Abstract
The plant hormone abscisic acid (ABA) and the jasmonic acid related-compound 12-oxo-phytodienoic acid (OPDA) play crucial roles in seed development, dormancy, and germination. However, a lack of suitable techniques for visualising plant hormones has restricted the investigation of their biological mechanisms. In the present study, desorption electrospray ionisation-imaging mass spectrometry (DESI-IMS), a powerful tool for visualising metabolites in biological tissues, was used to visualise ABA and OPDA in immature Phaseolus vulgaris L. seed sections. The mass spectra, peak values and chemical formulae obtained from the analysis of seed sections were consistent with those determined for ABA and OPDA standards, as were the precursor and major fragment ions observed in tandem mass spectrometry (MS/MS) imaging. Furthermore, the precursor and fragment ion images showed similar distribution patterns. In addition, the localisation of ABA and OPDA using DESI-IMS was confirmed using liquid chromatography-MS/MS (LC-MS/MS). The results indicated that ABA was mainly distributed in the radical and cotyledon of the embryo, whereas OPDA was distributed exclusively in external structures, such as the hilum and seed coat. The present study is the first to report the visualisation of plant hormones using IMS, and demonstrates that DESI-IMS is a promising technique for future plant hormone research.
Plant hormones comprise chemically diverse compounds that occur at low concentrations and regulate growth, development, and responses to external stimuli1. In seeds, abscisic acid (ABA) plays a key role in the induction of primary dormancy during seed development and acts as a germination repressor2,3,4,5,6,7,8. Recently, 12-oxo-phytodienoic acid (OPDA), a jasmonic acid (JA)-related compound, was reported to act with ABA in the regulation of seed dormancy and germination and to have a role in a proper development of an embryo9,10,11,12.
Conventional procedures, such as liquid chromatography tandem mass spectrometry (LC-MS/MS), is commonly used to analyse plant hormones8,10,11,12,13. However, since this requires the extraction of target compounds from plant tissues, it is difficult to investigate their distributions inside plant tissues. Therefore, a technique allowing the visualisation of ABA and OPDA inside plant tissues would be useful, but to date, one has yet to be fully established.
Imaging mass spectrometry (IMS) is a powerful tool for direct visualisation of biomolecules in biological tissues. The use of IMS for visualising small molecules in the tissues or cells of animals and plants has primarily been by matrix-assisted laser desorption ionisation (MALDI)14,15,16,17,18,19,20,21,22. However, the detection of small molecules using MALDI-IMS is often complicated by interfering low-mass matrix ions generating a crowded low mass region. Although ultra-high mass resolution in orbitrap19,20, MS/MS in quadrupole-time-of-flight (Q-TOF)14. and ion trap17,21,22 systems have been used to circumvent this problem, avoiding the use of the matrix altogether is desirable. Desorption electrospray ionisation (DESI) is a growing ambient ionisation technique23,24,25,26,27,28,29, that is initiated when charged droplets impact the biological tissues directly23,24. Unlike MALDI, DESI does not require the application of a matrix. Therefore, DESI-IMS is often considered more suitable than MALDI-IMS for the visualisation of low molecules.
The DESI ion source is commonly used with orbitrap28 or Q-TOF29 mass spectrometers. Orbitraps are useful for visualising metabolites, owing to its ultra-high mass resolution and accuracy; however, its sensitivity depends on the stability of these ions during ion cyclotron resonance. Q-TOFs have high sensitivity, mass resolution and mass accuracy in both the MS and MS/MS modes, which permits the accurate calculation of the chemical formulae for small molecules and their derived fragments.
The common bean, Phaseolus vulgaris L., which is one of the most important protein and micronutrient sources for both humans and livestock worldwide30. In 2014, the genome of P. vulgaris L. became available, which has provided a unique opportunity to elucidate the role of plant hormones at the molecular level. In addition, the number of genes and number of members within each gene family in P. vulgaris L. does not significantly differ from that of Arabidopsis, the most popular model plant species30. As a result, P. vulgaris L. is increasingly being used as a model legume.
The present study describes and quantifies the distributions of ABA and OPDA in immature P. vulgaris L. seed sections using DESI-IMS coupled to a Q-TOF mass spectrometer and LC-ESI-MS/MS analysis.
Results and Discussion
Preparation of immature seed sections
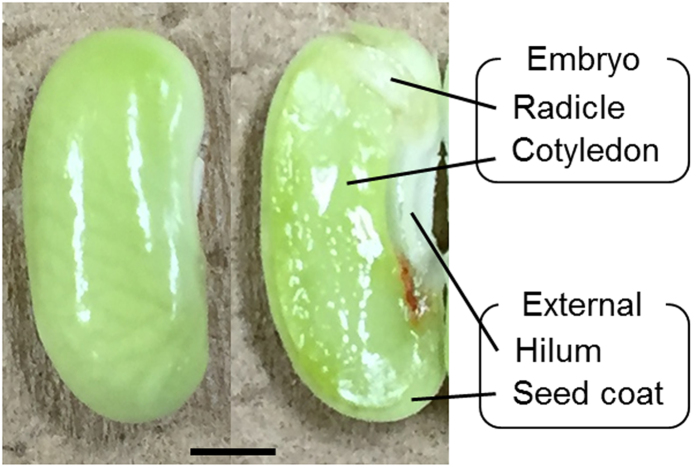
Figure 1 shows the cross-sectional structures of typical immature seeds. The ABA and OPDA contents of immature seeds are thought to change during development8,10,12. Using LC-ESI-MS/MS analysis, we quantified both ABA and OPDA in the immature seeds weighting between 30–210 mg. The analysis revealed that the levels of ABA and OPDA increased markedly in seeds weighting more than approximately 120 mg, and that high amounts of the ABA and OPDA were maintained in heavier seeds (data not shown). Therefore, 150– to 190-mg immature seeds were used for DESI-IMS.
Figure 1. Structure of immature Phaseolus vulgaris L. seeds.
Optical image of immature seed. Scale bar = 2 mm.
Because efficient sectioning is crucial for successful DESI-IMS analysis31, we also compared a few preparation techniques. When frozen in liquid nitrogen and sectioned at 20-μm thickness, the external structures of immature seeds, such as the hilum and seed coat, were destroyed. However, CMC freeze-embedding allowed for good quality sections of immature seeds with structures remaining intact and both the embryo and external structures clearly observed (Fig. 2b).
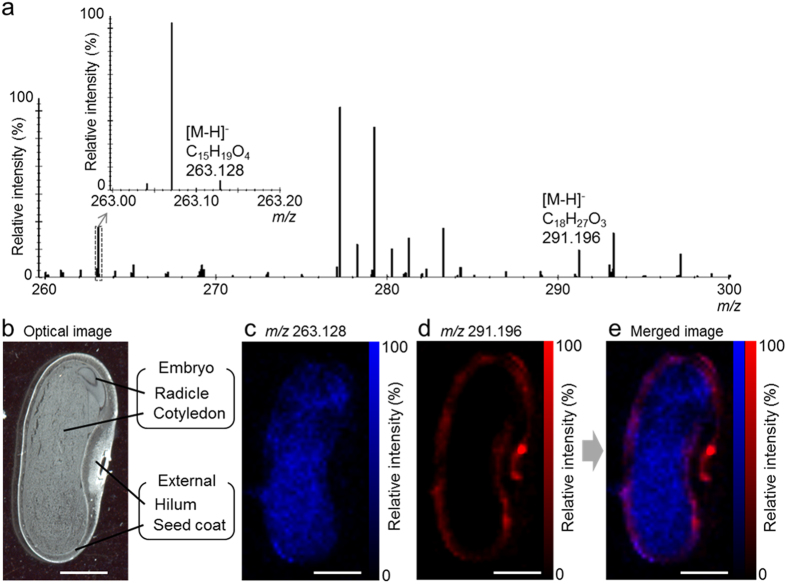
Figure 2. Desorption electrospray ionisation-imaging mass spectrometry (DESI-IMS) of immature Phaseolus vulgaris L. seed.
Sections were prepared from a 163.7-mg seed. Spatial resolution was set at 200 μm. Internal mass calibration was performed post-acquisition using the exact m/z of the palmitic acid [M-H]− ion (m/z 255.233). (a) Mass spectrum at the m/z 260–300. The chemical formulae are shown at the top of the respective peaks. (b) Optical image of a seed section after DESI-IMS. (c) Ion image at m/z 263.128. (d) Ion image at m/z 291.196. (e) Merged ion image of (c and d). Scale bar = 2 mm.
DESI-IMS of immature seed sections
Using the Q-TOF, we initially measured ABA and OPDA standards using DESI-MS in the negative ion mode at 30 000 full width at half maximum (FWHM) mass resolution. Both ABA and OPDA were detected as [M-H]− ions. This was followed by DESI-IMS analysis of immature seed sections (Fig. 2a). Peaks corresponding to the ABA [M-H]− and OPDA [M-H]− ions were detected at mass/charge (m/z) 263.128 and m/z 291.196, respectively (Table 1). The differences between the exact and detected m/z values were −3.4 ppm for ABA and −2.1 ppm for OPDA. The chemical formulae for the peaks were consistent with those generated from ABA and OPDA standards (Fig. 2a). The ion images at m/z 263.128 (Fig. 2c) and m/z 291.196 (Fig. 2d), together with the merged image of these ions (Fig. 2e), indicated that ABA was mainly distributed in the embryo, including the radicle and cotyledon, whereas OPDA exclusively distributed in external structures, such as the hilum and seed coat. DESI-IMS coupled to an orbitrap was also performed at 100 000 FWHM mass resolution. The OPDA [M-H]− ion was detected; however, the ABA [M-H]− ion was not (Supplementary Fig. S1). Therefore, the DESI-IMS orbitrap setup was not further used in this study. We hypothesized that the main reason ABA was not detected in the orbitrap because of the poor stability of the ABA ion during ion cyclotron resonance. Fortunately, the Q-TOF did not exhibit this problem, and we were also able to improve the stability and sensitivity of the ABA and OPDA signals using a new DESI sprayer design using a fixed fused silica emitter32.
Table 1. Desorption electrospray ionisation-imaging mass spectrometry (DESI-IMS) of abscisic acid (ABA) and 12-oxo-phytodienoic acid (OPDA).
| Analyte | Chemical Formula | Exact Mass (Da) | Exact m/z [M-H]− | Detected m/z [M-H]− | Error (ppm) |
|---|---|---|---|---|---|
| ABA | C15H20O4 | 264.1362 | 263.1289 | 263.128 | −3.4 |
| OPDA | C18H28O3 | 292.2038 | 291.1966 | 291.196 | −2.1 |
DESI-IMS/MS of immature seed sections
In order to further increase the specificity of the imaging experiment, we analysed the immature seed sections using DESI-IMS/MS Q-TOF21,26. The precursor and fragment ions of both ABA and OPDA were monitored.
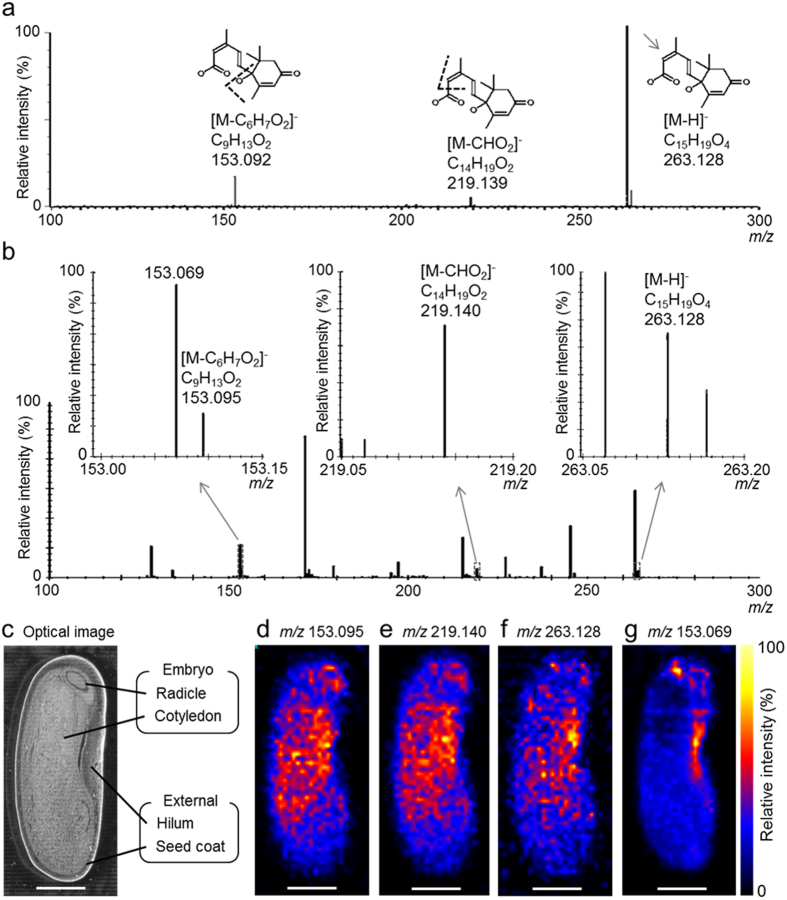
DESI-MS/MS of the ABA standard precursor [M-H]− ion at m/z 263.128 generated two major fragment ions at m/z 153.092 and m/z 219.139 (Fig. 3a). The associated chemical formulae and structures are shown on top of each peak. Subsequent DESI-IMS/MS of the immature seed sections resulted in the same precursor (m/z 263.128) and fragment ions (m/z 153.095 and m/z 219.140) (Fig. 3b). Another peak was observed at m/z 153.069 close to the ABA-derived fragment ion at m/z 153.095 (Fig. 3b). We found that at least 20 000 FWHM mass resolution was required to distinguish between the peaks at m/z 153.069 and m/z 153.095 (data not shown).
Figure 3. Desorption electrospray ionisation - tandem imaging mass spectrometry (DESI-IMS/MS) of abscisic acid (ABA) in immature Phaseolus vulgaris L. seed.
Sections were prepared from a 163.7-mg seed. (a) Tandem mass spectrum (MS/MS) of ABA standard. The chemical formulae and structures are shown at the top of the respective peaks. (b) MS/MS spectrum of the immature seed section at m/z 262–265. Internal calibration of the MS/MS spectrum was performed post-acquisition using the exact m/z of ABA [M-H]− ion (m/z 263.1289). (c) Optical image of seed section after DESI-IMS/MS. (d) Ion image at m/z 153.095. (e) Ion image at m/z 219.140. (f) Ion image at m/z 263.128. Scale bar = 2 mm.
Images derived from the two major ABA fragment ions (Fig. 3d,e) and the ABA precursor ion (Fig. 3f) exhibited identical distribution patterns. In contrast, a different distribution pattern was observed for the m/z 153.069 ion (Fig. 3g). Furthermore, the ABA distribution patterns obtained using DESI-IMS/MS (Fig. 3d–f) were similar to those obtained using DESI-IMS (Fig. 2c), thereby confirming the precise localization of ABA in immature P. vulgaris L. seeds.
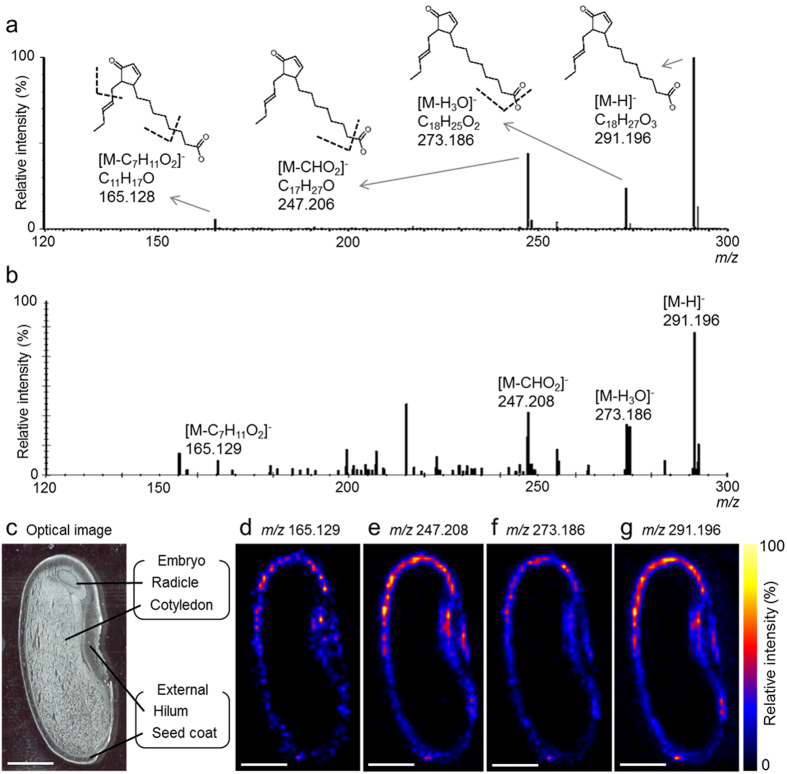
Meanwhile, the DESI-MS/MS of the OPDA standard precursor [M-H]− ion at m/z 291.196 resulted in three fragment ions at m/z 165.128, m/z 247.206, and m/z 273.186 (Fig. 4a). The associated chemical formulae and structures are shown on the top of each peak. Subsequent DESI-IMS/MS of immature seed sections resulted in the same precursor ion (m/z 291.196) and major fragment ions (m/z 165.129, m/z 247.208, and m/z 273.186; Fig. 4b). The distribution patterns obtained using DESI-IMS/MS for the OPDA precursor ion (Fig. 4g) and the three major OPDA fragment ions (Fig. 4d–f) were similar to those obtained using DESI-IMS (Fig. 2d), thereby confirming the precise localization of OPDA in immature P. vulgaris L. seeds.
Figure 4. Desorption electrospray ionisation-tandem imaging mass spectrometry (DESI-IMS/MS) of 12-oxo-phytodienoic acid (OPDA) in immature Phaseolus vulgaris L. seed.
Sections were prepared from a 163.7-mg seed. (a) Tandem mass spectrum (MS/MS) of the OPDA standard. The chemical formulae and structures are shown at the top of the respective peaks. (b) MS/MS spectrum of the immature seed section at m/z 290–293. Internal calibration of the MS/MS spectrum was performed post-acquisition using the exact m/z of OPDA [M-H]− ion (m/z 291.1966). (c) Optical image of a seed section after DESI-IMS/MS. (d) Ion image at m/z 165.129. (e) Ion image at m/z 247.208. (f) Ion image at m/z 273.186. (g) Ion image at m/z 291.196. Scale bar = 2 mm.
LC-ESI-MS/MS of ABA and OPDA in immature seeds
In order to confirm the ABA and OPDA distribution patterns observed in the DESI-IMS analysis, ABA and OPDA were quantified in the 80% methanol extracts of the embryo and external structures of four different immature seeds using LC-ESI-MS/MS (Table 2). The mean concentration of ABA in the embryo (1844 ± 478 pg/mg fresh weight (FW)) was approximately 18 times higher than that of the external structures (103 ± 52 pg/mg FW). In contrast, the mean concentration of OPDA in the external structures (8164 ± 1549 pg/mg FW) was approximately 154 times higher than that in the embryos (53 ± 24 pg/mg FW). These results agreed with those of the DESI-IMS analysis.
Table 2. Abscisic acid (ABA) and 12-oxo-phytodienoic acid (OPDA) concentrations in the embryo and external structures of immature Phaseolus vulgaris L. seeds.
| Tissue | Weight (mg FW) | OPDA (pg/mg FW) | ABA (pg/mg FW) |
|---|---|---|---|
| Embryo | 113 ± 7 | 53 ± 24 | 1844 ± 478 |
| External structures | 59 ± 1 | 8164 ± 1549 | 103 ± 52 |
ABA and OPDA in aqueous methanol (80%) extracts of embryo and external structures separated from four different immature seeds were quantified by liquid chromatography-electrospray ionisation-tandem mass spectrometry (LC-ESI-MS/MS) analysis. Concentrations of ABA and OPDA in embryo and external structures are shown per mg of fresh weight (FW). Each value is represented by the mean ± standard deviation (n = 4).
Goetz et al.12 reported that, in tomato, OPDA occurs almost exclusively in the seed coat tissues. They further reported that the female-sterile phenotype of the JA-insensitive mutants of tomato (jai1) could be rescued by the wound-induced synthesis of OPDA, which suggests that OPDA, rather than JA, is involved in embryo development. Similarly, the present study revealed that OPDA is localised in the seed coat of P. vulgaris L.
Meanwhile, using Arabidopsis mutants disrupted in peroxisomal β-oxidation, Dave et al.9,10 demonstrated that OPDA, rather than JA or JA-isoleucine, is involved in the repression of seed germination and that its action is synergistic with ABA. Dave et al.11 showed that the synergistic action observed in Arabidopsis results from the accumulation of OPDA in response to ABA and a dormancy-promoting factor, MOTHER-OF-FT-AND-TFL1. In addition to its inhibitive roles in seed germination, ABA is important in the regulation of embryo growth7. Therefore, ABA and OPDA constitute indispensable counterparts in the regulation of seed growth and germination. However, there is limited reliable information on the distribution of ABA in seeds. To the best of our knowledge, the present work using DESI-IMS Q-TOF is the first to report that ABA is mainly localized in the embryo including the radicle and cotyledon. In addition, the DESI-IMS Q-TOF could simultaneously visualise the distribution of ABA and OPDA in seed tissues. Furthermore, DESI-IMS had a great advantage over conventional procedures, such as LC-MS/MS, in that it can directly visualize with finer spatial precision the gradients of the ABA and OPDA concentrations inside plant tissues (Figs 2c–e, 3d–g and 4d–g).
In conclusion, it was found that the DESI-IMS is a valuable tool for the visualisation of plant hormones and investigations of their biological roles.
Methods
Reagents
cis-12-Oxo-phytodienoic acid (OPDA) was purchased from Cayman Chemical Company (Ann Arbor, MI, USA). (+)-Abscisic acid (ABA), methanol, and water were purchased from Sigma Aldrich (St. Louis, MO, USA). D6-ABA was purchased from Icon Isotopes (Summit, NJ, USA). D5-OPDA was purchased from OlChemlm Ltd (Olomouc, Czech Republic). Carboxymethyl cellulose (CMC) sodium salt was purchased from Wako Chemicals (Tokyo, Japan). Superfrost Plus microscope glass slides were purchased from Thermo Scientific (San José, CA, USA). All reagents and solvents used in the present study were of analytical grade.
Seeds
Phaseolus vulgaris L. were purchased from a local supermarket, and immature seeds were removed from the pods, weighed, flash frozen in liquid nitrogen, and preserved at −80 °C until used.
Sample preparation for LC–electrospray ionisation (ESI)–MS/MS analysis
Frozen immature seeds weighting 30–210 mg were homogenized using a disposable homogenizer, and 1 ml cold aqueous methanol (80%) was immediately added. After mixing for 30 min, the suspensions were centrifuged at 3 000 rpm for 10 min. After adding of D6-ABA and D5-OPDA in the supernatants (final concentrations were 100 ng/mL) as internal standards, the solutions were subsequently analysed using LC-ESI-MS/MS.
For the quantification of ABA and OPDA in the embryo and external structures of the immature seeds, tissues were separated from four different seeds weighting 150–190 mg, weighed, and then subjected to the same extraction procedure.
LC–ESI–MS/MS analysis
One μl of each tissue extract was injected into an Agilent 1200 high performance liquid chromatography system coupled to an Agilent 6430 triple quadrupole mass spectrometer equipped with an ESI source (all Agilent Technologies, Palo Alto, CA, USA). An Agilent ZORBAX Eclipse XDB-C18 reversed-phase column (50 × 2.1 mm, 1.8 μm particle size; Agilent Technologies) was used. The solvents used were water (solvent A) and acetonitrile (solvent B), each containing 0.05% acetic acid. The flow rate and LC gradient conditions were 3%–70% B from 0–22 min at a flow rate of 0.2 ml/min, 98% B from 22–27 min at a flow rate of 0.3 ml/min, and 3% B from 27–37 min at a flow rate of 0.2 ml/min.
The MS instrument was operated in the negative ion mode using a drying gas temperature of 300 °C (5.0 l/min). ABA, D6-ABA, OPDA, and D5-OPDA were monitored using multi-reaction monitoring (MRM) transitions at m/z 263.1 > 153.1, m/z 269.1 > 159.1, m/z 291.1 > 165.1, and m/z 296.2 > 170.1 respectively. The collision energy (CE) was optimized for these MRM transitions (CE = 2 for ABA and D6-ABA, and CE = 16 for OPDA and D5-OPDA). D6-ABA and D5-OPDA were used as internal standards for quantification. Both Q1 (MS1) and Q3 (MS2) were maintained at 0.7 m/z FWHM during the LC-ESI-MS/MS experiments. Data analysis was performed using the MassHunter software (Agilent Technologies). The concentrations of ABA and OPDA in the embryo and external structures of four different immature seeds were calculated per mg of fresh weight (FW). Each value is represented by the mean ± standard deviation (n = 4).
DESI-MS/MS analysis
One μl of aqueous methanol (50%) containing 1 ng/μl of the ABA or OPDA standards were placed on a glass slide, and allowed to dry. The MS/MS spectrum of each compound was measured using a Xevo G2-XS Q-TOF instrument equipped with a 2D DESI ion source (all Waters, Milford MA, USA). The DESI spray solvent (95% methanol:5% water, v/v) was delivered at a flow rate of 3 μl/min using a Harvard syringe pump. A voltage of 3 kV was applied to the DESI sprayer, and nitrogen was delivered at 0.4 MPa from an external gas cylinder. The optimised CE voltages for the generation of major fragment ions were 10 eV for ABA and 20 eV for OPDA. External calibration was performed by 10 mmol/L sodium formate solution in 90% 2-propanol (v/v) prior to measurement. The chemical formulae and structures of the major fragment ions were calculated using the MassFragment software (Waters).
Preparation of seed sections for DESI-IMS
Seed sections were prepared from a 163.7-mg seed using CMC freeze-embedding31. Briefly, the immature seed was immersed in 2% CMC and frozen in liquid nitrogen. Subsequently, 20-μm sections were consecutively prepared using a CM 1860 cryostat (Leica Microsystems, Wetzlar, Germany). The sections were then mounted onto Superfrost microscope glass slides (Thermo Scientific), which were dried in a vacuum desiccator for 10 min. The sections were placed in 15-ml centrifuge tubes that contained silica gel for drying, and were preserved at −80 °C until DESI-IMS analysis.
DESI-IMS analysis
DESI-IMS was performed using a Xevo G2-XS Q-TOF instrument equipped with a 2D DESI ion source (Waters). The DESI solvent (95% methanol, v/v) was delivered at a flow rate of 3 μl/min using a Harvard syringe pump. A voltage of 3 kV was applied to the DESI sprayer, and nitrogen was delivered at 0.4 MPa from an external gas cylinder. The seed sections were scanned in horizontal lines at 200 μm/s, and collecting a mass spectrum every second. Scanning rows were separated by vertical steps of 200 μm, resulting in a lateral resolution of 200 μm. Signals at m/z 200–400 were measured in the negative ion mode at 30 000 FWHM mass resolution. External calibration was performed by 10 mmol/L sodium formate solution in 90% 2-propanol (v/v) prior to measurement. Internal calibration was performed post-acquisition in the HDImaging software (Waters) using the exact m/z of the palmitic acid [M-H]− ion (m/z 255.233). Peak picking was performed in order of the most intense peaks from the entire m/z range at 30 000 FWHM mass resolution, and ion images were constructed using the HDImaging software (Waters). Chemical formulae and structures were calculated using the MassFragment and Elemental Composition software (Waters).
DESI-tandem imaging mass spectrometry (IMS/MS) was performed at m/z 262–265 for ABA and m/z 290–293 for OPDA, respectively. The CE was set at 10 eV for ABA and 20 eV for OPDA. Internal calibration was performed post-acquisition in the HDImaging software (Waters) using the exact m/z of the precursor [M-H]− ions (m/z 263.1289 for ABA and m/z 291.1966 for OPDA). The major fragment ions from ABA and OPDA were used to construct ion images using the HDImaging software (Waters). The chemical formulae and structures were calculated using the MassFragment and Elemental Composition software (Waters).
Ultra-high mass resolution imaging was also performed using the Exactive (Thermo Scientific) orbitrap mass spectrometer equipped with a 2D DESI ion source (Prosolia Inc. Indianapolis, IN, USA). Methanol (95%) was used as the spray solvent. Spatial resolution was set to 150 μm, and mass resolution was set at 100 000 FWHM (@m/z 400). Signals at m/z 200–400 were measured in the negative ion mode. Image files (IMG files) were created from the acquired data using the FireFly software, and processed to generate images using the BioMAP software (Novartis, Basel, Switzerland). Chemical formulae were calculated using the Xcalibur software (Thermo Scientific).
Statistical analysis
The concentrations of ABA and OPDA in the embryo and external structures are the means of four different seeds ± standard deviation. Statistical differences between the means were evaluated using Student’s t-test (P < 0.05).
Additional Information
How to cite this article: Enomoto, H. et al. Visualisation of abscisic acid and 12-oxo-phytodienoic acid in immature Phaseolus vulgaris L. seeds using desorption electrospray ionisation-imaging mass spectrometry. Sci. Rep. 7, 42977; doi: 10.1038/srep42977 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Satoshi Uehara for his assistance in performing DESI-IMS with an orbitrap mass spectrometer. We would also like to thank Editage (www.editage.jp) for English language editing. This work was supported by JSPS KAKENHI (26292144) and by the Ministry of Education, Culture, Sports, Science, and Technology-Supported Program for the Strategic Research Foundation at Private Universities (2013–2017) [project number S131052A01].
Footnotes
The authors declare no competing financial interests.
Author Contributions H.E. and H.Y. designed the research. H.E., T.S., K.S., F.S., E.Y., K.M., M.A., and H.Y. performed the experiments. H.E., T.Y., and H.Y. analysed the data. H.E. wrote the manuscript. H.E., T.P., T.Y., and H.Y. edited the manuscript. H.E., T.P., M.A., K.M., T.Y., and H.Y. reviewed the manuscript. All authors have read and approved the manuscript.
References
- Davies P. J. The plant hormones: Their nature, occurrence, and function in Plant hormones: Biosynthesis, Signal transduction, Action! (ed. Davies P. J.) 1–15 (Springer, 2010). [Google Scholar]
- Chernys J. T. & Zeevaart J. A. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 124, 343–353 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. Salt and drought stress signal transduction in plants. Ann. Rev. Plant Biol. 53, 247–273 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T., Park Y. & Hwang I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 58, 29–48 (2015). [DOI] [PubMed] [Google Scholar]
- McCarty D. R. Genetic control and integration of maturation and germination pathways in seed development. Ann. Rev. Plant Physiol. Plant Mol. Biol. 46, 71–93 (1995). [Google Scholar]
- Oishi M. Y. & Bewley J. D. Premature drying, fluridone-treatment, and embryo isolation during development of maize kernels (Zea mays L.) induce germination, but the protein synthetic responses are different. Potential regulation of germination and protein synthesis by abscisic acid. J. Exp. Bot. 43, 759–767 (1992). [Google Scholar]
- Cheng Z. J. et al. Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell. 26, 1053–1068 (2014). [DOI] [PMC free article] [PubMed]
- Kanno Y. et al. Comprehensive hormone profiling in developing Arabidopsis seeds: Examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 51, 1988–2001 (2010). [DOI] [PubMed] [Google Scholar]
- Dave A. & Graham I. A. Oxylipin signaling: A distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA). Front. Plant Sci. 3, 140–144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A. et al. 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell. 23, 583–599 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A., Vaistij F. E., Gilday A. D., Penfield S. D. & Graham I. A. Regulation of Arabidopsis thaliana seed dormancy and germination by 12-oxo-phytodienoic acid. J. Exp. Bot. 67, 2277–2284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S. et al. Role of cis-12-oxo-phytodienoic acid in tomato embryo development. Plant Physiol. 158, 1715–1727 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiwocha S. D. et al. A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: An analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J. 35, 405–417 (2003). [DOI] [PubMed] [Google Scholar]
- Reyzer M. L., Hsieh Y., Ng K., Korfmacher W. A. & Caprioli R. M. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 38, 1081–1092 (2003). [DOI] [PubMed] [Google Scholar]
- Caprioli R. M., Farmer T. B. & Gile J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 69, 4751–4760 (1997). [DOI] [PubMed] [Google Scholar]
- Setou M. et al. Developments and applications of mass microscopy. Med. Mol. Morphol. 43, 1–5 (2010). [DOI] [PubMed] [Google Scholar]
- Yoshimura Y. et al. Visualization of anthocyanin species in rabbiteye blueberry Vaccinium ashei by matrix-assistedlaser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 403, 1885–1895 (2012). [DOI] [PubMed] [Google Scholar]
- Enomoto H., Sugiura Y., Setou M. & Zaima N. Visualization of phosphatidylcholine, lysophosphatidylcholine and sphingomyelin in mouse tongue body by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 400, 1913–1921 (2011). [DOI] [PubMed] [Google Scholar]
- Römp A. & Spengler B. Mass spectrometry imaging with high resolution in mass and space. Histochem. Cell Biol. 139, 759–783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf R. R., Conaway M. C. P., Garrett T. J., Stacpoole P. W. & Yost R. A. Imaging of lipids in spinal cord using intermediate pressure matrix-assisted laser desorption-linear ion trap/Orbitrap MS. Anal. Chem. 81, 8488–8495 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler D. M. et al. Utility of imaging mass spectrometry (IMS) by matrix-assisted laser desorption ionization (MALDI) on an ion trap mass spectrometer in the analysis of drugs and metabolites in biological tissues. J. Pharmacol. Toxicol. Methods. 55, 279–288 (2007). [DOI] [PubMed] [Google Scholar]
- Sugiura Y., Zaima N., Setou M., Ito S. & Yao I. Visualization of acetylcholine distribution in central nervous system tissue sections by tandem imaging mass spectrometry. Anal. Bioanal. Chem. 403, 1851–1861 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolta N. et al. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 306, 471–473 (2004). [DOI] [PubMed] [Google Scholar]
- Cooks R. G., Ouyang Z., Takats Z. & Wiseman J. M. Ambient mass spectrometry. Science. 311, 1566–1570 (2006). [DOI] [PubMed] [Google Scholar]
- Wiseman J. M., Ifa D. R., Song Q. & Cooks R. G. Tissue Imaging at Atmospheric Pressure Using Desorption Electrospray Ionization (DESI) Mass Spectrometry. Angewandte Chem. Int. 45, 7188–7192 (2006). [DOI] [PubMed] [Google Scholar]
- Wiseman J. M. et al. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. Proc. Natl. Acad. Sci. USA 105, 18120–18125 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T. et al. Direct Plant Tissue Analysis and Imprint Imaging by Desorption Electrospray Ionization Mass Spectrometry. Anal. Chem. 83, 5754–5761 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicke N. E., Dill A. L., Ifa D. R. & Cooks R. G. High resolution tissue imaging on an orbitrap mass spectrometer by desorption electro-spray ionization mass spectrometry (DESI-MS). J. Mass Spectrom. 45, 223–226 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Škrášková K. et al. Enhanced capabilities for imaging gangliosides in murine brain with matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry coupled to ion mobility separation. Methods. 104, 69–78 (2016). [DOI] [PubMed] [Google Scholar]
- Castro-Guerrero N. A., Isidra-Arellano M. C., Mendoza-Cozatl D. G. & Valdés-López O. Common bean: A legume model on the rise for unraveling responses and adaptations to iron, zinc, and phosphate deficiencies. Front. Plant Sci. 7, 600 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y. et al. Sample preparation for mass spectrometry imaging of plant tissues: A review. Front. Plant Sci. 10, 7–60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillner J. et al. Investigation of the impact of desorption electrospray ionization sprayer geometry on its performance in imaging of biological tissue. Anal Chem. 88, 4808–4816 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.