2013). The MYC regulon controls a pleiotropic physiological and developmental response including the repression of SA-dependent transcriptional output (Kazan and Manners, 2013).
At least two Psy type III effectors, HopX1 and HopZ1a, and the phytotoxin coronatine can activate the JA pathway. Coronatine is a structural mimic of JA-Ile (Katsir et al., 2008). HopX1 is a cysteine protease that eliminates JAZ proteins by cleaving their central ZIM domain (Gimenez-Ibanez et al., 2014). HopZ1a is an acetyltransferase that acetylates soybean and Arabidopsis JAZ proteins, promoting COI1-dependent JAZ turnover (Jiang et al., 2013). Both HopX1 and HopZ1a were identified from Psy strains deficient in coronatine biosynthesis and each can rescue the growth defects of a Psy mutant unable to synthesize coronatine (Gimenez-Ibanez et al., 2014; Jiang et al., 2013). Effects of HopX1 or HopZ1a action on the global JA-activated transcriptional landscape have not been defined, though each causes de-repression of a few tested JA response genes (Gimenez-Ibanez et al., 2014; Jiang et al., 2013).
Here, we provide a mechanistic model for how one of the many TCP14-targeting effectors suppresses defense by manipulating the host defense hormone network to promote Psy virulence. Our data demonstrate that the previously unstudied bacterial type III effector, HopBB1, alters sub-sets of targets from two heretofore unlinked transcriptional regulons, TCP14 and MYC, to de-repress a subset of JA responses and promote virulence while avoiding pleiotropic effects associated with full misregulation of either regulon.
Results
TCP14 is a negative regulator of JA signaling
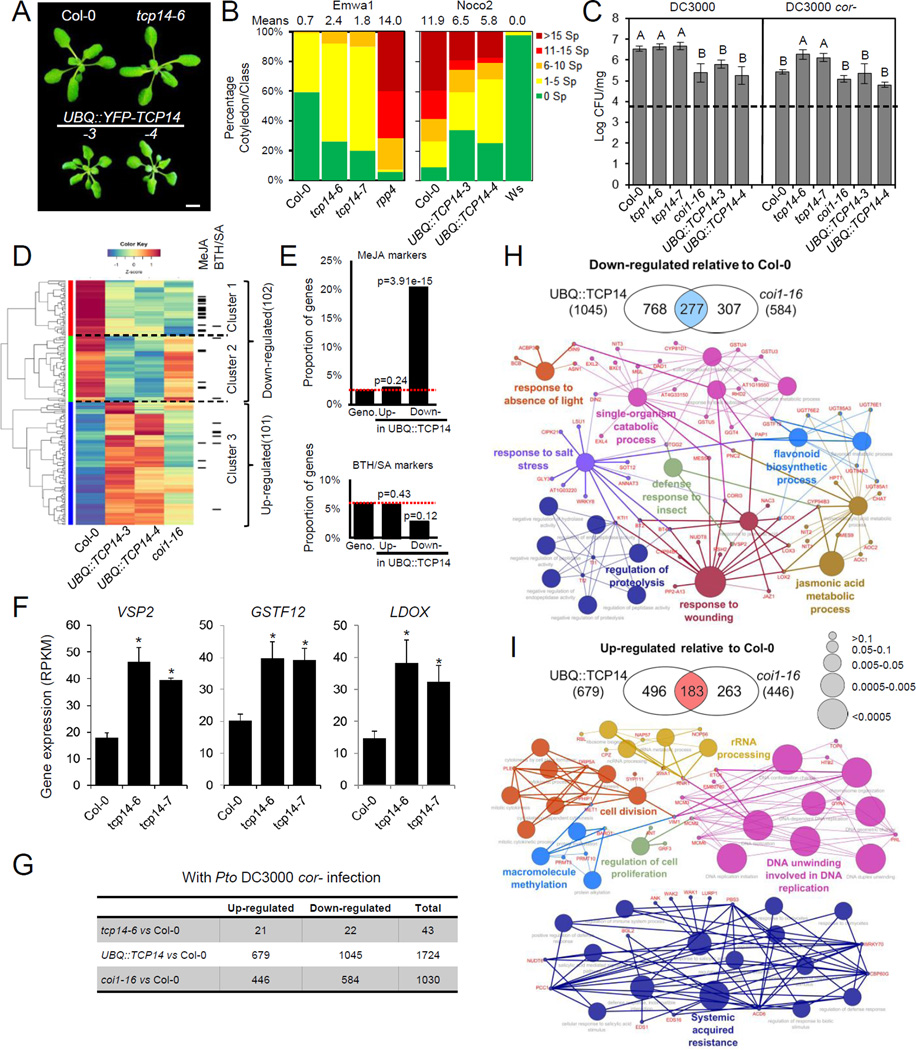
We showed that a tcp14 mutant enhanced susceptibility to the avirulent Emwa1 isolate of the oomycete pathogen, Hpa (Mukhtar et al., 2011; Wessling et al., 2014). We confirmed and extended this result using a second tcp14 allele and transgenic Arabidopsis over-expressing YFP-TCP14 from the UBQ promoter (Figures 1A and 1B). These plants were modestly smaller than wild-type at the same developmental stage (Figures 1A and S1A). These plants displayed enhanced disease resistance when challenged with the virulent Hpa isolate Noco2 (Figure 1B). We examined the in planta growth of P. syringae pv. tomato strain DC3000 (Pto DC3000, hereafter DC3000) and a coronatine-deficient mutant, Pto DC3000 cor- (hereafter DC3000 cor-) on tcp14 mutants and TCP14 over-expression lines. Plants overexpressing TCP14 displayed enhanced resistance to DC3000 at the same levels as coi1 mutants (Figure 1C). The growth of DC3000 cor- was the same on Col-0 and plants overexpressing TCP14 (Figure 1C). tcp14 mutants were unaltered in their response to DC3000, but rescued the growth defects of DC3000 cor- (Figure 1C). These disease phenotypes suggest that TCP14 regulates immune system output by suppressing the JA response.
Figure 1. TCP14 represses JA response and promotes disease resistance.
(A) Three-week-old plants of Col-0, tcp14-6, UBQ::YFP-TCP14-3 and UBQ::YFP-TCP14-4. Bar=5mm
(B) tcp14 mutants are more susceptible to Hpa Emwa1 than Col-0. Overexpressing TCP14 in Arabidopsis enhances disease resistance against Hpa Noco2. The means represent the numbers of sporangiophores (sp) on each cotyledon (n>50).
(C) Mutation in TCP14 enhances the virulence of DC3000 cor-. Two-week-old plants were dip inoculated with indicated bacteria strains at OD600=0.05. CFU: Bacterial colony formation units. Error bars represent ±SD.
(D) JA-responsive genes are repressed in two-week-old TCP14-overexpressing plants. Hierarchical clustering of the 203 genes identified as differentially expressed in the pairwise comparison between Col-0 and UBQ::YFP-TCP14-3, UBQ::YFP-TCP14-4 and the coi1-16 mutant were also included. MeJA or BTH/SA-responsive genes (Table S1) are indicated on the right. Cluster 1: genes repressed in UBQ::YFP-TCP14 and coi1-16; Cluster 2: genes repressed in UBQ::YFP-TCP14 but not in coi1-16; Cluster 3: genes upregulated in UBQ::YFP-TCP14.
(E) Genes down-regulated in UBQ::YFP-TCP14-3 are enriched for JA markers. Dashed lines represent the proportion of genes that belong to each group in the Arabidopsis genome. Geno. (Genome): Expected proportion of MeJA (or BTH/SA) markers in Arabidopsis genome. Up- /Down- : Proportion of MeJA (or BTH/SA) markers in genes up-regulated or down-regulated by UBQ::YFP-TCP14.
(F) Elevated expression of JA-responsive genes in steady-state tcp14 mutants. *: FDR<0.0001
(G) Summary of transcriptional alterations in tcp14-6, UBQ::YFP-TCP14 overexpression line and in coi1-16 plants 24h after DC3000 cor- infection. The table depicts the number of differentially expressed genes in each line relative to infected wild-type plants.
(H) Gene Ontology terms (biological processes) enriched in the set of 277 genes down-regulated in both UBQ::YFP-TCP14 and coi1-16 when compared to wild-type plants after infection. Note the prevalence of terms associated with the JA pathway. Gene names are labeled in red.
(I) Gene Ontology terms (biological processes) enriched in the set of 183 genes up-regulated in both UBQ::YFP-TCP14 and coi1-16 when compared to wild-type plants after infection. Note the prevalence of terms associated with the SA pathway. Gene names are labeled in red. The complete GO enrichment results for (H) and (I) are shown in Table S2. See also Figure S1 and Tables S1 and S2.
To test the hypothesis that TCP14 is a negative regulator of the JA pathway, we defined a comprehensive set of marker genes for the JA and SA responses (Figures S1B–S1F; Table S1) and analyzed the transcriptomes of Col-0, tcp14 mutants and transgenic plants overexpressing TCP14 in two-week old seedlings; the time point when altered infection phenotypes were observed. A total of 203 genes were differentially expressed in TCP14-overexpressing seedlings compared to Col-0 (Figures 1D; Table S2). Genes down-regulated by TCP14 overexpression were significantly enriched for genes that are activated by JA treatment (26/102; p=2.19e-19, hypergeometric test; cluster 1, Figures 1D and 1E). Indeed, many of these down-regulated genes were also weakly expressed in the coi1–16 mutant (Figure 1D; Table S2). In contrast, only 6 of the 101 genes that were up-regulated in the UBQ::YFP-TCP14-3 line are markers of the SA response (Figure 1D), suggesting that TCP14-driven repression of the JA pathway was not a consequence of activated SA response. No global transcriptome changes were observed in tcp14 mutants relative to Col-0 in non-infected plants (Table S2). However, some JA-responsive genes, including VSP2 and those required for anthocyanin biosynthesis, were up-regulated in the mutants (Figure 1F).
We also examined transcriptional alterations in these plants 24h after infection with DC3000 cor-. When compared to infected Col-0, both coi1–16 and the UBQ::YFP-TCP14-4 line showed weaker activation of JA-responsive genes (Figures 1G and 1H; Table S2). Although the suppression of the JA response is an obvious transcriptional alteration in either wild type or infected UBQ::YFP-TCP14 plants (Figures 1D and 1H), TCP14 may also participate in the regulation of other sectors of the plant immune system. Indeed, the enhanced disease phenotype in UBQ::YFP-TCP14 also correlates with suppression of ABA-responsive genes and responses related to ABA signaling (Figure 1I, Table S2). Moreover, TCP14-overexpressing lines displayed enhanced activation of SA-responsive genes after infection (Figure 1I; Table S2), consistent with their enhanced resistance to both DC3000 and Hpa isolate Noco2 (Figures 1B and 1C). However, only 43 genes were differentially expressed in the infected tcp14-6 mutant relative to wild-type plants (Figure 1G; Table S2). We do not know if the effects of tcp14 mutation would be more dramatic at different time points in the response. Overall, this transcriptional profile supports the conclusion that TCP14 contributes to plant immunity as a negative regulator of subsets of the JA response.
The P. syringae effector HopBB1 interacts with TCP14 in vivo
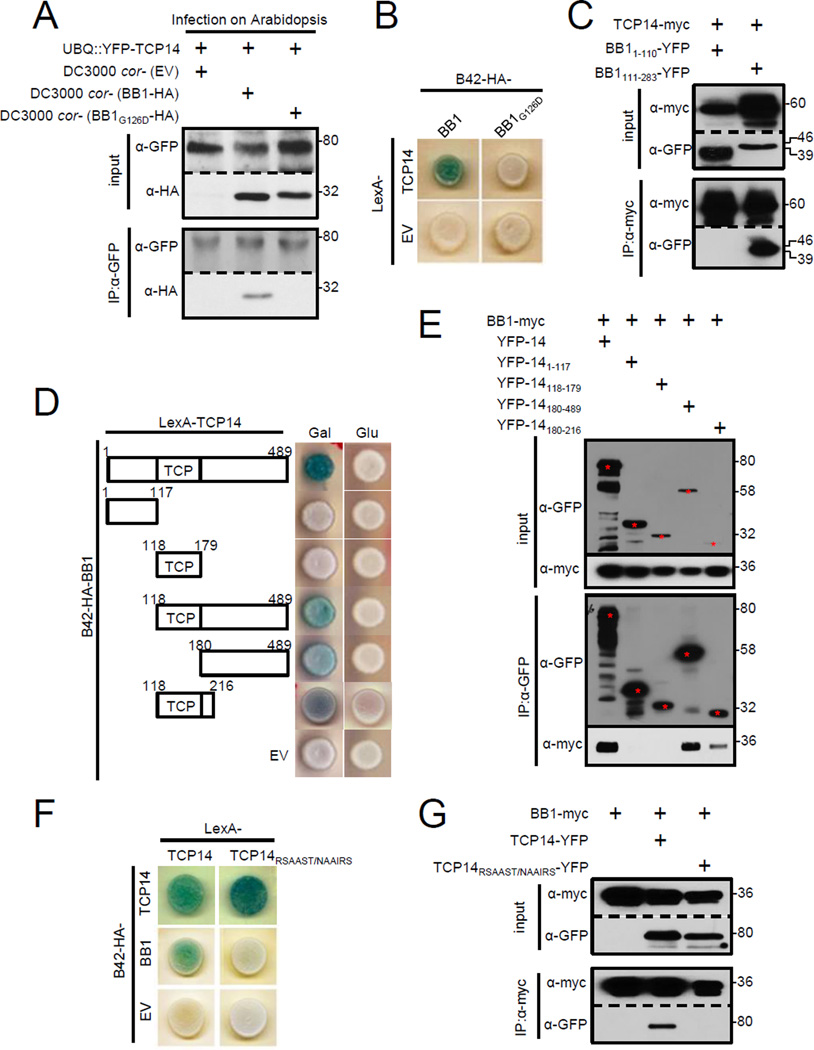
To demonstrate how effectors modulate TCP14 function, we focused first on an uncharacterized TCP14-interacting Psy type III effector, HopBB1 (Mukhtar et al., 2011). In yeast, HopBB1 selectively interacts with a subset of 24 Arabidopsis TCP family members (Figure S2A). We validated the interactions between HopBB1 and TCP14 in planta by inoculating YFP-TCP14 overexpressing Arabidopsis with DC3000 cor-expressing HopBB1-HA at native levels. We observed that HopBB1-HA was co-immunoprecipitated with YFP-TCP14 (Figure 2A), demonstrating that these two proteins associate in vivo during Psy infection. We used random mutagenesis to isolate a HopBB1 mutant, HopBB1G126D that lost interaction with TCP14 in yeast-two-hybridization (Y2H) and failed to associate with TCP14 in planta (Figures 2A, 2B and S2B). HopBB1111–283 that contains G126, but has no annotated function, was sufficient for association with TCP14 (Figure 2C). TCP14180–489 downstream of the conserved TCP DNA binding domain was sufficient for interaction with HopBB1 in yeast and in planta (Figures 2D, 2E and S2C). TCP14180–216 co-immunoprecipitated with HopBB1 (Figure 2E). We replaced every six amino acids in this region with a structurally flexible sequence (NAAIRS; (Wilson et al., 1985)), and revealed that the TCP14 sequence motif 204-RSAAST-209 is necessary for interaction between full length TCP14 and HopBB1 (Figures 2F, 2G and S2D). Collectively, these data are consistent with the hypothesis that HopBB1 associates with TCP14 in vivo.
Figure 2. HopBB1 interacts with TCP14 in planta.
(A) Pto-delivered HopBB1, but not HopBB1G126D associates with TCP14 in Arabidopsis. DC3000 cor- with empty vector (EV), HA-tagged HopBB1 or HopBB1G126D were hand-infiltrated at OD600=0.05 into leaves of four-week-old transgenic Arabidopsis expressing YFP-TCP14. Leaves were harvested 24 hrs after inoculation.
(B) HopBB1G126D loses interaction with TCP14 in yeast.
(C) The C-terminus (111–283) of HopBB1 is sufficient to associate with TCP14 in N. benthamiana.
(D) HopBB1 interacts with TCP14(aa180-216) in yeast.
(E) HopBB1 associates with TCP14180–216 in N. benthamiana.
(F) TCP14RSAAST/NAAIRS fails to interact with HopBB1 in yeast, but retains homodimerization.
(G) TCP14RSAAST/NAAIRS fails to associate with HopBB1 in N. benthamiana.
Proteins were transiently expressed in N. benthamiana from a 35S promoter for (C), (E) and (G). See also Figure S2.
HopBB1 de-represses JA response
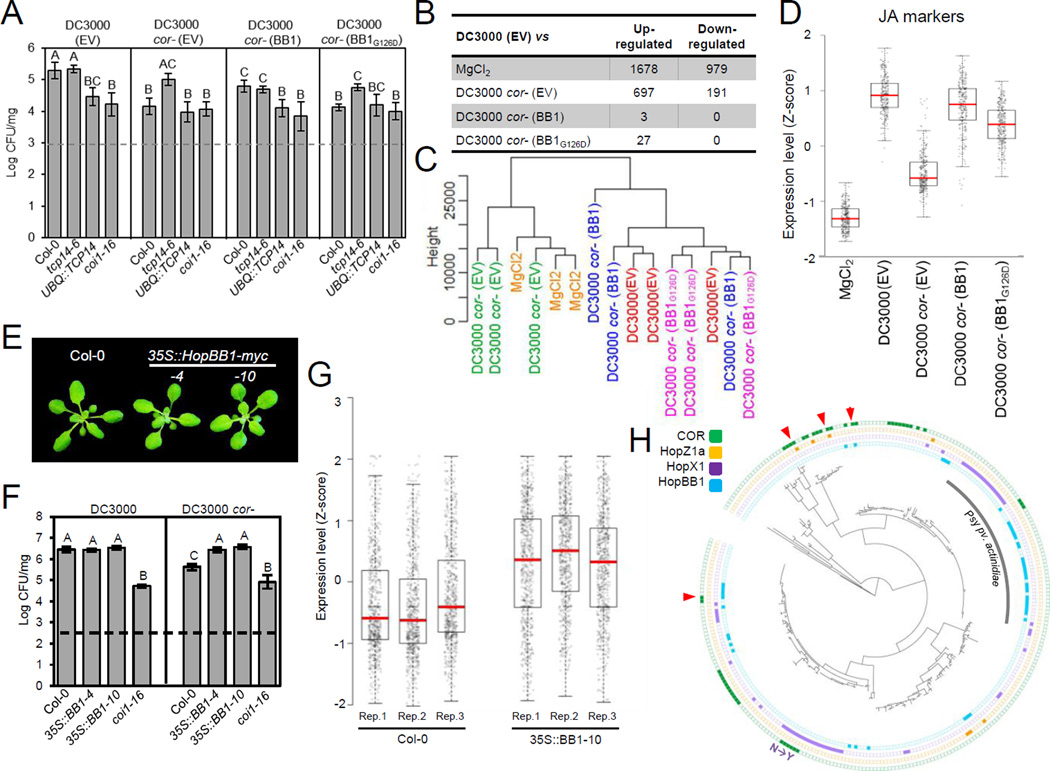
We tested the hypothesis that HopBB1 targets TCP14 to manipulate plant JA response. Following delivery of native levels via type III secretion, HopBB1, but not HopBB1G126D, partially rescued the growth defects of DC3000 cor- on Col-0 plants (Figure 3A and S3A). Growth promotion of DC3000 cor- contributed by HopBB1 was suppressed by over-expression of TCP14 and in coi1 (Figure 3A). These observations indicate that HopBB1 partially complements the defects of coronatine deficiency, that this can be modulated by TCP14 and requires COI1.
Figure 3. HopBB1 promotes bacteria growth and activates JA response.
(A) Bacterial-delivered HopBB1 promotes the growth of DC3000 cor- in Col-0. Two-week-old plants were spray inoculated with a bacteria suspension at OD600=0.2. CFU: Bacterial colony formation units. Error bars represent ±SD.
(B) Summary of transcriptional changes in Col-0 plants 24h after the treatment with DC3000 (EV), MgCl2 and the coronatine-deficient mutant strains carrying the empty vector (EV), HopBB1 or HopBB1G126D. Numbers represent the differentially expressed genes relative to the DC3000 treatment.
(C) Dendrogram constructed based on the entire transcriptome showing that the transcriptional signature of DC3000 cor- (EV), treated plants resembles that of the mock treatment; DC3000 cor- expressing either HopBB1 or HopBB1G126D trigger similar transcriptional responses as DC3000 (EV).
(D) A set of 253 JA marker genes are activated by DC3000 (EV) and/or DC3000 cor-(HopBB1); HopBB1G126D has reduced ability to activate these genes.
(E) Transgenic Arabidopsis plants expressing HopBB1 are morphologically indistinguishable from Col-0 wild-type. Bar=5mm
(F) Plants expressing HopBB1 complement the growth defects of DC3000 cor-.
(G) JA-responsive genes are activated in transgenic plants expressing HopBB1-myc. The z-score transformed expression of 672 JA responsive marker genes is shown for three biological replicates of Col-0 and transgenic plants expressing HopBB1-myc.
(H) The distribution of HopBB1, HopX1, HopZ1a and coronatine biosynthesis pathway in 287 sequenced Pseudomonas syringae genomes. Arrowhead: genomes contain two JA-activating tools. N→Y: A polymorphism (N→Y) exists in the HopX1 allele. See also Figure S3 and Tables S3 and S4.
We then investigated the effect of HopBB1 on the transcriptome of wild-type plants 24h after the infection. As expected, the transcriptome of plants infected with DC3000 was significantly different from those sprayed with either a mock or DC3000 cor- (EV) (Figures 3B and 3C). The set of 697 genes that were more strongly induced by DC3000 than by DC3000 cor- (EV) was enriched in biological processes related to JA and ABA responses (Table S3). Remarkably, infection with DC3000 cor- (HopBB1) resulted in a global transcriptional signature that resembled infection with DC3000 (Figure 3C), supporting our conclusion that HopBB1 can rescue the impaired ability of DC3000 cor-to establish infection. A total of 129 of the 672 (19%) JA-responsive genes were expressed to higher levels in plants infected with DC3000 or DC3000 cor- (HopBB1) compared to DC3000 cor- (EV) treatment. Although the transcriptional changes induced by DC3000 cor- (HopBB1G126D) qualitatively resembled that induced by DC3000 (Figure 3C), these JA-responsive genes were less activated (p-value = 2.2e-16, Student’s t-test), indicating that interaction with TCP14 is required for the full virulence function of HopBB1 (Figure 3D).
To exclude the possibility that the transcriptome change induced by bacteria-delivered HopBB1 may be confounded by other effectors that may influence JA signaling, we defined the transcriptome of transgenic plants expressing only HopBB1. As expected if HopBB1 potentiates JA responses, these plants were hypersensitive to JA-mediated inhibition of root elongation (Figure S3C). In addition, DC3000 cor- is more virulent on HopBB1 transgenic plants than on wild-type Col-0, demonstrating that heterologous HopBB1 complements this strain’s coronatine deficiency (Figure 3F), analogous to tcp14 (Figure 1C). We compared the transcriptome of HopBB1 expressing plants to Col-0 at steady state and identified 628 differentially expressed genes (593 up- and 35 down-regulated) (Table S3). Many of our JA response marker genes (93/672; p=3.41e-47; hypergeometric test) were up-regulated in the HopBB1 expressing plants (Table S3), and the average expression of all 672 JA-responsive genes was higher in these transgenic plants (Figure 3G). JA response genes were enriched in the overlap between HopBB1-upregulated and TCP14-suppressed genes: out of the 102 genes that were down-regulated by steady state TCP14 overexpression (Figure 1D; Table S4), 12 were up-regulated in HopBB1 transgenic plants and 10 of these are JA markers (p=2.26e-17; hypergeometric test) (Table S4). Genes specific to BTH/SA response were also enriched in the HopBB1 up-regulated genes (139/2096; p=2.53e-33; hypergeometric test). However, genes that are typically associated with SA-mediated defense responses (e.g., PR-1, PR-5, ICS1, WRKYs) were not differentially expressed, suggesting that the SA response activated in HopBB1-expressing plants is likely to be insufficient for robust defense.
As expected, the JA response genes defined in our study were enriched for MYC2 binding motifs in their promoters (Figure S3D). In fact, these genes were enriched for co-occurrence of MYC2 and TCP binding sites (Franco-Zorrilla et al., 2014; Kosugi and Ohashi, 2002). Out of the 88 JA response genes that contain consensus MYC and TCP motifs in their promoters, 22 (25%) were also up-regulated by HopBB1 expression (Figure S3E). Interestingly, neither constitutive nor conditional overexpression of HopBB1 caused the chlorotic leaf phenotype observed previously after either coronatine treatment or HopX1 expression (Figure S3F) (Gimenez-Ibanez et al., 2014; Kloek et al., 2001). Consistent with this observation, the expression of MYC-dependent and JA-responsive photosynthetic genes (Qi et al., 2015) was not altered in HopBB1 expressing plants (Figure S3G). In sum, our transcriptome data are consistent with our pathology data and support the hypothesis that HopBB1 activates a sector of the overall JA response that is co-regulated by TCPs and MYC.
We surveyed the genomic distribution of HopBB1, coronatine biosynthetic genes, HopX1 and HopZ1a in 287 Psy genomes. Only four (1.3%) genomes contain two presumably functional JA-activating virulence factors (Figure 3H, Table S5). Nearly 50% (141) of Psy genomes carry one, and only one, functional version of these four JA-activating virulence factors (Figure 3H; Table S5). Strikingly, in a few additional cases where HopX1 and coronatine biosynthetic genes co-exist in a single strain, the HopX1 alleles have mutations in functionally essential residues (Nimchuk et al., 2007). The phylogeny of the 141 Psy isolates suggests that independent gene gain/loss occurred in each lineage (Figure 3H; Table S5). This is particularly true across otherwise very closely related strains from the Psy pathovar actinidae, currently responsible for epidemic disease outbreaks that threaten the kiwi industries of New Zealand and Italy (Table S5) (McCann et al., 2013).
HopBB1-mediated degradation of TCP14 requires SCFCOI1
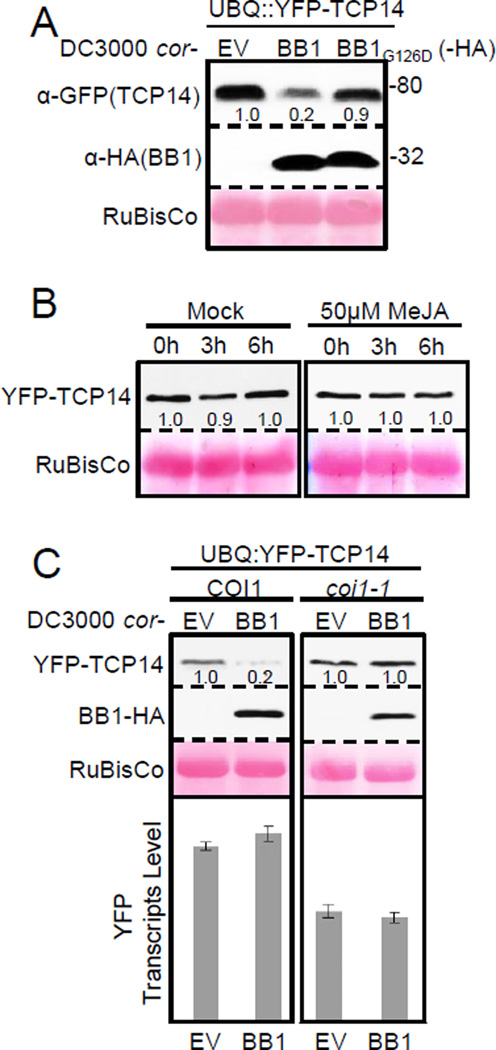
Given that HopBB1 interacts with TCP14 and de-represses JA responses during infection, we investigated how HopBB1 disrupts TCP14 function. Delivery of native levels of HopBB1, but not HopBB1G126D, led to reduced TCP14 protein accumulation (Figure 4A). MeJA treatment alone did not alter the accumulation of TCP14, indicating that the HopBB1-induced turnover of TCP14 was not a consequence of activated JA response (Figure 4B). TCP14 degradation in these experiments required the SCFCOI1 complex, since it was blocked in coi1-1 plants expressing UBQ::YFP-TCP14 (Figure 4C). Thus, HopBB1 promotes the degradation of TCP14 via the SCFCOI1 degradation pathway during infection, ultimately facilitating the activation of JA responses.
Figure 4. HopBB1-mediated degradation of TCP14 requires SCFCOI1 pathway.
(A) Bacterial-delivered HopBB1, but not HopBB1G126D, induced turnover of TCP14 during infection on Arabidopsis. Bacteria were hand-inoculated into leaves of four-week-old plants at an OD600=0.05. Samples were harvested 24 hrs after inoculation.
(B) YFP-TCP14 is not subject to JA-mediated degradation in the absence of HopBB1 in Arabidopsis. Two-week old seedlings expressing UBQ::YFP-TCP14 were sprayed with mock or 50µM MeJA solution, and sampled at the indicated time. Each sample was pooled from eight seedlings.
(C) Pto-delivered HopBB1 reduces TCP14 protein level in wild-type Col-0, but not in coi1-1 mutant. Experiments were performed as Figure 4A. YFP-TCP14 transcripts were quantified using real-time PCR. Error bars indicate ±SD
HopBB1 interacts with JAZ3
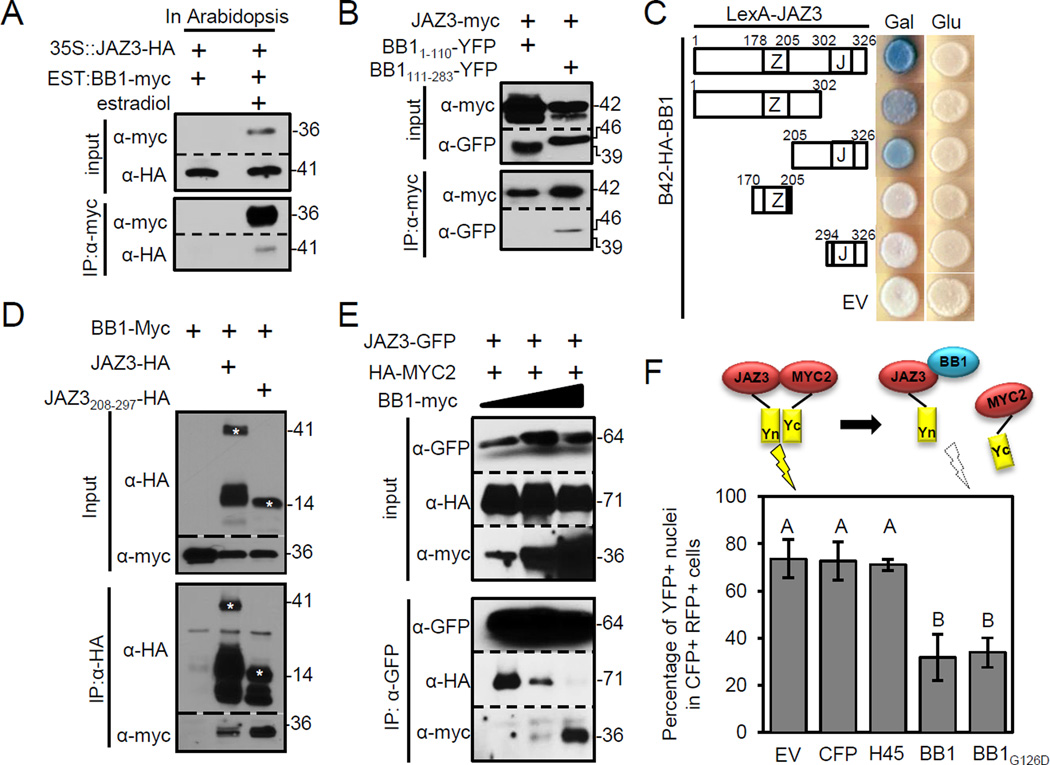
We sought to define which component(s) of the SCFCOI1 pathway mediated TCP14 degradation. We confirmed the finding that JAZ3 interacted with HopBB1 (Mukhtar et al., 2011; Wessling et al., 2014) (Figure S4A). Conditionally expressed HopBB1 co-immunoprecipitated JAZ3 in both transgenic Arabidopsis and transiently expressing N. benthamiana leaves (Figure 5A and Figure S4D). HopBB1111–283 was also sufficient for association with JAZ3 (Figure 5B). However, HopBB1G126D retained interaction with JAZ3 (Figures S4C and S4D), suggesting that a different association surface within the HopBB1 C-terminus is required. JAZ3208–297 is required for HopBB1-interaction, supported by association analyses in yeast and N. benthamiana (Figure 5C, 5D).
Figure 5. HopBB1 interacts with JAZ3 and disrupts its association with MYC2.
(A) HopBB1 interacts with JAZ3 in Arabidopsis. HopBB1-myc conditionally expressed from an estradiol-inducible promoter was transformed into Arabidopsis constitutively expressing JAZ3-HA from a 35S promoter. Three-week-old seedlings were induced with 50µM estradiol and sampled 6 hrs after induction.
(B) The C-terminus (111–283) of HopBB1 is sufficient to associate with JAZ3 in N. benthamiana.
(C) HopBB1 interacts with an uncharacterized JAZ3 domain (206–302) in yeast.
(D) HopBB1 associates with an uncharacterized JAZ3 domain (206–302) in N. benthamiana.
(E) HopBB1 reduces the association between JAZ3 and MYC2 in planta. Proteins were transiently co-expressed in N. benthamiana. HopBB1 was induced using a gradient of estradiol (0.01µM, 1µM, 10µM) 6 hrs after Agrobacteria infiltration. Samples were harvested 18 hrs after induction.
(F) HopBB1 disrupts the BiFC signal generated from the association between JAZ3 and MYC2. The co-expression of HopBB1 or HopBB1G126D reduces the efficiency from 80% to 20%. RFP-positive cells were examined for the presence of CFP and YFP signal in nuclei. For each combination, the ratios were calculated by counting approximately 200 nuclei collected from four independent samples. Error bars indicate ±SD.
See also Figure S4.
In contrast to HopX1 and HopZ1a, HopBB1 expression was not sufficient to alter JAZ3 accumulation (Figure S4E) (Gimenez-Ibanez et al., 2014; Jiang et al., 2013). However, increasing HopBB1 levels did reduce the amount of MYC2 associated with JAZ3 in a competitive co-immunoprecipitation assay in N. benthamiana, indicating that HopBB1 interferes with the interaction of MYC2 and JAZ3 (Figure 5E). We developed a bimolecular fluorescence complementation (BiFC)-based assay to examine this disassociation in vivo. We co-infiltrated Agrobacterium strains carrying either a BiFC construct expressing JAZ3-nYFP, cYFP-MYC2 and mRFP as a co-expression reporter, or a second construct carrying an estradiol inducible HopBB1-CFP, to test the ability of HopBB1 co-expression to block JAZ3-MYC2 interaction mediated YFP reconstruction (Figure 5F and S4F, S4G). Co-expression of HopBB1-CFP or HopBB1G126D-CFP dramatically reduced the percentage of re-constituted YFP signal in CFP-and RFP-positive nuclei. Neither CFP nor CFP tagged with HaRxL45 [an Hpa effector that interacts with TCP14, but not JAZ3 (Wessling et al., 2014)] altered the BiFC efficiency (Figures 5F and S4H). These observations support our contention that HopBB1-JAZ3 association interferes with the interaction between JAZ3 and MYC2 in vivo.
TCP14 is subject to JA-mediated degradation in the presence of HopBB1 and JAZ3
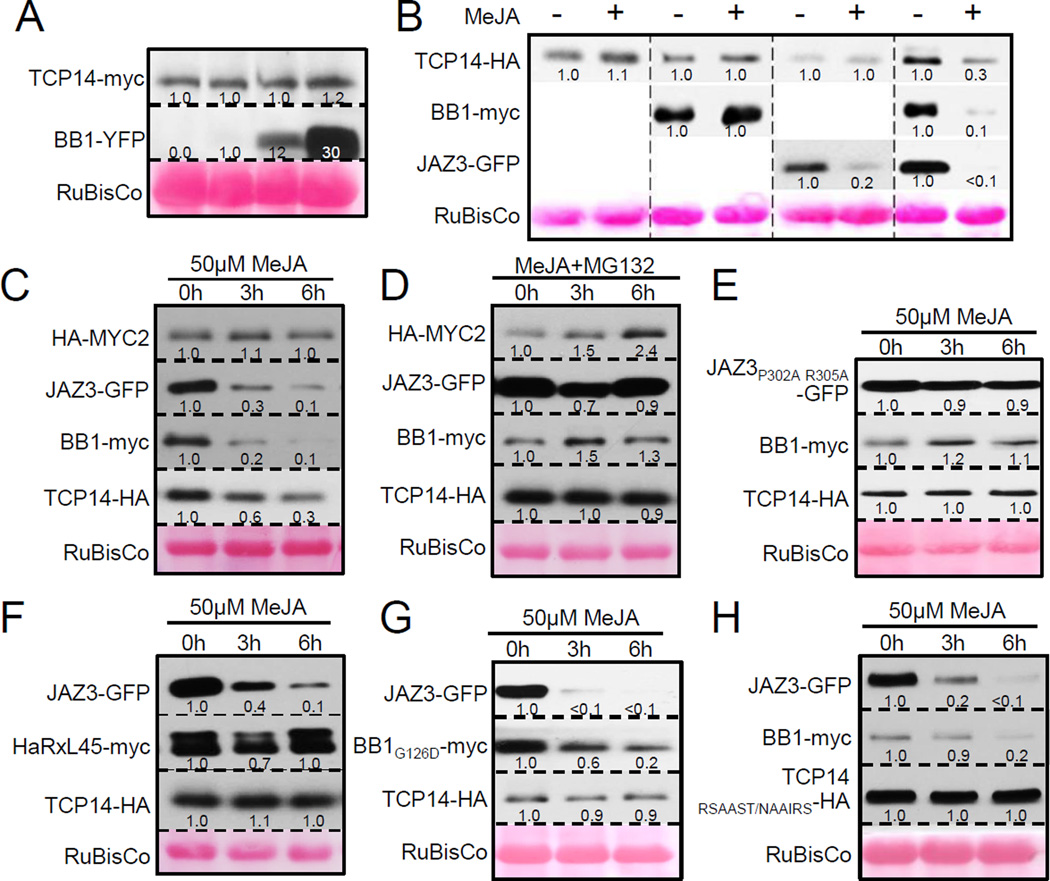
Since JAZ proteins are subject to SCFCOI1-mediated degradation, we tested how HopBB1 and JAZ3 influence the degradation of TCP14 in a reconstructed degradation system in N. benthamiana. TCP14 was not subject to JA-mediated protein degradation when transiently expressed (Figures 4B and 6B) or when co-expressed with either HopBB1 or JAZ3 (Figures 6A and 6B). Importantly, when TCP14, JAZ3 and HopBB1 were co-expressed, levels of all three proteins were dramatically reduced upon MeJA treatment (Figure 6B). As anticipated, MYC2 levels were not reduced in these experiments (Figure 6C). Thus, HopBB1-mediated TCP14 turnover requires JA and JAZ3.
Figure 6. TCP14 is subject to JA-mediated degradation in the presence of HopBB1 and JAZ3.
(A) HopBB1 alone does not trigger TCP14 degradation without MeJA. Leaves were co-infiltrated with Agrobacteria delivering 35S::TCP14-myc or EST::HopBB1-YFP-HA genes.
(B) TCP14 is subject to JA-mediated degradation in the presence of HopBB1 and JAZ3. Agrobacteria carrying vectors expressing each protein under 35S constitutive promoter were co-infiltrated into N. benthamiana leaves. 50µM of MeJA was hand-infiltrated into leaves 24 hours post inoculation. Numbers below western signal indicate the relative signal intensity. The same method was applied to (B–H).
(C) MYC2 is not co-degraded with TCP14.
(D) HopBB1-mediated degradation of TCP14 is blocked by the 26S proteasome inhibitor, MG132. 50µM MeJA and 50µM MG132 were co-infiltrated 24 hours post inoculation.
(E) The recruitment of JAZ3 to SCFCOI1 is required for HopBB1-mediated degradation of TCP14.
(F) HaRxL45 cannot mediate TCP14 degradation with the presence of JAZ3 and MeJA.
(G) HopBB1G126D cannot mediate TCP14 degradation with the presence of JAZ3 and MeJA.
(H) TCP14RSAAST/NAAIRS is not subject to HopBB1-mediated degradation.
See also Figure S5.
The protease inhibitor MG132 can block JA-triggered degradation of JAZ proteins (Chini et al., 2007; Thines et al., 2007); it also blocked HopBB1-mediated degradation of TCP14 (Figure 6D). We generated JAZ3P302A R305A, an allele that cannot interact with COI1 and is thus MeJA resistant (Figures 6E and S5A, S5B). Importantly, JAZ3P302A R305A still interacted with HopBB1 in yeast, suggesting that its overall structure is not altered (Figure S5C). When we co-expressed JAZ3P302A R305A with TCP14 and HopBB1, MeJA-induced degradation of HopBB1 and TCP14 was blocked (Figure 6E). This observation suggested that SCFCOI1-dependent degradation of JAZ3 is required for HopBB1-mediated, JA-dependent turnover of TCP14. TCP14 turnover was not driven by HaRxL45 co-expression in the presence of JAZ3 (Figure 6F), implying that effectors interacting with TCP14 modulate its activity by at least two different mechanisms. Remarkably, HopBB1G126D failed to mediate TCP14 turnover, but was still degraded with JAZ3 in the presence of MeJA (Figure 6G). Conversely, the TCP14RSAAST/NAAIRS mutant that fails to interact with HopBB1 was also resistant to HopBB1-mediated degradation (Figure 6H). Thus, HopBB1-mediated degradation of TCP14 requires its interactions with TCP14 and the degradation of JAZ3 through SCFCOI1.
HopBB1 recruits TCP14 into a JAZ3-containing sub-nuclear structure
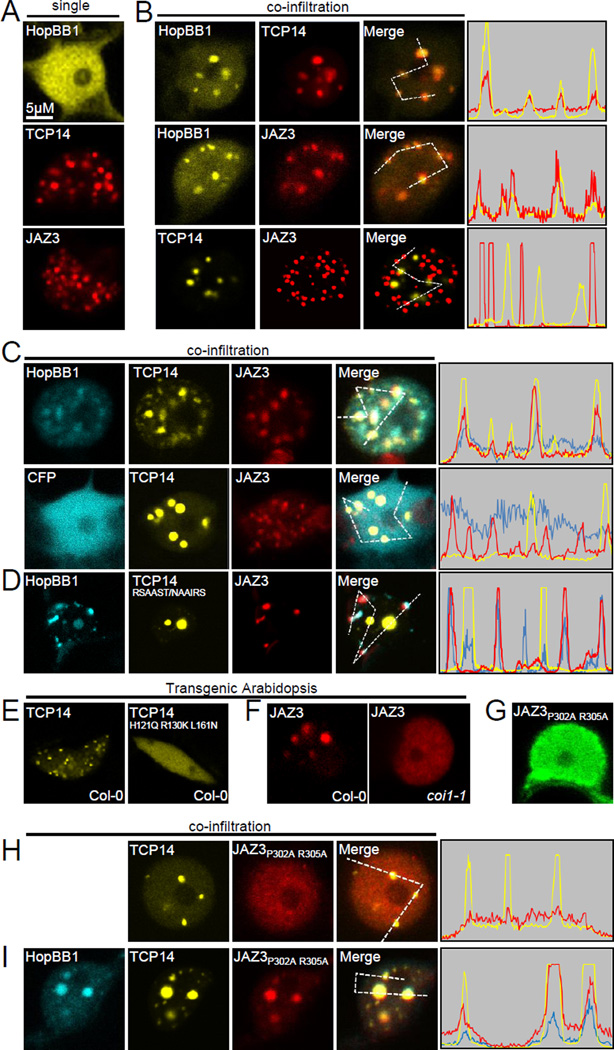
We previously demonstrated that TCP14 re-locates HopBB1 into an uncharacterized sub-nuclear structure (Figures 7A, 7B top and S6A) (Wessling et al., 2014). Similarly, we noted that JAZ3 also forms sub-nuclear foci (Figure 7A). We found that HopBB1 can also be re-localized into JAZ3-foci (Figures 7B middle and S6B). Strikingly, JAZ3-foci and TCP14-foci did not overlap when co-expressed, implying that they represent different structures (Figures 7B bottom and S6C). We then co-expressed a CFP-tagged HopBB1 together with TCP14 and JAZ3. In nuclei with HopBB1-CFP (but not CFP alone), the CFP, YFP and RFP signals co-localized in the same foci (Figure 7C and S6D), suggesting that HopBB1 re-distributed TCP14 and JAZ3 from separate sub-nuclear foci into the same structure. Although HopBB1G126D failed to associate with TCP14 in both Y2H and co-IP assay, it was still re-localized by TCP14, suggesting that these assays report different interaction affinities (Figure S6E). For this reason, we could not specifically test the requirement of HopBB1-TCP14 interaction in redistributing JAZ3 and TCP14. However, TCP14RSAAST/NAAIRS, which still forms nuclear foci but cannot interact with HopBB1, also cannot be fully re-distributed to the JAZ3-containing foci in the presence of HopBB1 (Figure 7D). This result confirms that redistribution of TCP14 to JAZ3-containing foci requires its association with HopBB1.
Figure 7. HopBB1 recruits TCP14 to a JAZ3-containing degradation site.
(A) Localization of HopBB1, TCP14 and JAZ3 in nuclei. HopBB1 is evenly distributed in nuclei, while TCP14 and JAZ3 form subnuclear foci. Bar=5µM Proteins were transiently expressed in N. benthamiana for (A)-(D), (G)-(I).
(B) HopBB1 was re-localized to subnuclear foci by TCP14 (top) and JAZ3 (middle). However, TCP14 and JAZ3 localize in distinct nuclear foci (bottom). Histograms represent the intensity of fluorescent signal on the pathway of the lines in the “Merged” panel.
(C) HopBB1 (top), but not CFP (bottom), drives TCP14 and JAZ3 into the same sub-nuclear foci.
(D) HopBB1 cannot co-localize TCP14RSAAST/NAAIRS -YFP into the same foci as JAZ3-RFP.
(E) The formation of TCP14 foci depends on its binding DNA ability. YFP-TCP14, but not TCP14H121Q R130K L161N, expressed under UBQ promoter forms subnuclear foci in transgenic Arabidopsis.
(F) The formation of JAZ3 foci depends on its ability to associate with COI1. JAZ3-RFP expressed from a constitutive 35S promoter forms subnuclear foci in transgenic Arabidopsis Col-0, but not coi1-1 mutant. Images in (E) and (F) were taken from cotyledon epidermal cells in transgenic Arabidopsis.
(G) JAZ3P302A R305A cannot form subnuclear foci in N. benthamiana.
(H) TCP14 cannot re-localize JAZ3P302A R305A.
(I) TCP14 re-located JAZ3P302AR305A to nuclear foci in the presence of HopBB1.
See also Figure S6.
We utilized TCP14 and JAZ3 mutants to examine functional requirements for the formation of sub-nuclear foci. Amino acids H121, R130 and L161 in the TCP domain are conserved in all 24 Arabidopsis TCP proteins (Figure S6F). Single mutation in any of these abolished TCP4 binding to DNA (Aggarwal et al., 2010). TCP14H121Q R130K L161N almost completely abolished the formation of sub-nuclear foci in transgenic Arabidopsis, although it was still exclusively localized in nuclei and retained its ability to homodimerize and associate with HopBB1 (Figures 7E, and S6G–6I). Thus, the formation of the TCP14 nuclear foci is dependent on TCP14 DNA binding activity.
We tested whether the JAZ3 nuclear foci represent a structure for its degradation. We observed that JAZ3-RFP formed sub-nuclear foci in Col-0, but not in coi1-1 (Figure 7F), a COI1 allele without detectable protein accumulation (He et al., 2012). JAZ3P302A R305A was unable to form nuclear foci (Figure 7G). Thus, formation of JAZ3 nuclear foci requires COI1. Importantly, JAZ3P302A R305A was re-localized into TCP14 subnuclear foci only in the presence of HopBB1 (Figures 7H and 7I). In sum, these data are consistent with a model where HopBB1 links template DNA-bound TCP14 to a degradation complex containing JAZ3.
Discussion
We demonstrate that the HopBB1 type III effector protein modulates subsets of two Arabidopsis transcriptional regulons, those negatively regulated by TCP14 and activated by MYC2, leading to a fine-tuned perturbation in plant defense output that facilitates bacterial pathogen proliferation. Expressing HopBB1 from bacteria or in Arabidopsis rescues the virulence defect of a pathogenic Psy strain lacking the JA-Ile structural mimic, coronatine, suggesting its role as a regulator of host JA response (Figure 3). HopBB1 has dual functions in de-repressing the JA signaling pathway: HopBB1 facilitates the degradation of TCP14 and possibly other TCPs through SCFCOI1 by connecting JAZ3 to it (Figures 4, 6 and 7). HopBB1 disrupts the inhibitory association between JAZ3 and MYC2, leading to MYC2-dependent transcriptional activation of JA responses (Figure 5), which may contribute to the residual function of HopBB1G126D in activating JA response (Figure 3D). However, only subsets of either the TCP14 or MYC regulons are transcriptionally perturbed in the presence of HopBB1 (Figures 1D and S3E). Thus, we propose that HopBB1 has evolved to minimize pleiotropic negative effects on host physiology generated by wholesale de-repression of the JA response output (defined here by MeJA treatment), while maintaining the ability to modulate defense hormone signaling to the pathogen’s advantage.
Four Psy virulence factors, coronatine, HopX1, HopZ1a and HopBB1, activate the JA-response at different steps in the signaling pathway. Coronatine and HopX1 stimulate an overlapping spectrum of JA-related phenotypes including activation of a few tested JA responsive genes, promotion of stomatal opening and induction of chlorotic symptoms in infected plants. This pleiotropy is likely attributable to the ability of HopX1 to directly cleave almost all members of the JAZ family; this is functionally analogous to coronatine action (Gimenez-Ibanez et al., 2014; Kloek et al., 2001). In contrast, we conclude that HopBB1 expression specifically activates a subset of JA-mediated responses (Figure 3D and S3E). This conclusion is supported by several observations. First, expressing HopBB1 in Arabidopsis activates only about 18% (168 of 933) of JA responsive genes (Figure S3E). Second, HopBB1 manipulates JA response by dissociating the JAZ3-MYC2 complex, leading to SCFCOI1-dependent degradation of the JAZ3-HopBB1-TCP14 complex (Figures 6 and 7). Third, HopBB1 is apparently more selective than HopX1 or the action of coronatine, since it only interacts with a small subset of JAZ proteins and TCP14 to pinpoint a sector of JA regulon. Fourth, the chlorotic phenotypes observed in HopX1 transgenic plants and coronatine treated plants are not visible following either constitutive or conditional expression of HopBB1 (Figures 3E and S3F). Importantly, coronatine-induced chlorosis can be decoupled from bacterial growth promotion and repression of SA-dependent responses (Kloek et al., 2001). We suggest that HopBB1 fine-tunes JA response by targeting a sub-group of JAZ proteins leading to transcriptional activation of genes enriched in those co-regulated by TCP and MYC.
Modulation of plant JA responses is an important virulence strategy for phytopathogenic bacteria (Gimenez-Ibanez et al., 2014; Jiang et al., 2013; Zheng et al., 2012). The evolutionary mechanism driving the mutual exclusivity of JA-modulating virulence factors in Psy genomes (Figure 3H and Table S5) is unknown, but is consistent with negative frequency-dependent selection driven by the centrality of JA response manipulation to Psy virulence, balanced by host immune recognition. This particular arms race is evident in various plants. The ZAR1 NLR innate immune receptor in Arabidopsis recognizes the acetylation activity of HopZ1a on the ZED1 pseudokinase (Lewis et al., 2013;). Alleles of HopX are recognized by the as yet uncloned R2 disease resistance gene in beans (Mansfield et al., 1994). Additionally, plants can evolve JAZ proteins that are resistant to COI1-mediated degradation (Chung and Howe, 2009; Shyu et al., 2012). These JAZ proteins might antagonize coronatine function. Although a host surveillance mechanism recognizing HopBB1 has not been discovered, it could be achieved by monitoring an as yet unknown activity on TCP14 or JAZ3, or on the relevant interacting domains integrated into recently described decoy fusion NLR proteins (Cesari et al., 2014).
TCP14 is targeted by effectors from three evolutionarily divergent pathogens (Wessling et al., 2014). Our results demonstrated that TCP14 contributes to disease resistance against Psy as a negative regulator of JA signaling (Figure 1). JA responsive genes are repressed in seedlings overexpressing TCP14 (Figure 1D) and tcp14 mutants rescue the growth defects of DC3000 cor- (Figure 1C). However, TCP14 may regulate other defense pathways against different pathogens at different developmental stages or in different tissues. Indeed, Kim et al suggested that TCP8, TCP14 and TCP15 are part of a transcriptional complex involved in NLR-mediated immune system signaling (Kim et al., 2014). Thus, we speculate that the TCP14-interacting effectors identified from Psy, Hpa and Go will manipulate TCP14 via different mechanisms to facilitate proliferation of pathogens with different life cycles and infection strategies. It is therefore noteworthy that an Hpa-derived TCP14-interacting effector, HaRxL45, fails to activate degradation of TCP14 in the presence of JA, indicating that this effector modulates TCP14 in a manner mechanistically different from that of HopBB1. Future studies will explore the different sectors of the immune response that are under direct control of TCP14.
Experimental Procedures
Transient protein expression in N. benthamiana
N. benthamiana plants were grown at 24°C (day) / 20°C (night) unde r a 16-h light / 8-h dark cycle. Agrobacteria were collected and re-suspended in 2 ml resuspension buffer (10 mM MES pH5.6, 10 mM MgCI2 and 200 µM acetosyringone) to a final concentration of the OD600=0.2. To reach equal protein accumulation, the final concentration of Agrobacteria expressing HopBB1 and HopBB1G126D was OD600 (0.02) and OD600 (0.2), respectively. GV3101 carrying 35S promoter-driven p19 protein was co-infiltrated at OD600=0.05 in each experiment to prevent the onset of post-transcriptional gene silencing and improve the efficiency of transient expression. Agrobacterium tumefaciens GV3101 (pMP90) transformed with mixtures of binary vector constructs were infiltrated into N. benthamiana leaves using a needleless syringe. Samples were harvested 24 hours after infiltration unless otherwise indicated.
Immunoblot and co-immunoprecipitation analyses
Leaf tissues were ground in liquid nitrogen, and extracted with 150–200 µL of grinding buffer (50mM Tris pH8.0, 1% SDS, 1mM EDTA) also containing 1ul/ml β-mercaptoethanol and 1× protease inhibitor (Sigma-Aldrich). The lysates were centrifuged at 12,000 rpm for 10 min at 4°C. Supernatants were collected and the protein concentration was determined with the BioRad Bradford quantification method (BioRad).
For co-immunoprecipitation analyses, proteins were extracted from 0.5 g of fresh tissue using 2 ml extraction buffer (50 mM HEPES [pH 7.5], 50 mM NaCl, 10 mM EDTA [pH 8.0], 0.5% Triton X-100, 5 mM DTT, and 1× Plant protease inhibitor cocktail from Sigma-Aldrich). Magnetic labeling and separation of tagged proteins was performed using mMACS Epitope Tag Protein Isolation Kit (Miltenyi Biotec). Protein samples were separated by SDS-PAGE. Immunoblots were performed with a 1:1,000 dilution of α-HA (Roche),1:1,000 dilution of α-GFP (Roche), 1:1,000 dilution of α-myc and 1:1,000 dilution of α-FLAG. Blots were detected by ECL prime (GE Healthcare).
Mutagenesis
JAZ3P302A R305A and TCP14H121Q R130K L161N were generated using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent). Random mutagenesis of HopBB1 was performed using the GeneMorph II EZClone Domain Mutagenesis Kit. A pJG4–5-HopBB1 construct was mutagenized according to the manufacture’s protocol. The library was transformed into yeast strain RFY206. Each RFY206 (pJG4–5-HopBB1) clone was mated with yeast EGY48 strain carrying pEG202-TCP14 or pEG202-JAZ3. HopBB1 clones that lost interaction with either TCP14 or JAZ3, but not both, were sequenced. If multiple mutations were present in one clone, single mutations were introduced into wild-type HopBB1 and confirmed by re-testing.
Real time PCR
Total RNAs were extracted with the RNeasy Plant Mini kit (Qiagen). cDNAs were sytnthesized using SuperScript® III Reverse Transcriptase (Invitrogen). qPCR was performed using SYBR green master mix (Applied Biosystems) with cycle: 95°C for 3min, 40 cycles of 95°C for 15 secs, 58°C for 15 secs and 72°C for 20secs. Expression levels were normalized to multiple endogenous controls including UBQ5 (AT3G62250), TUB (AT5G62690) and SAND (AT2G28390).
Statistics for in planta bacterial growth
In Figures 1C, 3A and 3F, error bars represent ±SD. Statistics were performed using one-way ANOVA test with Tukey-Kramer HSD with 95% confidence. In each case, the result displayed is one of three independent analyses giving similar results.
Supplementary Material
Highlights.
The transcriptional regulator TCP14 represses JA response to promote disease resistance
The Pseudomonas syringae type III effector HopBB1 interacts with TCP14
HopBB1 activates TCP14-repressed JA response genes and promotes bacterial virulence
HopBB1 targets TCP14 for SCFCOI1-dependent degradation by connecting it to JAZ3
Acknowledgments
We thank Dangl-Grant lab members for fruitful discussions and criticisms, Drs. Freddy Monteiro and Farid El Kasmi for tissue harvest, the UNC-CH High Throughput Sequencing Center for assistance and Prof. Sarah Grant for critical comments on the manuscript. We thank Prof.. Sheng Yang He, Michigan State University, for the gift of JAZ3 fragments, entry clones of JAZ family members and 35S::JAZ3-HA plants and Prof. Steve A. Kay for entry clones of TCP family members. This work was funded by grants to JLD from National Institutes of Health (1RO1 GM107444), the Gordon and Betty Moore Foundation (GBMF3030), and the HHMI. JLD is an Investigator of the Howard Hughes Medical Institute. LY was funded in part by the Gordon and Betty Moore Foundation through Grant GBMF 2550.02 to the Life Sciences Research Foundation. PJPLT was supported in part by a fellowship from the Pew Latin American Fellows Program in the Biomedical Sciences. YH was supported by a Distinguished Guest Professorship, Eberhard-Karls-Universität, Tübingen, Germany to JLD. Omri M. Finkel is supported by National Institutes of Health grant F32 GM117758-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
LY, PJPLT, PE and JD designed the study. LY, PJPLT, PE, SB, YH, ISG, OMF, MEE and PM performed experiments. LY, PJPLT and JD wrote the paper.
References
- Aggarwal P, Das Gupta M, Joseph AP, Chatterjee N, Srinivasan N, Nath U. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell. 2010;22:1174–1189. doi: 10.1105/tpc.109.066647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Yang L, Hetzel J, Dangl JL, Chory J. The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem. Sci. 2014;39:447–456. doi: 10.1016/j.tibs.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentham A, Burdett H, Anderson PA, Williams SJ, Kobe B. Animal NLRs provide structural insights into plant NLR function. Ann Bot. 2016 doi: 10.1093/aob/mcw171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- Cesari S, Bernoux M, Moncuquet P, Kroj T, Dodds PN. A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front. Plant Sci. 2014;5:606. doi: 10.3389/fpls.2014.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21:131–145. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreze M, Carvunis A-R, Charloteaux B, Galli M, Pevzner SJ, Tasan M, Ahn Y-Y, Balumuri P, Barabási A-L, Bautista V. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Lopez-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Boter M, Fernandez-Barbero G, Chini A, Rathjen JP, Solano R. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 2014;12:e1001792. doi: 10.1371/journal.pbio.1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Chintamanani S, Chen Z, Zhu L, Kunkel BN, Alfano JR, Tang X, Zhou JM. Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 2004;37:589–602. doi: 10.1111/j.1365-313x.2003.01986.x. [DOI] [PubMed] [Google Scholar]
- He Y, Chung EH, Hubert DA, Tornero P, Dangl JL. Specific missense alleles of the arabidopsis jasmonic acid co-receptor COI1 regulate innate immune receptor accumulation and function. PLoS Genet. 2012;8:e1003018. doi: 10.1371/journal.pgen.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Yao J, Ma KW, Zhou H, Song J, He SY, Ma W. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 2013;9:e1003715. doi: 10.1371/journal.ppat.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Lyons R. Intervention of Phytohormone Pathways by Pathogen Effectors. Plant Cell. 2014;26:2285–2309. doi: 10.1105/tpc.114.125419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. MYC2: the master in action. Mol. Plant. 2013;6:686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 2011;68:147–158. doi: 10.1111/j.1365-313X.2011.04674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Son GH, Bhattacharjee S, Kim HJ, Nam JC, Nguyen PDT, Hong JC, Gassmann W. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector - triggered immunity. Plant J. 2014;78:978–989. doi: 10.1111/tpj.12527. [DOI] [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN. Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 2001;26:509–522. doi: 10.1046/j.1365-313x.2001.01050.x. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002;30:337–348. doi: 10.1046/j.1365-313x.2002.01294.x. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Lee AH, Hassan JA, Wan J, Hurley B, Jhingree JR, Wang PW, Lo T, Youn JY, Guttman DS, et al. The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc. Natl. Acad. Sci. USA. 2013;110:18722–18727. doi: 10.1073/pnas.1315520110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LA, Polanski K, de Torres-Zabala M, Jayaraman S, Bowden L, Moore J, Penfold CA, Jenkins DJ, Hill C, Baxter L. Transcriptional dynamics driving MAMP-triggered immunity and pathogen effector-mediated immunosuppression in Arabidopsis leaves following infection with pseudomonas syringae pv tomato DC3000. Plant Cell. 2015;27:3038–3064. doi: 10.1105/tpc.15.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JA, Sun Y, Blair PB, Mukhtar MS. TCP three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci. 2015;20:238–245. doi: 10.1016/j.tplants.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Mansfield J, Jenner C, Hockenhull R, Bennett MA, Stewart R. Characterization of avrPphE, a gene for cultivar-specific avirulence from Pseudomonas syringae pv. phaseolicola which is physically linked to hrpY, a new hrp gene identified in the halo-blight bacterium. Mol. Plant Microbe Interact. 1994;7:726–739. doi: 10.1094/mpmi-7-0726. [DOI] [PubMed] [Google Scholar]
- McCann HC, Rikkerink EH, Bertels F, Fiers M, Lu A, Rees-George J, Andersen MT, Gleave AP, Haubold B, Wohlers MW, et al. Genomic analysis of the Kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog. 2013;9:e1003503. doi: 10.1371/journal.ppat.1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Fisher EJ, Desveaux D, Chang JH, Dangl JL. The HopX (AvrPphE) family of Pseudomonas syringae type III effectors require a catalytic triad and a novel N-terminal domain for function. Mol. Plant Microbe Interact. 2007;20:346–357. doi: 10.1094/MPMI-20-4-0346. [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Perez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Wang J, Huang H, Liu B, Gao H, Liu Y, Song S, Xie D. Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between bHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell. 2015;27:1634–1649. doi: 10.1105/tpc.15.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resentini F, Felipo-Benavent A, Colombo L, Blazquez MA, Alabadi D, Masiero S. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant. 2015;8:482–485. doi: 10.1016/j.molp.2014.11.018. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Alborn HT, O’Donnell P, Sammons M, Toshima H, Tumlinson JH. Simultaneous analysis of phytohormones, phytotoxins, and volatile organic compounds in plants. Proc. Natl. Acad. Sci. USA. 2003;100:10552–10557. doi: 10.1073/pnas.1633615100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu C, Figueroa P, Depew CL, Cooke TF, Sheard LB, Moreno JE, Katsir L, Zheng N, Browse J, Howe GA. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell. 2012;24:536–550. doi: 10.1105/tpc.111.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell. 2012;24:96–108. doi: 10.1105/tpc.111.093518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Kingdom HN, MacLean AM, Grieve VM, Hogenhout SA. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA. 2011;108:E1254–E1263. doi: 10.1073/pnas.1105664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA, Kamoun S. From Guard to Decoy: a new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111:1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessling R, Epple P, Altmann S, He Y, Yang L, Henz SR, McDonald N, Wiley K, Bader KC, Glasser C, et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe. 2014;16:364–375. doi: 10.1016/j.chom.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Haft DH, Getzoff ED, Tainer JA, Lerner RA, Brenner S. Identical short peptide sequences in unrelated proteins can have different conformations: a testing ground for theories of immune recognition. Proc. Natl. Acad. Sci. USA. 1985;82:5255–5259. doi: 10.1073/pnas.82.16.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Yao J, Ke J, Zhang L, Lam VQ, Xin XF, Zhou XE, Chen J, Brunzelle J, Griffin PR, et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature. 2015;525:269–273. doi: 10.1038/nature14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe. 2012;11:587–596. doi: 10.1016/j.chom.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.